Abstract
The molecular mechanism by which neural progenitor cells commit to a specified lineage of the central nervous system remains unknown. We show that HDAC1 and HDAC2 redundantly control neuronal development and are required for neuronal specification. Mice lacking HDAC1 or HDAC2 in neuronal precursors show no overt histoarchitectural phenotypes, whereas deletion of both HDAC1 and HDAC2 in developing neurons results in severe hippocampal abnormalities, absence of cerebellar foliation, disorganization of cortical neurons, and lethality by postnatal day 7. These abnormalities in brain formation can be attributed to a failure of neuronal precursors to differentiate into mature neurons and to excessive cell death. These results reveal redundant and essential roles for HDAC1 and HDAC2 in the progression of neuronal precursors to mature neurons in vivo.
Keywords: cerebellum, hippocampus, neurogenesis, neuronal precursors
Histone acetyltransferases (HATs) and histone deacetylases (HDACs) provide the enzymatic basis for transcriptional activation and repression, respectively, through alterations of the chromatin landscape (1). Transcription factors recruit HDACs, either individually, or in repressive complexes to deacetylate lysine residues on histone tails, resulting in chromatin condensation and repression of gene expression (2). There are 4 classes of HDACs that coordinate proper gene regulation for numerous cellular processes: class I (HDAC1, -2, -3, and -8), class II (HDAC4, -5, -6, -7, -9, and -10), sirtuin class III, and class IV (HDAC11) (3). Although much has been learned through in vitro and inhibitor studies, little is known about the biological function of these individual enzymes in vivo (4). We have shown that the class I HDACs, HDAC1 and HDAC2, redundantly regulate cardiac growth and morphogenesis (5), however, the functions of HDAC1 and HDAC2 in other tissues remain unknown.
HDAC inhibitors have shown significant potential for therapeutic use in a variety of disorders, including those of the central nervous system (CNS), such as neurodegenerative disease, motor neuron disease, and a number of other neurological disease states (6). Furthermore, it has been shown that HDAC inhibitors induce differentiation of both embryonic and adult cortical neuronal progenitor cells to neurons specifically (7–9). The wide expression pattern of a number of HDACs in the developing brain suggests specific roles for individual HDACs in neuronal development (10), however, the broad inhibition of classical HDAC inhibitors has precluded the analysis of individual HDACs pharmacologically. Additionally, the early lethality associated with global deletion of class I HDACs in knockout mice has compounded the difficulties in analyzing the functions of these enzymes during specific stages of neurogenesis in vivo (5, 11).
To further investigate the specific roles of HDAC1 and HDAC2 in neuronal development, we generated conditional deletions of HDAC1 and HDAC2 in the central nervous system. Here, we show both HDAC1 and HDAC2 are required for multiple aspects of neuronal development. Deletion of both HDAC1 and HDAC2 results in major abnormalities of cortical, hippocampal, and cerebellar development, whereas deletion of HDAC1 or HDAC2 individually has no apparent effect on neuronal development. Neuronal precursors lacking HDAC1 and HDAC2 are unable to differentiate into mature neurons specifically and undergo cell death, resulting in defects in superficial cortical layering and death by postnatal day 7. These results reveal HDAC1 and HDAC2 as critical and redundant regulators of neuronal differentiation during neocortical, hippocampal, and cerebellar development.
Results
Neuronal Abnormalities upon CNS Deletion of HDAC1 and HDAC2.
To generate mice that lack HDAC1 and HDAC2 in the central nervous system, we crossed HDAC1loxP/loxP and HDAC2loxP/loxP mice to transgenic mice expressing Cre recombinase under the control of the human glial fibrillary acidic protein promoter, GFAP-Cre, which is widely expressed in the central nervous system, including neural stem cells (5, 12). HDAC1 and HDAC2 expression during brain development has been extensively studied and show relatively contrasting expression patterns throughout the central nervous system. HDAC1 is highly expressed in glia, whereas HDAC2 shows high expression in mature neurons, however, both are expressed in neuronal stem cells/progenitors during embryonic development (13). HDAC1loxP/loxP; GFAP-Cre and HDAC2loxP/loxP; GFAP-Cre mice were born at normal Mendelian ratios and showed normal brain histoarchitecture, behavior, motor skills, and lifespan (supporting information (SI) Fig. S1). HDAC1loxP/loxP; HDAC2loxP/loxP; GFAP-Cre mice were also born at normal Mendelian ratios, but died within the first week, accompanied by a reduced brain size and compacted cerebellum (Fig. 1). Immunohistochemistry of HDAC1 and HDAC2 showed efficient deletion of HDAC1 and HDAC2 in the ventricular zone and cortical plate regions of the neocortex (Fig. S2). One copy of a wild-type allele of HDAC1 or HDAC2 was sufficient for neural development and viability, further indicating the redundant functions of these genes.
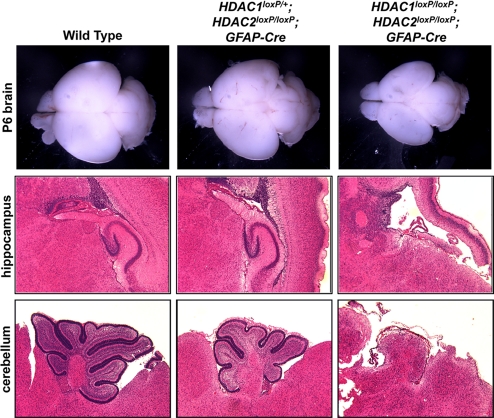
Fig. 1.
HDAC1 and HDAC2 deletion causes severe hippocampal and cerebellar abnormalities. Gross brain, hippocampus, and cerebellum from wild-type, HDAC1loxP/+; HDAC2loxP/loxP; GFAP-Cre, and HDAC1loxP/loxP; HDAC2loxP/loxP; GFAP-Cre mice stained with hematoxylin and eosin showing loss of hippocampus and loss of foliation in cerebellum of double HDAC1 and HDAC2 mutants. One copy of a HDAC1 allele is sufficient for development and viability.
To delineate the defects associated with loss of HDAC1 and HDAC2 in the developing CNS, we performed histological analysis on 6-day-old pups. Histology on brains from wild-type, HDAC1loxP/+; HDAC2loxP/loxP; GFAP-Cre and HDAC1loxP/loxP; HDAC2loxP/loxP; GFAP-Cre mice showed dramatic defects localized to the cortex, hippocampus, and cerebellum of the double mutant animals. Deletion of HDAC1 and HDAC2 together resulted in severe deficiencies in cortical laminar organization, a loss of all hippocampal structures, and a lack of foliation in the cerebellum (Fig. 1). Mice with only 1 wild-type HDAC1 allele showed reduced cerebellar foliation (Fig. 1), however, these mice survived to adulthood and showed no obvious motor defects. Histological sections of embryos obtained from timed matings showed the neuronal defects to originate between E14.5 and E16.5, a time of maximal neurogenesis and neuronal migration.
Abnormal Purkinje Cell Migration in HDAC1/2 Double-Mutant Mice.
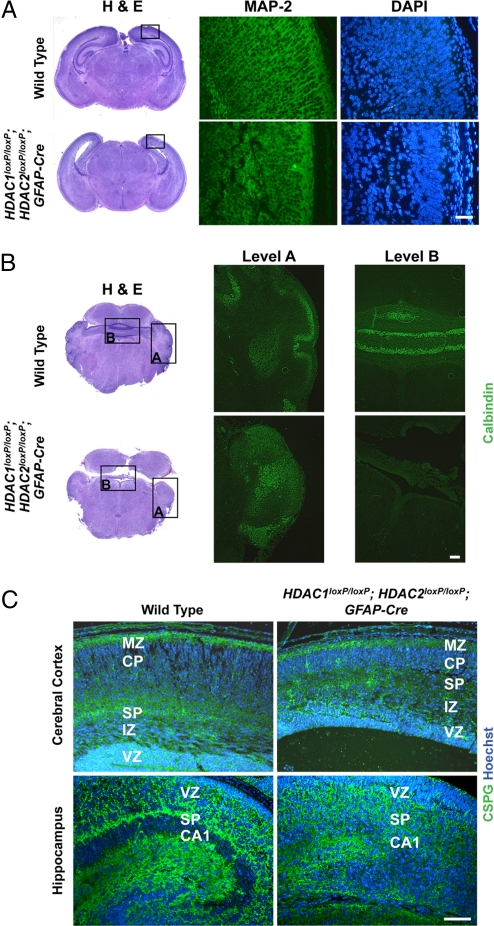
To further investigate the cortical and cerebellar abnormalities resulting from HDAC1/2 deletion, we performed immunohistochemistry at postnatal day 1 to detect microtubule-associated protein 2 (MAP-2), a neuronal marker highly expressed in dendritic extensions, and calbindin, a marker of Purkinje cells in the cerebellum. In wild-type mice, MAP-2 staining of the neocortex showed proper dendritic extensions between cortical layers, whereas HDAC1loxP/loxP; HDAC2loxP/loxP; GFAP-Cre mice showed highly disorganized cortical neurons with a complete absence of dendritic extensions (Fig. 2A). Similarly, calbindin staining of wild-type and HDAC1loxP/loxP; HDAC2loxP/loxP; GFAP-Cre cerebellum revealed distinct neuronal malformations (Fig. 2B). At postnatal day 1, cerebellar development is ongoing and Purkinje cells have not yet formed the tight single-cell layer present in later stages. Consistently, wild-type mice showed proper organization of Purkinje cells and normal foliation, whereas Purkinje cells failed to migrate and remained within the deep nuclei of the cerebellum in HDAC1loxP/loxP; HDAC2loxP/loxP; GFAP-Cre mice. The lack of Purkinje cell migration is detrimental to the development of the cerebellum, because the overlaying granule cell layer requires proliferative signals from the Purkinje cells for proper cerebellar growth (14).
Fig. 2.
Neuronal defects in HDAC1loxP/loxP; HDAC2loxP/loxP; GFAP-Cre mice. (A) Immunohistochemical staining of MAP-2 on the neocortex of wild-type and mutant mice. Mutant mice show a lack of dendritic extensions compared with control. Scale bar = 40 μm. (B) Immunohistochemical staining for calbindin on wild-type and mutant cerebellum. Mutant cerebellum shows Purkinje cells have failed to migrate and remain among the deep cerebellar nuclei. Scale bar = 40 μm. (C) Immunohistochemistry of CSPGs (green) and Hoechst (blue) of wild-type and mutant cerebral cortex and hippocampus. Scale bar = 40 μm. Deletion of HDAC1 and HDAC2 results in disorganized molecular layers. Hippocampal structures are unidentifiable in mutant mice. MZ, marginal zone; CP, cortical plate; SP, subplate; IZ, intermediate zone; VZ, ventricular zone.
Defective Cortical Laminar Organization from Loss of HDAC1 and HDAC2 in CNS.
To further characterize the defects in cortical laminar organization, we performed immunohistochemistry to detect chondroitin sulfate proteoglycans (CSPGs), which serve as molecular markers for the subplate and marginal zone of the cerebral cortex and the subplate of the hippocampus (15). In wild-type mice, by postnatal day 1, the cortical plate neurons have split the preplate into 2 layers, the superficial marginal zone and the subplate layer (16) (Fig. 2C). Immunohistochemical staining for CSPGs on HDAC1loxP/loxP; HDAC2loxP/loxP; GFAP-Cre mice also showed 2 distinct layers for the subplate and marginal zone, albeit much less pronounced than in normal mice. Additionally, the intermittent cortical plate was greatly reduced in the cortex of HDAC1/2 double mutant mice, suggesting either disorganization or compaction of the cortical plate neurons or a reduced number of cortical plate neurons. Similarly, CSPG staining on the hippocampus of HDAC1loxP/loxP; HDAC2loxP/loxP; GFAP-Cre mice showed a complete disorganization of the hippocampal subplate region compared with wild-type controls (Fig. 2C).
Neuronal Migration Is Intact in CNS Deletion of HDAC1 and HDAC2.
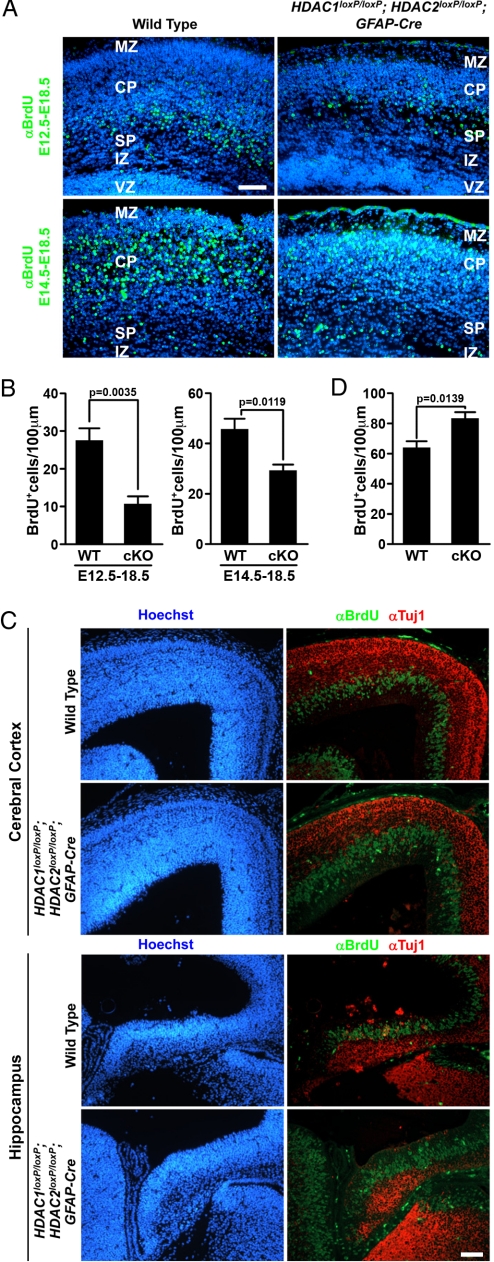
Because HDACs are involved in regulation of the cell cycle (17) and have been shown to control cell migration (18, 19), we speculated that the observed phenotypes could be because of inefficient neuronal migration and/or defective progenitor specification. Development of the cerebral cortex is an intricate process that involves sequential neurogenesis and migration. As the cortex develops, newly generated neurons migrate past the older, more mature neurons to populate the cortical plate just below the marginal zone in a classical inside-out pattern (16). To examine neuronal migration, we performed neuronal birth date analyses at E12.5 and E14.5. Pregnant females were injected with a single i.p. injection of bromodeoxyuridine (BrdU) at either stage E12.5 or E14.5, and embryos were harvested at E18.5. In wild-type mice, immunohistochemistry against BrdU detected neurons born at E12.5 at the base of the cortical plate (Fig. 3A). HDAC1loxP/loxP; HDAC2loxP/loxP; GFAP-Cre mice also showed BrdU labeled neurons at the base of the cortical plate, indicating proper migration of neurons, however, the number of mutant neurons born at E12.5 was significantly diminished compared with wild-type controls (Fig. 3B). Consistently, BrdU labeled neurons born at E14.5 were visible in the cortical plate in both wild-type and HDAC1loxP/loxP; HDAC2loxP/loxP; GFAP-Cre mice, but again, the mutant mice showed a significant reduction in the number of BrdU-positive neurons (Fig. 3 A and B). These findings suggest that although neuronal migration of mutant neurons is intact, significantly fewer postmitotic neurons exist in HDAC1loxP/loxP; HDAC2loxP/loxP; GFAP-Cre mice.
Fig. 3.
Neuronal birth date and proliferation analysis of HDAC1loxP/loxP; HDAC2loxP/loxP; GFAP-Cre mice. (A) Immunohistochemistry of BrdU (green) and Hoechst (blue) of wild-type and mutant cerebral cortex injected with BrdU at E12.5 or E14.5. Deletion of HDAC1 and HDAC2 results in normal neuronal migration but results in fewer differentiated neurons. (B) Quantification of BrdU+ cells from (A) showing a statistically significant reduction in neurons from double mutant (cko) mice. Scale bar = 40 μm. (C) Immunohistochemistry at E14.5 detecting Hoechst (blue), S-phase neuronal precursors labeled by BrdU (green), and Tuj1 (red) on wild-type and mutant cerebral cortex (Top) and developing hippocampus (Bottom). Scale bar = 40 microns. (D) Quantification of BrdU+ cells at E14.5 to assess proliferation. Deletion of HDAC1 and HDAC2 results in increased proliferation of neuronal precursors at the ventricular zone and reduced number of Tuj1-expressing neurons at E14.5. MZ, marginal zone; CP, cortical plate; SP, subplate; IZ, intermediate zone; VZ, ventricular zone; cKO, double conditional knockout.
CNS Deletion of HDAC1 and HDAC2 Results in Increased Proliferation of Neuronal Progenitors.
Because of the reduced number of BrdU-positive neurons in HDAC1/2 double mutant mice, we suspected the developing neurons either failed to differentiate into neurons and/or underwent apoptosis. To determine whether neuronal proliferation/differentiation was affected in mutant mice, we injected pregnant females with a single i.p. injection of BrdU at E14.5 and killed the embryos 1 hour later. Compared with wild-type mice, HDAC1loxP/loxP; HDAC2loxP/loxP; GFAP-Cre mice showed a significant increase in BrdU-positive neuronal precursors in the ventricular zone of the neocortex (Fig. 3 C and D) and in the developing hippocampus (Fig. 3 C and D). Additionally, Tuj1-positive neurons appeared to be decreased in mutant mice compared with wild-type littermates, especially in the cortical plate region where neurons born at this time point would normally be migrating (Fig. 3C). Surprisingly, the increased proliferation of neuronal precursors was restricted to E14.5 as the number of S-phase neuronal precursors at E13.5 was similar between wild-type and mutant mice, whereas E15.5 mutant S-phase cells were significantly decreased compared with wild-type mice, likely because of the overall reduced number of neurons in the mutant cortical plate (Fig. S3).
Deletion of HDAC1 and HDAC2 in Neuronal Progenitors Blocks Neuronal Differentiation.
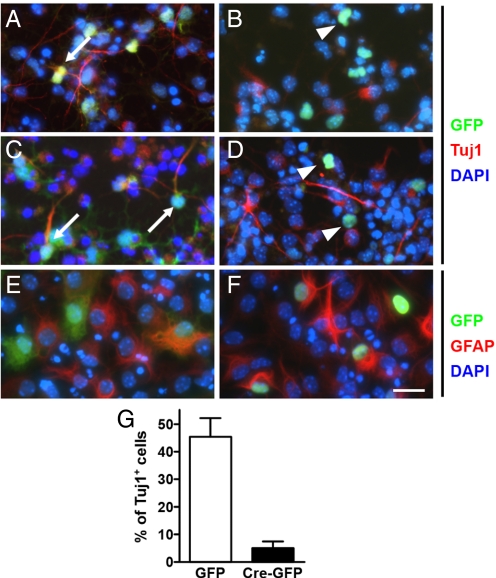
To quantify the potential defect in cell fate specification, we prepared cortical neuronal precursors from E14.5 HDAC1loxP/loxP; HDAC2loxP/loxP mice and infected them with a retrovirus expressing Cre-GFP or GFP alone. Two days after infection, we withdrew FGF and added differentiation media. Surprisingly, deletion of HDAC1 and HDAC2 resulted in a nearly complete block to neuronal differentiation, in response to Isoxazole-9, retinoic acid plus forskolin, or valproic acid (Fig. 4 A–D), as shown by the lack of Tuj1-positive cells expressing Cre-GFP. In contrast a significant number of GFP-alone cells expressed Tuj1. The blockade of differentiation by deletion of HDAC1 and HDAC2 was robust, showing only 5% Tuj1-positive cells of the total GFP-positive cells in the Cre-GFP condition compared with 45% Tuj1-positive cells of the total GFP-positive cells in the GFP alone control (Fig. 4G). Surprisingly, the blockade of differentiation was restricted to neurons. Cre-mediated deletion of HDAC1 and HDAC2 followed by addition of differentiation media to drive an astrocytic fate resulted in similar levels of differentiation between wild type and mutant cells (Fig. 4 E and F).
Fig. 4.
E14.5 neuronal precursors lacking HDAC1 and HDAC2 cannot differentiate into neurons in vitro. Neuronal precursors expressing control, GFP (A, C, and E) or Cre-GFP (B, D, and F), in growth (FGF) (A and B), neuronal (Isoxazole-9) (C and D), and astrocyte (LIF+BMP) (E and F) conditions in 4-day cultures. Control GFP cells colabeled with Tuj1 (white arrows), whereas Cre-GFP cells are notably absent for Tuj1 (white arrowheads) in growth (A and B) and neuronal conditions (C and D). Both control GFP and Cre-GFP colabeled with GFAP in astrocyte conditions (E and F). (G) Quantification of Tuj1+ cells from total GFP or Cre-GFP cells to assess neuronal differentiation. (Scale bar: 15 μm.) Each experiment represents replicates from 2 independent experiments.
Aberrant Cell Death from Loss of HDAC1 and HDAC2 in Neuronal Progenitors.
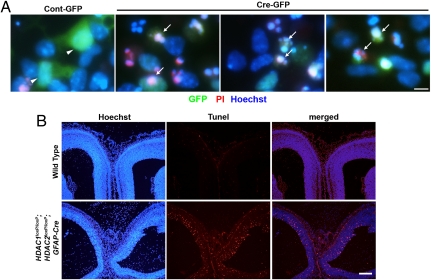
We next investigated whether the defects in neuronal differentiation were because of aberrant cell death. HDAC1loxP/loxP; HDAC2loxP/loxP neuronal precursors were infected with Cre-GFP or GFP alone for 2 days and then switched to neuronal differentiation conditions. After 24 h, cells were stained with propidium iodide (PI) to assess dead and dying cells and Hoechst to assess living cells. HDAC1loxP/loxP; HDAC2loxP/loxP neuronal precursors infected with GFP alone were largely PI-negative and had smooth, uniform nuclei, whereas those infected with Cre-GFP were PI-positive and contained fragmented nuclei indicative of cell death (Fig. 5A).
Fig. 5.
Deletion of HDAC1 and HDAC2 during neuronal differentiation induces cell death. (A) Live-staining of neuronal precursors after 24 h in neuronal differentiation media. Scale bar = 5 microns. Control-GFP are propidium iodide (PI)-negative, whereas Cre-GFP cells are PI-positive and show fragmented nuclei indicative of cell death. (B) TUNEL analysis of E15.5 wild-type and mutant mice shows significant lethality in the ventricular zone of mutant mice, whereas no cell death is observed in wild-type littermates. Scale bar: 100 microns.
To confirm whether similar mechanisms occurred in vivo, we performed Terminal deoxynucleotidyl Transferase biotin-dUTP Nick End Labeling (TUNEL) analysis on the hippocampus and neocortical regions of wild-type and HDAC1loxP/loxP; HDAC2loxP/loxP; GFAP-Cre embryos. Consistently, robust cell death was seen throughout the ventricular zone of mutant neocortex at E15.5, whereas no apoptosis was noticeable in wild-type embryos (Fig. 5B). The neuronal cell death extended throughout the ventricular zone of the hippocampus of mutant embryos, but was largely absent from wild-type hippocampus (Fig. 5B). The apoptosis of neuronal precursors remained confined to a specified time point as no cell death was seen at E14.5 or E16.5, suggesting HDAC1 and HDAC2 are specifically required for proper neuronal specification during neocortical and hippocampal development.
Discussion
In the present study, we provide genetic support that HDAC1 and HDAC2 act as redundant regulators during tissue development. Deletion of either HDAC1 or HDAC2 individually in the central nervous system does not evoke an overt developmental phenotype, however, deletion of both HDAC1 and HDAC2 resulted in robust neural abnormalities, including obliteration of the hippocampus, disorganization of the laminar structure of the neocortex, and loss of foliation of the cerebellum.
HDAC1 and HDAC2 as Regulators of Tissue Development.
We showed that HDAC1 and HDAC2 act redundantly in the regulation of cardiac development (5). Through these studies, it is apparent the redundancy between HDAC1 and HDAC2 extends beyond cardiac cell types, and includes the specific differentiation of neuronal precursors into neurons. Mice lacking HDAC1 or HDAC2 in the CNS showed no obvious histoarchitectural or developmental phenotypes and lived to adulthood with lifespan and mating patterns similar to wild-type littermates. However, the double deletion of HDAC1 and HDAC2 in the CNS resulted in severe neuronal death and animal lethality by 7 days. The specific time point at which the phenotypic abnormalities appeared (E14.5–15.5) is interesting because both neuronal differentiation and neuronal migration are high during this time. Additionally, the hGFAP-Cre line is not highly expressed in the frontal cortex until E13.5 (12), consistent with the time point at which the phenotypic abnormalities appeared.
Development of the cerebral cortex is a dynamic process that involves coordinated neurogenesis and neuronal migration. Neurogenesis occurs in the ventricular zone and new postmitotic neurons migrate outwardly along radial glial fibers to populate the preplate region around E10.5. Newly generated and migrating neurons split the preplate into the superficial marginal zone and the subplate. As the cortex develops, all newly generated neurons migrate past the older neurons to populate the cortical plate just below the marginal zone in a classical inside-out pattern (16). Given the known roles of HDAC1 and HDAC2 in regulating cell cycle and migration (17–19), it is interesting to see loss of HDAC1 and HDAC2 not overtly affecting migration. Mutant migrating neurons along radial glial fibers showed a moderate displacement compared with wild-type controls, however, the most pronounced phenotype was the overall lack of mature neurons after deletion of HDAC1 and HDAC2, especially those born at E14.5. The lack of a pronounced migratory phenotype in the cerebral cortex contrasts greatly with that seen in the cerebellum. Especially intriguing is the fact that hGFAP-Cre is not expressed in Purkinje cells of the cerebellum during development (12), suggesting a non-cell-autonomous role for HDAC1 and HDAC2 in regulating cerebellar Purkinje cell migration.
The mechanistic redundancy between HDAC1 and HDAC2 in the control of neuronal differentiation leads to the question of whether these 2 enzymes control some specified program of differentiation that extends beyond cell-type, or whether the deletion of these 2 master regulators of transcriptional repression is so deleterious that cell death is rampant and ultimate organ failure occurs. It is well known that HDAC1 and HDAC2 regulate certain modulators of cellular proliferation and/or differentiation such as the cyclin-dependent kinase inhibitor p21, however, it remains to be determined whether these targets directly regulate processes across cell-types in an HDAC-dependent manner. To investigate whether there is a common program regulated by HDAC1 and HDAC2 across cell-types, we examined microarray data from brain and cardiac tissue where both HDAC1 and HDAC2 had been deleted. Surprisingly, when the dysregulated transcripts were plotted against one another, there was essentially no distinct overlap between up-regulated transcripts, suggesting the alterations in transcript levels after deletion of HDAC1 and HDAC2 occur in a tissue- and cellular process-specific manner. This is additionally supported by the fact that astrocytic differentiation was not affected by loss of HDAC1 and HDAC2 in progenitor cells. Cells lacking HDAC1 and HDAC2 together were able to differentiate into astrocytes at similar levels as wild-type controls, suggesting the robust phenotype from loss of HDAC1 and HDAC2 is specific to cellular function and not to an overall cellular catastrophe from loss of 2 master regulators of transcriptional repression.
Contrasting Phenotypes Between Inhibitors and Genetic Deletion.
The robust phenotype from deletion of HDAC1 and HDAC2 in the progression of neuronal precursors to neurons is surprising given prior studies on HDAC inhibitors and neurogenesis. HDAC inhibitor treatment of adult or embryonic neuronal precursors specifically induces the differentiation of neurons (7–9), whereas we show deletion of HDAC1 and HDAC2 induces cell death during differentiation to neurons specifically. Whereas it is obvious pharmacological inhibition does not equate with genetic deletion, it is interesting given current thought on how HDAC inhibitor treatment preferentially targets class I HDACs (20, 21). It is likely the transient and incomplete nature of pharmacological inhibition allows for the survival of neuronal precursors; and the derepression of proneurogenic genes allows for the specific induction of neurogenesis, whereas the genetic loss of HDAC1 and HDAC2 is so robust, differentiation is blocked and cellular death ensues. A similar phenotype is seen in lower organisms such as zebrafish, where loss-of-function mutants for hdac1 show a blockade of neuronal differentiation in vivo through the activation of the neurogenic repressor, Hes, and show significant apoptosis in hindbrain neural progenitor cells (22–24). Additionally, neuronal precursors transduced with a mutant HDAC1 that lacks catalytic activity show a fifty percent reduction in neuronal specification (25). It is also interesting that HDAC inhibitors do not induce differentiation of astrocytes or oligodendrocytes, but are specific to the neuronal lineage (7).
Taken together, these data show HDAC1 and HDAC2 are specific and critical components of the neuronal differentiation program and are dramatically sensitive to alterations in gene dosage, whereby transient inhibition induces neuronal differentiation whereas complete loss of function yields robust cellular death. A further difference between pharmacological inhibition and genetic deletion of HDACs that might account for differences in phenotypic outcomes of these 2 approaches is that HDAC proteins remain integrated within transcriptional complexes in the presence of HDAC inhibitors, whereas the proteins are absent in cells in which the HDAC genes have been deleted. Given the myriad protein–protein interactions in which HDACs participate, the complete absence of HDAC proteins may perturb protein–protein interactions required for modulation of transcriptional programs that are otherwise insensitive to HDAC inhibitors.
Therapeutic Implications.
The specific role of HDACs in neuronal differentiation is of particular interest given recent reports that HDAC inhibitors enhance learning and memory and show promising therapeutic applications (26). Further dissection of the specificity of HDAC inhibitors for each HDAC isoform, and understanding the molecular mechanisms by which HDACs control neurogenesis, will be crucial to the development of highly specific therapeutics for the many disease states of the CNS, and other tissues in which HDAC inhibitors are being tested.
Materials and Methods
Animal Studies.
Generation of HDAC1 and HDAC2 conditional alleles has been described (5). GFAP-Cre has been described (12). For BrdU studies, pregnant females were injected with a single i.p. injection of BrdU (Roche) (100 μg/g BW) at E12.5 or E14.5. For neuronal birth date analysis, females were killed at E18.5. For proliferation studies, pregnant females were killed 1 h after injection, embryos fixed in 4% paraformaldehyde, and sectioned for immunostaining. All animal experimental procedures were reviewed and approved by the Institutional Animal Care and Use Committees at the University of Texas Southwestern Medical Center.
Histology and Immunohistochemistry.
Tissues were fixed in 4% paraformaldehyde, embedded in paraffin, and sectioned at 5-μm intervals. Sections were stained with hematoxylin and eosin by using standard procedures (27). TUNEL assays were performed according to a standard protocol (Roche). Immunohistochemistry was performed with the following primary antibodies: rabbit anti-HDAC1 (1:100, Abcam), rabbit anti-HDAC2 (1:100, Abcam), mouse anti-MAP2 (1:200; Sigma), mouse anti-calbindin (1:200; Sigma), mouse anti-CSPG (Sigma), and rabbit anti-Tuj1 (1:5,000, Covance), according to a protocol described (28). Immunohistochemistry using mouse anti-BrdU (Roche) was performed according to manufacturer's instructions.
In Vitro Differentiation Analysis.
Neuronal cortices were isolated from HDAC1loxP/loxP and HDAC2loxP/loxP mice at E14.5 as described in ref. 29 and neuronal differentiation was induced as described in ref. 30. Briefly, neuronal precursors were isolated and placed in insulin-containing N2-supplemented (Invitrogen) DME:Ham's F-12 (Omega Scientific) medium containing 20 ng/mL FGF-2 (PeproTech, Inc.) and 20 ng/ml EGF (Peprotech, Inc.). Retrovirus containing Cre-GFP or GFP alone was infected for 2 days. After 2 days of infection, FGF-2 was withdrawn and neuronal differentiation media (N2 medium plus 5 μM Isoxazole-9 (31) or astrocyte differentiation media (N2 medium plus 50 ng/mL LIF and 50 ng/mL BMP-2) was added. After 4 days, cells were fixed in 4% paraformaldehyde and immunostained for Tuj1 as described in ref. 30. In some cultures, 1 μg/mL propidium iodide (Molecular Probes) or 1 μg/mL Hoechst 22242 (Sigma) were added to label dead cells or all cells, respectively.
Supplementary Material
Acknowledgments.
We thank Jose Cabrera for graphics, Jennifer Brown for editorial assistance, and Cheryl Nolen for technical assistance. We are grateful to Ed Seto and Tso-Pang Yao for comments on the manuscript. This work was supported by grants from the National Institutes of Health, the D.W. Reynolds Clinical Cardiovascular Research Center, and the Robert A. Welch Foundation.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/cgi/content/full/0902750106/DCSupplemental.
References
- 1.Kouzarides T. Chromatin modifications and their function. Cell. 2007;128:693–705. doi: 10.1016/j.cell.2007.02.005. [DOI] [PubMed] [Google Scholar]
- 2.Grozinger CM, Schreiber SL. Deacetylase enzymes: Biological functions and the use of small-molecule inhibitors. Chem Biol. 2002;9:3–16. doi: 10.1016/s1074-5521(02)00092-3. [DOI] [PubMed] [Google Scholar]
- 3.Gregoretti IV, Lee YM, Goodson HV. Molecular evolution of the histone deacetylase family: Functional implications of phylogenetic analysis. J Mol Biol. 2004;338:17–31. doi: 10.1016/j.jmb.2004.02.006. [DOI] [PubMed] [Google Scholar]
- 4.Haberland M, Montgomery RL, Olson EN. The many roles of histone deacetylases in development and physiology: Implications for disease and therapy. Nat Rev Genet. 2009;10:32–42. doi: 10.1038/nrg2485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Montgomery RL, et al. Histone deacetylases 1 and 2 redundantly regulate cardiac morphogenesis, growth, and contractility. Genes Dev. 2007;21:1790–1802. doi: 10.1101/gad.1563807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kazantsev AG, Thompson LM. Therapeutic application of histone deacetylase inhibitors for central nervous system disorders. Nat Rev Drug Discov. 2008;7:854–868. doi: 10.1038/nrd2681. [DOI] [PubMed] [Google Scholar]
- 7.Hsieh J, Nakashima K, Kuwabara T, Mejia E, Gage FH. Histone deacetylase inhibition-mediated neuronal differentiation of multipotent adult neural progenitor cells. Proc Natl Acad Sci USA. 2004;101:16659–16664. doi: 10.1073/pnas.0407643101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Shaked M, et al. Histone deacetylases control neurogenesis in embryonic brain by inhibition of BMP2/4 signaling. PLoS ONE. 2008;3:e2668. doi: 10.1371/journal.pone.0002668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Balasubramaniyan V, et al. Effects of histone deacetylation inhibition on neuronal differentiation of embryonic mouse neural stem cells. Neuroscience. 2006;143:939–951. doi: 10.1016/j.neuroscience.2006.08.082. [DOI] [PubMed] [Google Scholar]
- 10.Broide RS, et al. Distribution of histone deacetylases 1–11 in the rat brain. J Mol Neurosci. 2007;31:47–58. doi: 10.1007/BF02686117. [DOI] [PubMed] [Google Scholar]
- 11.Lagger G, et al. Essential function of histone deacetylase 1 in proliferation control and CDK inhibitor repression. EMBO J. 2002;21:2672–2681. doi: 10.1093/emboj/21.11.2672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhuo L, et al. hGFAP-cre transgenic mice for manipulation of glial and neuronal function in vivo. Genesis. 2001;31:85–94. doi: 10.1002/gene.10008. [DOI] [PubMed] [Google Scholar]
- 13.MacDonald JL, Roskams AJ. Histone deacetylases 1 and 2 are expressed at distinct stages of neuro-glial development. Dev Dyn. 2008;237:2256–2267. doi: 10.1002/dvdy.21626. [DOI] [PubMed] [Google Scholar]
- 14.Chizhikov V, Millen KJ. Development and malformations of the cerebellum in mice. Mol Genet Metab. 2003;80:54–65. doi: 10.1016/j.ymgme.2003.08.019. [DOI] [PubMed] [Google Scholar]
- 15.Sheppard AM, Pearlman AL. Abnormal reorganization of preplate neurons and their associated extracellular matrix: An early manifestation of altered neocortical development in the reeler mutant mouse. J Comp Neurol. 1997;378:173–179. doi: 10.1002/(sici)1096-9861(19970210)378:2<173::aid-cne2>3.0.co;2-0. [DOI] [PubMed] [Google Scholar]
- 16.Hatten ME. Central nervous system neuronal migration. Annu Rev Neurosci. 1999;22:511–539. doi: 10.1146/annurev.neuro.22.1.511. [DOI] [PubMed] [Google Scholar]
- 17.Wang C, et al. Histone acetylation and the cell-cycle in cancer. Front Biosci. 2001;6:D610–D629. doi: 10.2741/1wang1. [DOI] [PubMed] [Google Scholar]
- 18.Whetstine JR, et al. Regulation of tissue-specific and extracellular matrix-related genes by a class I histone deacetylase. Mol Cell. 2005;18:483–490. doi: 10.1016/j.molcel.2005.04.006. [DOI] [PubMed] [Google Scholar]
- 19.Zinovyeva AY, Graham SM, Cloud VJ, Forrester WC. The C. elegans histone deacetylase HDA-1 is required for cell migration and axon pathfinding. Dev Biol. 2006;289:229–242. doi: 10.1016/j.ydbio.2005.10.033. [DOI] [PubMed] [Google Scholar]
- 20.Kramer OH, et al. The histone deacetylase inhibitor valproic acid selectively induces proteasomal degradation of HDAC2. EMBO J. 2003;22:3411–3420. doi: 10.1093/emboj/cdg315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Trivedi CM, et al. Hdac2 regulates the cardiac hypertrophic response by modulating Gsk3 beta activity. Nat Med. 2007;13:324–331. doi: 10.1038/nm1552. [DOI] [PubMed] [Google Scholar]
- 22.Cunliffe VT. Histone deacetylase 1 is required to repress Notch target gene expression during zebrafish neurogenesis and to maintain the production of motoneurones in response to hedgehog signalling. Development. 2004;131:2983–2995. doi: 10.1242/dev.01166. [DOI] [PubMed] [Google Scholar]
- 23.Cunliffe VT, Casaccia-Bonnefil P. Histone deacetylase 1 is essential for oligodendrocyte specification in the zebrafish CNS. Mech Dev. 2006;123:24–30. doi: 10.1016/j.mod.2005.10.005. [DOI] [PubMed] [Google Scholar]
- 24.Stadler JA, et al. Histone deacetylase 1 is required for cell cycle exit and differentiation in the zebrafish retina. Dev Dyn. 2005;233:883–889. doi: 10.1002/dvdy.20427. [DOI] [PubMed] [Google Scholar]
- 25.Humphrey GW, et al. Complementary roles for histone deacetylases 1, 2, and 3 in differentiation of pluripotent stem cells. Differentiation. 2008;76:348–356. doi: 10.1111/j.1432-0436.2007.00232.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fischer A, Sananbenesi F, Wang X, Dobbin M, Tsai LH. Recovery of learning and memory is associated with chromatin remodelling. Nature. 2007;447:178–182. doi: 10.1038/nature05772. [DOI] [PubMed] [Google Scholar]
- 27.Shelton JM, Lee MH, Richardson JA, Patel SB. Microsomal triglyceride transfer protein expression during mouse development. J Lipid Res. 2000;41:532–537. [PubMed] [Google Scholar]
- 28.Xin M, et al. A threshold of GATA4 and GATA6 expression is required for cardiovascular development. Proc Natl Acad Sci USA. 2006;103:11189–11194. doi: 10.1073/pnas.0604604103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Groszer M, et al. Negative regulation of neural stem/progenitor cell proliferation by the Pten tumor suppressor gene in vivo. Science. 2001;294:2186–2189. doi: 10.1126/science.1065518. [DOI] [PubMed] [Google Scholar]
- 30.Hsieh J, et al. IGF-I instructs multipotent adult neural progenitor cells to become oligodendrocytes. J Cell Biol. 2004;164:111–122. doi: 10.1083/jcb.200308101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Schneider JW, et al. Small-molecule activation of neuronal cell fate. Nat Chem Biol. 2008;4:408–410. doi: 10.1038/nchembio.95. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.