Abstract
MicroRNAs (miRNAs) inhibit the translation of target mRNAs and affect, directly or indirectly, the expression of a large portion of the protein-coding genes. This study focuses on miRNAs that are expressed in the mouse cochlea and vestibule, the 2 inner ear compartments. A conditional knock-out mouse for Dicer1 demonstrated that miRNAs are crucial for postnatal survival of functional hair cells of the inner ear. We identified miRNAs that have a role in the vertebrate developing inner ear by combining miRNA transcriptome analysis, spatial and temporal expression patterns, and bioinformatics. Microarrays revealed similar miRNA profiles in newborn-mouse whole cochleae and vestibules, but different temporal and spatial expression patterns of six miRNAs (miR-15a, miR-18a, miR-30b, miR-99a, miR-182, and miR-199a) may reflect their roles. Two of these miRNAs, miR-15a-1 and miR-18a, were also shown to be crucial for zebrafish inner ear development and morphogenesis. To suggest putative target mRNAs whose translation may be inhibited by selected miRNAs, we combined bioinformatics-based predictions and mRNA expression data. Finally, we present indirect evidence that Slc12a2, Cldn12, and Bdnf mRNAs may be targets for miR-15a. Our data support the hypothesis that inner ear tissue differentiation and maintenance are regulated and controlled by conserved sets of cell-specific miRNAs in both mouse and zebrafish.
Keywords: cochlea, deafness, Dicer, mouse, zebrafish
The inner ear is one of the most complex tiny organs in the vertebrate body and is responsible for both hearing and balance. The mammalian cochlear labyrinth is a coiled duct, containing 1 sensory epithelium (SE), referred to as the organ of Corti. The mammalian vestibular SE is comprised of 3 cristae and 2 otolith organs (maculae), the saccule, and the utricle. In both the cochlea and the vestibule, SE is composed of mechanosensory hair cells (HCs) submerged in a matrix of nonsensory supporting cells (SC). All HCs contain actin elongations, named stereocilia, which are responsible for mechanoelectrical transduction to provide an electrical signal that is transferred to the brain. Nevertheless, auditory and vestibular HCs have morphological and physiological differences (1). Interestingly enough, in mammals, cochlear HCs are unable to regenerate but a limited amount of HC regeneration may occur in the vestibular SE (2, 3).
Hereditary deafness and vestibular dysfunction often involve inner ear developmental defects or degeneration of the sensory neuroepithelial HCs. While over 100 loci have been mapped and 44 protein-coding genes with mutations have been cloned (Hereditary Hearing Loss Homepage; http://webh01.ua.ac.be/hhh/), many cases of hereditary hearing loss remain a mystery. Approximately 98% of RNAs in mammalian cells do not code for proteins and epigenetic factors are associated with hearing loss (4), raising the possibility that noncoding RNAs, such as microRNAs (miRNAs), may also be involved in inner ear development and hearing loss. miRNAs are 17 to 25 nt RNAs produced from stem-loop precursors (pre-miRNAs) that are encoded by miR genes (also known as Mirn genes). miRNAs were recently discovered as a major form of regulation in the cell and associated with many developmental processes and diseases (5). Hundreds of animal miR genes have been discovered and their sequences are listed in the miRBase database (6). The most understood functions of mature miRNAs in animals are translational suppression of mRNAs with imperfect complementary sequences in their 3′ untranslated region (3′UTR) and cleavage of mRNAs with a perfect match (7). It has been suggested that at least 60% of the vertebrate protein-coding genes is directly regulated by miRNAs (8, 9), and that miRNAs affect, directly or indirectly, virtually all cellular and organismal processes. miRNAs are differentially expressed in different tissues (10) and a triad of miRNAs (miR-183, miR-96, and miR-182), clustered within 4 kb and transcribed in the order shown, is expressed specifically in the mechanosensory HCs in the vertebrate inner ear (11, 12). In parallel to our study, 2 other groups linked point mutations in the seed region of the Mirn96 gene with hearing loss in both human and mouse (13, 14). In addition, a conditional knockout of Dicer mainly in the brain and the early otic placode, under the control of the Pax2 promoter, led to abnormal development and innervation of the inner ear and to death prior to birth (15).
To study the roles of inner ear SE miRNAs, we conditionally knocked out Dicer only in the inner ear SE hair and SCs after their normal differentiation from progenitor cells. Our study demonstrates that Dicer activity is required for the survival of normal and functional HCs in the cochlear SE. Removal of Dicer from the SE, which initially develops normally, causes abnormal growth and subsequent degeneration of HCs, leading to deafness. We also provide Tables S1–S5 of miRNAs that are newly predicted to have a role in the mammalian inner ear SE. Antisense morpholino experiments in zebrafish reveal that silencing of miR-15a-1 and miR-18a leads to loss of mechanosensory HCs, as well as other inner ear defects. Therefore, miRNAs have an essential role in inner ear development and function.
Results
Dicer1 Is Essential for Survival of Functional HCs in the Dicer-PCKO Mouse Cochlea.
The type III ribonuclease Dicer is required for the production of mature and functional miRNAs from stem-loop pre-miRNAs (16). Dicer1 mutant mice arrest miRNA processing and die at an early embryonic stage (17). To study miRNA loss in a specific tissue, Dicer1 may be knocked out conditionally in mice using the Cre-loxP recombination system (18). The Pou4f3/Brn3c gene is expressed, in the inner ear, specifically in HCs (19, 20). Using mice that express Cre downstream to the Pou4f3 promoter (21) and Dicerflox/flox mice (18), we created a mutant Dicer-PCKO (Pou4f3-Cre induced Conditional Knock-Out) mouse in which the Dicer1 gene is conditionally knocked out in the inner ear HCs. In the postnatal day 6 (P6) mouse inner ear, Pou4f3-Cre is expressed in both HCs and some of the SCs in the cochlear SE, and only in HCs in the vestibule (21). The Pou4f3-Cre is expressed in many organs in addition to the inner ear as well (D. Vetter, unpublished work), and Dicer-PCKO mice had many defects in addition to inner ear defects (see SI Results and Discussion and Fig. 1A and Fig. S1); here, we focus on the inner ear phenotype. Auditory brainstem responses (ABR) confirmed that at 38 days Dicer-PCKO mice are deaf, with no response to auditory stimuli (Fig. 1B). On the other hand, the mice exhibited a mild vestibular phenotype, with a partial reaching response and no circling behavior.
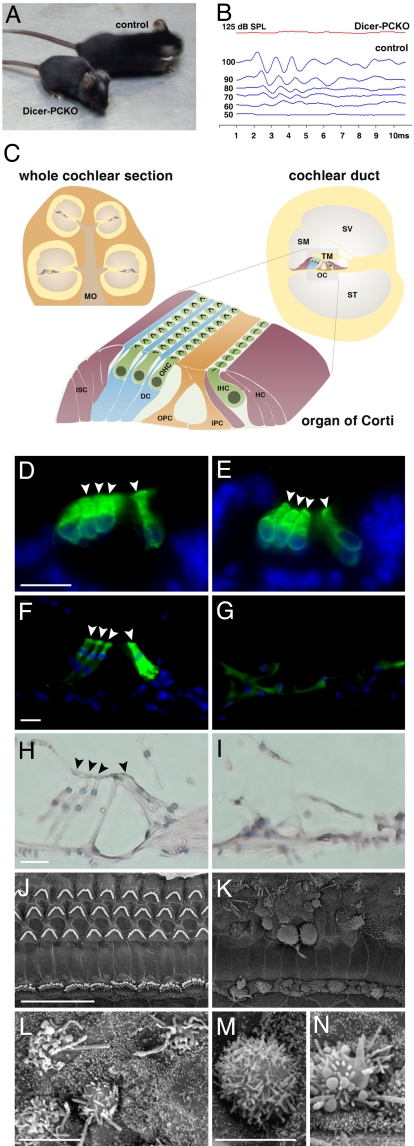
Fig. 1.
P38 Dicer-PCKO mutant mice were deaf and developed aberrant hair bundle and stereocilia defects in the cochlea. (A) P38 mutant mice were smaller than heterozygote siblings and exhibited a distinct shape, including typical alopecia (see also Fig. S1). (B) ABR demonstrated complete loss of hearing in mutant (red) compared to control (blue) mice (8 kHz stimuli). (C) Schematic representation of the organ of Corti: (green) sensory HCs; (other colors) SCs. DC, Deiters' cells; HC, Hensen cells; IHC, inner hair cells; IPC, inner pillar cells; ISC, inner sulcus cells; MO, modiolus; OC, organ of Corti; OHC, outer hair cells; OPC, outer pillar cells; SM, scala media; ST, scala tympani; SV, scala vestibuli; TM, tectorial membrane. (D–G) Immunofluorescence with the inner ear hair cell marker myosin VI (green) in P0 (D and E) and P38 (F and G) control (D, F) and mutant (E, G) mice. Cell nuclei stained with DAPI (blue). (H and I) H&E staining of the P38 organ of Corti from control (H) and mutant (I) mice. (D–I) Sections from the base of the cochlea. (J–N) SEM of inner ear hair cell stereocilia in P38 control (J) and mutant (K–N) mice. The hair cells are observed from the scala media. (L–N) Higher magnification. [Scale bars: (D, F, H, J), 20 μm; (L, M, N), 5 μm.) Control mice are littermate heterozygotes.
The Pou4f3 promoter is known to be expressed from embryonic day 12.5 (E12.5) in the mouse vestibule and from E14.5 in the mouse cochlea (22). The cochlear SE, the organ of Corti, is illustrated in Fig. 1C. The Dicer–PCKO inner ear SE exhibited normal morphology at E18 (data not shown) and P0 (Fig. 1 D and E). Because there was no abnormal phenotype at P0, inner ears were examined after maturation. At P38, some of the cochlear HCs had an aberrant shape and myosin VI expression (a HC marker) was lost (Fig. 1 F–I). The severity of the HC phenotype varied longitudinally along the cochlea, with HCs in the base showing more severe malformations than those in the apex. At the same time, in the vestibular maculae, myosin VI-expressing HCs still existed (data not shown). Inner ear HCs have specialized organized stereocilia that are crucial for mechanotransduction (23). Scanning electron microscopy at P38 revealed that in the cochlea, many HCs lost their stereocilia, and the apical surfaces of residual inner and outer HCs became uniformly rounded and visibly reduced. In other cases, HCs displayed disorganized bundles of thin, microvilli-like stereocilia of uniform length, and sometimes adjacent stereocilia fused together to form giant protrusions (Fig. 1 J–N). In the vestibule, HCs were present and still had stereocilia, although these stereocilia were abnormally organized or fused together, which correlated with the partially preserved vestibular function (data not shown).
miRNA Transcriptomes of WT Mouse Whole Cochlea and Vestibule.
Microarray scanning of small (<40 nt) RNAs isolated from newborn (P0) mouse cochleae and vestibules detected 105 miRNAs expressed in the cochlea and 114 miRNAs expressed in the vestibule, with average intensities higher than twice the global background, out of 206 included in the arrays (Fig. 2A and Table S1). Only 24 miRNAs were found to have different levels of expression in these whole organs, and these differences were mild (15–40%) (Fig. 2B). Two complementary approaches were used to choose candidate miRNAs predicted to be expressed specifically in the inner ear SE. To identify miRNAs that may be expressed in inner ear SE we searched for miR genes in introns of mouse protein-coding genes that are expressed in the cochlear and vestibular SE at P2. These miR genes may be transcribed together with the hosting protein-coding gene, using the same promoter (see SI Results and Discussion and Table S2). To identify miRNAs that may be hearing-related, we looked for miR genes in chromosomal loci that were linked with nonsyndromic hereditary hearing loss in humans, but their responsible genes have not been identified yet (see SI Results and Discussion and Table S3). The intersection between miRNA microarray results and list of predictions is shown (Fig. 2C and Tables S4 and S5). Because the microarrays contained probes for only 206 miRNAs, while the input for predictions included 488 known mouse miR genes [miRBase release 12.0, September 2008 (6)], at least half of the predicted miRNAs may not be included in the microarray results. miRNAs that are expressed in the whole inner ear according to microarrays and are predicted to be expressed in the SE and to be hearing-related are proposed candidates for further study.
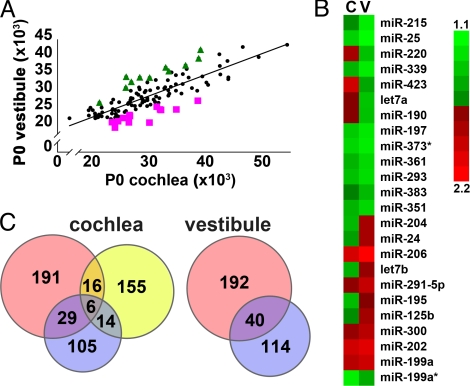
Fig. 2.
Identification of miRNAs expressed differentially in the auditory and vestibular organs in newborn mice. (A) Scatter-plot representation of the distribution of miRNAs expressed in P0 mouse vestibule versus cochlea. Results were averaged from 3 individual arrays, including 8 to 12 spots per each miRNA. Only miRNAs with an average intensity higher than twice the global background intensity, at least in 1 of the organs, are plotted (for details, see Table S1). miRNAs that are over-expressed in the vestibule (triangles) or cochlea (squares) by at least 15% than their expected expression, according to the linear regression of all expressed miRNAs, are labeled. (B) Heat map representation of the 24 differentially expressed miRNAs in P0 cochleae and vestibules (at least 1.15-fold difference) that are labeled in (A). (C) Intersection of microarray results (light blue) with predicted SE-related miRNAs. Left Venn diagram: miR genes included in protein-coding genes expressed in mouse P2 cochlear SE (pink), miR genes in human deafness-related loci with unidentified genes (yellow) and miRNAs expressed in whole P0 cochleae (blue). Right Venn diagram: miR genes included in protein-coding genes expressed in mouse P2 vestibular SE (pink) and miRNAs expressed in whole P0 vestibules (blue).
Expression Patterns of Selected miRNAs in the WT Mouse Inner Ear.
Five miRNAs that are similarly expressed in whole P0 cochleae and vestibules and have orthologues in zebrafish were selected for further study. They are listed from the most abundant to the less abundant expression in P0 mouse inner ears: miR-199a, miR-30b, miR-18a, miR-15a, and miR-99a. While miR-199a was one of the highest expressed (together with the other miR-199a-2 cluster member, miR-214), miR-99a had a very low or no expression in the mouse inner ear, according to microarray data. miR-15a and miR-199a were predicted to be expressed in inner ear SE and their genes are included in human deafness-related loci (see Tables S2 and S3). miR-182 expression was used as a positive control, as it is known to be expressed specifically in vertebrate inner ear HCs (11, 12). The miRNAs evaluated in detail have a similar global level in whole cochleae and vestibules, according to microarray results.
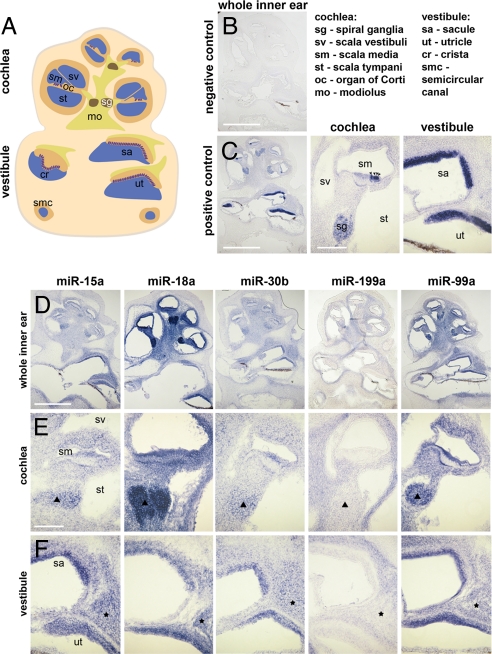
miRNAs that are most highly expressed in the inner ear before it is functional may have a specific role in its development. The newborn-mouse inner ear is not yet functional, while at P30 the mouse inner ear is fully developed and miRNAs that are highly expressed at this age may participate in its survival and maintenance. Quantitative real time RT-PCR (qRT-PCR) detected a gradual change in expression for most of the studied miRNAs during inner ear development from E16.5 to P30. miR-182 and miR-199a were specifically expressed in the inner ear and very low in the brain, and presented a similar expression pattern during inner ear development, with highest expression at P0. miRNAs 99a, 15a, 18a, and 30b were expressed at lower levels in the inner ear as compared to the P0 brain at most time-points. For most time-points and miRNAs, the cochlear and vestibular expression levels were similar, but some exceptions were detected (Fig. S2). In situ hybridization (ISH) suggested differences in miRNA functions at P0 because they differed in their spatial expression patterns in the inner ear (Fig. 3). miR-182 is specifically expressed in HCs of the cochlear and vestibular SE, as well as in the spiral ganglion (see Fig. 3C) (11). miRNAs 15a, 30b, and 99a were expressed in both SCs and HCs in the cochlear SE, as well as in the spiral ganglion neurons and the basilar membrane (see Fig. 3 D and E). However, in the vestibular SE, their expression was restricted to HCs only (see Fig. 3 D and F). Most interesting, miR-199a and miR-99a expression patterns are mirrored. Almost all cochlear cells, including those of the modiolus, expressed miR-199a, except for the SE and the other cells surrounding the scala media. In the vestibule, miR-199a was not expressed in SE, neither in HCs nor the SCs at their basal side. miR-99a was expressed in almost all cell types in the cochlea, but mainly in the cells surrounding the scala media (not only in HCs), the basilar membrane, and the spiral ganglion. In the vestibule, miR-99a was expressed mainly in HCs (but not in the basilar SCs). miR-18a had a similar expression pattern to miR-99a, but its expression in the spiral ganglion neurons was much more prominent, and in the vestibular SE it was expressed in both HCs and SCs (see Fig. 3 D and F). The ISH specificity was confirmed by the nonidentical hybridization of miR-15a and miR-99a probes, which differ in only 2 bases, whose cochlear expression patterns are different.
Fig. 3.
Distinct patterns of spatial expression of miRNAs 15a, 18a, 30b, 99a, and 199a in the newborn-mouse inner ear. (A) Schematic illustration of a P0 whole inner ear section: (blue) fluid-filled lumens, (orange) epithelial tissues, (violet) hair cells, (brown spots) spiral ganglion, (yellow) cochlear modiolus and vestibular nerve fibers; vestibular orange areas, SE SCs. (B–F) Cell-specific expression patterns were demonstrated for miRNAs by ISH in whole-mount inner ears, followed by cryosectioning. (B) A scrambled probe was used as a negative control. (C) A probe for miR-182 was used as a positive control and confirmed solution penetration and staining throughout. Whole inner ear section (Left) and magnifications of cochlear (Middle) and vestibular (Right) areas are shown: (arrowheads) cochlear HCs. (D) Whole inner ear sections enable comparison of the expression pattern of each miRNA in the cochlea and vestibule. (E) Magnified cochlea figures present the epithelial cells surrounding the scala media and the spiral ganglion (triangles). (F) Magnified vestibule figures include the SE of the saccule and utricle, as well as nerve fibers (asterisks). [Scale bars: (B, C Left, D), 500 μm; (C Middle, E), 100 μm.]
Zebrafish Inner Ear Development Is Dependent on miR-15a-1 and miR-18a.
The zebrafish provides a useful and robust model for functional analysis of individual miRNAs and their conserved roles in vertebrates (24, 25). The zebrafish inner ear resembles the mammalian vestibule and was chosen to evaluate the contribution of miRNAs 15a-1 and 18a to HC and otic development. At 48 h postfertilization (hpf), canal pillars protrude into the otocyst and will fuse to create the 3 semicircular canals (26). At this stage, the maculae of the saccule (hearing) and utricle (balance) have ≈20 HCs, while the 3 cristae (angular acceleration) have 1 to 4 HCs each (27). At 48 hpf, miR-15a and miR-18a are expressed throughout the head, as reported previously for 72-hpf fish (12); after sectioning the signals are particularly strong in the otocysts (Fig. 4A).
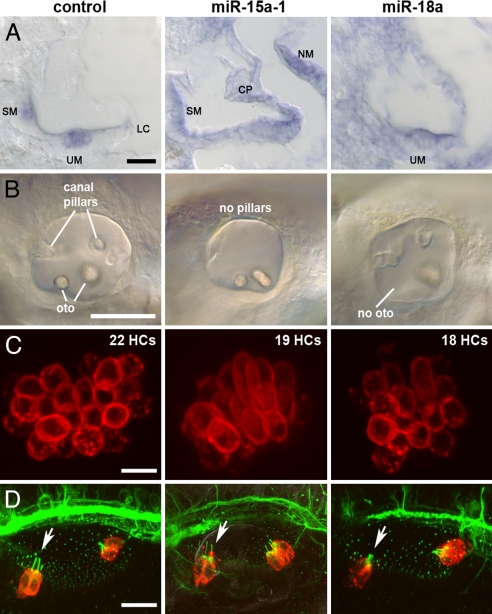
Fig. 4.
miRNAs 15a and 18a are expressed in the zebrafish inner ear and miR-15a-1 or miR-18 morphant phenotypes include inner ear defects and reduced HC numbers. (A) ISH shows expression at 48 hpf: miR-183 served as a positive control (Left) for expression in HCs in the saccular macula (SM), utricular macula (UM), and lateral crista (LC). miR-15a was expressed in lateral line neuromasts (NM) and throughout the inner ear, including SCs and canal pillars (CP), and miR-18a expression was strong in the UM and adjacent cells. Dorsal is up, lateral is right. (B–D) Otocysts shown after treatment with MMO-miR-15a-1 or MMO-miR-18a (see Fig. S3). Injection with a standard MO that is not expected to target any known zebrafish RNA serves as a negative control (Left) to emphasize defects in canal pillars, otoliths (oto), and number of HCs in the other morphants. (B) Otocysts at 55 hpf. Anterior is left, dorsal is up. (C) UMs at 55 hpf. Confocal image stacks of HCs viewed en face from the ventral side. HC numbers are listed for each sample. Anterior is left, medial is up. (D) Otocysts at 30 hpf. Confocal image stacks show that morphants have differentiated the earliest HCs, called tether cells (red), at the anterior (arrow) and posterior poles. Primary cilia, labeled with acetylated tubulin, project into the lumen of the otocyst (green dots) but are substantially elongated for tether cells; axons also stain with this antibody. Anterior is left, dorsal is up. [Scale bars: (A), 20 μm; (B), 100 μm; (C), 10 μm; (D), 20 μm.]
The rapid development of the inner ear in zebrafish embryos enables antisense-based silencing of individual miRNAs following injection of morpholinos (MOs) into 1-celled embryos (Fig. S3). Reductions of the targeted miRNAs, miR-15a-1 and miR-18a, were confirmed by ISH with specific probes at 30 hpf (Figs. S4 and S5). Treatment with at least 2 distinct MOs against each miRNA resulted in similar defects in overall otic morphogenesis, with a third morpholino to pre-miRNA sequences showing reduced ability to phenocopy (Figs. S4–S6). miR-15a-1 morphants lacked canal pillars, and the otoliths that rest above each sensory macula were often abnormal, being small, extra, fused, or untethered to a macula (see Fig. 4B and Fig. S4). miR-18a morphants consistently showed small or missing anterior otoliths (see Fig. 4B and Fig. S5), although the initial pair of anterior (and posterior) HCs that tether each otolith were present and normally differentiated at 30 hpf (Fig. 4D). At 54 to 56 hpf, both miR-15a-1 and miR-18a morphants had significantly reduced numbers of lateral line neuromasts and mechanosensory HCs (see Fig. 4C and Table S6). The spatial organization of sensory organs, the statoacoustic ganglion, and some other cranial ganglia were impaired to varying degrees in both classes of morphants (see Fig. S6 C–E). For more details, see SI Results and Discussion.
Potential Targets of Inner Ear miRNAs in Mice.
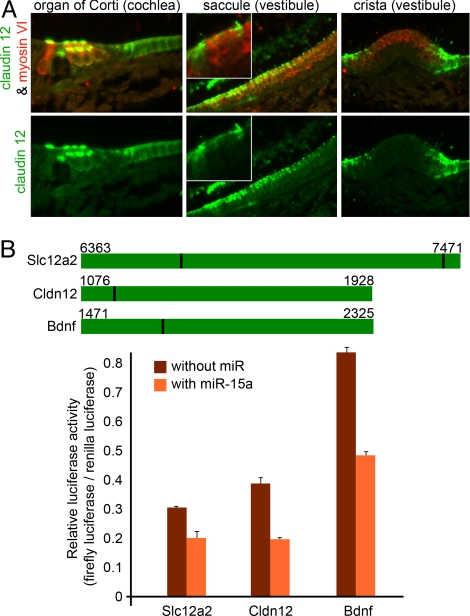
miRNAs down-regulate the stability and translation of target mRNAs that contain a complementary sequence to the miRNA seed (nucleotides 2–8 of the miRNA) in their 3′ UTR. The unique expression pattern of each miRNA may be used for searching for its targets. To suggest target mRNAs for miRNAs that are expressed in the newborn-mouse inner ear SE (miRNAs 15a, 18a, 30b, and 99a) (see Fig. 3), we looked for predicted targets, based on 3 Web-based target prediction algorithms: TargetScan (28), miRanda (29), and PicTar (30), and intersected the predictions with information regarding the expression of these putative targets in inner ear SE at P2 (see Tables S7–S10). To identify biologically relevant targets for miR-15a, 3 putative targets that are expressed in the inner ear SE (as mRNAs) were selected: Slc12a2/Nkcc1, Cldn12, and Bdnf (details in SI Results and Discussion). For example, claudin 12 protein was detected in P0 cochlear HCs, which also express miR-15a. In the P0 vestibule, claudin 12 was detected only in SCs that do not express miR-15a, but not in vestibular HCs that express this miRNA (Fig. 5A). While the 3′-UTR of Slc12a2 mRNA contains 2 putative binding sites for miR-15a, Cldn12 and Bdnf 3′-UTRs contain a single binding site for this miRNA. The dual luciferase assay confirmed that miR-15a can inhibit the expression of a gene upstream to ≈1kb 3′-UTR fragments surrounding these 4 miR-15a binding sites (Fig. 5B) and that miR-15a can reduce the translation of Slc12a2, Cldn12, and Bdnf mRNAs when each is coexpressed with this miRNA.
Fig. 5.
Slc12a2, Cldn12 and Bdnf mRNAs are putative targets for miR-15a in the inner ear SE. (A) Immunolabeling of Claudin 12 (Cldn12) in P0 cochlea and vestibule (green). Myosin VI-specific immunostaining detects HCs (red). (B) The dual luciferase assay was used to measure the ability of miR-15a to reduce the expression of the firefly luciferase gene that is located upstream to putative target 3′ UTRs. Fragments of about 1 kb around miR-15a binding sites from Slc12a2, Bdnf, and Cldn12 3′-UTRs were cloned into pGL3 plasmids, downstream to firefly luciferase cDNA, and luciferase activity was measured in transiently transfected HEK-293T cells. (Top) Illustrations of the 3′-UTR fragments cloned into pGL3 plasmids. Base numbers of beginning and end of each cloned fragment are presented for Slc12a2 (NM_009194), Cldn12 (NM_022890), and Bdnf (NM_007540) mRNAs. Binding sites for miR-15a labeled dark green. (Bottom) Relative luciferase activity in cells in the absence or presence of miR-15a. Cells were cotransfected with pRL-TK plasmids that include the renilla luciferase gene, and relative firefly/renilla luciferase activity is presented.
Discussion
miRNAs are required for growth and survival of inner ear neurosensory epithelia in both mouse and zebrafish. We have shown that normal function and survival of the postmitotic HCs depend on miRNA maturation. When mouse inner ear HCs were deprived of miRNAs during their differentiation by a conditional knockout of Dicer1, degeneration of HCs, hair-bundle malformations, and subsequent hearing loss were observed several weeks later. Pou4f3 expression in the mouse inner ear HCs begins 6.5 to 8.5 days before birth (22), suggesting that the Pou4f3-promoter-induced knockout of Dicer1 should initiate at this time. Yet the Dicer-PCKO mice exhibited no HC phenotypes through the day of birth, perhaps because residual Dicer1 and mature miRNAs before onset of Cre expression may still be active for a short period. Alternatively, the regulation of key miR targets may be minimal during early differentiation events but more essential during later phases of HC maturation, perhaps to mediate stereociliary stabilization and HC survival. miRNAs were found to be required for the survival of neuronal tissues, including forebrain cortical neurons (31), postmitotic cerebellar Purkinje cells (32), and midbrain dopaminergic neurons (33) in mice, when Dicer1 was knocked out conditionally in these cells. Similarly, a reduction in miRNAs may induce apoptosis in neurosensory HCs in our Dicer-PCKO mice, causing them to die prematurely.
The cochlear and vestibular HCs may differ in their sensitivity to miRNA loss, as a faster deterioration of the cochlear HCs occurred, although Pou4f3 is expressed in vestibular HCs 2 days before its onset in cochlear HCs (22). The massive loss of cochlear stereocilia and the milder vestibular phenotype at P38 may explain why these mice are deaf but do not suffer from significant vestibular symptoms. The more severe cochlear phenotype may be because of the fact that after birth, Pou4f3-Cre is expressed only in HCs in the vestibule, but in both HCs and some SCs in the cochlea (21). Thus, miRNAs in SE SCs may be more important for survival of functional HCs than miRNAs in the HCs themselves. The P38 cochleae of our mutant mice exhibited a gradient phenotype, when HCs in the cochlea base were more deformed than in the apex. This finding correlates with the published observation of a graded expression of miR-182 from the base to apex in mouse cochlear HCs, with highest expression at the base (11).
We aimed at identifying the repertoire of miRNAs expressed in the auditory and vestibular systems of the inner ear. From the over 100 miRNAs detected in the cells of these inner ear compartments, a subset was chosen for further evaluation. miRNAs were found to be expressed differentially in diverse cells, suggesting they have cell-specific regulatory roles. For example, miR-182 is expressed only in cells that do not express miR-199a. miR-99a is expressed in the SCs of cochlear but not vestibular SE. Interestingly, we found 2 miRNAs, miR-99a and miR-199a, with a complementary pattern of expression in P0 mouse inner ears, suggesting that they may participate in a negative feedback loop to down-regulate each other. An indirect mechanism, via down-regulated target mRNAs, appears most plausible for such an interaction.
miRNA levels in the mouse inner ear change during development, as was previously shown (11), suggesting that the roles of these miRNAs are specific to particular development stages. We were unique in comparing temporal expression of selected miRNAs in separated cochleae and vestibules from embryonic to fully developed inner ears. Both vestibular and cochlear expression of an miRNA may be similar at certain time-points and different at others, suggesting they regulate diverse targets and are thus responsible for distinctive functions (e.g., miR-30b, miR-99a).
What targets do miRNAs control in the inner ear? Our data suggests that Slc12a2, Cldn12, and Bdnf, all proteins known for their essential function in the inner ear, are genuine targets of miR-15a. Our target-prediction approach is supported by a report describing miR-30a-5p that has an identical seed sequence to miR-30b as down-regulating Bdnf mRNA in the human prefrontal brain cortex (34). Interestingly, both the mRNAs and proteins of Bdnf and Cldn12 are highly expressed in the newborn-mouse cochlear HCs that express miR-15a as well, while the luciferase assay in proliferating (HEK-293) cells showed that miR-15a inhibits the translation of these mRNAs when they are coexpressed. However, the HCs in the postnatal cochleae are differentiated and held in mitotic arrest (i.e., quiescent). Two recent studies proposed that while miRNAs inhibit the translation of their target mRNAs in proliferating cells, in quiescent cells they have the reverse effect and may up-regulate the translation of the same targets (35, 36). This may indeed be the case in quiescent auditory and vestibular HCs.
The mutant-mouse phenotype and identity of subsequentmiRNAs found to differ in their expression between auditory and vestibular cells, such as miR-99a, suggest that differences in control rendered by miRNAs may provide fine-tuning of regulation of protein translation in the many cell types of the inner ear.
Taking all these results into account, how do miRNAs regulate HC growth and maintenance? Studies from other systems can serve as a guide to decipher the functions in the inner ear of the miRNAs that we identified in the current screen. Some of these are up-regulated in cancer and may induce proliferation and angiogenesis [e.g., miR-18a (37)], while others are down-regulated following exposure to carcinogens [miR-99a (38) and miR-199a (39)]. miR-15a is known to have anticarcinogenic and apoptotic effects (40). Recently, miR-15a has been linked to pancreas regeneration (41). A knockout of the miR-199a-2 cluster affected normal musculoskeletal development and growth (42).
Our work is unique in demonstrating that miRNAs are essential for the development and survival of sensory HCs in the vertebrate inner ear. Moreover, we identified many miRNAs that may have a role in the vertebrate developing inner ear, in addition to the previously known miR-182 cluster (11, 12), by combining miRNA transcriptome analysis, spatial and temporal expression patterns, and bioinformatics. Indeed, 2 of these miRNAs were confirmed to be essential for normal morphogenesis of the zebrafish inner ear. We provide Tables S1–S5 with the many additional miRNAs whose role in the inner ear deserves further study. Some of these may be responsible for differences between the cells of the hearing and balance systems, such as miR-211, expressed in vestibular SE but not in comparable cells in the cochlea. The comparison of organ- and cell-specific miRNAs will help to further define the fine-tuning of regulation and expression in the inner ear.
Materials and Methods
Generation of Dicer-PCKO Mice.
Mice homozygote for Dicerflox [provided by C. Tabin (18)] and mice harboring a Pou4f3-Cre allele [developed by D. Vetter, Tufts, and Z.-Y. Chen, Harvard (21)] were bred in accordance with Tel Aviv University Institutional Animal Care and Use Committee (M-06–042) and National Institutes of Health Guide for the Care and Use of Laboratory Animals.
Assays for Inner Ear Phenotype in Mice.
ABR thresholds for click (≈4 kHz) and 8 kHz stimuli were measured to assess mouse hearing (control, n = 3; Dicer-PCKO, n = 3). Paraffin sections of inner ears were analyzed by H&E staining, immunohistochemistry (antibodies listed in SI Materials and Methods), and scanning electron microscopy, as previously described (43).
miRNA Microarrays and qRT-PCR.
Microarrays were printed in-house with the mirVana miRNA Probe Set version 1 and hybridized with labeled cochlea and vestibule RNA samples. For qRT-PCR, specific miRNAs or U6B RNA (as endogenous control) were reverse transcribed from <200 nt RNA fractions, and their expression was measured using the TaqMan MicroRNA kits and the ABI Prism 7000 or 7900 PCR machine (Applied Biosystems).
In Situ Hybridization of miRNAs.
At least 3 independent ISH experiments were performed with each probe, and at least 4 inner ears were included in each experiment. ISH was performed using antisense 3′-digoxygenin (DIG)-labeled miRCURY LNA Detection probes (Exiqon) and anti-DIG-AP (alkaline phosphatase conjugated) antibodies (44). Mouse inner ears and zebrafish embryos were cryosectioned to 8- to 10- and 30-μm sections, respectively.
MO Injections to Zebrafish Embryos and Phenotypic Evaluation.
All protocols were approved by the Purdue University Animal Care and Use Committee.
Dual Luciferase Assay.
Fragments of ≈1 kb from the 3′-UTRs of Slc12a2, Cldn12, and Bdnf mRNAs were cloned into the pGL3 Luciferase Reporter Vector (Promega) as 3′UTRs of the firefly luciferase cDNA. miR-15a vector was provided by R. Agami (45). Three experiments were performed, each in triplicate.
Further details of the methods used may be found in the SI Materials and Methods.
Supplementary Material
Acknowledgments.
We thank Douglas Vetter and Zheng-Yi Chen for Pou4f3-Cre mice and for sharing unpublished data, Kursad Turksen for anti-Cldn12 antibodies, Haim Sohmer and Cahtia Adelman for aid with ABR measurements, Betsy Tao for assistance with zebrafish, Cliff Tabin, and Reuven Agami. This work was supported by the Israel Science Foundation Grant 1486/07 (to K.B.A.), European Commission FP6 Integrated Projects EuroHear LSHG-CT-20054–512063 and Eumodic 037188 (to K.B.A.), the National Organization for Hearing Research (to T.S.) the Deafness Research Foundation (to T.S.), and National Institutes of Health Grants R01 DC005641 (to K.B.A.) and R21 DC008997 (to D.M.F.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The data reported in this paper have been deposited in the Gene Expression Omnibus (GEO) database, www.ncbi.nlm.nih.gov/geo (accession no. GSE15496).
This article contains supporting information online at www.pnas.org/cgi/content/full/0812446106/DCSupplemental.
References
- 1.Kelley MW. Regulation of cell fate in the sensory epithelia of the inner ear. Nat Rev Neurosci. 2006;7:837–849. doi: 10.1038/nrn1987. [DOI] [PubMed] [Google Scholar]
- 2.Warchol ME, Lambert PR, Goldstein BJ, Forge A, Corwin JT. Regenerative proliferation in inner ear sensory epithelia from adult guinea pigs and humans. Science. 1993;259:1619–1622. doi: 10.1126/science.8456285. [DOI] [PubMed] [Google Scholar]
- 3.Kawamoto K, Izumikawa M, Beyer LA, Atkin GM, Raphael Y. Spontaneous hair cell regeneration in the mouse utricle following gentamicin ototoxicity. Hear Res. 2009;247:17–26. doi: 10.1016/j.heares.2008.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Provenzano MJ, Domann FE. A role for epigenetics in hearing: Establishment and maintenance of auditory specific gene expression patterns. Hear Res. 2007;233:1–13. doi: 10.1016/j.heares.2007.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhang B, Wang Q, Pan X. MicroRNAs and their regulatory roles in animals and plants. J Cell Physiol. 2007;210:279–289. doi: 10.1002/jcp.20869. [DOI] [PubMed] [Google Scholar]
- 6.Griffiths-Jones S, Grocock RJ, van Dongen S, Bateman A, Enright AJ. miRBase: microRNA sequences, targets and gene nomenclature. Nucleic Acids Res. 2006;34:D140–D144. doi: 10.1093/nar/gkj112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 8.Miranda KC, et al. A pattern-based method for the identification of microRNA binding sites and their corresponding heteroduplexes. Cell. 2006;126:1203–1217. doi: 10.1016/j.cell.2006.07.031. [DOI] [PubMed] [Google Scholar]
- 9.Friedman RC, Farh KK, Burge CB, Bartel DP. Most mammalian mRNAs are conserved targets of microRNAs. Genome Res. 2009;19:92–105. doi: 10.1101/gr.082701.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Landgraf P, et al. A mammalian microRNA expression atlas based on small RNA library sequencing. Cell. 2007;129:1401–1414. doi: 10.1016/j.cell.2007.04.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Weston MD, Pierce ML, Rocha-Sanchez S, Beisel KW, Soukup GA. MicroRNA gene expression in the mouse inner ear. Brain Res. 2006;1111:95–104. doi: 10.1016/j.brainres.2006.07.006. [DOI] [PubMed] [Google Scholar]
- 12.Wienholds E, et al. MicroRNA expression in zebrafish embryonic development. Science. 2005;309:310–311. doi: 10.1126/science.1114519. [DOI] [PubMed] [Google Scholar]
- 13.Mencía A, et al. Mutations in the seed region of human miR-96 are responsible for non-syndromic progressive hearing loss. Nat Genet. 2009 doi: 10.1038/ng.355. [DOI] [PubMed] [Google Scholar]
- 14.Lewis M, et al. An ENU-induced mutation of miR-96 associated with progressive hearing loss in mice. Nat Genet. 2009 doi: 10.1038/ng.369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Soukup GA, et al. Residual microRNA expression dictates the extent of inner ear development in conditional Dicer knockout mice. Dev Biol. 2009 doi: 10.1016/j.ydbio.2009.01.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bernstein E, Caudy AA, Hammond SM, Hannon GJ. Role for a bidentate ribonuclease in the initiation step of RNA interference. Nature. 2001;409:363–366. doi: 10.1038/35053110. [DOI] [PubMed] [Google Scholar]
- 17.Bernstein E, et al. Dicer is essential for mouse development. Nat Genet. 2003;35:215–217. doi: 10.1038/ng1253. [DOI] [PubMed] [Google Scholar]
- 18.Harfe BD, McManus MT, Mansfield JH, Hornstein E, Tabin CJ. The RNaseIII enzyme Dicer is required for morphogenesis but not patterning of the vertebrate limb. Proc Natl Acad Sci USA. 2005;102:10898–10903. doi: 10.1073/pnas.0504834102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Erkman L, et al. Role of transcription factors Brn-3.1 and Brn-3.2 in auditory and visual system development. Nature. 1996;381:603–606. doi: 10.1038/381603a0. [DOI] [PubMed] [Google Scholar]
- 20.Xiang M, et al. Essential role of POU-domain factor Brn-3c in auditory and vestibular hair cell development. Proc Natl Acad Sci USA. 1997;94:9445–9450. doi: 10.1073/pnas.94.17.9445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sage C, et al. Essential role of retinoblastoma protein in mammalian hair cell development and hearing. Proc Natl Acad Sci USA. 2006;103:7345–7350. doi: 10.1073/pnas.0510631103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Xiang M, Gao WQ, Hasson T, Shin JJ. Requirement for Brn-3c in maturation and survival, but not in fate determination of inner ear hair cells. Development. 1998;125:3935–3946. doi: 10.1242/dev.125.20.3935. [DOI] [PubMed] [Google Scholar]
- 23.Frolenkov GI, Belyantseva IA, Friedman TB, Griffith AJ. Genetic insights into the morphogenesis of inner ear hair cells. Nat Rev Genet. 2004;5:489–498. doi: 10.1038/nrg1377. [DOI] [PubMed] [Google Scholar]
- 24.Choi PS, et al. Members of the miRNA-200 family regulate olfactory neurogenesis. Neuron. 2008;57:41–55. doi: 10.1016/j.neuron.2007.11.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kloosterman WP, Lagendijk AK, Ketting RF, Moulton JD, Plasterk RH. Targeted inhibition of miRNA maturation with morpholinos reveals a role for miR-375 in pancreatic islet development. PLoS Biol. 2007;5:e203. doi: 10.1371/journal.pbio.0050203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Haddon C, Lewis J. Early ear development in the embryo of the zebrafish, Danio rerio. J Comp Neurol. 1996;365:113–128. doi: 10.1002/(SICI)1096-9861(19960129)365:1<113::AID-CNE9>3.0.CO;2-6. [DOI] [PubMed] [Google Scholar]
- 27.Bever MM, Fekete DM. Atlas of the developing inner ear in zebrafish. Dev Dyn. 2002;223:536–543. doi: 10.1002/dvdy.10062. [DOI] [PubMed] [Google Scholar]
- 28.Lewis BP, Shih IH, Jones-Rhoades MW, Bartel DP, Burge CB. Prediction of mammalian microRNA targets. Cell. 2003;115:787–798. doi: 10.1016/s0092-8674(03)01018-3. [DOI] [PubMed] [Google Scholar]
- 29.John B, et al. Human MicroRNA targets. PLoS Biol. 2004;2:e363. doi: 10.1371/journal.pbio.0020363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Krek A, et al. Combinatorial microRNA target predictions. Nat Genet. 2005;37:495–500. doi: 10.1038/ng1536. [DOI] [PubMed] [Google Scholar]
- 31.Davis TH, et al. Conditional loss of Dicer disrupts cellular and tissue morphogenesis in the cortex and hippocampus. J Neurosci. 2008;28:4322–4330. doi: 10.1523/JNEUROSCI.4815-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Schaefer A, et al. Cerebellar neurodegeneration in the absence of microRNAs. J Exp Med. 2007;204:1553–1558. doi: 10.1084/jem.20070823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kim J, et al. A microRNA feedback circuit in midbrain dopamine neurons. Science. 2007;317:1220–1224. doi: 10.1126/science.1140481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mellios N, Huang HS, Grigorenko A, Rogaev E, Akbarian S. A set of differentially expressed miRNAs, including miR-30a–5p, act as post-transcriptional inhibitors of BDNF in prefrontal cortex. Hum Mol Genet. 2008;17:3030–3042. doi: 10.1093/hmg/ddn201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Vasudevan S, Tong Y, Steitz JA. Switching from repression to activation: microRNAs can up-regulate translation. Science. 2007;318:1931–1934. doi: 10.1126/science.1149460. [DOI] [PubMed] [Google Scholar]
- 36.Vasudevan S, Tong Y, Steitz JA. Cell-cycle control of microRNA-mediated translation regulation. Cell Cycle. 2008;7:1545–1549. doi: 10.4161/cc.7.11.6018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mendell JT. miRiad roles for the miR-17–92 cluster in development and disease. Cell. 2008;133:217–222. doi: 10.1016/j.cell.2008.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Izzotti A, et al. Downregulation of microRNA expression in the lungs of rats exposed to cigarette smoke. Faseb J. 2008;23:806–812. doi: 10.1096/fj.08-121384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kalscheuer S, Zhang X, Zeng Y, Upadhyaya P. Differential expression of microRNAs in early-stage neoplastic transformation in the lungs of F344 rats chronically treated with the tobacco carcinogen 4-(methylnitrosamino)-1-(3-pyridyl)-1-butanone. Carcinogenesis. 2008;29:2394–2399. doi: 10.1093/carcin/bgn209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Cimmino A, et al. miR-15 and miR-16 induce apoptosis by targeting BCL2. Proc Natl Acad Sci USA. 2005;102:13944–13949. doi: 10.1073/pnas.0506654102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Joglekar MV, Parekh VS, Mehta S, Bhonde RR, Hardikar AA. MicroRNA profiling of developing and regenerating pancreas reveal post-transcriptional regulation of neurogenin3. Dev Biol. 2007;311:603–612. doi: 10.1016/j.ydbio.2007.09.008. [DOI] [PubMed] [Google Scholar]
- 42.Watanabe T, et al. Dnm3os, a non-coding RNA, is required for normal growth and skeletal development in mice. Dev Dyn. 2008;237:3738–3748. doi: 10.1002/dvdy.21787. [DOI] [PubMed] [Google Scholar]
- 43.Hertzano R, et al. Transcription profiling of inner ears from Pou4f3(ddl/ddl) identifies Gfi1 as a target of the Pou4f3 deafness gene. Hum Mol Genet. 2004;13:2143–2153. doi: 10.1093/hmg/ddh218. [DOI] [PubMed] [Google Scholar]
- 44.Kloosterman WP, Wienholds E, de Bruijn E, Kauppinen S, Plasterk RH. In situ detection of miRNAs in animal embryos using LNA-modified oligonucleotide probes. Nat Methods. 2006;3:27–29. doi: 10.1038/nmeth843. [DOI] [PubMed] [Google Scholar]
- 45.Voorhoeve PM, et al. A genetic screen implicates miRNA-372 and miRNA-373 as oncogenes in testicular germ cell tumors. Cell. 2006;124:1169–1181. doi: 10.1016/j.cell.2006.02.037. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.