Abstract
The outer membranes (OMs) of Gram-negative bacteria have an asymmetric lipid distribution with lipopolysaccharides at the outer leaflet and phospholipids (PLs) at the inner leaflet. This lipid arrangement is essential for the barrier function of the OM and for the viability of most Gram-negative bacteria. Cells with OM assembly defects or cells exposed to harsh chemical treatments accumulate PLs in the outer leaflet of the OM and this disrupts lipopolysaccharide organization and increases sensitivity to small toxic molecules. We have identified an ABC transport system in Escherichia coli with predicted import function that serves to prevent PL accumulation in the outer leaflet of the OM. This highly conserved pathway, which we have termed the Mla pathway for its role in preserving OM lipid asymmetry, is composed of at least 6 proteins and contains at least 1 component in each cellular compartment. We propose that the Mla pathway constitutes a bacterial intermembrane PL trafficking system.
Keywords: Mla pathway, phospholipid, pldA, YrbC, VacJ
Gram-negative bacteria are generally more resistant than Gram-positive bacteria to antibiotics, detergents, and other toxic chemicals because of the sophisticated organization and composition of the outer membrane (OM). The OM is separated from the inner membrane (IM) phospholipid (PL) bilayer by an aqueous periplasm and a thin peptidoglycan layer. The lipids of the OM are asymmetrically distributed, with lipopolysaccharides (LPS) in the outer leaflet and PLs in the inner leaflet (1).
In Escherichia coli K12 strains, LPS is composed of a lipid A moiety conjugated to a core oligosaccharide (2). Lipid A is typically acylated with 6 saturated fatty acids that anchor the molecule in the outer leaflet of the OM (3). The hydrophobicity of lipid A together with the strong lateral interactions between LPS molecules contribute to the effectiveness of the OM barrier (2). LPS organization is disrupted by OM assembly defects or by exposure to antimicrobial peptides and chelating agents such as EDTA, which displace divalent cations needed to reduce repulsive charges between LPS molecules (2). As a consequence, much of the LPS layer is shed (4) and PLs from the inner leaflet are forced to migrate into the breached areas of the outer leaflet. Packing disruptions at the interface of PL and LPS molecules and the formation of PL bilayer patches at the OM reduce barrier function (5). Therefore appreciable levels of outer leaflet PLs are detrimental to the cell.
Surface-exposed PLs are only detectable in stressed cells (1) where they can be modified by 1 of 2 OM β-barrel enzymes, PldA or PagP. PldA is an OM phospholipase that hydrolyzes a wide range of PL substrates and can remove the sn-1 and sn-2 fatty acid side chains from the glycerophosphodiester backbone of both PLs and lysophospholipids (lyso-PLs) (6). PldA normally exists as an inactive monomer in the OM, but the presence of PLs and lyso-PLs in the outer leaflet of the OM initiates the formation of a catalytically active PldA dimer (6) that sequesters and destroys the invading lipid substrates. Thus the enzyme's proposed function is to maintain lipid asymmetry of the OM under stress conditions when enough substrate is available to potentiate dimerization (6).
PagP also utilizes PLs in the outer leaflet as a substrate (3), but the proposed function of PagP is fundamentally different from that of PldA. Like pldA, pagP is expressed at low levels and its protein product is dormant in unstressed cells (7). However, pagP is inducible by the PhoP/Q stress response, which senses the limitation of divalent cations (8). PagP cleaves a palmitate moiety from the sn-1 position of a suitable PL and transfers it to lipid A, forming a hepta-acylated LPS molecule of increased hydrophobicity and a lyso-PL by-product that cannot be further degraded by this enzyme (3). Because lyso-PLs can destabilize the OM, it is presumably the lipid A modification that improves the quality of the OM when divalent cations are limiting (9), but the level to which PagP's limited phospholipase activity alters the levels of cell-surface PLs is not known. Moreover, the contribution of PagP activity to barrier function is context dependent and can negatively influence OM's properties under noninducing conditions (9).
Here, we characterize a third mechanism that impacts cell-surface PLs. We describe an ABC (ATP-binding cassette) transport system with predicted import function (10) that actively prevents PL accumulation at the cell surface in the absence of obvious extracellular stress. We have renamed the genes encoding components of this pathway as mlaA (vacJ), mlaB (yrbB), mlaC (yrbC), mlaD (yrbD), mlaE (yrbE), and mlaF (yrbF), based on their role in maintenance of OM lipid asymmetry. Core components of the Mla pathway are conserved in Gram-negative bacteria and in the chloroplasts of plants (10). The orthologous ABC transport system of Arabidopsis thaliana, the TGD pathway, functions to deliver phosphatidic acid from the OM to the IM of chloroplasts (11); while a paralogous Mce4 pathway restricted to Actinobacteria imports exogenous cholesterol (12, 13). We propose that the Mla pathway constitutes an analogous intermembrane transport system, but is functionally distinct as its purpose is to prevent surface exposure of PLs. The intriguing relationship between cell-surface PLs and microbial pathogenesis is also discussed.
Results
Candidate Genes of the mla Pathway.
Homologs of the E. coli mlaF, -E, and -D genes have been implicated in membrane transport in chloroplasts (11) and in Actinobacteria (12, 13), but the role of this conserved ABC transport pathway in most Gram-negative bacteria is unknown. To address this question, we first set out to identify candidate genes of the putative mla pathway.
In bacterial systems, operon structure often reflects a shared biological function among the protein products of coexpressed genes. In E. coli, mlaF, -E, and -D, are colinear and lie upstream of mlaC and mlaB (Fig. 1A), and all are predicted to be part of the same operon (14). The colocalization of the mlaF-B subset of genes is commonly preserved among Gram-negative bacteria and is occasionally interspersed with the mlaA gene (also annotated as vacJ) (10), although in E. coli mlaA is not genetically linked to the mlaF-B locus (Fig. 1A).
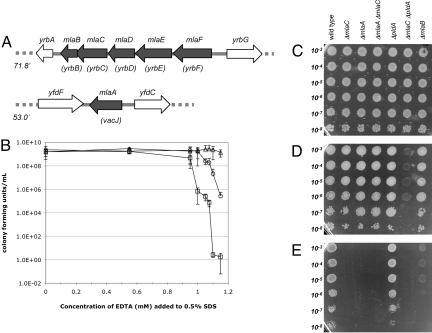
Fig. 1.
The mla genes and the SDS-EDTAS profiles of various strains. (A) The relative chromosomal positions of the mlaFEDCB operon and the mlaA gene are given in minutes, and the original gene names are indicated in parentheses. (B) CFU/mL on agar media containing 0.5% SDS and varying concentrations of EDTA (0–1.15 mM). Wild-type (triangles), ΔmlaB (circles), and ΔmlaC (squares) strains are shown. Data for ΔmlaC is representative of results for ΔmlaF/E/D/A. Serial culture dilutions were spotted onto solid medium: (C) LB, (D) LB + 0.5% SDS, 0.55 mM EDTA, (E) LB + 0.5% SDS, 1.1 mM EDTA. Approximate dilutions are given on the Left.
Two of the candidate gene products, MlaA and MlaC, are secreted proteins. MlaA is an OM lipoprotein of unknown function (15, 16), although MlaC resides in the periplasm (17) and is a predicted substrate-binding protein (18). At the IM, the ABC transport machine is likely to consist of the MlaF, -E, -D, and -B proteins. MlaF is a classic ABC transport nucleotide-binding component (17, 18), whereas MlaE is an integral IM protein (19) and predicted permease (18) with a signature sequence similar to those typically found in prokaryotic ABC import pathways (10). MlaD is a putative substrate-binding protein (18) and localizes to the periplasmic face of the IM by an uncleaved signal sequence (Fig. S1, SI Text). Finally, MlaB is a predicted cytoplasmic protein with a STAS domain (10, 20), a protein fold thought to bind NTPs (20). In sulfate-transport systems, a STAS domain is fused to a transmembrane transport domain and removal of the STAS portion influences the localization and kinetic activity of its associated transport domain (21). The mlaE sequence is fused to the mlaB sequence in several Gram-negative bacteria and may indicate that the 2 gene products interact in E. coli in a manner conceptually analogous to STAS and transport domains of sulfate transporters (10).
Loss-of-Function mla Mutations Result in Increased SDS-EDTA Sensitivity.
We would expect that loss-of-function mutations in any of the genes that encode an ABC transport pathway would lead to similar phenotypes, because presumably the encoded proteins would work together to execute a single task. Our next goal was to identify a phenotype for cells lacking the Mla pathway and to determine which of the candidate proteins are essential for function.
We constructed single-deletion mutations in each of the mla genes and none displays obvious growth defects in standard growth media. However, the deletion of mla orthologs in other organisms increases OM permeability (22–24). To test whether E. coli mla mutants have a defective OM, we analyzed the levels of LPS and OM β-barrel proteins and assayed their sensitivity to antimicrobial compounds. We detected no reduction of LPS or OM proteins compared to wild-type cells by Western blot, nor could we detect an increased sensitivity to erythromycin, rifampicin, bacitracin, and novobiocin by disc diffusion assays (data not shown). Next, we grew each mla mutant on agar media containing 0.5% SDS with varying amounts of EDTA, a chelator of divalent cations that destabilizes LPS interactions (2, 4). We found that each of the ΔmlaF/E/D/C/A single mutants displayed an identical SDS-EDTA sensitivity (SDS-EDTAS) profile, with a steep and dramatic increase in sensitivity between 0.95 and 1.1 mM EDTA (Fig. 1B). In addition, the ΔmlaA mutation in combination with any of the ΔmlaF/E/D/C mutations also exhibited an SDS-EDTAS profile identical to all of the single mutants (Fig. 1 D and E, ΔmlaA and ΔmlaC are a representative combination).
The SDS-EDTA resistance (SDS-EDTAR) threshold of ΔmlaB cells was higher than that of the other mla mutants (Fig. 1 B–E). Nevertheless, the growth of ΔmlaB cells was severely compromised between 0.95 and 1.1 mM EDTA, forming “dust-like” colonies in this range (Fig. 1E). In the case of the ΔmlaA ΔmlaB double mutant, the ΔmlaA sensitivity was epistatic to the less severe ΔmlaB phenotype and identical to that of the ΔmlaA single mutant (data not shown).
The similar SDS-EDTA phenotypes displayed by the mla mutants indicate that they may be part of a singular pathway. Furthermore, the lack of an additive OM sensitivity phenotype between the mlaA mutation and any of the other mla mutations is additional evidence that a single function is disrupted in all of the double mutants.
Increased PldA Levels Suppress the mla− OM Defect.
To understand the function of the Mla system, we searched for spontaneous suppressor mutations that repair the OM permeability defect of mla loss-of-function mutants in the presence of SDS-EDTA (0.5%/1.15 mM). As the phenotypes of most of our mutants are identical, we chose to employ the initial selection with a single representative mutant strain, ΔmlaC.
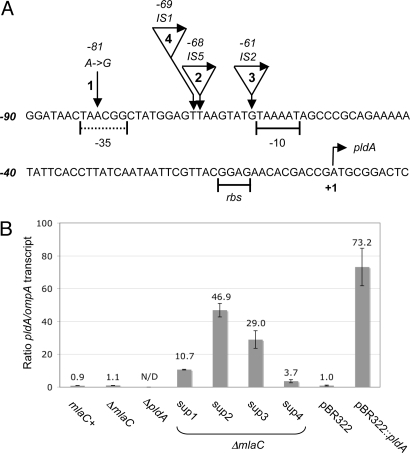
Suppressors of ΔmlaC SDS-EDTAS at the concentration used in our selection were extremely rare; they arose at a frequency of approximately 1 per 109 cells on selective media. We isolated 51 stable suppressor mutants from 6 individual cultures and found that each contained 1 of 2 general alterations of the pldA locus (Fig. 2A). Of the 51 suppressors, 35 contained an A → G transition mutation at position −81, a mutation that arose independently at least 5 times. The second suppressor class is made up of insertion sequence (IS) element transpositions upstream of the pldA promoter (Fig. 2A). Of the 16 IS suppressors, 3 different IS elements were identified, at least 1 of which arose twice independently.
Fig. 2.
Suppressors of ΔmlaC SDS-EDTAS. (A) Each suppressor class is labeled 1–4. The pldA translation start is indicated as +1. The predicted −10 hexamer and the ribosomal binding site (rbs) are underlined (41), the inferred −35 region is underscored with a dashed line. (B) Fold induction of pldA relative to the ompA internal control as determined by qRT-PCR. Sups 1–4 correspond to the suppressor classes as defined in A. N/D, not detected
We used quantitative real time-PCR (qRT-PCR) to determine the level of pldA expression for each of our strains. We found that pldA transcript levels remained unchanged in the ΔmlaC strain compared with wild type, but was increased in all of our ΔmlaC suppressor mutants (Fig. 2B) and correlated with the degree of suppression as determined by the number of colony forming units (CFUs) on the selection medium (data not shown). In addition, we constructed a plasmid-borne copy of pldA, pBR322::pldA, which yielded pldA transcript levels at ≈70 times that of wild-type cells (Fig. 2B). The pBR322::pldA plasmid was an effective suppressor of the ΔmlaC OM defect and fully restored viability on selective media (Fig. S2).
Although our initial selection was performed with a ΔmlaC mutant, subsequent selections for SDS-EDTAR using several other mla mutants yielded the same low frequency of suppressor mutants, and the same suppressor categories as described above (data not shown). We believe our selection is saturated. The simplest explanation for our genetic analysis is that the candidate members of the mla pathway execute a common function that is required for OM integrity and the loss of this function can only be compensated for by an increase in pldA transcription.
PldA Is Not a General Suppressor of SDS-EDTAS.
We wanted to determine whether increased levels of PldA could act as a general suppressor for mutations in other pathways that result in SDS-EDTAS. First, we introduced pBR322::pldA into the lptD4213 strain, which is defective in the assembly of LPS (25, 26). The lptD4213 cells carrying pBR322::pldA did not exhibit a reduction in the zones of inhibition in antibiotic disc diffusion assays compared to the pBR322 control (data not shown). Next, we examined the effects of pBR322::pldA in SDS-EDTAS mutants of the Bam complex that are mildly defective in the assembly of OM β-barrel proteins (27). Multicopy pldA conferred no detectable SDS-EDTAR to the bamD4 mutant and only partially improved the quality of the ΔbamC OM (Fig. S2). On the other hand, the pBR322::pldA conferred full SDS-EDTAR to ΔmlaC, with CFUs comparable to that of the wild-type strain (Fig. S2).
We conclude that increased pldA expression does not suppress all SDS-EDTAS mutants because it compensates poorly or not at all for defects in the Bam and Lpt OM biogenesis pathways. Furthermore, PldA-mediated suppression is not simply limited to cells with modest increases in OM permeability, as ΔbamC cells are clearly more SDS-EDTAR than the ΔmlaC cells, yet increased PldA fully suppresses the permeability defect associated with ΔmlaC and not ΔbamC cells (Fig. S2).
Mla Pathway Function Is Distinct from, but Related to, the Function of PldA.
PldA-mediated suppression of the Δmla OM defects suggests 1 of 2 scenarios. First, increased PldA may be a bypass suppressor of Δmla cells, whereby PldA functions downstream of the Mla components but as part of the same pathway. If so, then cells lacking PldA and cells disrupted in the Mla pathway should have similar phenotypes. However, this is not the case. The ΔmlaC mutant is SDS-EDTAS (0.5%/1.1 mM) whereas ΔpldA cells are fully SDS-EDTAR (Fig. 1 C–E). Moreover, if we reduce the concentration of EDTA in the SDS plates to 0.55 mM, the ΔmlaC cells and the ΔpldA cells each grow as well as wild type, but the double ΔmlaC ΔpldA mutant grows poorly (Fig. 1D).
The alternative explanation for our genetic results is that PldA and the Mla transport system achieve the same end result under our selection conditions using 2 separate pathways. Loss-of-function phenotypes for the 2 pathways are distinct, i.e., the function of the Mla transport system is not simply to induce or activate PldA. The synergistic SDS-EDTAS phenotype displayed by the ΔpldA ΔmlaC double mutant and the fact that increased expression of pldA transcript suppresses the OM permeability defect of mla mutants (Fig. S2) agree with this model and suggest that the 2 pathways are distinct but functionally related. Therefore, we predict that both systems function independently to prevent PL accumulation at the cell surface.
Mutations in the Mla Pathway Lead to PL Accumulation in the Outer Leaflet of the OM.
Although we suspected a relationship between Mla pathway function and PLs, we saw no dramatic differences in the total PL extracts prepared from the Δmla mutants and our wild-type strain, as assayed by thin layer chromatography (TLC) (data not shown). However, our genetic data predict that the Mla pathway would affect PL accumulation in the outer leaflet of the OM, which represents only a small fraction of total PLs even in stressed cells (1). Therefore, we sought to specifically measure the levels of surface-exposed PLs in each of our mutants. The enzymatic activity of PagP can serve as a sensitive, in vivo reporter of the relative levels of cell-surface PLs (28, 29), as the PagP-mediated conversion of lipid A from the hexa- to hepta-acylated form only occurs when PLs are located in the outer leaflet of the OM (7).
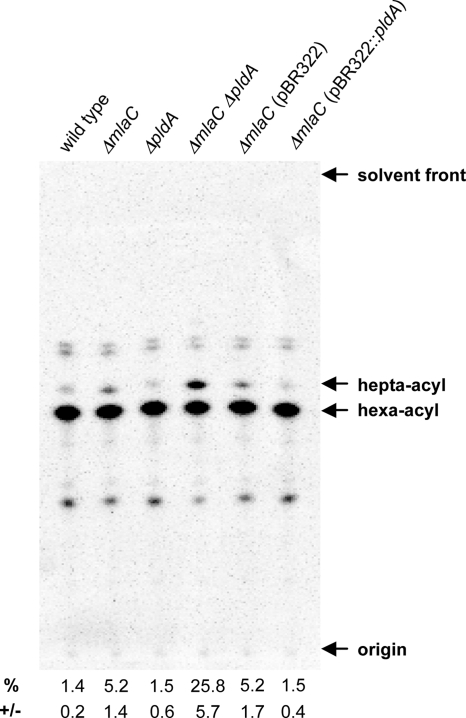
We extracted radiolabeled lipid A from our strains of interest and separated the lipid species using TLC (Fig. 3). As expected, our wild-type sample exhibited little-to-no activation of PagP, as demonstrated by the low percentage of hepta-acylated lipid A (Fig. 3, lane 1). However, the ΔmlaC mutant contained approximately 4-fold more hepta-acylated lipid A than wild type (Fig. 3, lanes 1 and 2). Deletion of the pldA gene had no effect on the level of lipid A palmitoylation in otherwise wild-type cells (Fig. 3, lane 3), although the double ΔpldA ΔmlaC mutant had much higher levels of hepta-acylated lipid A than either of the single mutants (Fig. 3, lanes 1–4). Finally, expression of the pBR322::pldA construct reduced the level of palmitoylated lipid A in the ΔmlaC mutant to wild-type levels (Fig. 3, lanes 5 and 6).
Fig. 3.
TLC analysis of radiolabeled lipid A. The percentage of palmitoylation of lipid A (%) and the percentage of error (+/−) are shown below each lane. Data were quantified from triplicate experiments.
Although the increased lipid A palmitoylation is most likely because of increased PLs at the cell surface, high levels of PagP can also lead to increased lipid A palmitoylation even in wild-type cells (7). Thus, an alternative explanation for our results is that the OM defect in the mutants triggers an increase in pagP expression, regulated by both the PhoP/Q (8) and EvgA/S systems in E. coli (30). However, we did not detect any increase in pagP transcript levels in the single mutants by qRT-PCR and detected at most a 2-fold increase in pagP transcript in the ΔmlaC ΔpldA double mutant compared to wild type (Fig. S3). Therefore, the increased level of lipid A palmitoylation in the ΔmlaC mutant is not an indirect effect of increased PagP production; rather, it is a direct reflection of increased PLs in the outer leaflet of the OM.
This data correlates well with our genetic observations: ΔpldA cells are SDS-EDTAR and do not display an OM defect (Fig. 1E), consistent with the idea that unstressed cells have minimal surface-exposed PL (2); the ΔmlaC cells are SDS-EDTAS and have more hepta-acylated lipid A relative to wild type, and both phenotypes are suppressed by increased pldA expression (Fig. S2 and Fig. 3); and the ΔmlaC ΔpldA double mutant has a more severe OM permeability defect (Fig. 1 D and E) and higher levels of hepta-acylated lipid A (Fig. 3) than either of the single mutants. The ΔmlaC and ΔpldA mutations are synergistic. Because pldA expression does not change in a ΔmlaC strain (Fig. 2B), this synergy suggests that the absence of the mla pathway is partially offset by an increase in PldA activity, a reaction that is regulated in part by the amount of PL substrate in the outer leaflet of the OM (6).
Discussion
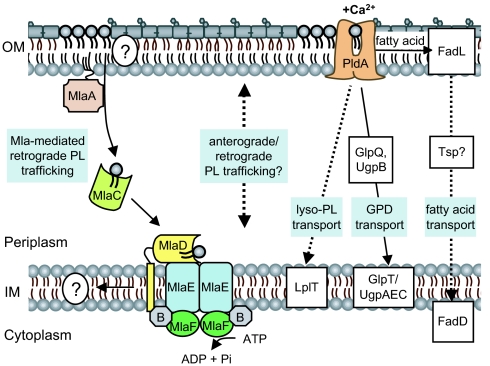
We have identified a highly conserved ABC transport system of predicted import function (10), the Mla pathway, which prevents PL accumulation in the outer leaflet of the OM. There are functional components of this pathway in each and every cellular compartment (Fig. 4). When we remove any one of these components, PLs accumulate in the outer leaflet of the OM and this results in increased OM permeability. This OM defect can only be suppressed by increasing the levels of the OM protein PldA, a phospholipase that destroys surface-exposed PLs (3). The crystal structure of the Ralstonia solanacearum ortholog of the periplasmic component of the pathway, MlaC, has been solved with bound PE (Protein Data Bank ID: 2QGU; Northeast Structural Genomics Consortium), an observation that strongly suggests that pathway substrates are PLs. Therefore, we conclude that the function of the Mla system is to maintain lipid asymmetry in the OM and we think it likely does so by retrograde trafficking of PLs from the OM to the IM (Fig. 4).
Fig. 4.
Model of the Mla and PldA PL turnover pathways. The Mla pathway removes PL from the OM (either from the inner leaflet before surface migration or directly from the outer leaflet) and delivers it to the MlaFEDB complex at the IM via the periplasmic substrate binding protein MlaC. The fate of the PL is unknown, but may be reintroduced at the IM. Other retrograde PL trafficking systems likely exist, and the mode of bulk anterograde and retrograde PL trafficking is still unknown. Increased levels of PldA suppress mutants of the Mla pathway by destroying surface exposed PL, resulting in the production of free fatty acids, lyso-PLs, and glycerophosphodiesters (GPDs). Each of these molecules can be taken up from exogenous or intramembrane sources and delivered to the IM by various pathways (42–45) but it is unclear whether these same pathways are used to clear PldA breakdown products from the outer leaflet. Dashed lines represent unknown modes of transport across the periplasm, and proteins depicted in white boxes have not been directly proven to be part of the PldA turnover pathway.
The presence of PLs in the outer leaflet of the OM increases permeability to hydrophobic molecules such as bile salts, and therefore many Gram-negative bacteria, especially the enterics, have mechanisms that destroy or remove PLs from the cell surface. The Mla pathway represents a novel mechanism for maintaining lipid asymmetry in E. coli, and we think it is the most important. Loss-of-function mla mutations result in increased OM permeability, whereas loss-of-function mutations in pagP or pldA, which specify 2 OM enzymes that share varying degrees of phospholipase activity (3), do not. The Mla system prevents the accumulation of PLs at the cell surface in the absence of PldA but the converse is not true. Indeed, it is the activity of the Mla pathway that inhibits activation of the phospholipases in unstressed, wild-type cells.
We demonstrated that increased expression of pldA compensates for the loss of the mla pathway, but interestingly, pagP does not. In fact, we found that increased pagP expression in fact conferred a modest increase in the SDS-EDTAS phenotype of Δmla cells (data not shown). Nor were there any synthetic phenotypes when ΔpagP was introduced into the ΔmlaC and ΔpldA single mutants or the ΔmlaC ΔpldA double mutant (data not shown). We think these results reflect the different functions of PldA and PagP. PldA functions to completely remove PLs from the outer leaflet of the OM, a function it shares with the Mla pathway. This shared function in maintaining lipid asymmetry is accomplished by different mechanisms: the Mla pathway likely removes PLs from the outer leaflet; PldA destroys them. In contrast, the function of PagP is to modify LPS by adding a 7th acyl chain. It is likely that increased PagP cannot suppress the OM defects in Δmla mutants because the enzyme does not completely destroy the PL. Furthermore, the fate of the lyso-PLs released into the OM as a result of the PagP reaction and their contribution to OM destabilization is unknown. Alternatively, the lipid A palmitoylation mediated by PagP may not be beneficial to the LPS network in the context of our growth conditions (9) and could counteract any benefit stemming from partial PL hydrolysis in cells that lack the Mla pathway.
Our model for Mla pathway function (Fig. 4) provides a plausible explanation for the many reports of pleiotropic phenotypes of mla mutants such as increased OM permeability (22–24) and loss of virulence in E. coli and other Gram-negative organisms (15, 22, 31). Orthologs of mlaA, mlaC, mlaE, and/or mlaF affect the virulence of enteroinvasive E. coli and Shigella flexneri (15, 22), causative agents of bacillary dysentery, and of Burkholderia pseudomallei (31), the agent that causes melioidosis. These pathogens infect nonphagocytic eukaryotic host cells via a sequence of cell invasion, intracellular replication, and spreading into neighboring cells. Intercellular spread into adjacent cells requires actin-based motility followed by escape from host-derived membranes that surround the invading microbe (32). Mutations in orthologs of the Mla pathway do not affect the earlier steps in pathogenesis, but the Mla pathway is required, together with other factors (33, 34), for escape from the host double membrane (15). Thus, the Mla pathway is an important virulence factor. We suggest that the avirulent phenotypes of mla mutants are the consequence of increased PLs in the outer leaflet of the OM. It will be interesting to determine whether the prevention of intercellular spread is the direct result of increased OM permeability, the inhibition of a primary virulence factor as an indirect consequence of disrupting the OM, or whether retrograde PL transport mediates downstream signaling functions critical to the persistence of host infection.
PLs are synthesized in the IM and the mechanism(s) that traffics them between the IM and the OM remain mysterious. Nonetheless, it has been established by several groups that PL transport occurs rapidly and in a reversible fashion from the OM to the IM (35–38). We attempted to demonstrate the contribution of the Mla pathway to retrograde PL transport by performing experiments similar to those previously described (36, 37). Briefly, these experiments enrich the cellular content of phosphatidylserine (PS), a trace PL species in E. coli, using either genetic or biochemical means. PS transport from the OM to the IM is then assayed via the conversion of PS to PE by an IM enzyme, PS decarboxylase (Psd). Using these methods, we could not detect a significant difference in the rate of PS to PE conversion in our Mla mutants compared to wild type (data not shown). One explanation may be that the Mla pathway cannot recognize PS. Nevertheless, the lack of significant changes to the overall PL profiles of the Mla mutants compared to wild type suggests that the Mla pathway is not responsible for the bulk retrograde PL flow. Given the subtle phenotypes of the loss-of-function mla mutants under laboratory conditions, we suspect that the Mla pathway targets a relatively minor population of OM PL molecules (i.e., those that ultimately reach the outer leaflet of the OM). Therefore it is likely that Mla-mediated retrograde PL transport cannot be distinguished biochemically from the major PL transport routes in these assays.
Finally, the identification of Mla function necessitates a reevaluation of our current understanding of OM permeability defects. Cells defective in the assembly of OM β-barrel proteins or LPS are thought to accumulate PLs in their outer leaflets to compensate for discontinuities in their OMs, thus rendering these cells sensitive to small toxic molecules. In contrast, mutants of the Mla pathway are relatively impermeable to most compounds with the exception of SDS-EDTA, and they exhibit no additional defects in LPS or OM protein levels; their only membrane defect is an increase in surface-exposed PLs. Interestingly, increased expression of pldA compensates poorly or not at all for defects in the Bam or Lpt OM biogenesis pathways (Fig. S2). One explanation is that the OM protection offered by increased PldA production is limited to OMs with modest PL accumulation and that the Bam and Lpt SDS-EDTAS mutants exceed this threshold. However, ΔbamC cells are initially more SDS-EDTAR than the ΔmlaC cells, yet pldA-mediated suppression confers complete resistance only to the ΔmlaC cells (Fig. S2). Thus, the barrier defect of the Mla mutants is specifically because of PL accumulation in the outer leaflet, whereas additional OM alterations contribute to the increased permeability phenotypes of Lpt and Bam mutants.
Materials and Methods
Bacterial Strains and Growth Conditions.
A list of the bacterial strains, phages, plasmids, and primers are given in Tables S1 and S2. Luria Bertani (LB) medium was purchased from BD Difco. SDS and EDTA, purchased from Sigma-Aldrich, were prepared as filter sterilized stock solutions of 10% SDS and 50 mM EDTA (pH 7.5).
Quantification of CFUs.
Cultures grown overnight in LB broth at 37 °C were added to a 96-well microtiter dish in duplicate, and 6 to 8 10-fold dilutions were made in fresh LB medium. A 48-pin manifold was used to transfer ≈2 μL of culture from the dish to solid LB medium with or without SDS-EDTA at the indicated concentrations. To quantify CFUs for strains with viability below the detection level (≈103 cells), 1–5 mL of overnight culture was concentrated and spread onto selection media. Colonies were counted between 12 and 15 h of incubation.
Isolation of ΔmlaC SDS-EDTAR Suppressors.
Six independent ΔmlaC cultures were grown to saturation in 5 mL of LB broth, pelleted, and individually resuspended in 200 μL of LB before plating onto LB-agar medium (0.5% SDS/1.15 mM EDTA). Fifty-one stable SDS-EDTAR suppressors were obtained and one was selected for mapping. The ΔmlaC suppressor was infected with λNK1323 to generate random miniTn10 chromosomal insertions (39) and selected on LB agar containing 10 μg/mL of tetracycline (Tet). Approximately 1,000 TetR colonies were pooled into 5 mL of LB + 50 mM Na-citrate and washed in an equal volume of LB + Na-citrate. One hundred microliters were added to 5 mL of LB and used to prepare a P1 vir lysate (40). This lysate was transduced (40) into a ΔmlaC recipient and plated onto LB + Tet. TetR colonies were pooled in LB + Na-citrate and dilutions plated onto SDS/EDTA (0.5%/1.15 mM). Several SDS/EDTAR colonies were purified and the positions of the miniTn10 insertion mutations were identified using arbitrary PCR and DNA sequence analysis (see SI). Standard genetic mapping techniques narrowed the approximate location of the suppressor mutation and subsequent PCR analysis of regions flanking the pldA promoter confirmed suppressor identity.
Analysis of Lipid A.
Cultures were grown overnight at 37 °C in LB broth (+Tet where appropriate) and diluted 1:100 in 5 mL of fresh LB without antibiotics so as to avoid potential artifacts from drug treatment. We added 5 μCi/mL of 32PO4 to the cultures, which were incubated in a 37 °C water bath shaker for 2.5 h. We used the mild-acid hydrolysis procedure to quantify lipid A modifications as described by the Bishop laboratory (7). Radiolabeled lipid samples were spotted onto a Silica 60 Å pore TLC plate (Merck). The mobile phase consisted of chloroform, pyridine, 88% formic acid, and water (50:50:16:5, vol/vol). After developing, plates were dried and exposed to a phosphor screen overnight. Samples were visualized with a GE Storm PhosphorImager and bands were quantified using ImageQuant TL image analysis software (Amersham Biosciences). Values are averaged from 3 independent experiments.
Supplementary Material
Acknowledgments.
We thank the members of the Silhavy lab, especially Natividad Ruiz and Annie Hiniker, for their helpful discussions and reading of the manuscript, and Valerie Carabetta and Joe Sklar for technical assistance. We are especially grateful to Daniel Kahne and Luisa Gronenberg for their help and suggestions during early stages of this project and to Russell Bishop for his generous guidance with the lipid A analysis experiment. This work was supported by National Institute of General Medical Sciences Grant GM34821 (T.J.S.) and a Ruth L. Kirschstein National Research Service Award GM073494 (J.C.M.).
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/cgi/content/full/0903229106/DCSupplemental.
References
- 1.Kamio Y, Nikaido H. Outer membrane of Salmonella typhimurium: Accessibility of phospholipid head groups to phospholipase c and cyanogen bromide activated dextran in the external medium. Biochemistry. 1976;15(12):2561–2570. doi: 10.1021/bi00657a012. [DOI] [PubMed] [Google Scholar]
- 2.Nikaido H. Molecular basis of bacterial outer membrane permeability revisited. Microbiol Mol Biol Rev. 2003;67(4):593–656. doi: 10.1128/MMBR.67.4.593-656.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bishop RE. Structural biology of membrane-intrinsic β-barrel enzymes: Sentinels of the bacterial outer membrane. Biochim Biophys Acta. 2008;1778(9):1881–1896. doi: 10.1016/j.bbamem.2007.07.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Leive L. Release of lipopolysaccharide by EDTA treatment of E. coli. Biochem Biophys Res Commun. 1965;21(4):290–296. doi: 10.1016/0006-291x(65)90191-9. [DOI] [PubMed] [Google Scholar]
- 5.Nikaido H. Restoring permeability barrier function to outer membrane. Chem Biol. 2005;12(5):507–509. doi: 10.1016/j.chembiol.2005.05.001. [DOI] [PubMed] [Google Scholar]
- 6.Dekker N. Outer-membrane phospholipase A: Known structure, unknown biological function. Mol Microbiol. 2000;35(4):711–717. doi: 10.1046/j.1365-2958.2000.01775.x. [DOI] [PubMed] [Google Scholar]
- 7.Jia W, et al. Lipid trafficking controls endotoxin acylation in outer membranes of Escherichia coli. J Biol Chem. 2004;279(43):44966–44975. doi: 10.1074/jbc.M404963200. [DOI] [PubMed] [Google Scholar]
- 8.Groisman EA. The pleiotropic two-component regulatory system PhoP-PhoQ. J Bacteriol. 2001;183(6):1835–1842. doi: 10.1128/JB.183.6.1835-1842.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Murata T, Tseng W, Guina T, Miller SI, Nikaido H. PhoPQ-mediated regulation produces a more robust permeability barrier in the outer membrane of Salmonella enterica serovar typhimurium. J Bacteriol. 2007;189(20):7213–7222. doi: 10.1128/JB.00973-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Casali N, Riley L. A phylogenomic analysis of the Actinomycetales mce operons. BMC Genomics. 2007;8(1):60. doi: 10.1186/1471-2164-8-60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Benning C. A role for lipid trafficking in chloroplast biogenesis. Prog Lipid Res. 2008;47(5):381–389. doi: 10.1016/j.plipres.2008.04.001. [DOI] [PubMed] [Google Scholar]
- 12.Mohn WW, et al. The actinobacterial mce4 locus encodes a steroid transporter. J Biol Chem. 2008;283(51):35368–35374. doi: 10.1074/jbc.M805496200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pandey AK, Sassetti CM. Mycobacterial persistence requires the utilization of host cholesterol. Proc Natl Acad Sci USA. 2008;105(11):4376–4380. doi: 10.1073/pnas.0711159105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gama-Castro S, et al. RegulonDB (version 6.0): Gene regulation model of Escherichia coli K-12 beyond transcription, active (experimental) annotated promoters and Textpresso navigation. Nucleic Acids Res. 2008;36:D120–D124. doi: 10.1093/nar/gkm994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Suzuki T, et al. Identification and characterization of a chromosomal virulence gene, vacJ, required for intercellular spreading of Shigella flexneri. Mol Microbiol. 1994;11(1):31–41. doi: 10.1111/j.1365-2958.1994.tb00287.x. [DOI] [PubMed] [Google Scholar]
- 16.Juncker AS, et al. Prediction of lipoprotein signal peptides in Gram-negative bacteria. Protein Sci. 2003;12(8):1652–1662. doi: 10.1110/ps.0303703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lopez-Campistrous A, et al. Localization, annotation, and comparison of the Escherichia coli K-12 proteome under two states of growth. Mol Cell Proteomics. 2005;4(8):1205–1209. doi: 10.1074/mcp.D500006-MCP200. [DOI] [PubMed] [Google Scholar]
- 18.Linton KJ, Higgins CF. The Escherichia coli ATP-binding cassette (ABC) proteins. Mol Microbiol. 1998;28(1):5–13. doi: 10.1046/j.1365-2958.1998.00764.x. [DOI] [PubMed] [Google Scholar]
- 19.Daley DO, et al. Global topology analysis of the Escherichia coli inner membrane proteome. Science. 2005;308(5726):1321–1323. doi: 10.1126/science.1109730. [DOI] [PubMed] [Google Scholar]
- 20.Aravind L, Koonin EV. The STAS domain: A link between anion transporters and antisigma-factor antagonists. Curr Biol. 2000;10(2):R53–R55. doi: 10.1016/s0960-9822(00)00335-3. [DOI] [PubMed] [Google Scholar]
- 21.Shibagaki N, Grossman AR. The role of the STAS domain in the function and biogenesis of a sulfate transporter as probed by random mutagenesis. J Biol Chem. 2006;281(32):22964–22973. doi: 10.1074/jbc.M603462200. [DOI] [PubMed] [Google Scholar]
- 22.Hong M, Gleason Y, Wyckoff EE, Payne SM. Identification of two Shigella flexneri chromosomal loci involved in intercellular spreading. Infect Immun. 1998;66(10):4700–4710. doi: 10.1128/iai.66.10.4700-4710.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kim K, Lee S, Lee K, Lim D. Isolation and characterization of toluene-sensitive mutants from the toluene-resistant bacterium Pseudomonas putida GM73. J Bacteriol. 1998;180(14):3692–3696. doi: 10.1128/jb.180.14.3692-3696.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Endo R, Ohtsubo Y, Tsuda M, Nagata Y. Identification and characterization of genes encoding a putative ABC-type transporter essential for utilization of gamma-hexachlorocyclohexane in Sphingobium japonicum UT26. J Bacteriol. 2007;189(10):3712–3720. doi: 10.1128/JB.01883-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ruiz N, Falcone B, Kahne D, Silhavy TJ. Chemical conditionality: A genetic strategy to probe organelle assembly. Cell. 2005;121:307–317. doi: 10.1016/j.cell.2005.02.014. [DOI] [PubMed] [Google Scholar]
- 26.Bos MP, Tefsen B, Geurtsen J, Tommassen J. Identification of an outer membrane protein required for the transport of lipopolysaccharide to the bacterial cell surface. Proc Natl Acad Sci USA. 2004;101(25):9417–9422. doi: 10.1073/pnas.0402340101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wu T, et al. Identification of a multicomponent complex required for outer membrane biogenesis in Escherichia coli. Cell. 2005;121(2):235–245. doi: 10.1016/j.cell.2005.02.015. [DOI] [PubMed] [Google Scholar]
- 28.Ruiz N, Gronenberg LS, Kahne D, Silhavy TJ. Identification of two inner-membrane proteins required for the transport of lipopolysaccharide to the outer membrane of Escherichia coli. Proc Natl Acad Sci USA. 2008;105:5537–5542. doi: 10.1073/pnas.0801196105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wu T, et al. Identification of a protein complex that assembles lipopolysaccharide in the outer membrane of Escherichia coli. Proc Natl Acad Sci USA. 2006;103(31):11754–11759. doi: 10.1073/pnas.0604744103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Eguchi Y, et al. Signal transduction cascade between EvgA/EvgS and PhoP/PhoQ two-component systems of Escherichia coli. J Bacteriol. 2004;186(10):3006–3014. doi: 10.1128/JB.186.10.3006-3014.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cuccui J, et al. Development of signature-tagged mutagenesis in Burkholderia pseudomallei to identify genes important in survival and pathogenesis. Infect Immun. 2007;75(3):1186–1195. doi: 10.1128/IAI.01240-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ogawa M, Sasakawa C. Bacterial evasion of the autophagic defense system. Curr Opin Microbiol. 2006;9(1):62–68. doi: 10.1016/j.mib.2005.12.007. [DOI] [PubMed] [Google Scholar]
- 33.Page AL, Ohayon H, Sansonetti PJ, Parsot C. The secreted IpaB and IpaC invasins and their cytoplasmic chaperone IpgC are required for intercellular dissemination of Shigella flexneri. Cell Microbiol. 1999;1(2):183–193. doi: 10.1046/j.1462-5822.1999.00019.x. [DOI] [PubMed] [Google Scholar]
- 34.Rathman M, et al. The development of a FACS-based strategy for the isolation of Shigella flexneri mutants that are deficient in intercellular spread. Mol Microbiol. 2000;35(5):974–990. doi: 10.1046/j.1365-2958.2000.01770.x. [DOI] [PubMed] [Google Scholar]
- 35.Kol MA, et al. Uptake and remodeling of exogenous phosphatidylethanolamine in E. coli. Biochim Biophys Acta. 2004;1636(2–3):205–212. doi: 10.1016/j.bbalip.2004.01.002. [DOI] [PubMed] [Google Scholar]
- 36.Jones NC, Osborn MJ. Translocation of phospholipids between the outer and inner membranes of Salmonella typhimurium. J Biol Chem. 1977;252(20):7405–7412. [PubMed] [Google Scholar]
- 37.Langley KE, Hawrot E, Kennedy EP. Membrane assembly: Movement of phosphatidylserine between the cytoplasmic and outer membranes of Escherichia coli. J Bacteriol. 1982;152(3):1033–1041. doi: 10.1128/jb.152.3.1033-1041.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Cohen LK, Lueking DR, Kaplan S. Intermembrane phospholipid transfer mediated by cell-free extracts of Rhodopseudomonas sphaeroides. J Biol Chem. 1979;254(3):721–728. [PubMed] [Google Scholar]
- 39.Kleckner N, Bender J, Gottesman S. Uses of transposons with emphasis on Tn10. Methods Enzymol. 1991;204:139–180. doi: 10.1016/0076-6879(91)04009-d. [DOI] [PubMed] [Google Scholar]
- 40.Silhavy TJ, Berman ML, Enquist LW. Experiments with Gene Fusions. Plainview, NY: Cold Spring Harbor Lab Press; 1984. [Google Scholar]
- 41.Brok RG, et al. Molecular characterization of enterobacterial pldA genes encoding outer membrane phospholipase A. J Bacteriol. 1994;176(3):861–870. doi: 10.1128/jb.176.3.861-870.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hearn EM, Patel DR, Lepore BW, Indic M, van den Berg B. Transmembrane passage of hydrophobic compounds through a protein channel wall. Nature. 2009;458(7236):367–370. doi: 10.1038/nature07678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Azizan A, Black PN. Use of transposon TnphoA to identify genes for cell envelope proteins of Escherichia coli required for long-chain fatty acid transport: The periplasmic protein Tsp potentiates long-chain fatty acid transport. J Bacteriol. 1994;176(21):6653–6662. doi: 10.1128/jb.176.21.6653-6662.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ohshima N, et al. Escherichia coli cytosolic glycerophosphodiester phosphodiesterase (UgpQ) requires Mg2+, Co2+, or Mn2+ for its enzyme activity. J Bacteriol. 2008;190(4):1219–1223. doi: 10.1128/JB.01223-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Harvat EM, et al. Lysophospholipid flipping across the Escherichia coli inner membrane catalyzed by a transporter (LplT) belonging to the major facilitator superfamily. J Biol Chem. 2005;280(12):12028–12034. doi: 10.1074/jbc.M414368200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.