Abstract
Adaptation to stress in vertebrates occurs via activation of hormonal and neuronal signaling cascades in which corticotropin-releasing hormone (CRH) plays a central role. Expression of brain CRH is subject to strong, brain-region specific regulation by glucocorticoid hormones and neurogenic intracellular signals. We hypothesized that Steroid Receptor Coactivator 1 (SRC-1), a transcriptional coregulator of the glucocorticoid receptor, is involved in the sensitivity of CRH regulation by stress-related factors. In the brains of SRC-1 knockout mice we found basal CRH mRNA levels to be lower in the central nucleus of the amygdala. Hypothalamic CRH up-regulation after chronic (but not acute) stress, as well as region-dependent up- and down-regulation induced by synthetic glucocorticoids, were significantly attenuated compared with wild type. The impaired induction of the crh gene by neurogenic signals was corroborated in AtT-20 cells, where siRNA and overexpression experiments showed that SRC-1 is necessary for full induction of a CRH promoter reporter gene by forskolin, suggestive of involvement of transcription factor CREB. In conclusion, SRC-1 is involved in positive and negative regulation of the crh gene, and an important factor for the adaptive capacity of stress.
Keywords: adaptation, amygdala, HPA axis, neuroendocrinology, transcription
Brain corticotropin-releasing hormone (CRH) plays a pivotal role in the mammalian response to stress. CRH synthesized by neurons in the central nucleus of the amygdala (CeA) mediates the effect of stress on emotional states, including fear and anxiety (1, 2). From the paraventricular nucleus of the hypothalamus (PVN) CRH coordinates autonomic outflow and the activity of the hypothalamus-pituitary-adrenal (HPA) axis. The latter leads to secretion of the adrenal glucocorticoid hormones, which in turn orchestrate the adaptation to stress of virtually all tissues in the body (3–5).
As part of adaptation, CRH expression itself is strongly regulated in response to stress-induced elevations of glucocorticoids and neurogenic signals. In the core of the HPA-axis, activation of the glucocorticoid receptor (GR) can repress transcription from the crh gene as part of negative feedback, whereas stress-related noradrenergic and glutamatergic excitatory signals can activate the gene, in part via activation of the transcription factor CREB. In the CeA, both GR activation and excitatory signals can lead to an increased CRH expression (6, 7). These modulations are considered crucial for stress adaptation, and a considerable amount of research has been devoted to understand how transcriptional signals are integrated at the level of the CRH promoter, both in vitro and in vivo (8, 9). Nevertheless, the regulation of the crh gene in vivo is far from understood.
Transcriptional coregulators constitute an expanding class of molecules that act as mediators and integrators of signals carried by nuclear receptors, such as GR (10). The p160 coregulator Steroid Receptor Coactivator (SRC-1) is strongly expressed in brain (11), can affect neuronal differentiation (12), and mediates effects of androgens and estrogens in brain areas involved in sexual behavior (13–15). SRC-1 is a multimodal protein that can also interact with nonnuclear receptor transcription factors (16, 17), but to date these have not been shown to be relevant in the brain. We have recently shown that the splice variant SRC-1A can increase the sensitivity of the crh promoter to repression via GR in cultured cells (18). We therefore hypothesized that this coactivator is involved in regulation of the crh gene by stress and glucocorticoids in vivo. We tested the hypothesis by examining the effects of stress and glucocorticoids in SRC-1 knockout mice (19).
Results
Basal Expression of CRH mRNA Is Changed in SRC-1 Knockout Mice.
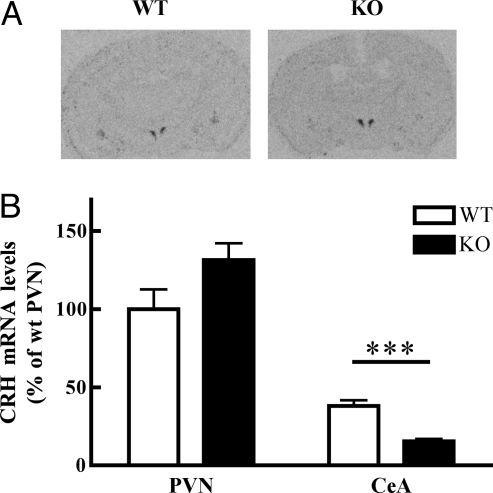
Under basal conditions CRH mRNA expression in the PVN of SRC-1 knockout mice did not significantly differ from controls (Fig. 1). Consistent with this finding, basal levels of the HPA axis hormones ACTH and corticosterone did not differ between genotypes (Fig. S1). However, CRH expression levels in the CeA were 2.5-fold lower in SRC-1 knockout mice (n = 6) compared with wild type mice (n = 8) (Fig. 1). Expression of the two types of corticosteroid receptors that may affect CRH expression, the mineralocorticoid receptor (MR) and GR, did not differ between genotypes in either nucleus (Fig. S1).
Fig. 1.
SRC-1 affects basal CRH mRNA expression in a region specific manner. (A) Autoradiograms showing CRH mRNA signal in the brain at the level of PVN and CeA. (B) CRH mRNA is lower in the CeA in SRC-1 knockout mice under basal conditions (n = 6–8). Two-way ANOVA indicates a significant effect for brain region (F1,26 = 130.1; P < 0.0001) and an interaction for genotype x brain region (F1,26 = 9.60; P < 0.01). **, P < 0.01, comparison with vehicle treated group.
SRC-1 Knockout Mice Are Resistant to both Up- and Down-Regulation of CRH by GCs.
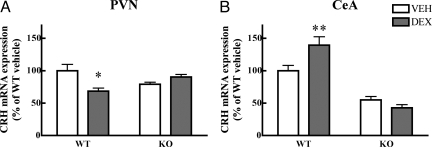
A unique feature of the crh gene is its opposite regulation by glucocorticoids in PVN and CeA. Both transactivating (e.g., induction of the tyrosine amino transferase gene in liver) and transrepressive effects (repression of the POMC gene in the pituitary) of GR activation may depend on SRC-1 in vivo (20). We therefore tested whether dexamethasone, a potent synthetic glucocorticoid, was able to regulate CRH mRNA expression in absence of SRC-1. In wild type mice, 5 day treatment with dexamethasone (5 mg/kg) led to the expected down-regulation of CRH mRNA in PVN and an up-regulation in CeA. However, dexamethasone induced up-, and down regulation were absent in SRC-1 knockout animals (Fig. 2). Despite resistance of crh and pomc (20) genes to dexamethasone suppression, the treatment led to effective suppression of plasma corticosterone in both genotypes (Fig. S2).
Fig. 2.
CRH expression is resistant to dexamethasone treatment in SRC-1 knockout mice. (A) Down-regulation of CRH expression is observed in the PVN of wild type mice but not in SRC-1 knockout animals (n = 3–5). Two-way ANOVA indicates a significant interaction between genotype and treatment (F1,13 = 6.63; P < 0.05). (B) Up-regulation in the CeA occurs only in wild type mice. Two-way ANOVA indicates a significant effect of genotype and an interaction effect (F1,13 = 7.74; P < 0.05). Data are expressed as percentage of vehicle treated wild type mice. *, P < 0.05 and **, P < 0.01, comparisons with vehicle treated group.
Modulation of CRH Expression Is Impaired in SRC-1 Knockout Mice.
To test the role of SRC-1 in the regulation of CRH expression by stress, we subjected mice to 10 days of predator stress. Corticosterone was taken as the relevant readout of the HPA axis, in view of its potential to regulate CRH expression. Once or twice daily a rat was placed on the mouse cage for one hour, at various starting times. This led to a robust stress response that did not differ between genotypes (Fig. 3A). After 10 days of rat stress, the mice displayed a habituated response to the rat exposure, but SRC-1 knockout mice had modestly higher corticosterone levels in response to stress, compared with wild type controls (Fig. 3B). Basal plasma corticosterone levels did not change as a consequence of the stress paradigm.
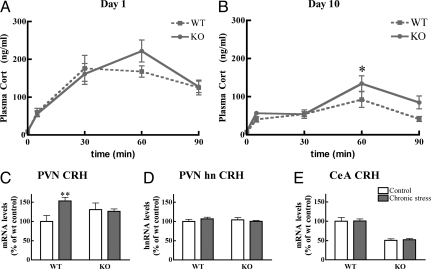
Fig. 3.
Adaptation to chronic rat stress depends on SRC-1. (A) Acute rat exposure leads to a robust corticosterone response, which is not different between the genotypes (n = 7–10). Two-way ANOVA, time effect: F3,54 = 13.87; P < 0.0001. (B) After 10 days the mice had habituated to the stressor but still showed a significant response to the stressor of rat exposure. Two-way ANOVA indicates a significant effect for time (F3,54 = 9.67; P < 0.0001) and for genotype (F1,54 = 5.42; P < 0.05). (C) CRH mRNA expression in the PVN is increased after chronic stress in wild type but not knockout mice (n = 7–19). Data are expressed as percentage of nonstressed wild type mice. Two-way ANOVA shows significance for treatment (F1,41 = 5.42; P < 0.05) and an interaction between treatment and genotype (F1,41 = 8.85; P < 0.01). (D) In PVN, the CRH hnRNA response to acute restraint is not affected by prior rat stress or genotype. (E) Rat stress does not lead to significant changes in expression of CRH in CeA in either genotype. Two-way ANOVA only shows the genotype effect also observed in untreated animals because of >2-fold lower expression of CRH mRNA in SRC-1 knockout mice (F1,41 = 93.93; P < 0.001; see also Fig. 1).
To further test changes in the stress system, we subjected mice to the heterotypic stressor of 30 min restraint stress, one day after the last rat exposure. The corticosterone response at 5 min after the onset of restraint was 2-fold higher in chronically stressed compared with naive SRC-1 knockout mice (60 ± 5 vs. 31 ± 7 ng/ml; P < 0.001), but not in wild type controls. Peak corticosterone levels at 30 min of restraint stress were similar between naive and chronically stressed mice (wt naive 189 ± 12 ng/ml; wt rat stress 179 ± 9 ng/ml), and this was independent of genotype. ACTH levels at 30 min after restraint did not differ between groups (Fig. S3). Thus, at the level of adrenal output the SRC-1 knockout mice show a mild difference in adjustment to chronic stress.
More pronounced changes were observed in brain, where, in accordance with different adaptation to stress in the two genotypes, the chronic stress paradigm led to increased CRH mRNA expression in the PVN of wild type, but not of SRC-1 knockout mice (Fig. 3C). The acute transcriptional response of the crh gene in the PVN as measured by an intronic probe (21), 30 min after restraint, did not depend on genotype or prior stress history (Fig. 3D). Stressors had no effect in the CeA of either genotype (Fig. 3E); independent of stress history, CeA CRH mRNA was >2-fold lower in SRC-1 knockout mice.
SRC-1 Can Modulate the Induction of the CRH Promoter by Forskolin.
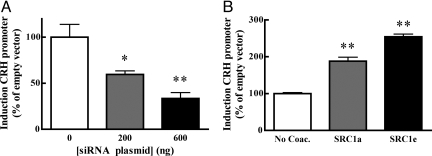
The SRC-1 knockout mice data showed that both basal and chronic stress-induced CRH expression is to some extent dependent on SRC-1. This suggests that on the CRH promoter the activity of transcription factors other than GR depend on SRC-1, particularly because there are no major differences between genotypes in circulating corticosterone levels, and expression of MR and GR. To directly test this hypothesis we used a reporter construct containing the proximal promoter of the human CRH gene (22) in AtT-20 cells, a well established model to study modulation of CRH expression. Treatment of these cells with forskolin leads to a robust increase in reporter activity, which depends on an intact CRE in the reporter (9). Expression of a siRNA directed against SRC-1 (23) dose dependently reduced the induction of the CRH promoter (Fig. 4A). The splice variants SRC-1a and 1e differentially affect steroid receptor signaling at multiple promoters, including GR-mediated repression of the CRH promoter (24, 25). We therefore studied the effect of both splice variants on forskolin-stimulated induction of the CRH promoter by overexpression. SRC-1a and 1e potentiated the effect of forskolin to a similar extent (Fig. 4B).
Fig. 4.
Forskolin-stimulated induction of the CRH promoter depends on SRC-1. (A) Knockdown of SRC-1 with siRNA dose-dependently attenuates induction of the hCRH promoter by forskolin, as tested by 1-way ANOVA (F2,8 = 11.53; P < 0.005). *, P < 0.05, **, P < 0.01, compared with control condition, Tukey's posthoc test. (B) The 2 main SRC-1 splice variants both increase forskolin-stimulated induction of the hCRH promoter as indicated by 1-way ANOVA (F2,13 = 156.1, P < 0.001). **, significantly different from control condition, P < 0.01, Tukey's posthoc test.
Discussion
The crh gene is arguably the primary GR target gene in brain with the best-understood function, i.e., coordination of the stress response. In the present study we show that the nuclear receptor coactivator SRC-1 is involved in regulation of the crh gene by multiple stimuli in two different brain regions that are crucial for regulation of stress responses. CRH mRNA induction by glucocorticoids in the CeA was abolished in SRC-1 knockout mice. CRH repression in the PVN was also affected by the absence of SRC-1. More surprisingly, our data also suggest SRC-1 involvement in likely nonGR dependent positive regulation of the CRH gene, as indicated by the strong reduction in basal expression levels in the CeA, and the lack of chronic stress-induced up-regulation of CRH expression in the PVN. We corroborated the latter effect by overexpression and knockdown of SRC-1 in the context of forskolin-stimulated activity of the proximal promoter of the crh gene.
Our data confirm earlier results that showed that the SRC-1 knockout mice have normal basal corticosterone levels, and a mild adrenal hyperactivity in response to restraint stress that is accompanied by increased adrenal size and enhanced expression of melanocortin 2 receptors (20). However, in relation to rat stress, SRC-1 knockout mice showed an enhanced adrenal output only after chronic exposure, indicating involvement in the adaptation to this stressor. In relation to regulation of the crh gene, it is relevant that corticosterone and receptor levels were not dramatically different between genotypes during the chronic stress paradigm.
Given the large degree of gene and cell specificity of coregulator involvement in nuclear receptor signaling, the extent of SRC-1 involvement with the crh gene is striking. A role in GR-mediated CRH mRNA induction in CeA is in line with the traditional view of SRC action, based on agonist-induced coactivator interactions with the transactivation function-2 domain of steroid receptors (26). Although studies in cell lines have shown that SRC-1 is recruited directly to the crh promoter via the estrogen receptor (27), it is not known whether there is a functional positive glucocorticoid response element (GRE) in the gene. The involvement of SRC-1 does support a GRE-dependent mechanism for GR-induction of CRH mRNA. However, not all GRE-mediated signaling in brain is affected in SRC-1 knockout mice because GR-mediated induction of the ubiquitous GR target gene gilz (28) is unaffected in the brain of these mice. Other stress-related neuropeptides can also be influenced by glucocorticoids, e.g., vasopressin that is coexpressed with CRH in the PVN (8), and it will be interesting to evaluate such effect in relation to SRC-1 and other coregulators.
The lack of dexamethasone-induced down-regulation of CRH mRNA in the PVN of SRC-1 knockout mice followed our hypothesis, based on earlier data on SRC-1 involvement in repression of the pomc gene (20), and the effects of SRC-1A overexpression on CRH gene repression in cell lines (18). The mechanism of the corepressor effect of SRC-1 remains elusive, and may involve the SRC-1A specific NR-interaction box and repression function (24) or an alternative NR interaction domain (29). For both crh and pomc genes, valid arguments are available for repression via GR that is DNA-binding dependent (22, 30) or independent (31, 32). Irrespective, it is striking that GR's negative feedback-related regulation of the two main secretagogues of the HPA-axis depends on SRC-1 (20), in two tissues with a large bias toward SRC-1A expression (11). However, direct evidence for specific involvement of particular splice variants is lacking.
SRC-1 also drives crh gene expression under basal conditions in the CeA, and chronic stress-induced increased CRH mRNA expression in PVN. Also, a trend toward higher CRH expression was present in the PVN of naive mice (Figs. 1 and 3). This may have failed to reach significance because of lack of statistical power. These effects likely do not involve decreased GR function; complete absence of GR activity induced by adrenalectomy was reported to have only a marginal effect of CRH expression in the CeA, and in PVN would lead to the opposite effect of what is observed (33). Other nuclear receptors may be involved in these differences. In particular ER and TR have been shown to affect CRH mRNA levels during development (34) and in adulthood (35, 36), and to depend in part on SRC-1 (19, 37).
Alternatively, other transcription factors are involved, such as CREB; the CRH promoter contains a well characterized cAMP response element, that mediates hypothalamic transactivation after acute stress (38). Our hnRNA data suggest that SRC-1 is involved in the response to chronic, rather than acute stress, which may be relevant in relation to the different demands on the stress system under acute and chronic conditions. The identical hnRNA induction argues against coactivation of CREB by SRC-1 in the acute situation. However, our results in AtT-20 cells show that SRC-1 can affect stimulation of the CRH promoter by forskolin in some cellular contexts, potentially via CREB signaling (39). Recent data suggest that occupation of this element by phosphorylated CREB is by itself not sufficient to drive CRH expression, but that additional transcription factors and coregulators are necessary (9). If the in vitro effect underlies some of our in vivo data, SRC-1 may act as a coactivator for a CRE-associated transcription factor, but only in the context of chronic stressors (40).
Given the importance of SRC-1 for the regulation of crh pomc genes, the phenotype of SRC-1 knockout mice may seem surprisingly mild. However, earlier studies with these mice have shown that increased SRC-2 expression can compensate and therefore mask certain phenotypes, such as involvement of SRC-1 in estrogen receptor signaling in the hypothalamus (13, 19). The complexity of the HPA axis, and the number of check points that lie between mRNA expression of the secretagogues CRH and ACTH and actual secretion of biologically active peptides (41, 42) may allow genomic compensation that is almost complete under basal conditions. The escape from compensation of the crh gene may be due to the involvement of SRC-1 splice variant specific functions, or interactions of SRC-1 with nonnuclear receptors, which may display a lesser degree of redundancy that the classical transactivating properties of the p160 SRC family on nuclear receptors.
In conclusion, we identified SRC-1 as pivotal for regulation of the crh gene in response to GR-related and other, possibly nonnuclear receptor dependent stimuli. This coactivator was known to play a role in transferring states of different sexual responsiveness, and now also seems to play an important role in (re)setting the central responses to stress via actions within and outside the hypothalamus. This hypothesis will have to be further studied by using transient knockdown techniques, in which compensatory mechanisms may be less prominent. Future studies will have to include the role of SRC-1 in the interplay with other transcription factors and coregulators known to impact on the crh gene (43). For now it is an open question what other genes in stress-related circuitry depend on SRC-1. In addition, it will be important to study behavioral consequences of transient SRC-1 knockdown, or otherwise altered activity of the coactivator, e.g. by posttranslation modifications (44). Nevertheless, given the central role of CRH in adaptation to stress, the present data merit the conclusion that SRC-1 can be considered an important factor for the “adaptive capacity” to stress.
Materials and Methods
Animals.
SRC-1 knockout mice (c57bl/6 background) and wild type littermates (19) were bred in Houston (dex experiment) and Leiden (stress experiments), and after weaning were housed at 2–3 per cage under standard lab conditions, at a 12:12-h light dark cycle, lights on at 7:00 AM, and chow available ad lib. 2–4 months old male mice were used. The experiments were carried out in accordance with European Communities Council Directive 86/609/EEC and the Dutch law on animal experiments, and were approved by the Leiden University animal ethical commission (protocol 02136), or the BCM Animal Care and Use Committee, respectively.
For determination of basal parameters, mice were decapitated between 8 and 10 AM, trunk blood and organs were collected and stored. For experiment 2, mice were treated with dexamethasone phosphate (Sigma) at 5 mg/kg body weight, i.p. in saline, or vehicle for 5 consecutive days in the morning. 3 h after the last injection the animals were decapitated and blood and organs were collected.
In the third experiment, wild type and SRC-1 knockout mice were exposed to the predator stress of rat exposure if left undisturbed. In the first week, the “rat stress” groups were exposed to rats on 5 consecutive days for 1–3 h a day at semirandom times, resulting in a total exposure of 10 h in the first week. In the 2nd week, two exposures took place of 1 h each. Adult male Sprague–Dawley rats were placed in cages with a grid floor on top of the mouse cages, so that animals could see, hear and smell, but not touch each other. During rat exposure, food and water were not present. The mice were kept in another cage than their home cage, but always the same cage was used for the same group of mice. The position in the rat stress room was different for each exposure, as were the rats used per cage. After rat stress, mice were put back in their home cage and in the same room than the “nonrat stress” animals.
To follow the stress response of the different groups, blood samples were taken by tail cuts at different time points: 3 days before the start of stress paradigm, for an (AM) basal level. On day 0 (first rat exposure: acute phase), the rat stress groups were divided into two different groups and two blood samples were taken via a tail cut; one group had tail cut after 5 and 60 min of rat stress, the second group at 30 and 90 min after rat stress. On day 9, a sample was collected in all animals to measure basal AM level. On day 11 (last rat exposure: chronic phase), the same procedure as on day 0 was performed.
The day after the last rat exposure, all 4 groups of mice were stressed by restraint inside a plastic tube (internal diameter 25 mm). The mice had one tail cut after 5 min restraint and were decapitated 30 min after restraint. Trunk blood was collected, and different tissues were dissected.
Tissue Measurements.
In situ hybridizations were performed as described in ref. 41 by using 35S-UTP labeled riboprobes against mouse MR exon 2, GR exon 2 (courtesy Dr. T. Cole), a 1 kb fragment of the CRH gene (courtesy Dr. G. Dent), and a 505-bp fragment of the mouse CRH intron (courtesy of Dr. A. Seasholtz). Final stringency of posthybridization washes was at 0.1 x standard sodium citrate at 65 °C. Signal was visualized with exposure of Kodak Biomax MR films, and 14C microscales (GE Healthcare) were used to calibrate the signal. Films were scanned and quantified by using Image J software (National Institutes of Health). PVN and CeA CRH mRNA was measured bilaterally in two sections per animal, a rostral section located between 0.58 and 0.94 mm caudal from Bregma, and a caudal section located between 0.94 and 1.06 mm from Bregma (45). Values represent the average value for the 2 levels.
For plasma corticosterone measurement, tail cut and trunk blood was collected into precooled EDTA-coated microfuge tubes, centrifuged for 15 min at 13000 rpm at 4 °C, and plasma was stored at −20 °C. Corticosterone and ACTH levels were determined by 125I RIA kits (ImmuChem, MP Biochemicals). Detection limits were 5 ng/ml (corticosterone) and 10 pg/ml (ACTH).
Transfection Studies.
The pSuperSRC-1 plasmid codes a shRNA specifically targeting SRC-1 (courtesy Dr. C. Massaad) (23). The CRH-LUC plasmid contains 900 bp of the 5′flanking region of the human CRH gene fused to luciferase (courtesy Dr. R. Dorin) (46). Expression vectors for SRC-1A and -1E were described in ref. 25. AtT-20/D-16V mouse tumor cells were used as described in ref. 18. For transfections we used 0.1 × 106 cells, 1.6 μl of Lipofectamine 2000 (Invitrogen Life Technologies) and 0.8 μg of total plasmid per well. In all assays 200 ng of CRH-LUC reporter plasmid was used. Knock-down was performed by using varying amounts of pSuperSRC-1 (23) 48 h before treatment. To induce the CRH-promoter, the cells were treated overnight with 10 μM forskolin (Calbiochem). For overexpression studies 400 ng of expression vector plasmid was used per well. The following day, the cells were treated for 4 h with 10 μM FSK. The cells were harvested and assayed according to the luciferase kit instructions (Promega) by using a luminometer (LUMAT LB 9507, Berthold).
Statistical Analysis.
All data are presented as means ±SEM. Statistical analysis was determined by one-way or two-way ANOVA as appropriate, followed by Tukey or Bonferroni posthoc tests, respectively, by using Prism GraphPad. P values <0.05 were considered statistically significant.
Supplementary Material
Acknowledgments.
This work was supported by Netherlands Organisation for Health Research and Development Vidi Grant 016.036.381 (to O.C.M.), National Alliance for Research on Schizophrenia and Depression (O.C.M.), the Royal Netherlands Academy of Arts and Sciences (E.R.d.K. and O.C.M.), National Institute of Child Health and Human Development/National Institutes of Health (NIH) through cooperative agreement U54 HD07495 Project One (E.M.A.) as part of the Specialized Cooperative Centers Program in Reproduction Research (E.M.A.), and NIH Grant CA112403 (to J.X.). We thank Drs. C. Massaad (University Paris Descartes, Paris), R. Dorin (University of New Mexico School of Medicine, Albuquerque, NM), and A. Seasholtz (University of Michigan Medical School, Ann Arbor, MI) for sharing plasmids.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0812062106/DCSupplemental.
References
- 1.Gray TS. Amygdaloid CRF pathways. Role in autonomic, neuroendocrine, and behavioral responses to stress. Ann N Y Acad Sci. 1993;697:53–60. doi: 10.1111/j.1749-6632.1993.tb49922.x. [DOI] [PubMed] [Google Scholar]
- 2.Heinrichs SC, Koob GF. Corticotropin-releasing factor in brain: A role in activation, arousal, and affect regulation. J Pharmacol Exp Ther. 2004;311:427–440. doi: 10.1124/jpet.103.052092. [DOI] [PubMed] [Google Scholar]
- 3.Herman JP, et al. Central mechanisms of stress integration: Hierarchical circuitry controlling hypothalamo-pituitary-adrenocortical responsiveness. Front Neuroendocrinol. 2003;24:151–180. doi: 10.1016/j.yfrne.2003.07.001. [DOI] [PubMed] [Google Scholar]
- 4.Bale TL, Vale WW. CRF and CRF receptors: Role in stress responsivity and other behaviors. Annu Rev Pharmacol Toxicol. 2004;44:525–557. doi: 10.1146/annurev.pharmtox.44.101802.121410. [DOI] [PubMed] [Google Scholar]
- 5.de Kloet ER, Joels M, Holsboer F. Stress and the brain: From adaptation to disease. Nat Rev Neurosci. 2005;6:463–475. doi: 10.1038/nrn1683. [DOI] [PubMed] [Google Scholar]
- 6.Kalin NH, Takahashi LK, Chen FL. Restraint stress increases corticotropin-releasing hormone mRNA content in the amygdala and paraventricular nucleus. Brain Res. 1994;656:182–186. doi: 10.1016/0006-8993(94)91382-x. [DOI] [PubMed] [Google Scholar]
- 7.Swanson LW, Simmons DM. Differential steroid hormone and neural influences on peptide mRNA levels in CRH cells of the paraventricular nucleus: A hybridization histochemical study in the rat. J Comp Neurol. 1989;285:413–435. doi: 10.1002/cne.902850402. [DOI] [PubMed] [Google Scholar]
- 8.Watts AG. Glucocorticoid regulation of peptide genes in neuroendocrine CRH neurons: A complexity beyond negative feedback. Front Neuroendocrinol. 2005;26:109–130. doi: 10.1016/j.yfrne.2005.09.001. [DOI] [PubMed] [Google Scholar]
- 9.Liu Y, Kamitakahara A, Kim AJ, Aguilera G. Cyclic adenosine 3′,5′-monophosphate responsive element binding protein phosphorylation is required but not sufficient for activation of corticotropin-releasing hormone transcription. Endocrinology. 2008;149:3512–3520. doi: 10.1210/en.2008-0052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lonard DM, O'Malley BW. The expanding cosmos of nuclear receptor coactivators. Cell. 2006;125:411–414. doi: 10.1016/j.cell.2006.04.021. [DOI] [PubMed] [Google Scholar]
- 11.Meijer OC, Steenbergen PJ, De Kloet ER. Differential expression and regional distribution of steroid receptor coactivators SRC-1 and SRC-2 in brain and pituitary. Endocrinology. 2000;141:2192–2199. doi: 10.1210/endo.141.6.7489. [DOI] [PubMed] [Google Scholar]
- 12.Nishihara E, et al. SRC-1 null mice exhibit moderate motor dysfunction and delayed development of cerebellar purkinje cells. J Neurosci. 2003;23:213–222. doi: 10.1523/JNEUROSCI.23-01-00213.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Apostolakis EM, Ramamurphy M, Zhou D, Onate S, O'Malley BW. Acute disruption of select steroid receptor coactivators prevents reproductive behavior in rats and unmasks genetic adaptation in knockout mice. Mol Endocrinol. 2002;16:1511–1523. doi: 10.1210/mend.16.7.0877. [DOI] [PubMed] [Google Scholar]
- 14.Auger AP, Tetel MJ, McCarthy MM. Steroid receptor coactivator-1 (SRC-1) mediates the development of sex- specific brain morphology and behavior. Proc Natl Acad Sci USA. 2000;97:7551–7555. doi: 10.1073/pnas.97.13.7551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Charlier TD, Ball GF, Balthazart J. Inhibition of steroid receptor coactivator-1 blocks estrogen and androgen action on male sex behavior and associated brain plasticity. J Neurosci. 2005;25:906–913. doi: 10.1523/JNEUROSCI.3533-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lee SK, et al. Steroid receptor coactivator-1 coactivates activating protein-1-mediated transactivations through interaction with the c-Jun and c-Fos subunits. J Biol Chem. 1998;273:16651–16654. doi: 10.1074/jbc.273.27.16651. [DOI] [PubMed] [Google Scholar]
- 17.Lee SK, Kim HJ, Kim JW, Lee JW. Steroid receptor coactivator-1 and its family members differentially regulate transactivation by the tumor suppressor protein p53. Mol Endocrinol. 1999;13:1924–1933. doi: 10.1210/mend.13.11.0365. [DOI] [PubMed] [Google Scholar]
- 18.van der Laan S, Lachize SB, Vreugdenhil E, de Kloet ER, Meijer OC. Nuclear receptor coregulators differentially modulate induction and glucocorticoid receptor-mediated repression of the corticotropin-releasing hormone gene. Endocrinology. 2008;149:725–732. doi: 10.1210/en.2007-1234. [DOI] [PubMed] [Google Scholar]
- 19.Xu J, et al. Partial hormone resistance in mice with disruption of the steroid receptor coactivator-1 (SRC-1) gene. Science. 1998;279:1922–1925. doi: 10.1126/science.279.5358.1922. [DOI] [PubMed] [Google Scholar]
- 20.Winnay JN, Xu J, O'Malley BW, Hammer GD. Steroid receptor coactivator-1-deficient mice exhibit altered hypothalamic-pituitary-adrenal axis function. Endocrinology. 2006;147:1322–1332. doi: 10.1210/en.2005-0751. [DOI] [PubMed] [Google Scholar]
- 21.Rozeboom AM, Akil H, Seasholtz AF. Mineralocorticoid receptor overexpression in forebrain decreases anxiety-like behavior and alters the stress response in mice. Proc Natl Acad Sci USA. 2007;104:4688–4693. doi: 10.1073/pnas.0606067104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Malkoski SP, Dorin RI. Composite glucocorticoid regulation at a functionally defined negative glucocorticoid response element of the human corticotropin-releasing hormone gene. Mol Endocrinol. 1999;13:1629–1644. doi: 10.1210/mend.13.10.0351. [DOI] [PubMed] [Google Scholar]
- 23.Grenier J, et al. Differential recruitment of p160 coactivators by glucocorticoid receptor between Schwann cells and astrocytes. Mol Endocrinol. 2006;20:254–267. doi: 10.1210/me.2005-0061. [DOI] [PubMed] [Google Scholar]
- 24.Kalkhoven E, Valentine JE, Heery DM, Parker MG. Isoforms of steroid receptor co-activator 1 differ in their ability to potentiate transcription by the oestrogen receptor. EMBO J. 1998;17:232–243. doi: 10.1093/emboj/17.1.232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Meijer OC, et al. Steroid receptor coactivator-1 splice variants differentially affect corticosteroid receptor signaling. Endocrinology. 2005;146:1438–1448. doi: 10.1210/en.2004-0411. [DOI] [PubMed] [Google Scholar]
- 26.Onate SA, Tsai SY, Tsai MJ, O'Malley BW. Sequence and characterization of a coactivator for the steroid hormone receptor superfamily. Science. 1995;270:1354–1357. doi: 10.1126/science.270.5240.1354. [DOI] [PubMed] [Google Scholar]
- 27.Lalmansingh AS, Uht RM. Estradiol regulates corticotropin-releasing hormone gene (crh) expression in a rapid and phasic manner that parallels estrogen receptor-alpha and -beta recruitment to a 3′,5′-cyclic adenosine 5′-monophosphate regulatory region of the proximal crh promoter. Endocrinology. 2008;149:346–357. doi: 10.1210/en.2007-0372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.van der Laan S, et al. Chromatin immunoprecipitation (ChIP) scanning identifies glucocorticoid receptor binding regions in the proximal promoter of a ubiquitously expressed glucocorticoid target gene in brain. J Neurochem. 2008;106:2515–2523. doi: 10.1111/j.1471-4159.2008.05575.x. [DOI] [PubMed] [Google Scholar]
- 29.Bevan CL, Hoare S, Claessens F, Heery DM, Parker MG. The AF1 and AF2 domains of the androgen receptor interact with distinct regions of SRC1. Mol Cell Biol. 1999;19:8383–8392. doi: 10.1128/mcb.19.12.8383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Reichardt HM, et al. DNA binding of the glucocorticoid receptor is not essential for survival. Cell. 1998;93:531–541. doi: 10.1016/s0092-8674(00)81183-6. [DOI] [PubMed] [Google Scholar]
- 31.Martens C, Bilodeau S, Maira M, Gauthier Y, Drouin J. Protein–protein interactions and transcriptional antagonism between the subfamily of NGFI-B/Nur77 orphan nuclear receptors and glucocorticoid receptor. Mol Endocrinol. 2005;19:885–897. doi: 10.1210/me.2004-0333. [DOI] [PubMed] [Google Scholar]
- 32.Yamamori E, et al. Molecular mechanisms for corticotropin-releasing hormone gene repression by glucocorticoid in BE(2)C neuronal cell line. Mol Cell Endocrinol. 2007;264:142–148. doi: 10.1016/j.mce.2006.11.001. [DOI] [PubMed] [Google Scholar]
- 33.Makino S, Gold PW, Schulkin J. Effects of corticosterone on CRH mRNA and content in the bed nucleus of the stria terminalis; comparison with the effects in the central nucleus of the amygdala and the paraventricular nucleus of the hypothalamus. Brain Res. 1994;657:141–149. doi: 10.1016/0006-8993(94)90961-x. [DOI] [PubMed] [Google Scholar]
- 34.Seale JV, Wood SA, Atkinson HC, Lightman SL, Harbuz MS. Organizational role for testosterone and estrogen on adult hypothalamic-pituitary-adrenal axis activity in the male rat. Endocrinology. 2005;146:1973–1982. doi: 10.1210/en.2004-1201. [DOI] [PubMed] [Google Scholar]
- 35.Shi ZX, Levy A, Lightman SL. Thyroid hormone-mediated regulation of corticotropin-releasing hormone messenger ribonucleic acid in the rat. Endocrinology. 1994;134:1577–1580. doi: 10.1210/endo.134.3.8119200. [DOI] [PubMed] [Google Scholar]
- 36.Seale JV, Wood SA, Atkinson HC, Harbuz MS, Lightman SL. Gonadal steroid replacement reverses gonadectomy-induced changes in the corticosterone pulse profile and stress-induced hypothalamic-pituitary-adrenal axis activity of male and female rats. J Neuroendocrinol. 2004;16:989–998. doi: 10.1111/j.1365-2826.2004.01258.x. [DOI] [PubMed] [Google Scholar]
- 37.Weiss RE, et al. Mice deficient in the steroid receptor co-activator 1 (SRC-1) are resistant to thyroid hormone. EMBO J. 1999;18:1900–1904. doi: 10.1093/emboj/18.7.1900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Shepard JD, Liu Y, Sassone-Corsi P, Aguilera G. Role of glucocorticoids and cAMP-mediated repression in limiting corticotropin-releasing hormone transcription during stress. J Neurosci. 2005;25:4073–4081. doi: 10.1523/JNEUROSCI.0122-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sands WA, Palmer TM. Regulating gene transcription in response to cyclic AMP elevation. Cell Signal. 2008;20:460–466. doi: 10.1016/j.cellsig.2007.10.005. [DOI] [PubMed] [Google Scholar]
- 40.Herman JP, Flak J, Jankord R. Chronic stress plasticity in the hypothalamic paraventricular nucleus. Prog Brain Res. 2008;170:353–364. doi: 10.1016/S0079-6123(08)00429-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Karssen AM, Meijer OC, Berry A, Sanjuan Pinol R, de Kloet ER. Low doses of dexamethasone can produce a hypocorticosteroid state in the brain. Endocrinology. 2005;146:5587–5595. doi: 10.1210/en.2005-0501. [DOI] [PubMed] [Google Scholar]
- 42.John CD, Gavins FN, Buss NA, Cover PO, Buckingham JC. Annexin A1 and the formyl peptide receptor family: Neuroendocrine and metabolic aspects. Curr Opin Pharmacol. 2008;8:765–776. doi: 10.1016/j.coph.2008.09.005. [DOI] [PubMed] [Google Scholar]
- 43.Aguilera G, Kiss A, Liu Y, Kamitakahara A. Negative regulation of corticotropin releasing factor expression and limitation of stress response. Stress. 2007;10:153–161. doi: 10.1080/10253890701391192. [DOI] [PubMed] [Google Scholar]
- 44.Han SJ, Lonard DM, O'Malley BW. Multi-modulation of nuclear receptor coactivators through posttranslational modifications. Trends Endocrinol Metab. 2009;20:8–15. doi: 10.1016/j.tem.2008.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Franklin KBJ, Paxinos G. The Mouse Brain in Stereotaxic Coordinates. San Diego: Academic; 1997. [Google Scholar]
- 46.Malkoski SP, Handanos CM, Dorin RI. Localization of a negative glucocorticoid response element of the human corticotropin-releasing hormone gene. Mol Cell Endocrinol. 1997;127:189–199. doi: 10.1016/s0303-7207(96)04004-x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.