Abstract
By using a whole-genome oligonucleotide microarray, designed based on known and predicted indica rice genes, we investigated transcriptome profiles in developing leaves and panicles of superhybrid rice LYP9 and its parental cultivars 93-11 and PA64s. We detected 22,266 expressed genes out of 36,926 total genes set collectively from 7 tissues, including leaves at seedling and tillering stages, flag leaves at booting, heading, flowering, and filling stages, and panicles at filling stage. Clustering results showed that the F1 hybrid's expression profiles resembled those of its parental lines more than that which lies between the 2 parental lines. Out of the total gene set, 7,078 genes are shared by all sampled tissues and 3,926 genes (10.6% of the total gene set) are differentially expressed genes (DG). As we divided DG into those between the parents (DGPP) and between the hybrid and its parents (DGHP), the comparative results showed that genes in the categories of energy metabolism and transport are enriched in DGHP rather than in DGPP. In addition, we correlated the concurrence of DG and yield-related quantitative trait loci, providing a potential group of heterosis-related genes.
Keywords: heterosis, hybrid rice, transcriptome, quantitative trait loci, differentially expressed genes
Extensive sequence diversity at the microstructural level has been demonstrated in a number of plant species (1), and such diversity can extend even to allelic regions (2). These intraspecific allelic variations should have impacts on gene expressions that lead to phenotypic variation, perhaps including hybrid vigor as a beneficial trait used in crop breeding. In a hybrid, in which 2 different alleles of a gene are often brought together, the combined allelic expression may deviate from that of either parent or the midparent predictions (3). In maize, both allelic diversity and expression variation were found between inbred parents and their hybrid (4). In maize hybrids, not only the allelic variation in gene expression but also different responses to extrinsic stimuli supported the presence of allelic expression variation in the same genetic context (5). Large-scale transcriptome profiling has been used for heterosis studies in maize (6), Arabidopsis (7), and wheat (8). In rice, an investigation of a yield-related quantitative trait locus (QTL) resulted in a discovery of allelic variation that affected the expression of a leucine-rich repeat receptor kinase gene cluster (9). Another survey with a cDNA microarray concerning 9,188 expressed sequence tags on expression polymorphism between an elite rice hybrid and its parental varieties revealed significant heterotic expression for 141 expressed sequences (10).
We have recently focused our heterosis research on Liang-You-Pei-Jiu (LYP9), a superhybrid rice strain from a cross between the maternal inbred PA64s, a photothermosensitive male sterile line, and the paternal inbred 93-11, an elite indica variety, after we sequenced the 2 parental genomes (11, 12). Two-dimensional electrophoresis analysis among 93-11, PA64s, and LYP9 revealed significant numbers of different embryo protein spots, many of which were shown to display mirrored relationships between parents and the first filial generations (13). Further analysis on mature embryos of this hybrid triad identified 54 differentially expressed proteins involved in major biological processes including nutrient reservoir, response to stress, and metabolism. Among these embryos, most of the storage proteins exhibit overdominance and stress-induced proteins display additivity (14). We also carried out transcriptome profiling for the hybrid and its parents using both sequencing-based (15–17) and hybridization-based methods (18). We now report a rather large-scale comparative transcriptome analysis of the triad, concerning 7 tissues sampled across developmental times and different tissues. We expect this genome-wide transcriptome comparison to be an initial step forward in understanding the causative mechanism of the altered gene expression in the hybrid and the molecular mechanism underlying heterosis.
Results
The Rice Whole-Genome Microarrays Are of Satisfactory Quality.
Our 70-mer oligonucleotide microarray, with 36,926 unique features identified, was designed based on known and predicted gene models of the indica rice 93-11 genome (18). We calibrated our microarray by doing 4 preliminary tests. First, a self-hybridization experiment was conducted, detecting only 9 false differentially expressed genes (DG) with marginal intensity above the background. Second, we conducted hybridizations between the seedling shoot and the filling panicle and discovered >5,000 DG with correlation coefficients of 0.85 between duplication and correlation coefficients of 0.81 in dye-swapping experiments. Third, to better define the background and fold changes we introduced a polyubiquitin gene as positive control, the fold changes of which are both consistent and always below the threshold (Fig. S1). We acquired at least 3 independent replicates for each sample pair in general and a total of 48 raw datasets (96 slides) for 7 tissues from the triad (collective correlation coefficient among all replicates >0.8). Finally, we validated our microarray results with semiquantitative RT-PCR, and out of 25 primer pairs with amplification products, 20 (80%) DG showed consistent results compared with those obtained from the microarray data (Fig. S2). Collectively, these results demonstrated the satisfactory quality of our experimental procedures and data.
Transcriptome Profiles of LYP9 and Its Parents Revealed Consistent Trends with Phenotypic Observations.
Our data were derived from 7 tissues of the LYP9 hybrid triads, including seedling shoot, leaf at tillering stage, flag leaf at booting stage, flag leaf at heading stage, flag leaf at flowering stage, flag leaf at filling stage, and panicle at filling stage, out of which we identified 11,448–14,592 genes expressed in each pairwise comparison (Table S1). Our analysis revealed 7,078 genes expressed in all studies tissues and 22,266 genes expressed collectively.
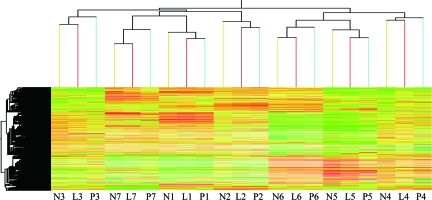
We used a cluster analysis method to investigate correlations among transcriptome profiles. The results revealed that tissues from different cultivars at the same developmental stage always formed the primary groups (Fig. 1). In a broader spectrum, the transcriptome profiles of LYP9 are similar to PA64s (maternal) at the early developmental stages but closer to 93-11 (paternal) at the later stages. Both are consistent with the morphological appearances or characteristics of the hybrid plant at corresponding stages, observed empirically in the field as either 93-11-like or PA64s-like. A distinct result was found in the cluster of the panicle at filling stage, where the profile of LYP9 is more similar to that of 93-11 because PA64s is a photothermosensitive male sterile rice line (19), and many of its genes may not express appropriately or at levels comparable to those of 93-11 and LYP9.
Fig. 1.
Hierarchical clustering analysis of all gene models based on expression data. Normalized expression values for the microarray (37K) clustered with Genespring (Silicon Genetics). Each horizontal line refers to a gene. The color represents the logarithmic intensity of the expressed genes. N, L, and P stand for 93-11, LYP9, and PA64s, respectively. Numbers 1–7 denote samples from the following tissues in order: seedling shoot, leaf at tillering stage, flag leaf at booting stage, flag leaf at heading stage, flag leaf at flowering stage, flag leaf at filling stage, and panicle at filling stage.
When we looked at universally expressed genes, some are undoubtedly housekeeping genes whereas the molecular category of structure was found overrepresented (Fig. S3). We also noticed that among structure molecules, genes encoding cytoplasmic (60S/40S) protein and plastid ribosomal (50S/30S) protein have a synergistic expression profile except in the filling stage panicle where the former are up-regulated and the latter are down-regulated (Fig. S4) as compared with those in other tissues. This result is consistent with the fact that the number of chloroplasts in panicles is significantly lower than that found in leaf tissues.
DG and Their Functional Analysis.
We defined DG between the parental lines as DGPP and those between the hybrid and its parents as DGHP. DGPP only denote the differences between 2 inbred lines, but DGHP may underlie heterosis because differences in expression between hybrid and parents should underlie their phenotypic differences. DGHP can be divided into 2 classes—i.e., those shared by DGPP and DGHP (DGO) and those uniquely belonging to DGHP (DGHPU). We found 3,926 (10.6%) DG observed at least once among all sample pairs (Dataset S1), and the numbers of DGHPU are larger than DGO in all 7 tissues investigated (Table 1). By comparing DG between the hybrid and its parents, we found that the great majority of DG are close to either their maternal or their paternal parent and that a minority of them are close to neither parent. To further understand the function of DG, we classified these genes according to their functional categories and relatedness. For instance, DGPP are enriched in 16 out of 161 categories as compared with DGHP, which are enriched in 25 function categories (Table S2 and S3). Since DGHP are composed of DGO and DGHPU, we expected that heterosis-related genes may be enriched in DGHPU rather than in DGO. Indeed, the DGHPU identified in this study are enriched mostly in the categories of energy metabolism and transport (Table 2).
Table 1.
Number and classification of DG
| Sample | DGPP | DGHP |
||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| L/N | L/P | DGHPU | DGO | H2P | CHP | B2P | CLP | L2P | ||
| S1 | 305 | 243 | 167 | 215 | 161 | 19 | 190 | 21 | 142 | 4 |
| S2 | 312 | 309 | 266 | 328 | 201 | 17 | 247 | 46 | 208 | 11 |
| S3 | 472 | 323 | 412 | 424 | 272 | 14 | 465 | 42 | 174 | 1 |
| S4 | 389 | 345 | 313 | 447 | 180 | 36 | 324 | 17 | 235 | 15 |
| S5 | 342 | 337 | 333 | 401 | 208 | 57 | 315 | 40 | 182 | 15 |
| S6 | 331 | 313 | 323 | 347 | 203 | 36 | 222 | 53 | 199 | 40 |
| S7 | 383 | 405 | 451 | 505 | 289 | 24 | 321 | 11 | 400 | 38 |
| Total | 2132 | 1913 | 1898 | 2260 | 1280 | 196 | 1851 | 198 | 1316 | 108 |
N, L, and P refer to 93-11, LYP9, and PA64s, respectively. DGPP refers to DG between both parents, DGHP refers to DG between the hybrid and parent. DGHPU denotes the unique portion of DGHP, and DGO denotes the overlap between DGPP and DGHP. H2P, CHP, B2P, CLP, and L2P represent higher than both parents, close to higher parent, between both parents, close to lower parent, and lower than both parents, respectively.
Table 2.
Functional classification of unique portion of DGHPU
| Functional categories | S1 | S2 | S3 | S4 | S5 | S6 | S7 |
|---|---|---|---|---|---|---|---|
| Metabolism | |||||||
| Amino acid metabolism | 7 | 14* | 14 | 10 | 16 | 19** | 29** |
| Biosynthesis of polyketides and nonribosomal peptides | 0 | 1 | 0 | 1 | 0 | 1 | 1 |
| Biosynthesis of secondary metabolites | 7 | 15 | 23 | 15 | 19 | 19 | 20 |
| Carbohydrate metabolism | 19 | 28 | 39 | 37 | 47** | 37* | 40 |
| Energy metabolism | 9** | 14** | 17** | 11 | 11 | 29** | 39** |
| Glycan biosynthesis and metabolism | 0 | 0 | 6 | 3 | 3 | 6 | 9* |
| Lipid metabolism | 5 | 7 | 8 | 7 | 8 | 12* | 6 |
| Metabolism of cofactors and vitamins | 10 | 19 | 21 | 14 | 24 | 17 | 16 |
| Metabolism of other amino acids | 4 | 8* | 4 | 4 | 8* | 3 | 7 |
| Nucleotide metabolism | 6 | 5 | 5 | 8 | 8 | 2 | 10 |
| Xenobiotics biodegradation and metabolism | 9 | 19 | 24 | 20 | 21 | 19 | 12 |
| Genetic Information Processing | |||||||
| DNA metabolism | 6 | 1 | 3 | 3 | 9 | 2 | 0 |
| RNA metabolism | 4 | 15 | 26 | 26 | 26 | 13 | 24 |
| Cellular protein metabolism | 17 | 34 | 61** | 50 | 44 | 22 | 41 |
| Environmental Information Processing | |||||||
| Signal transduction | 1 | 6 | 12 | 17* | 17** | 5 | 3 |
| Transport | 11 | 27 | 43** | 39* | 42** | 33** | 38 |
| Cellular Processes | |||||||
| Cell motility | 0 | 1 | 1 | 1 | 1 | 0 | 0 |
| Cell cycle | 2 | 6 | 4 | 2 | 4 | 2 | 7 |
| Cell–cell signaling | 0 | 0 | 2 | 1 | 1 | 1 | 1 |
| Cell death | 5 | 4 | 1 | 2 | 5 | 5 | 3 |
| Cell growth | 0 | 1 | 1 | 1 | 0 | 0 | 0 |
| Other | 28* | 32 | 59** | 59** | 52** | 50** | 75** |
| Unknown | 123 | 181 | 193 | 229 | 155 | 152 | 249 |
| Total | 215 | 328 | 424 | 447 | 401 | 347 | 505 |
* and ** denote significant enrichment of DG among function category with P < 0.05 and P < 0.01, respectively.
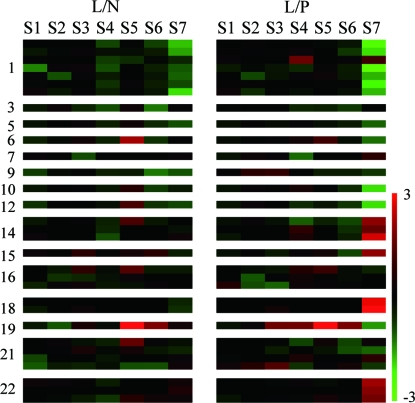
Since the most important trait of hybrid rice is grain yield, we analyzed the genes involved in carbohydrate biosynthesis (20, 21)—such as starch biosynthesis—and noticed that genes involved in starch synthesis have much higher expression in the panicle of LYP9 than PA64s at filling stage, including the key enzymes in starch biosynthesis such as sucrose synthase, ADP-glucose pyrophosphorylase, and starch synthase. The result is in agreement with the fact that starch biosynthesis cannot take place in the panicles of PA64s. In addition, rubisco, a key protein in the pathway, showed a lower expression level in LYP9 than in PA64s, thus supporting the fact that the panicle of PA64s remained green long after flowering was observed in the field. It is interesting that the genes taking part in sucrose and starch metabolism, such as ADP-glucose pyrophosphorylase, sucrose-P synthase, invertase, and branching enzyme, tend to be highly expressed in the hybrid (Fig. 2).
Fig. 2.
Expression profiles of DG between LYP9 and its parents in carbohydrate biosynthesis pathway. Genes involved in carbohydrate metabolism were identified according to their Enzyme Commission annotation, and those genes that differentially expressed at least once were shown. The log2-transformed ratio between the hybrid and either parent was used (L, LYP9; N, 9311; P, PA64s). Each row represents a single gene, and the number indicates a group of isoenzymes in the pathway according to its position in the path and order. Red and green colors denote up- and down-regulated genes, respectively. The genes are listed as follows: (1) ribulose-bisphosphate carboxylase, (3) glyceraldehyde-3-phosphate dehydrogenase, (5) fructose-bisphosphate aldolase, (6) fructose-bisphosphatase, (7) glucose-6-phosphate isomerase, (9) transketolase, (10) sedoheptulose-1,7-bisphosphatase (SBPase), (12) phosphoribulokinase, (14) ADP-glucose pyrophosphorylase, (15) UDP-glucose pyrophosphorylase, (16) sucrose-P synthase, (18) sucrose synthase, (19) invertase, (21) starch synthase, and (22) branching enzyme.
Nonadditive-Expressed Genes.
Concerning the relative level of gene expression among a hybrid–parent triad, we often expect 2 scenarios to come into play. In the first scenario, gene expression in the hybrid exhibits a cumulative mode, contributed by each allele from the respective parents. In the other scenario, the expression deviates from the midparental level. The former scenario is additive, indicating that alleles from both parents may contribute to gene expression in the hybrid, attributable mostly to a cis-regulation mechanism. The latter scenario is nonadditive, in which other regulators probably contribute to an altered expression of the corresponding alleles in the hybrid, attributable mostly to trans-regulation (3). In comparison with gene expression among the LYP9 triad, we detected 860 up-regulated and 1,095 down-regulated nonadditive genes (NAG). The number of NAG in each sampling triad ranged from 195 to 497 (Table 3); they composed 0.5–1.4% of the total gene set and 29.6–53.7% of DGHP identified at 7 tissues.
Table 3.
Nonadditive-expressed genes in LYP9
| Sample | Number of NAG |
Number of NAG in DGHP |
||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Up | Down | Total | a% | DGHP | b% | DGHPU | c% | DGO | d% | |
| S1 | 80 | 115 | 195 | 0.5 | 144 | 38.3 | 108 | 50.2 | 36 | 22.4 |
| S2 | 97 | 180 | 277 | 0.8 | 184 | 34.8 | 148 | 45.1 | 36 | 17.9 |
| S3 | 163 | 147 | 310 | 0.8 | 206 | 29.6 | 168 | 39.6 | 38 | 14.0 |
| S4 | 182 | 220 | 402 | 1.1 | 261 | 41.6 | 222 | 49.7 | 39 | 21.7 |
| S5 | 140 | 126 | 266 | 0.7 | 239 | 39.2 | 209 | 52.1 | 30 | 14.4 |
| S6 | 103 | 177 | 280 | 0.8 | 218 | 39.6 | 189 | 55.4 | 29 | 14.3 |
| S7 | 158 | 339 | 497 | 1.4 | 426 | 53.7 | 264 | 52.3 | 162 | 56.1 |
| Total | 860 | 1095 | 1846 | 5.0 | 1481 | 46.5 | 1245 | 55.1 | 488 | 38.1 |
a% denotes the percentage of NAG in the total gene set (36,926), b%, c%, and d% denote the percentage of NAG in total numbers of DGHP, DGHPU and DGO, respectively.
DGHP Are Enriched in Known QTLs.
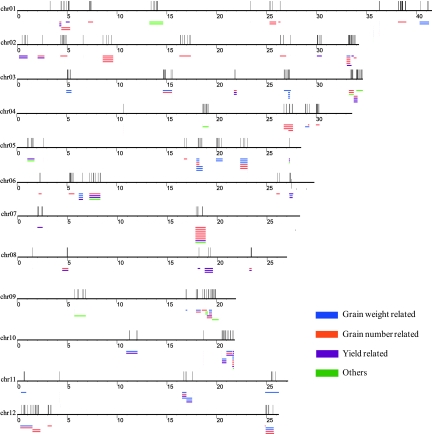
We were able to map 2,673 DGHP to 3,128 QTLs classified into 9 categories and 209 traits in the rice genome (www.gramene.org). One important piece of evidence supporting the correlation between the 2 types of data is the fact that the fraction of DGHP in the transcriptome profiles (36,926 expressed genes) is 8.6% as compared with the fractions of DGHP mapped to QTLs—10.1% and 11.8% in the QTL intervals that harbor less than 50 and 10 genes, respectively (Table S4). Among DGHP-related QTLs, many are well characterized, including 1000-seed weigh (e.g., AQCY015, CQAS23, AQAI076, and CQAS23), filled grain number (e.g., AQCY010, AQCY059, AQAK009, and AQAK011), grain number (CQB22, AQDR015, AQDR059, and AQED038), and grain yield per panicle (AQDR091, AQDR103, and AQDR104). The potential association between DGHP and QTLs were also suggested within many QTL regions, such as Starch synthase III (Os055024_01) to AQCY010 for filled grain number, putative sugar transporter (Os055048_01) to AQAI076 and AQEY022 for 1000-seed weight, and auxin response factor (Os016758_01) to CQK15 for panicle number. To help portray this DGHP-QTL correlation, we aligned DGHP over yield-related QTL regions covering less than 100 genes on rice chromosomes (Fig. 3).
Fig. 3.
Distribution of DG located in yield-category QTL of small intervals. Yield-category QTL of small intervals (number of genes ≤100) that harbor DGHP were aligned to TIGR's rice pseudochromosome version 5. The long horizontal lines represent 12 rice chromosomes, the short horizontal lines represent QTL intervals, and the short vertical lines represent DGHP.
Discussion
Complex Regulatory Mechanisms Probably Underlie Gene Expression Changes in Hybrid.
Transcriptomes are not only always specific to cell types but also are regulated at different levels, such as transcription and splicing, and through genetic or epigenetic mechanisms. Although in this current report we are unable to show detailed sequence comparisons and validations for different alleles of annotated DG, allelic sequence variation—especially those in the regulatory sequence/element—is undoubtedly one of the causes of gene-expression change in hybrids. We will certainly proceed in identifying these allelic differences of all DG in our dataset. Another class of gene regulators is trans-regulators, such as transcription factors (TFs). The dosage effect of such regulatory genes had been proposed to affect phenotypes in hybrids (22). We indeed found that 187 TFs exhibited differential expression in the hybrid compared with either parent. It is quite a coincidence that a recent study using seedling tissue of 2 hybrid triads, based on the genomic sequence of 93-11 and nipponbare, also suggested that altered gene expression caused by interactions between transcription factors and the allelic promoter region in the hybrids was one plausible mechanism underlying heterosis in rice (23).
Moreover, we noticed that among those differentially expressed TFs, the AP2-EREBP family—potential targets of miRNA (24)—is overrepresented. Noncoding RNAs are involved in epigenetic regulations, and other epigenetic mechanisms including DNA methylation, acetylation and deacetylation of histones, and chromatin remodeling. It had been reported that the degree of methylation in hybrids is different from that in inbred lines in Arabidopsis and rice (25, 26). A recent study reported that epigenetic regulation of a few regulatory genes (CCA1 and LHY in this case) induced cascade changes both in downstream genes (TOC1, GI, etc.) and in physiological pathways, and ultimately induced growth and development, which also indicates the presence of a general mechanism for the growth vigor and increased biomass commonly observed in hybrids (27). In the present survey, we also found that among DGHP there were many epigenesis-related genes, including methyltransferase, hydroxymethyltransferase, serine O-acetyltransferase, histone acetyltransferase, acetyl-CoA acyltransferases, and chromodomain helicase-DNA-binding protein 3. The expression of these genes is being verified experimentally as is their involvement in related biological pathways.
Gene Expression Variations in Hybrid Suggests Correlation to Genetic Mechanisms Responsible for Heterosis.
The dominance and overdominance hypotheses (28) were proposed to explain heterosis before the molecular concepts of genetics were formulated, and these hypotheses are not closely allied with molecular principles. We categorized DG between hybrid and parents (DGHP) into 5 basic categories: overdominance (H2P), underdominance (L2P), dominance (CHP and CLP), and midparent (B2P). We found that dominant expression was the most prevalent among DGHP (81.6–91.8%). Additive and nonadditive expression represent another possible genetic model for gene expression in hybrids (3). Whether or not a transcript shows nonadditive expression is most likely to be influenced by the contributions of cis- and trans-acting factors of this gene (29–32). In our data, the majority of genes in the hybrid showed additive expression, and the phenomenon suggests that cis-acting elements usually play a major role in the control of general gene expression. Nonadditively expressed genes in our entire dataset constituted only 0.5–1.4% of the total discovered in each sampled tissue but accounted for 29.6–53.7% of DGHP. A similar result was observed in a study of maize heterosis, in which the nonadditive-expressed genes were found to contain 2.2% of the total genes and 22% of DG (6). It should be noted that we were unable to detect those genes where the silencing of 1 allele was compensated by overexpression of the other, which might cause underestimation of nonadditive genes in hybrids, as mentioned previously (31). The analysis of nonadditive gene expression indicates that allelic expression in hybrids may not just be a combination of alleles from the 2 parents but is rather regulated by other genes or epistatic mechanisms. Nonadditive gene expression was also considered as midparent heterosis or heterotic expression (10, 23).
A study in gene expression in maize endosperm revealed heterochronic expression of 3 allele pairs (33). In the present study, 85% of DG were detected only once in 7 tested tissues. For those DG that appeared more than once, 63% (LYP9/PA64s) to 75% (LYP9/93-11) differed in the same direction; i.e., either up- or down-regulated. This trend indicates that their corresponding regulatory mechanisms may function in the same way in different tissues and under different conditions. In contrast, 25–39% of those genes follow a different trend; they differ in the opposite direction, so that a gene in the hybrid may be under a different control mechanism or the regulatory factors may function in a different way under variable conditions.
DG Are Candidates for Genes That Play an Important Role in Heterosis.
Microarray-based expression studies allowed us to identify genes that are differentially expressed between a hybrid and its parents, and these DG are often found to be expressed in a biased pattern in comparison with regular transcriptomes. For example, we found that the DGHP involved in the carbohydrate–metabolism pathway had a larger fraction of up-regulated genes than down-regulated genes, similar to the recent studies (23, 27). However, of the genes taking part in oxidative phosphorylation, there were more down-regulated genes identified in the hybrid than in the parental lines. In addition, heading stage is an important period for panicle development and grain-yield formation, and our previous serial analysis of gene expression (SAGE) analysis showed that genes related to protein biosynthesis and peptide transport were up-regulated in the panicle of the hybrid LYP9 (16). Based on our current data, a similar conclusion was reached in the analysis of gene expression in flag leaves of heading stage and flowering stage. It was interesting to find that sucrose-transport genes are up-regulated in LYP9 panicles as compared with those in 93-11 panicles, suggesting that the transportation of carbohydrate from the source to the sink in LYP9 is more efficient than in at least one of the parents.
An altered expression of the maize domestic gene tb1 was characterized as the cause of observed quantitative phenotypic changes by a fine-mapping approach (34), and a transcription activator was demonstrated to be responsible for the significant plant-height changes in an Arabidopsis hybrid (35). Recently, a major quantitative gene in rice, Ghd7, isolated by map-base cloning and encoding a CCT domain protein, was considered as a crucial factor for increasing productivity and adaptability of an elite hybrid cultivar, Shanyou 63, and some other indica varieties (36). In our current study, not only have we found many TFs in our DG collections, but we also mapped a high fraction of DG to the intervals of grain-yield-related QTLs. These results led us to believe that DG between the hybrid and parents may contribute in a significant way to heterosis. We also have constructed databases integrating heterosis-related genes among major crops and experimental plants, identifying altered sequences among differentially expressed alleles (37), and mapping relative DG to QTLs discovered in this study.
Materials and Methods
Rice Whole-Genome Oligonucleotide Array.
The whole-genome array was developed based on annotated and predicted genes from the genome assembly of indica rice 93-11 (11, 12). Oligonucleotides were arrayed onto 2 poly-L-lysine-coated microscope slides as a set with a SpotArray72 microarrayer (Perkin–Elmer) in the microarray laboratory at Beijing Genomic Institute, and the slides were processed according to a standard procedure (38).
Plant Materials and Data Processing.
LYP9 and its parental lines (93-11 and PA64s) were grown in a greenhouse for the seedling samples and in the rice field for all other samples. The plant tissues were collected and stored at −80 °C. RNA samples were isolated (39), quantitated by using a NanoDrop1000 spectrophotometer (Nanodrop Technologies), and labeled (40, 41). Each sample had at least 3 biological replications to minimize systematic errors. Separate tiff images of Cy5 and Cy3 channels were obtained by ScanArray Lite scanner (Perkin–Elmer), and spot intensities were quantified by using the Axon GenePix Pro 5.1 image analysis software.
We categorized our raw data with 3 simple criteria. First, features were flagged as “bad” either by using Genepix or by manual investigation, second, a false positive rate ≤5% in reference to the controls was found, and third, legitimate features were found in at least 2 of the 3 replicate sets or 3 of the 4 replicate sets. The processed data were normalized based on the mean of all expressed genes. The normalization of the 2-channel data for each array was done by using the intensity-based Loess method with R language. DG were defined by a log-scale ratio between paired samples with a P value <0.01 (Z test).
Functional Annotation.
For each gene identified, we performed detailed functional annotations by using standard tools, such as BLAST (42, 43) and HMMer (44), against public data, including (i) the The Institute for Genomic Research (TIGR) Rice Pseudomolecules and Genome Annotation database (release 5.0, http://rice.plantbiology.msu.edu); (ii) the knowledge-based Oryza Molecular Biological Encyclopedia (http://cdna01.dna.affrc.go.jp/cDNA/); (iii) the TIGR Rice Gene Index (http://compbio.dfci.harvard.edu/tgi/); and (iv) the UniProtKB/Swiss-Prot (www.ebi.uniprot.org). We also used the Kyoto Encyclopedia of Genes and Genomes (http://www.genome.jp/kegg/) and Gene Ontology databases (45) for protein annotation (E value <10−7). The HMMpfam program (http://hmmer.janelia.org/) was used to search Pfam hidden Markov models retrieved from Pfam release 18 (46) for structural domains E value (<0.001).
Because some categories are larger (i.e., involve more genes) than others, they tend to show more frequently in any set of genes; thus, it is essential to identify the statistically significant categories in a set of genes. We took the whole set of genes as the default background distribution and used the reported method (47) to decide the significance of DG in each category, with P value cutoff of 0.05 as the significance threshold.
Mapping DG to QTL.
Rice QTL data with physical positions on the TIGR release 5 genome were acquired from Gramene (www.gramene.org) and 2,685 DG were mapped to 2,729 rice QTL, covering 9 QTL categories and 211 QTL traits. For better demonstration of the relationship between DG and QTL, we classified yield-related QTL according to the number of genes in each chromosome region and performed an enrichment test according to the method described in ref. 47.
Supplementary Material
Acknowledgments.
The authors thank Dr. Chengzhi Liang (Cold Spring Harbor Laboratory, Cold Spring Harbor, NY) for his supply of mapping data of rice QTL to Tigr5 pseudogenome, and Xiaojun Tan and Dr. Xiting Yan for technical support of data analysis. This project was funded by Chinese Academy of Sciences Grants KSCX2-SW-306 (to L.Z.), KSCX1-SW-03 (to Z.Z.), and KSCX1-SW-03–01 (to J.Y.), National Natural Science Foundation of China Grants 90208001 and 30550005 (to L.Z.) and 30221004 (to J.Y. and G.L.), National Basic Research Program of China Grants 2004CB720406 (to Z.Z.) and 2006CB101706 (to G.L.), Ministry of Science and Technology Grants 2002AA229021 (to J.Y.) and 2006AA10A101 (to Z.Z.), and Ph.D. Programs Foundation of Ministry of Education of China Grant 20060533064 (to L.Y.).
Footnotes
This contribution is part of the special series of Inaugural Articles by members of the National Academy of Sciences elected in 2006.
The authors declare no conflict of interest.
Data deposition: The sequence reported in this paper has been deposited in the Gene Expression Omnibus (GEO Accession number GSE14729).
This article contains supporting information online at www.pnas.org/cgi/content/full/0902340106/DCSupplemental.
References
- 1.Bennetzen JL. Comparative sequence analysis of plant nuclear genomes: Microcolinearity and its many exceptions. Plant Cell. 2000;12:1021–1029. doi: 10.1105/tpc.12.7.1021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Fu H, Dooner HK. Intraspecific violation of genetic colinearity and its implications in maize. Proc Natl Acad Sci USA. 2002;99:9573–9578. doi: 10.1073/pnas.132259199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Birchler JA, Auger DL, Riddle NC. In search of the molecular basis of heterosis. Plant Cell. 2003;15:2236–2239. doi: 10.1105/tpc.151030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Song R, Messing J. Gene expression of a gene family in maize based on noncollinear haplotypes. Proc Natl Acad Sci USA. 2003;100:9055–9060. doi: 10.1073/pnas.1032999100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Guo M, et al. Allelic variation of gene expression in maize hybrids. Plant Cell. 2004;16:1707–1716. doi: 10.1105/tpc.022087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Swanson-Wagner RA, et al. All possible modes of gene action are observed in a global comparison of gene expression in a maize F1 hybrid and its inbred parents. Proc Natl Acad Sci USA. 2006;103:6805–6810. doi: 10.1073/pnas.0510430103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Vuylsteke M, van Eeuwijk F, Van Hummelen P, Kuiper M, Zabeau M. Genetic analysis of variation in gene expression in Arabidopsis thaliana. Genetics. 2005;171:1267–1275. doi: 10.1534/genetics.105.041509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yao Y, et al. Identification of differentially expressed genes in leaf and root between wheat hybrid and its parental inbreds using PCR-based cDNA subtraction. Plant Mol Biol. 2005;58:367–384. doi: 10.1007/s11103-005-5102-x. [DOI] [PubMed] [Google Scholar]
- 9.He G, et al. Haplotype variation in structure and expression of a gene cluster associated with a quantitative trait locus for improved yield in rice. Genome Res. 2006;16:618–626. doi: 10.1101/gr.4814006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Huang Y, et al. Heterosis and polymorphisms of gene expression in an elite rice hybrid as revealed by a microarray analysis of 9198 unique ESTs. Plant Mol Biol. 2006;62:579–591. doi: 10.1007/s11103-006-9040-z. [DOI] [PubMed] [Google Scholar]
- 11.Yu J, et al. A draft sequence of the rice genome (Oryza sativa L. ssp. indica) Science. 2002;296:79–92. doi: 10.1126/science.1068037. [DOI] [PubMed] [Google Scholar]
- 12.Yu J, et al. The genomes of Oryza sativa: A history of duplications. PLoS Biol. 2005;3:e38. doi: 10.1371/journal.pbio.0030038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Xie Z, et al. Pedigree analysis of an elite rice hybrid using proteomic approach. Proteomics. 2006;6:474–486. doi: 10.1002/pmic.200500227. [DOI] [PubMed] [Google Scholar]
- 14.Wang W, et al. Proteomic profiling of rice embryos from a hybrid rice cultivar and its parental lines. Proteomics. 2008;8:4808–4821. doi: 10.1002/pmic.200701164. [DOI] [PubMed] [Google Scholar]
- 15.Zhou Y, et al. Gene identification and expression analysis of 86,136 Expressed Sequence Tags (EST) from the rice genome. Genomics Proteomics Bioinformatics. 2003;1:26–42. doi: 10.1016/S1672-0229(03)01005-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bao J, et al. Serial analysis of gene expression study of a hybrid rice strain (LYP9) and its parental cultivars. Plant Physiol. 2005;138:1216–1231. doi: 10.1104/pp.105.060988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Song S, Qu H, Chen C, Hu S, Yu J. Differential gene expression in an elite hybrid rice cultivar (Oryza sativa, L) and its parental lines based on SAGE data. BMC Plant Biol. 2007;7:49. doi: 10.1186/1471-2229-7-49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ma L, et al. A microarray analysis of the rice transcriptome and its comparison to Arabidopsis. Genome Res. 2005;15:1274–1283. doi: 10.1101/gr.3657405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.JZ Yi, Xiao W. The production technology of the Liang-You-Pei-Jiu. 2000 (in Chinese) [Google Scholar]
- 20.Malkin R, Niyogi K. In: Biochemistry and Molecular Biology of Plants. Buchanan B, Gruissem W, Jones R, editors. Rockville, MD: American Society of Plant Biologists; 2000. pp. 610–619. [Google Scholar]
- 21.Dennis D, Blakeley S. In: Biochemistry and Molecular Biology of Plants. Buchanan B, Gruissem W, Jones R, editors. Rockville, MD: American Society of Plant Biologists; 2000. pp. 630–672. [Google Scholar]
- 22.Birchler JA, Riddle NC, Auger DL, Veitia RA. Dosage balance in gene regulation: Biological implications. Trends Genet. 2005;21:219–226. doi: 10.1016/j.tig.2005.02.010. [DOI] [PubMed] [Google Scholar]
- 23.Zhang H-Y, et al. A genome-wide transcription analysis reveals a close correlation of promoter INDEL polymorphism and heterotic gene expression in rice hybrids. Mol Plant. 2008;1:720–731. doi: 10.1093/mp/ssn022. [DOI] [PubMed] [Google Scholar]
- 24.Shigyo M, Hasebe M, Ito M. Molecular evolution of the AP2 subfamily. Gene. 2006;366:256–265. doi: 10.1016/j.gene.2005.08.009. [DOI] [PubMed] [Google Scholar]
- 25.Xiong LZ, Xu CG, Saghai Maroof MA, Zhang Q. Patterns of cytosine methylation in an elite rice hybrid and its parental lines, detected by a methylation-sensitive amplification polymorphism technique. Mol Gen Genet. 1999;261:439–446. doi: 10.1007/s004380050986. [DOI] [PubMed] [Google Scholar]
- 26.Madlung A, et al. Remodeling of DNA methylation and phenotypic and transcriptional changes in synthetic Arabidopsis allotetraploids. Plant Physiol. 2002;129:733–746. doi: 10.1104/pp.003095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ni Z, et al. Altered circadian rhythms regulate growth vigour in hybrids and allopolyploids. Nature. 2008;457:327–31. doi: 10.1038/nature07523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Crow JF. Alternative hypotheses of hybrid vigor. Genetics. 1948;33:477–487. doi: 10.1093/genetics/33.5.477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Doss S, Schadt EE, Drake TA, Lusis AJ. Cis-acting expression quantitative trait loci in mice. Genome Res. 2005;15:681–691. doi: 10.1101/gr.3216905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pastinen T, Hudson TJ. Cis-acting regulatory variation in the human genome. Science. 2004;306:647–650. doi: 10.1126/science.1101659. [DOI] [PubMed] [Google Scholar]
- 31.Stupar RM, Springer NM. Cis-transcriptional variation in maize inbred lines B73 and Mo17 leads to additive expression patterns in the F1 hybrid. Genetics. 2006;173:2199–2210. doi: 10.1534/genetics.106.060699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ronald J, Brem RB, Whittle J, Kruglyak L. Local regulatory variation in Saccharomyces cerevisiae. PLoS Genet. 2005;1:e25. doi: 10.1371/journal.pgen.0010025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Guo M, Rupe MA, Danilevskaya ON, Yang X, Hu Z. Genome-wide mRNA profiling reveals heterochronic allelic variation and a new imprinted gene in hybrid maize endosperm. Plant J. 2003;36:30–44. doi: 10.1046/j.1365-313x.2003.01852.x. [DOI] [PubMed] [Google Scholar]
- 34.Clark RM, Wagler TN, Quijada P, Doebley J. A distant upstream enhancer at the maize domestication gene tb1 has pleiotropic effects on plant and inflorescent architecture. Nat Genet. 2006;38:594–597. doi: 10.1038/ng1784. [DOI] [PubMed] [Google Scholar]
- 35.Su N, Sullivan JA, Deng XW. Modulation of F1 hybrid stature without altering parent plants through trans-activated expression of a mutated rice GAI homologue. Plant Biotechnol J. 2005;3:157–164. doi: 10.1111/j.1467-7652.2004.00107.x. [DOI] [PubMed] [Google Scholar]
- 36.Xue W, et al. Natural variation in Ghd7 is an important regulator of heading date and yield potential in rice. Nat Genet. 2008;40:761–767. doi: 10.1038/ng.143. [DOI] [PubMed] [Google Scholar]
- 37.Song S, et al. HRGD: A database for mining potential heterosis-related genes in plants. Plant Mol Biol. 2008;69:255–260. doi: 10.1007/s11103-008-9421-6. [DOI] [PubMed] [Google Scholar]
- 38.Eisen MB, Brown PO. DNA arrays for analysis of gene expression. Methods Enzymol. 1999;303:179–205. doi: 10.1016/s0076-6879(99)03014-1. [DOI] [PubMed] [Google Scholar]
- 39.Bachem CWB, Oomen RJFJ, Visser RGF. Transcript imaging with cDNA-AFLP: A step-by-step protocol. Plant Mol Biol Reporter. 1998;16:157–173. [Google Scholar]
- 40.Ma L, et al. Genomic evidence for COP1 as a repressor of light-regulated gene expression and development in Arabidopsis. Plant Cell. 2002;14:2383–2398. doi: 10.1105/tpc.004416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ma L, et al. Light control of Arabidopsis development entails coordinated regulation of genome expression and cellular pathways. Plant Cell. 2001;13:2589–2607. doi: 10.1105/tpc.010229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 43.Altschul SF, et al. Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Durbin R, Eddy SR, Krogh A, Mitchison GJ. Biological Sequence Analysis: Probabilistic Models of Proteins and Nucleic Acids. Cambridge, U.K.: Cambridge Univ Press; 1998. [Google Scholar]
- 45.Ashburner M, et al. Gene ontology: Tool for the unification of biology. The Gene Ontology Consortium. Nat Genet. 2000;25:25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Mistry J, et al. Pfam: Clans, web tools and services. Nucleic Acids Res. 2006;34:D247–D251. doi: 10.1093/nar/gkj149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Mao X, Cai T, Olyarchuk JG, Wei L. Automated genome annotation and pathway identification using the KEGG Orthology (KO) as a controlled vocabulary. Bioinformatics. 2005;21:3787–3793. doi: 10.1093/bioinformatics/bti430. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.