Abstract
Organisms that compete for limited resources within a common environment may evolve traits that allow them to exploit distinct ecological niches, thus enabling multiple species to coexist within the same habitat. The process of niche partitioning now has been captured at the molecular level, employing the method of continuous in vitro evolution. Mixed populations of 2 different “species” of RNA enzymes were made to compete for limited amounts of one or more substrates, with utilization of the substrate being necessary for amplification of the RNA. Evolution in the presence of a single substrate led to the extinction of one or the other enzyme, whereas evolution in the presence of 5 alternative substrates led to the accumulation of mutations that allowed each enzyme to exploit a different preferred resource. The evolved enzymes were capable of sustained coevolution within a common environment, exemplifying the emergence of stable ecological niche behavior in a model system. Biochemical characterization of the 2 evolved enzymes revealed marked differences in their kinetic properties and adaptive strategies. One enzyme reacted with its preferred substrate ≈100-fold faster than the other, but the slower-reacting species produced 2- to 3-fold more progeny per reacted parent molecule. The in vitro coevolution of 2 or more species of RNA enzymes will make possible further studies in molecular ecology, including the exploration of more complex behaviors, such as predation or cooperation, under controlled laboratory conditions.
Keywords: in vitro evolution, ligase, molecular ecology, ribozyme
One of the aims of laboratory evolution is to model biological evolution experimentally, with precise control of the key variables relevant to processes of selection, amplification, and mutation. Several laboratory evolution systems have been developed in which populations of bacteria, viruses, or macromolecules are challenged to adapt to various selection constraints, such as depletion of a critical nutrient or alteration of the environment (1–4). The use of catalytic RNA in such evolution experiments is of special interest because of the relevance of RNA to the origins of life and the simplicity of representing both genotype and phenotype within the same molecule.
One especially powerful system for the evolution of catalytic nucleic acids involves the continuous in vitro evolution of RNA enzymes with RNA ligase activity (5). In this system, RNA enzymes are challenged to ligate an oligonucleotide substrate containing the sequence of an RNA polymerase promoter element to their 5′ ends. A primer complementary to the 3′ end of the enzyme is extended by reverse transcriptase to generate complementary DNA (cDNA). The cDNAs derived from reacted RNAs are then transcribed by RNA polymerase to generate many progeny RNA enzymes. Through repeated rounds of these steps, all carried out within a common reaction mixture, the population expands exponentially. Growth can be continued indefinitely by performing a serial transfer procedure, whereby each transfer provides a fresh supply of reaction materials and the opportunity to modify the reaction environment.
Continuous in vitro evolution first was applied to a variant of the class I (CL1) RNA ligase enzyme, which has an unusually fast catalytic rate of >100 min−1 (6, 7). Several rounds of stepwise evolution first were carried out to adapt the CL1 ligase to the promoter-containing substrate and to the reaction conditions required by reverse transcriptase and RNA polymerase. The resulting RNA enzymes were then used to initiate continuous evolution performed by serial transfer. After 100 rounds of 1,000-fold growth and dilution, the enzymes had adapted to the conditions of continuous evolution, with a typical individual isolated from the population exhibiting a kcat of 21 min−1 and KM of 1.7 μM (measured in the presence of 25 mM Mg2+ at pH 8.5 and 37 °C) (8).
Variants of the CL1 RNA enzyme have been used to initiate additional continuous evolution experiments, which have aimed to discover new catalytic behaviors and to test hypotheses relevant to evolutionary biology. For example, CL1 enzymes have been evolved that can tolerate a lower concentration of Mg2+ (9), resist attack by an RNA-cleaving DNA enzyme (8), employ substantially reduced concentrations of substrate (10), or operate at either higher or lower pH (11). Other experiments involving multiple parallel lineages of continuous evolution have been used to investigate conditions that favor recurrent evolutionary endpoints starting from the same initial conditions (12) or that result in extinction through the progressive accumulation of deleterious mutations (13).
Recently a second RNA ligase enzyme, termed DSL (14, 15), was made to undergo continuous in vitro evolution (16). As with the CL1 ligase, it first was necessary to conduct stepwise evolution to adapt the DSL enzyme to the promoter-containing substrate and the conditions required for continuous evolution. The resulting RNA enzymes were then used to initiate evolution by serial transfer, ultimately giving rise to molecules with a kcat of 1.4 min−1 and KM of 0.14 μM (measured under the same conditions as above). The CL1 and DSL enzymes catalyze the same chemical reaction but have completely independent origins and no sequence relatedness other than in the template region of the enzyme that binds the oligonucleotide substrate.
Heretofore, continuous in vitro evolution has been used to study the biochemical adaptation of a particular RNA enzyme. Now that there are 2 distinct enzymes capable of undergoing continuous evolution, it should be possible to study their coadaptation within a common environment. In nature, adaptation and complexity can arise from interactions between different organisms that share a common habitat. Laboratory evolution involving 2 distinct “species” of RNA enzymes makes it possible to conduct studies in experimental molecular ecology, with carefully controlled parameters and detailed knowledge of the evolutionary progression of genotype and phenotype. In the present study, 2 coevolving RNA enzymes were presented with a mixture of several potential substrates to provide an opportunity for niche partitioning. Once each enzyme had adapted to a distinct niche, the two could persist in a common environment, demonstrating an evolutionarily stable strategy in the context of laboratory evolution.
Results
Coevolution Employing a Common Substrate.
The CL1 and DSL enzymes (Fig. 1) first were challenged to undergo continuous coevolution within a common reaction mixture, employing limited amounts of a single oligonucleotide substrate. This process quickly led to the extinction of one or the other enzyme, depending on which substrate was used. A substrate similar to that used in the evolutionary development of the CL1 enzyme was used more efficiently by that enzyme, and likewise for the DSL enzyme and its preferred substrate. This advantage could not be overcome by evolution before extinction of the disadvantaged enzyme occurred. By staggering the starting concentration of the 2 enzymes, it was possible to delay the onset of extinction. It became clear, however, that additional constraints would be required to prevent runaway growth of the more advantageous species.
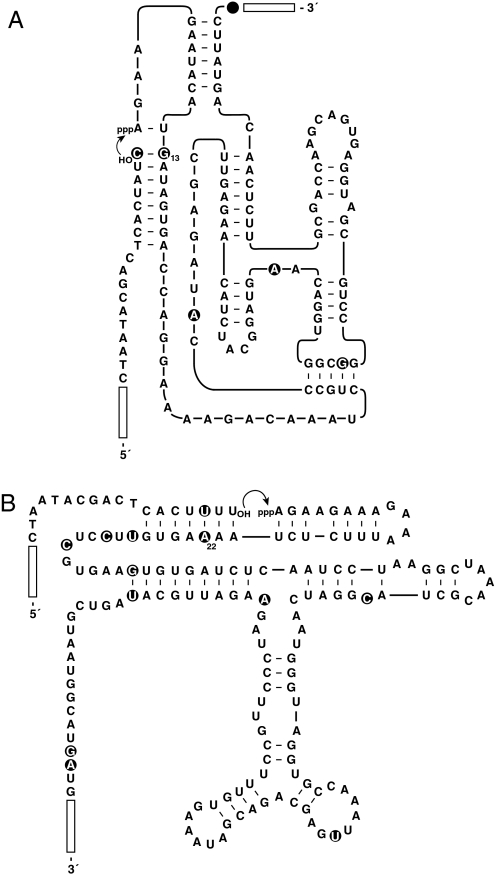
Fig. 1.
Sequence and secondary structure of the evolved CL1 (A) and DSL (B) enzymes, shown with substrates S4 and S5, respectively. Open rectangles indicate primer binding sites at the 5′ end of the substrate and 3′ end of the enzyme. Curved arrow indicates the site of ligation. Filled circles highlight mutations present in typical clones isolated after 50 transfers of coevolution (with 5 substrates) relative to the starting enzymes.
A serial transfer experiment was initiated, during which a 14-mer antisense oligodeoxynucleotide was used to inhibit the growth of the dominant ligase. Antisense oligos were designed to hybridize to a critical region of each of the 2 RNA enzymes, thus impairing their catalytic activity. Beginning with a population of 1 pmol each of randomized variants of the CL1 and DSL enzymes, 40 serial transfers of continuous coevolution were carried out, monitoring the concentration of each ligase (based on cDNA) before and after each transfer [supporting information (SI) Fig. S1]. To maintain diversity within the evolving population, error-prone PCR was performed on both enzymes after transfers 10, 20, and 30. Extinction was narrowly avoided during this coevolutionary process by employing the antisense oligos, but it required knowledge by the experimenter of which sequence to target and manipulation of the reaction conditions based on which enzyme required antisense inhibition. Furthermore, the enzymes began to develop resistance to the antisense oligos by acquiring escape mutations that destabilized the relevant base-pairing interactions. Prolonged use of this approach would require that the oligos be redesigned to maintain the desired effect.
Coevolution Employing 5 Different Substrates.
A second continuous coevolution experiment was initiated, employing a mixture of 5 different substrates (Table S1). Each substrate (S1–S5) contained a single nucleotide change within the promoter sequence compared with the substrate that was used previously (S0). This change reduced the efficiency of the promoter to 9%–63% relative to that of the wild-type promoter and created a single-nucleotide mismatch for each combination of enzyme and substrate.
Continuous coevolution was initiated with 10 pmol of the CL1 and 1 pmol of the DSL enzymes, which were transcribed from templates that had been subject to mutagenic PCR. The concentration of each ligase was measured before and after each transfer (Fig. 2), and the time between transfers was adjusted periodically to allow the enzymes to exhaust the supply of their corresponding substrates (Fig. S2). The concentrations of the CL1 and DSL enzymes were again staggered after transfers 30, 32, 37, 42, and 46 to compensate for their differential growth rates. Diversity was maintained within the evolving population by performing mutagenic PCR after transfers 5, 7, 10, 15, 20, 25, 30, 32, 37, 42, and 46.
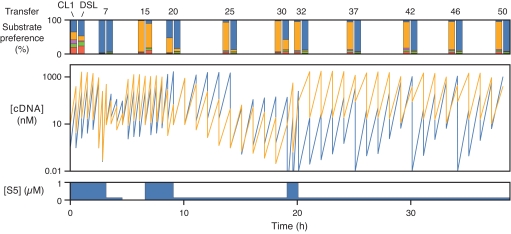
Fig. 2.
Time course of 50 transfers of continuous coevolution in the presence of 5 different substrates. The concentration of the CL1 (orange) and DSL (blue) enzymes was determined (based on their respective cDNAs) before and after each transfer. Paired bar graph at the Top indicates the substrate preference of each enzyme (CL1 at Left, DSL at Right) after various transfers when provided 1 μM S1 (red), S2 (green), S3 (purple), S4 (orange), or S5 (blue). Stepped graph at the Bottom indicates the concentration of S5 present during the coevolution experiment; all other substrates were present at 1 μM concentration throughout.
The initial reaction mixture contained 1 μM each of the 5 substrates. Assays were conducted before the start of the experiment and periodically thereafter to measure the ability of each enzyme to amplify in the presence of 1 μM of each substrate, tested individually (Fig. 2). Both enzymes showed a strong tendency to adapt to utilization of S5, and accordingly, the concentration of S5 was reduced to 0.1 μM or eliminated completely for intervals during the course of the experiment to provide selective advantage favoring the use of one of the other substrates.
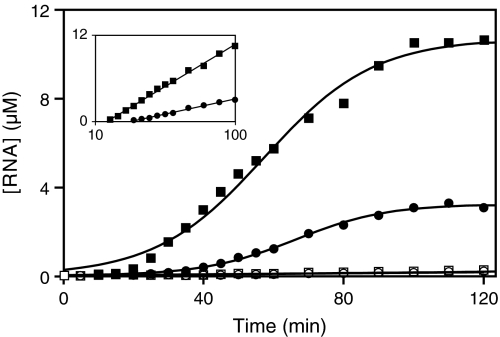
After 50 transfers of continuous coevolution, with an overall amplification of ≈10100-fold, individual CL1 and DSL molecules were cloned from the population and sequenced. A typical clone of each enzyme was chosen for more detailed analysis (Fig. 1). The evolved CL1 enzyme contained 5 mutations relative to the starting enzyme, whereas the evolved DSL enzyme contained 11 mutations. These mutations included compensatory changes within the portion of the enzyme that binds the substrate: A13→G for CL1 and U22→A for DSL, providing perfect complementarity with substrates S4 and S5, respectively. Amplification profiles were obtained for each of the evolved enzymes, demonstrating mutually exclusive use of S4 by CL1 and of S5 by DSL. The exponential growth rate of DSL appreciably exceeded that of CL1 when each enzyme was allowed to operate in the presence of its preferred substrate. After 30-min incubation, the DSL enzyme exhibited 150-fold amplification, whereas the CL1 enzyme exhibited only 13-fold amplification. In addition, DSL achieved a maximum extent of growth that was approximately 3-fold greater than that of CL1 (Fig. 3).
Fig. 3.
Amplification profiles of the CL1 (circles) and DSL (squares) enzymes operating in the presence of 1 μM of either S4 or S5. Filled symbols indicate behavior in the presence of the cognate substrate (CL1 with S4, DSL with S5); open symbols indicate behavior in the presence of the noncognate substrate. The concentration of RNA enzyme was determined at various times, and the data were fit to the logistic equation: [enzyme] = a/(1 + be−ct), where a is the maximum extent of amplification and c is the exponential growth rate. Curvilinear regression coefficients were 0.995 for CL1 with S4 and 0.998 for DSL with S5. Inset shows behavior during the linear phase of growth.
The compensatory mutations alone were installed in the CL1 and DSL enzymes. For CL1, the matched pairing between the enzyme and substrate resulted in growth characteristics similar to those of the fully evolved enzyme, suggesting that the other 4 mutations in the evolved enzyme are near-neutral with regard to fitness. For DSL, in contrast, the compensatory mutation alone did not provide the full growth rate of the evolved enzyme. DSL is a younger enzyme compared with CL1, having been subject to many fewer rounds of in vitro evolution, and thus is more likely to acquire mutations that enhance its fitness within the context of continuous in vitro evolution.
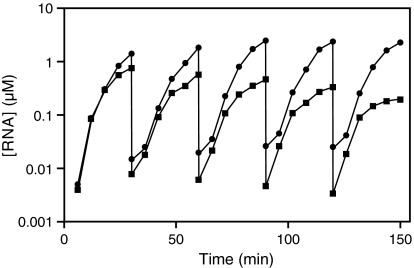
It is possible to balance the disparate growth rates of the evolved CL1 and DSL enzymes by adjusting the concentrations of their respective substrates (Fig. S3). The CL1 enzyme operating in the presence of 1 μM S4 has a nearly identical rate and maximum extent of growth compared with the DSL enzyme operating in the presence of 0.02 μM S5. Under these conditions, it is possible to carry out the sustained coevolution of CL1 and DSL within a common reaction mixture over the course of a serial transfer experiment (Fig. 4). In the absence of further evolutionary change, this behavior could be maintained indefinitely, demonstrating occupancy of distinct niches by the 2 different RNA enzymes.
Fig. 4.
Sustained coevolution of the CL1 (circles) and DSL (squares) enzymes. Five successive rounds of amplification and 100-fold dilution were performed over a period of 2.5 h. The concentration of each enzyme was determined based on incorporation of [α-32P]ATP into newly synthesized RNAs. The concentrations of S4 and S5 were 1 and 0.02 μM, respectively.
Kinetic and Transcriptional Analyses.
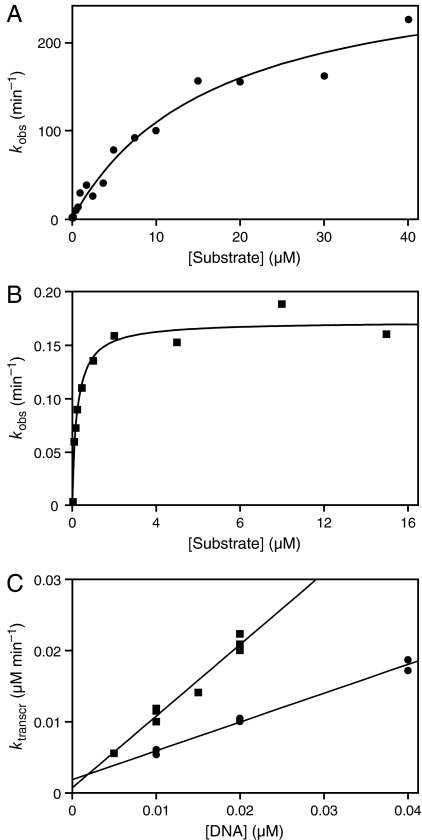
The catalytic activity of the evolved CL1 and DSL enzymes with their preferred substrates was examined under the conditions of continuous evolution (Fig. 5 A and B). CL1 demonstrated a kcat of 290 ± 30 min−1 and KM of 17 ± 4 μM. This catalytic rate is among the fastest ever measured for an RNA enzyme. In contrast, DSL exhibited a kcat of 0.17 ± 0.01 min−1 and KM of 0.24 ± 0.04 μM. Thus, the 2 enzymes exhibit markedly different kinetic parameters, with a ≈100-fold difference in observed rates in the presence of 1 μM of their respective substrates. Yet both enzymes are viable within the continuous evolution system.
Fig. 5.
Catalytic activity and transcription rate of the evolved CL1 (circles) and DSL (squares) enzymes. (A) Reaction of CL1 with S4. (B) Reaction of DSL with S5. Data were fit to the Michaelis–Menten equation; curvilinear regression coefficients were 0.984 and 0.981 for CL1 and DSL, respectively. (C) Transcription rates were measured by using various concentrations of RNA/cDNA heteroduplex templates. Linear regression coefficients were 0.994 and 0.983 for CL1 and DSL, respectively.
Interestingly, the slower DSL enzyme amplified more efficiently in the continuous evolution mixture, warranting investigation into other factors that might affect amplification rate. The rates of reverse transcription of the 2 ligated enzymes were measured and found to be 0.2 and 0.3 min−1 for CL1 and DSL, respectively. However, the rates of forward transcription (starting from cDNA) were more disparate, with DSL exhibiting a 2.2-fold faster rate of transcription over a range of cDNA concentrations equivalent to those present during continuous evolution. Synthetic DNA templates were prepared to validate the observed difference in transcription rates. The rate of transcription was linear over a range of DNA template concentrations, with a rate constant of 0.40 and 1.0 min−1 for the CL1 and DSL enzymes, respectively (Fig. 5C). This 2.5-fold difference is consistent with the measurements obtained starting from cDNAs that had been generated in the continuous evolution mixture.
Discussion
In nature, the competition for limited resources is a critical driving force for adaptation and evolutionary innovation, both among individuals of a given species and between species that occupy the same environment. Continuous in vitro evolution has proven to be a powerful method for evolving RNA molecules in the laboratory, enabling the selective amplification of individuals from a heterogeneous population over the course of hundreds of “generations” of sustained growth. This method had only been applied to 1 class of RNA enzymes at a time. In the present study, 2 unrelated catalytic species were made to undergo continuous evolution within a common environment, competing for limited amounts of substrate.
The competitive exclusion principle states that 2 species that compete for the exact same resource within the same environment cannot stably coexist (17). Such conflicts often are resolved in biological ecosystems by adaptation of the competing species to distinct ecological niches. In the context of continuous coevolution, when mixed populations of CL1 and DSL enzymes were challenged to compete for the same substrate, extinction of one species or the other could not be avoided. However, when the 2 enzymes were presented with 5 potential substrates, each enzyme adapted to use a different substrate, demonstrating what is termed “ecological character displacement” (18). Once differentiated in this way, the 2 enzymes were capable of sustained coevolution, which in principle could be continued indefinitely. Further evolutionary change might lead to extinction of one species if the other species usurped its substrate before an effective counterstrategy could be evolved. Barring such events, however, the current situation represents a stable strategy for coexistence.
The niche space (19) provided by the 5 substrates is well defined in molecular terms. These substrates differ by 1 or 2 nucleotides within the region of 8 nucleotides that is bound by the RNA enzyme (Table S1). The fitness landscape for each of the 2 coevolving enzymes reflects this niche space and potential interactions with the opposing enzyme. In the present study, those interactions were entirely competitive, but other types of interactions, such as commensalism or predation, might have emerged. The enzymes compete during the exponential growth phase for the finite supply of substrates, and as they reach the carrying capacity of the reaction mixture compete for other resources, such as the primer for cDNA synthesis and the NTPs required for transcription. These processes give rise to complex growth dynamics, which nonetheless could be described in precise molecular terms if one measured the detailed kinetic properties of each of the components of the continuous evolution system.
According to the r/K selection theory, a species will tend toward either of 2 general evolutionary outcomes when forced to adapt to selection pressures, where r refers to the reproduction rate and K refers to the carrying capacity of the local environment (20, 21). Although actual cases in biology usually are more complicated, r-selected species are exemplified by the rapid production of numerous offspring that are better suited to a fluctuating environment, whereas K-selected species have lower fecundity and more fully exploit the maximum carrying capacity of a stable environment. In broad terms, the evolved CL1 enzyme is a K-strategist that generates fewer copies per round of replication compared with DSL but more effectively utilizes the available substrate because of its very rapid rate of catalysis. Almost every CL1 molecule has reacted before it is reverse transcribed and, as a result, inactivated. The evolved DSL enzyme, in contrast, is an r-strategist that generates 2.5-fold more copies per round of replication compared with CL1 but, because of its slower catalytic rate, is more likely to be reverse transcribed. Both strategies are viable and at steady state result in a higher fraction of the CL1 molecules being in the reacted state compared with the DSL molecules.
The evolution of niche partitioning for a mixed population of 2 different catalytic RNAs broadens the scope of complex behaviors that have been realized in a molecular ecosystem. The disparate kinetic properties of the CL1 and DSL enzymes demonstrate that there is a wider range of adaptive solutions within the continuous evolution system than had previously been thought. The CL1 enzyme, with its extraordinary catalytic rate of 290 min−1, outruns reverse transcription (with an observed rate of 0.2 min−1), so that nearly every enzyme molecule is able to give rise to progeny. The DSL enzyme, with a catalytic rate of only 0.17 min−1, reacts slower than the observed rate of reverse transcription (0.3 min−1), but compensates for the reduced number of viable parents by the production of a higher number of progeny molecules per parent. Despite this enhanced transcription rate, the cognate substrate for the evolved DSL enzyme encodes a promoter that is 3-fold weaker than that encoded by the substrate for the evolved CL1 enzyme. This observation suggests that the enhanced transcriptional efficiency of the DSL enzyme derives from the sequence of the transcript itself, as has been observed for other RNA sequences (22, 23).
A longstanding goal of laboratory evolution has been to model complex population behaviors within a molecular system. Isothermal RNA-based amplification methods have been used to evolve noncatalytic molecules that mimic both predation and cooperation behaviors (24, 25). However, the absence of activity-based selection within such systems has led to parasitic side reactions, prohibiting long-term study of population dynamics. A prior study employing continuous in vitro evolution gave rise to RNA enzymes that were resistant to a toxicant, although unlike prey, the toxicant was not itself subject to evolution (8). The continuous coevolution of RNA enzymes provides a framework for the simultaneous evolution of multiple molecular species. This development, in conjunction with a better understanding of the biochemical parameters required for continuous evolution, will enable greater complexity to be achieved in future molecular ecology studies that employ diverse catalytic motifs.
Materials and Methods
Coevolution with 1 Substrate.
DNA templates for the starting pool were generated by performing error-prone PCR (26) on plasmid DNA encoding either a truncated form of the B16–19 CL1 RNA ligase (9) or the T100–1 form of the DSL ligase (16), and subsequently these PCR products were used as templates for in vitro transcription. The resulting RNAs were purified by denaturing polyacrylamide gel electrophoresis (PAGE). The truncated form of the B16–19 ligase had the sequence 5′-AGAAGAAUACAUAAUAGUGACCAGGAAAAGACAAAUCUGCCCUUAGAGCUUGAGAACAUCUUCGGAUGCACGGGAGGCAGCUCGCGAUGGAAGUGACGAACCAGCGUUCUCAACAGUAUUCACUGACUCCGUGCCAUCC-3′.
A starting population of 1 pmol of each ligase was challenged to ligate a chimeric DNA-RNA substrate (S0) having the sequence 5′-CTTGACGTCAGCCTGGACTAATACGACTCACUAUU-3′ (T7 promoter sequence in italics; RNA residues in bold). Continuous evolution was carried out as described in ref. 5, in a reaction mixture containing 5 μM substrate, 2.5 μM fluorescein-labeled cDNA primer having the sequence 5′-FAM-GGATGGCACGGAGTCAG-3′, 2 mM each NTP, 0.2 mM each dNTP, 25 mM MgCl2, 50 mM KCl, 4 mM DTT, 50 mM EPPS (pH 8.5), 10 U μL−1 SuperScript II reverse transcriptase (Invitrogen), 2.5 U μL−1 T7 RNA polymerase, and 0.001 U μL−1 inorganic pyrophosphatase, which was incubated at 37 °C for times decreasing from 30 to 12 min. At the end of each incubation, a small aliquot was transferred to the next reaction vessel, with the dilution increasing from 100-fold to 1,000-fold over the course of 40 transfers. Antisense oligos having the sequence 5′-CGAGCTGCCTCCCddG-3′ (complementary to CL1) and 5′-CTACCCATTGATCddC-3′ (complementary to DSL) were sometimes present within the reaction mixture: 5 μM anti-CL1 was present during reactions 6–10; 2 μM anti-DSL was present during reactions 16–20. The 3′-terminal dideoxynucleotide prevented these oligos from being extended by reverse transcriptase during continuous evolution. Error-prone PCR was performed after reactions 10, 20, and 30. The resulting mutagenized PCR products were transcribed, and the CL1 and DSL enzymes were purified by PAGE and then used to resume continuous coevolution.
Coevolution with 5 Substrates.
DNA templates for the starting pool were based on clonal isolates of the CL1 and DSL enzymes obtained after coevolution with a single substrate (Fig. S1), which were subject to error-prone PCR, followed by in vitro transcription and PAGE purification of the resulting RNA. One pmol of the CL1 and 10 pmol of the DSL enzymes were used to initiate coevolution in the presence of 1 μM each of 5 substrates having the sequence: S1, 5′-CTTGACGTCAGCCTGGACTAATACGACTCGCUAUU-3′; S2, 5′-CTTGACGTCAGCCTGGACTAATACGACTCACUGUU-3′; S3, 5′-CTTGACGTCAGCCTGGACTAATACGACTCACUACU-3′; S4, 5′-CTTGACGTCAGCCTGGACTAATACGACTCACUAUC-3′; and S5, 5′-CTTGACGTCAGCCTGGACTAATACGACTCACUUUU-3′ (T7 promoter sequence in italics; RNA residues in bold). All other reaction components were identical to those described above. Fifty successive transfers were carried out, with 20- to 60-min reaction times and 10- to 100-fold dilutions between transfers. The concentrations of S1–S4 were maintained at 1 μM each, whereas the concentration of S5 was varied as follows: 1 μM during reactions 1–7, 16–20, 31, and 32; 0.1 μM during reactions 8–10, 21–30, and 33–50; and none during reactions 11–15. Error-prone PCR was performed after reactions 5, 7, 10, 15, 20, 25, 30, 32, 37, 42, and 46, as described above. Reaction 11 was performed using stepwise rather than continuous in vitro coevolution, allowing 25 pmol each of CL1 and DSL to react for 30 min in the presence of 1 μM each of S1–S4.
Substrate utilization assays used 10 nM PAGE-purified CL1 or DSL RNA, isolated before initiation of continuous coevolution and at various times during the experiment. Each enzyme was tested separately for its ability to amplify in the presence of 1 μM S1, S2, S3, S4, or S5. The amount of cDNA generated after 30 min was determined, and the relative utilization of the various substrates was calculated, normalized to 100%.
Kinetic Analyses.
Ligation reactions catalyzed by the CL1 and DSL enzymes were performed under substrate-excess conditions, employing [α-32P]ATP-labeled enzyme and various concentrations of substrate that were in at least 10-fold excess over the concentration of enzyme. The reaction conditions were otherwise identical to those used in continuous evolution, except the protein enzymes were omitted. Reaction products were separated by PAGE and quantitated using a PharosFX Molecular Imager (BioRad). For DSL, the values for kobs were determined for each concentration of substrate based on at least 6 data points, which were fit to the equation: y = a (1 − e−kobst), where y is the fraction reacted and a is the maximum extent as determined from long time points. CL1 exhibited biphasic kinetics at all substrate concentrations, and for each concentration at least 8 data points were obtained and fit to the equation: y = a − be−k1t − ce−k2t, where b and k1 are the amplitude and rate of the initial fast phase and c and k2 are the amplitude and rate of the slow phase. The amplitude of the fast phase was typically ≈50% of the overall maximum extent. The rate k2 did not demonstrate saturation behavior under the substrate concentrations tested. Values for kcat and KM were obtained from a Michaelis-Menten plot of k1 or kobs versus substrate concentration for CL1 and DSL, respectively.
Measurement of the fast rate of the CL1 enzyme required use of a quench-flow apparatus (KinTek, model RQF-3) to achieve reaction times as short as 100 ms. Separate syringes of the apparatus were used to deliver 15 μL each of an enzyme and substrate solution, which both contained all of the other reaction components. A third syringe was used to deliver 87 μL of a quench solution containing 40 mM Na2EDTA after a precisely specified interval. The drive solution used to propel the other solutions through the reaction loop contained 25 mM MgCl2, 50 mM KCl, 4 mM DTT, and 50 mM EPPS (pH 8.5). For all concentrations of S4 <30 μM, each time point was collected and analyzed in duplicate.
The observed rates of reverse transcription were measured under continuous evolution conditions, excluding the substrate and T7 RNA polymerase, and employing 1 μM of ligated product resulting from the reaction of CL1 with S4 or the reaction of DSL with S5. After incubation for 0.75, 1, 1.5, or 2 min, the reaction products were separated by PAGE and the fraction of extended primer was determined. Linear regression coefficients were 0.997 and 0.998 for CL1 and DSL, respectively.
The products of reverse transcription were diluted into a continuous evolution mixture, but lacking reverse transcriptase and cDNA primer, to achieve a final template concentration of 0.01, 0.02, or 0.04 μM. Transcription rates were determined by measuring the incorporation of [α-32P]ATP into newly synthesized RNA. Similar experiments were performed using synthetic templates, which were assembled from equimolar amounts of PAGE-purified ligated RNAs and full-length cDNAs. The 2 strands were heated to 90 °C for 2 min in the presence of 50 mM KCl at pH 8.5, cooled to 50 °C over a period of 5 min, held at 50 °C for 5 min, and then cooled to room temperature over a period of 2 min, at which point MgCl2 was added to achieve a final concentration of 25 mM. Promoter strengths were measured by using templates that were generated in the context of continuous evolution and diluted to 0.01 μM concentration.
Supplementary Material
Acknowledgments.
This work was supported by National Aeronautics and Space Administration Grant NNX07AJ23G, National Science Foundation Grant MCB-0614614, and The Skaggs Institute for Chemical Biology.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/cgi/content/full/0903397106/DCSupplemental.
References
- 1.Bennett AF, Dao KM, Lenski RE. Rapid evolution in response to high-temperature selection. Nature. 1990;346:79–81. doi: 10.1038/346079a0. [DOI] [PubMed] [Google Scholar]
- 2.Elena SF, Cooper VS, Lenski RE. Punctuated evolution caused by selection of rare beneficial mutations. Science. 1996;272:1802–1804. doi: 10.1126/science.272.5269.1802. [DOI] [PubMed] [Google Scholar]
- 3.Hillis DM, Bull JJ, White ME, Badgett MR, Molineux IJ. Experimental phylogenetics: Generation of a known phylogeny. Science. 1992;255:589–592. doi: 10.1126/science.1736360. [DOI] [PubMed] [Google Scholar]
- 4.Johns GC, Joyce GF. The promise and peril of continuous in vitro evolution. J Mol Evol. 2005;61:253–263. doi: 10.1007/s00239-004-0307-1. [DOI] [PubMed] [Google Scholar]
- 5.Wright MC, Joyce GF. Continuous in vitro evolution of catalytic function. Science. 1997;276:614–617. doi: 10.1126/science.276.5312.614. [DOI] [PubMed] [Google Scholar]
- 6.Bartel DP, Szostak JW. Isolation of new ribozymes from a large pool of random sequences. Science. 1993;261:1411–1418. doi: 10.1126/science.7690155. [DOI] [PubMed] [Google Scholar]
- 7.Ekland EH, Szostak JW, Bartel DP. Structurally complex and highly active RNA ligases derived from random RNA sequences. Science. 1995;269:364–370. doi: 10.1126/science.7618102. [DOI] [PubMed] [Google Scholar]
- 8.Ordoukhanian P, Joyce GF. A molecular description of the evolution of resistance. Chem Biol. 1999;6:881–889. doi: 10.1016/s1074-5521(00)80007-1. [DOI] [PubMed] [Google Scholar]
- 9.Schmitt T, Lehman N. Non-unity molecular heritability demonstrated by continuous evolution in vitro. Chem Biol. 1999;6:857–869. doi: 10.1016/s1074-5521(00)80005-8. [DOI] [PubMed] [Google Scholar]
- 10.Paegel BM, Joyce GF. Darwinian evolution on a chip. PLoS Biol. 2008;6:900–906. doi: 10.1371/journal.pbio.0060085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kuhne H, Joyce GF. Continuous in vitro evolution of ribozymes that operate under conditions of extreme pH. J Mol Evol. 2003;57:292–298. doi: 10.1007/s00239-003-2480-z. [DOI] [PubMed] [Google Scholar]
- 12.Lehman N. Assessing the likelihood of recurrence during RNA evolution in vitro. Artif Life. 2004;10:1–22. doi: 10.1162/106454604322875887. [DOI] [PubMed] [Google Scholar]
- 13.Soll SJ, Díaz Arenas C, Lehman N. Accumulation of deleterious mutations in small abiotic populations of RNA. Genetics. 2007;175:267–275. doi: 10.1534/genetics.106.066142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ikawa Y, Tsuda K, Matsumura S, Inoue T. De novo synthesis and development of an RNA enzyme. Proc Natl Acad Sci USA. 2004;101:13750–13755. doi: 10.1073/pnas.0405886101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Horie S, Ikawa Y, Inoue T. Structural and biochemical characterization of DSL ribozyme. Biochem Biophys Res Commun. 2006;339:115–121. doi: 10.1016/j.bbrc.2005.11.007. [DOI] [PubMed] [Google Scholar]
- 16.Voytek SB, Joyce GF. Emergence of a fast-reacting ribozyme that is capable of undergoing continuous evolution. Proc Natl Acad Sci USA. 2007;104:15288–15293. doi: 10.1073/pnas.0707490104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hardin G. The competitive exclusion principle. Science. 1960;131:1292–1297. doi: 10.1126/science.131.3409.1292. [DOI] [PubMed] [Google Scholar]
- 18.Brown WL, Wilson EO. Character displacement. Syst Zool. 1956;5:49–64. [Google Scholar]
- 19.Hutchinson GE. Concluding remarks. Cold Spring Harbor Symp Quant Biol. 1957;22:415–427. [Google Scholar]
- 20.MacArthur RH, Wilson EO. Princeton, NJ: Princeton Univ Press; 1967. The Theory of Island Biogeography; pp. 145–180. [Google Scholar]
- 21.Pianka ER. On r-selection and K-selection. Am Nat. 1970;104:592–597. [Google Scholar]
- 22.Milligan JF, Groebe DR, Witherell GW, Uhlenbeck OC. Oligoribonucleotide synthesis using T7 RNA polymerase and synthetic DNA templates. Nucleic Acids Res. 1987;15:8783–8798. doi: 10.1093/nar/15.21.8783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Martin CT, Muller DK, Coleman JE. Processivity in early stages of transcription by T7 RNA polymerase. Biochemistry. 1988;27:3966–3974. doi: 10.1021/bi00411a012. [DOI] [PubMed] [Google Scholar]
- 24.Wlotzka B, McCaskill JS. A molecular predator and its prey: Coupled isothermal amplification of nucleic acids. Chem Biol. 1997;4:25–33. doi: 10.1016/s1074-5521(97)90234-9. [DOI] [PubMed] [Google Scholar]
- 25.Ellinger T, Ehricht R, McCaskill JS. In vitro evolution of molecular cooperation in CATCH, a cooperatively coupled amplification system. Chem Biol. 1998;5:729–741. doi: 10.1016/s1074-5521(98)90665-2. [DOI] [PubMed] [Google Scholar]
- 26.Cadwell RC, Joyce GF. Randomization of genes by PCR mutagenesis. PCR Meth Appl. 1992;2:28–33. doi: 10.1101/gr.2.1.28. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.