Abstract
Recent experimental evidence points to intermediates populated during the process of amyloid fibril formation as the toxic moieties primarily responsible for the development of increasingly common disorders such as Alzheimer's disease and type II diabetes. We describe here the application of a pulse-labeling hydrogen-deuterium (HD) exchange strategy monitored by mass spectrometry (MS) and NMR spectroscopy (NMR) to characterize the aggregation process of an SH3 domain under 2 different conditions, both of which ultimately lead to well-defined amyloid fibrils. Under one condition, the intermediates appear to be largely amorphous in nature, whereas under the other condition protofibrillar species are clearly evident. Under the conditions favoring amorphous-like intermediates, only species having no protection against HD exchange can be detected in addition to the mature fibrils that show a high degree of protection. By contrast, under the conditions favoring protofibrillar-like intermediates, MS reveals that multiple species are present with different degrees of HD exchange protection, indicating that aggregation occurs initially through relatively disordered species that subsequently evolve to form ordered aggregates that eventually lead to amyloid fibrils. Further analysis using NMR provides residue-specific information on the structural reorganizations that take place during aggregation, as well as on the time scales by which they occur.
Keywords: aggregation, HD exchange, misfolding intermediates, PI3-SH3
Protein aggregation into amyloid fibrils is associated with a wide range of increasingly prevalent disorders such as Alzheimer's disease, Creutzfeldt–Jakob disease, and type II diabetes (1, 2). Amyloid fibril formation appears to be a multistep process during which a series of intermediate aggregated states is sampled (3). Although early hypotheses proposed amyloid fibrils as the primary pathogenic agent, much recent evidence supports the view that smaller aggregates populated during the fibril assembly process represent the primary culprits (4, 5). The importance of the details of the aggregation process to the mechanism of disease makes characterization of the different species present during aggregation an issue of extreme importance. Of all of the species formed during aggregation, amyloid fibrils, despite their size and apparent intractability, are the easiest to characterize because of their long-lived nature and great regularity. Application of solid-state NMR (NMR) techniques (6), site-directed spin labeling in combination with electron paramagnetic resonance (EPR) (7), and hydrogen deuterium (HD) exchange experiments (8–10) have all made very significant contributions to the study of their structures. Prefibrillar intermediates, however, are very difficult to characterize because they are short-lived, are often heterogeneous, and may be present at low populations. Despite these problems, several techniques have been developed to study such species, notably photo-induced cross-linking of unmodified proteins (PICUP) (11) and various forms of mass spectrometry (12, 13). In addition, a conformation-specific antibody that recognizes soluble oligomers from many types of proteins, regardless of sequence, has been produced and has proved to be very useful in monitoring the kinetics of oligomer formation (14).
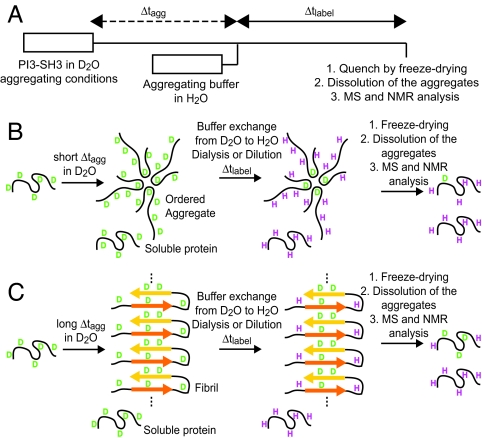
HD exchange experiments are based on solvent accessibilities; amide protons that normally undergo rapid exchange with solvent deuterons experience much slower exchange when involved in H-bonded structures and/or when sterically inaccessible to the solvent. HD exchange experiments can be applied to probe systems either at equilibrium or during the kinetic chain of reactions following perturbation from equilibrium conditions. In equilibrium experiments, the protein conformation under study does not change with time. In the context of protein aggregation, equilibrium HD exchange experiments have been used to probe the core structure of a range of amyloid fibrils (8–10) and protofibrils (15) and the dynamics of molecular recycling within an ensemble of fibrils (16). By contrast to such equilibrium studies, kinetic HD exchange experiments can monitor protein conformational changes as a function of time. Within this context, we have developed a pulse-labeling HD exchange experiment designed to gain both mechanistic information on the process of aggregation and structural information on the different species present during the course of aggregation (Fig. 1).
Fig. 1.
Schematic description of the pulse-labeling HD exchange experiment developed to study protein aggregation. (A) The experiment starts by incubating soluble protein under aggregation conditions in a deuterium based buffer. After a variable aggregation time, Δtagg, labeling takes place for a fixed period, Δtlabel, using protonated aggregation buffer. The magnitude of Δtlabel is chosen so that only unprotected amide deuterons will exchange significantly with the solvent. After the labeling pulse, freeze-drying is used to quench exchange. Different samples are prepared at defined Δtagg values, which are later solubilized into monomers by transfer to a DMSO solution and analyzed by NMR and ESI-MS. The figure illustrates hypothetical scenarios when the protein is left to aggregate for a short Δtagg (B) and a long Δtagg (C).
In this article, we describe the application of such pulse-labeling HD exchange experiments to the study of the aggregation process of the SH3 domain of the α-subunit of bovine phosphatidylinositol-3′-kinase (PI3-SH3). The PI3-SH3 domain is a protein that aggregates to form well-characterized fibrils in vitro, particularly at low pH (17, 18). Moreover, these fibrils and their precursors show structural and cytotoxic properties that are closely similar to those observed in many depositional disorders (19). The results described in this article provide important information about the aggregation process of a protein, such as the distribution and stability of the different aggregation states and the nature of structural reorganizations occurring during amyloid fibril formation.
Results
A Pulse-Labeling Strategy for HD Exchange Analysis of Aggregation Intermediates.
The pulse-labeling HD exchange experiment designed to probe the species present at different stages in the aggregation reaction involves first the incubation of soluble protein in deuterated buffer under specific conditions where aggregation occurs (Fig. 1). Variation in the period allowed for aggregation, Δtagg, permits the study of the species present at different times. Then, the solution is subjected to dilution or dialysis for a fixed labeling time, Δtlabel, in protonated aggregation buffer. Δtlabel is chosen so that deuterium atoms located in unprotected regions of the aggregates undergo isotope exchange, whereas deuterium atoms located in protected regions are retained. After the labeling pulse, the sample is freeze-dried to quench exchange. By using different Δtagg values and a fixed Δtlabel, the most protected regions of each of the aggregates under study are labeled. To analyze the deuterium incorporation in the aggregates, the latter are dissociated into monomers by transfer to a DMSO solution (9), and analyzed by a combination of electrospray ionization mass spectrometry (ESI-MS) (8, 16) and NMR (9, 10, 16). MS has the unique ability to detect and characterize populations of molecules with different degrees of exchange (20). NMR analysis complements the MS analysis by defining the average proton occupancy over the distribution of protein molecules on a residue-specific basis.
PI3-SH3 Amyloid Fibril Formation Proceeds Through Different Types of Intermediates Depending on the Solution Conditions.
We monitored the aggregation process of PI3-SH3 by using transmission electron microscopy (TEM), atomic force microscopy (AFM), and oligomer-specific immunoreactivity (14). Studies were carried out at pH 2.0, in accord with earlier HD exchange experiments (16), and at pH 1.5. The results show that even a small difference in pH can result in the population of different intermediate assemblies on the way to fibril formation.
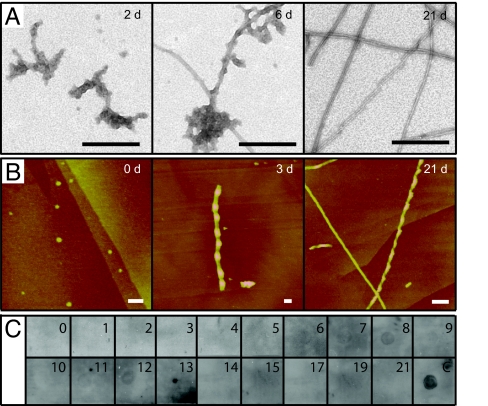
At pH 2.0, electron micrographs obtained for samples aggregated from 0 to 4 days show the dominant presence of amorphous aggregates. At intermediate times from 6 to 8 days, these structures coexist with fibrillar ones and from 10 days onward, electron micrographs reveal the presence of highly abundant and morphologically well-defined fibrils (Fig. 2A). Similar types of aggregates are observed by AFM (Fig. 2B), except for the amorphous ones, such aggregates may not adhere well to the graphite surface or they may be missed during imaging because of low tip adhesion. When we examined the binding of the oligomer-specific antibody A11 at different incubation times (14), no immunoreactivity was observed at any stage during the entire aggregation process (Fig. 2C), suggesting that under these conditions the type of aggregates recognized by this antibody are either absent or present only at low populations. From now on, we will refer to pH 2.0 conditions as AM, indicating that they favor the formation of amorphous-like intermediates.
Fig. 2.
Characterization of the PI3-SH3 aggregation process under AM conditions. (A) Electron micrographs obtained for Δtagg of 2, 6, and 21 days. (Scale bar, 200 nm.) (B) AFM images obtained for Δtagg of 0, 3, and 21 days. (Scale bar, 100 nm.) (C) Kinetics of oligomer-specific immunoreactivity. At the times indicated, aliquots were applied to a nitrocellulose membrane and probed with the oligomer-specific antibody, A11. The spot marked with a C corresponds to Aβ oligomers and was used as a positive control.
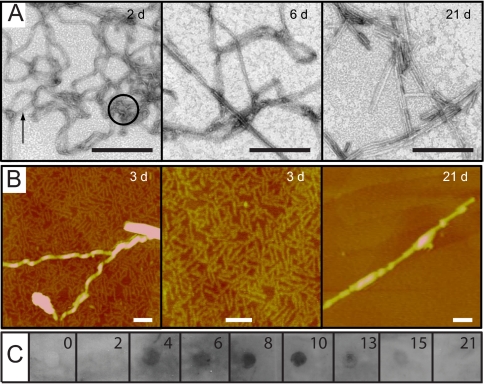
Electron micrographs obtained for samples aggregated from 0 to 4 days at pH 1.5 show the presence of short, thin protofibrils as well as flat-ribbon-like structures that can be seen to be formed from 2 fibril strands (21). After aggregation for 6 days, the flat-ribbon-like fibrils coexist with conventional amyloid-like fibrils, which become the major species after 8 days of aggregation (Fig. 3A). AFM analysis is highly complementary to that using TEM, because at early aggregation times AFM reveals the presence of abundant protofibrils (Fig. 3B), which are only rarely observed by TEM. These protofibrillar species have very small heights of 0.8–1.0 nm and, because of this, the TEM staining agent may not generate sufficient contrast for their observation. When these samples were examined by using the oligomer-specific antibody A11 (14), clear immunoreactivity can be observed in samples incubated for 4 to 10 days (Fig. 3C), indicating that oligomers recognized by this antibody are significantly populated. Throughout the article, we will refer to these conditions as PF because they favor protofibrillar-like intermediates.
Fig. 3.
Characterization of the PI3-SH3 aggregation process under PF conditions. (A) Electron micrographs obtained for Δtagg of 2, 6, and 21 days. The circle indicates a protofibril and the arrow shows a flat-ribbon-like fibril. (Scale bar, 200 nm.) (B) AFM images obtained for Δtagg of 3 and 21 days. (Scale bar, 100 nm.) (C) Kinetics of oligomer-specific immunoreactivity. At the times indicated, aliquots were applied to a nitrocellulose membrane and probed with the oligomer-specific antibody, A11.
The morphology of the mature fibrils generated under both conditions appears rather similar, the only clear differences being that the fibrils at pH 2.0 appear as individual, discrete structures, whereas those at pH 1.5 are shorter and generally appear as small bundles of 2 or 3 fibrils. Having 2 different conditions under which different types of intermediates are favored, amorphous and protofibrillar, while giving similar end products provides us with the opportunity for comparative studies by means of the pulse-labeling HD experiment discussed above and from which mechanistic and structural information on the aggregation process can be obtained.
PI3-SH3 Amyloid Fibril Formation Under Conditions Favoring Amorphous-Like Intermediates.
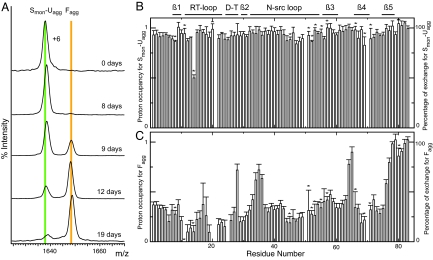
When the pulse-labeling experiment is used to study the aggregation process of PI3-SH3 under AM conditions, only 2 different HD exchange species are detected by ESI-MS throughout the entire aggregation process (Fig. 4A). From 0 to 8 days of aggregation, a single peak is observed at m/z 1637.1 (Fig. 4A). This peak corresponds in mass to the protein with no protected backbone amides, and as it may include soluble monomeric protein and/or aggregates with structural or dynamical properties that give no protection against HD exchange, we denote this population Smon-Uagg. After 9 days of aggregation, the peak representing the Smon-Uagg state remains visible, but a second peak at higher mass is evident with an m/z value of 1647.1 corresponding to species with an average of 52.2 ± 3.3 protected amides. The appearance of this second peak is concurrent with the appearance of well-defined fibrils in TEM micrographies (Fig. 2A); we therefore denote this peak as resulting from the fibrillar, Fagg, state. In agreement with this attribution, at longer aggregation times, the intensity of the Fagg species increases relative to that of the Smon-Uagg in the MS spectrum, such that Fagg is the dominant species after 12 days of aggregation (Fig. 4A).
Fig. 4.
PI3-SH3 pulse-labeled samples prepared under AM conditions. (A) ESI-MS mass spectra (+6 charge state). The spectra show the relative populations of Smon-Uagg and Fagg at the indicated Δtagg times. Peak intensities are normalized to the overall species population. (B) Exchange profile for Smon-Uagg obtained from NMR data of samples prepared after Δtagg of 8 days. (C) Exchange profile for Fagg obtained by deconvolution of the NMR data of samples prepared after Δtagg values of 10, 11, 12, 13, 17, and 19 days by using the distribution of populations obtained by ESI-MS analysis and the NMR exchange profile for the Smon-Uagg species. (B and C) The bars represent the proton occupancy and percentage of exchange for each residue. The locations of the β-sheet strands, turns, and loops in the native state of PI3-SH3 are indicated above the graph. An asterisk above a bar indicates a residue whose resonance is not fully resolved; the absence of a bar indicates that the resonance of the residue is not detectable or that the residue is proline that does not have an amide hydrogen (residues 50, 70, and 84).
The same samples were also analyzed by NMR spectroscopy to determine the proton occupancies at individual sites. A proton occupancy of 1.0 for a particular residue indicates that its amide backbone hydrogen has undergone complete HD exchange during the labeling pulse and it is thus located in a region of structure that does not provide any protection from solvent interactions. A value of 0.0 indicates that the amide hydrogen has not exchanged and it is thus located in a region of the structure able to offer protection. From 1 to 8 days of aggregation, NMR analysis shows a flat profile with proton occupancies close to 1.0, revealing no amide protection for any backbone amide [supporting information (SI) Fig. S1B], as expected from the MS results. Starting at 9 days and coinciding with the appearance of the Fagg peak by MS, the progressive protection of amide hydrogens is increasingly evident (Fig. S1C) and becomes more pronounced at longer aggregation times (Fig. S1D). NMR data, corresponding to aliquot samples where the Smon-Uagg and Fagg peaks were detected by ESI-MS, were deconvoluted by using both the population distribution obtained by ESI-MS analysis and the amide exchange profile measured by NMR after 8 days of incubation as the exchange profile for the Smon-Uagg state (Fig. 4B). This calculation allows an exchange profile for the PI3-SH3 fibrils to be obtained (Fig. 4C). The quantity of deuterium in the deconvoluted Fagg state, measured as 1 minus the measured proton occupancy and summed over all of the amide protons except for residues 50, 70, and 84 that are prolines, equals 53.3, a value very close to the number of amide deuterons, 52.2 ± 3.3, in the Fagg state determined by ESI-MS.
Examination of the PI3-SH3 fibril exchange profile reveals that the amide hydrogens from 7 residues at the C terminus (Ile 77 to Ser 83) and six (Gly 64 to Glu 65, Lys 34 to Ser 36, and Asp 28) residues distributed throughout the sequence exchange significantly faster than the average (≥0.6). The fact that these low-protected stretches of residues are preceded by, or contain, a glycine residue suggests that they are likely to include flexible loops and turns within the fibril structure. As far as the rest of the residues are concerned, the regions of the sequence comprising Ala 10 to Lys 16 and Glu 19 to Gly 27 show the lowest proton occupancies (≤0.2), whereas the remainder of the residues in the sequence exhibit proton occupancies of ≈0.35. This pattern of exchange suggests that residues Ala 10 to Lys 16 and Glu 19 to Gly 27 are likely to be present as part of a highly protected fibrillar core, and that the remainder of the residues are protected within the fibrillar structure and even perhaps within the core in such a way that their amide hydrogens are at least partially protected from exchange.
PI3-SH3 Amyloid Fibril Formation Under Conditions Favoring Protofibrillar-Like Intermediates.
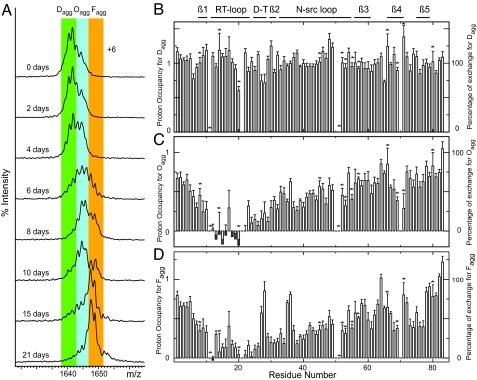
The aggregation process of PI3-SH3 under PF conditions was also studied by means of the pulse-labeling experiment. Under these conditions, ESI-MS analysis reveals that aggregation occurs as a multistate process with multiple species arising, coexisting, and decaying with time (Fig. 5A). A plot of the intensity of each peak as a function of the aggregation time, Δtagg, reveals that some species coexist for significant time periods (≈4 days) (Fig. S2). Thus, a first set of peaks with m/z values of 1639.1, 1640.3, and 1641.6, corresponding to a + 6 charge state and indicated with a green band in Fig. 5A, are dominant at early aggregation times, from 0 to 4 days. The peak with m/z of 1639.1 corresponds to a species in which all amide deuterons have exchanged with solvent within the labeling time, indicating that it corresponds to the species with the least persistent protective structure. The peaks at m/z 1640.3 and 1641.6 correspond, respectively, to species with average masses that have 11.3 ± 1.3 and 19.2 ± 1.7 amides protected. A second set of peaks with m/z values of 1643.1, 1644.5, and 1645.9, overlaid with a blue band in Fig. 5A, is present early in the aggregation reaction but become dominant at somewhat later times, from 6 to 10 days. These species correspond, respectively, to average masses indicative of 27.8 ± 1.8, 36.3 ± 1.5, and 44.9 ± 2.1 amides that are highly protected. Finally, a third set of 4 peaks, indicated by an orange band in Fig. 5A, correspond to the species that are most abundant late in the aggregation process. These peaks have m/z values of 1647.5, 1648.9, 1650.1, and 1651.8 corresponding, respectively, to average masses that indicate the presence of 54.6 ± 0.9, 62.7 ± 0.8, 70.2 ± 1.2, and 80.2 ± 1.2 protected amides. Because these species appear toward the end of the aggregation process and become the dominant species when electron micrographs show the presence of well-defined fibrils, we attribute these species to the fibrils, Fagg. The presence of several peaks from the aggregation reaction at pH 1.5, and the fact that in the analysis of the data several species coexist for significant periods, reveals that aggregation at this pH is a more heterogeneous process than at pH 2.0.
Fig. 5.
PI3-SH3 pulse-labeled samples prepared under PF conditions. (A) ESI-MS mass spectra (+6 charge state). The spectra show the relative populations of Dagg (green band), Oagg (blue band), and Fagg (orange band) at the indicated Δtagg times. Peak intensities are normalized to the overall species population. (B–D) Exchange profile for Dagg (B), Oagg (C), and Fagg (D). These profiles were obtained by multilinear regression analysis combining the NMR data of samples prepared after Δtagg equal to 0, 2, 6, 10, 13, and 15 days and the distribution of populations obtained by ESI-MS. The bars represent the proton occupancy and percentage of exchange for each residue. The locations of the β-sheet strands, turns, and loops in the native state of PI3-SH3 are indicated above the graph. An asterisk above a bar indicates a residue whose resonance is not fully resolved; the absence of a bar indicates that the resonance of the residue is not detectable or that the residue is proline that does not have an amide hydrogen (residues 50, 70, and 84).
ESI-MS results provide us with unique information about the distribution of species at different stages of the aggregation process. For a given value of Δtagg, NMR spectroscopy of a set of duplicate samples identical to those used for the MS analysis gives us the average proton occupancy on a residue-by-residue basis over the distribution of molecules detected by ESI-MS (Fig. S3). For a given Δtagg, we have combined the populations obtained by ESI-MS with NMR data (of the type indicated in Fig. S3) and have then used a multilinear regression analysis to obtain residue-specific information for each group of peaks detected by ESI-MS (Fig. 5 B–D).
The exchange profile defined by NMR spectroscopy for the protein molecules that correspond to the first set of peaks (see the green band in Fig. 5A) reveals that these species possess largely disordered conformations as all residues have similar proton occupancies with values close to 1.0 (Fig. 5B). We will therefore attribute these species to disordered aggregates lacking persistent structure, denoted Dagg. The NMR-based exchange profiles for the second set of species (corresponding to the blue band in Fig. 5A) reveal that residues Tyr 12 to Ile 22 are highly protected from exchange. The proton occupancies for the rest of the residues increase gradually as their positions in the sequence become more distant from the most protected ones (Fig. 5C). The fact that we are able to observe a clear pattern of protection within the sequences of these intermediate species suggests, therefore, that the initially disordered aggregates have reorganized to form conformations where residues Tyr 12 to Ile 22, in particular, are located in highly persistent structure; we refer to these species as ordered aggregates, Oagg. The deconvoluted exchange profiles for the species that make up the third group of peaks, the orange band in Fig. 5A, already assigned to fibrils, Fagg, reveal that the most protected region comprises residues Tyr 12 to Leu 26, similar to that of the Oagg species, but that further reorganization has taken place. Thus, in the Fagg state, some parts of the protein have become more protected (e.g., residues Tyr 73 to Ile 77), but others have become more exposed (e.g., residues Gly 27 and Glu 28, Gly 35 and Ser 36, and Gly 64) when compared with the Oagg species (Fig. 5D).
Discussion
On the Structure of the Fibrils.
We have previously carried out HD exchange experiments on PI3-SH3 amyloid fibrils (16). Under the conditions used, exchange was dominated by the continuous recycling of molecules within the ensemble of fibrils. Although our main aim in developing the pulse-labeling experiment has been to characterize the various species present at different stages of the aggregation process, the fact that use of short labeling times eliminates the contribution of recycling to exchange (Fig. S4) has allowed us to obtain the exchange patterns for the PI3-SH3 molecules incorporated within the fibrils (Figs. 4C and 5D). The pattern obtained for the fibrils produced under AM conditions (Fig. 4C) and under PF conditions (Fig. 5D) is very similar. Small differences, such a higher protection for residues at the N terminus of the sequence for fibrils produced under PF conditions could be explained by differences in the way the protofilaments interact within the higher-order structure of the mature fibrils.
It is interesting to compare our exchange data with reports in the literature. Work on a homologous protein of the PI3-SH3 domain, the α-spectrin SH3 domain, and several PI3-SH3 mutants has suggested that residues located in part of the RT loop and adjacent divergent turn (DT) in the native state, act as “mediators” or “facilitators” in the conversion of the soluble PI3-SH3 into amyloid fibrils (22). Other studies using molecular dynamics simulations to probe aggregation of the Src-SH3 domain concluded that the RT loop sequence constitutes the initial core structure for fibril formation (23). The RT loop and the DT region in the native structure of PI3-SH3 includes residues Ala 10 to Lys 16 and Glu 19 to Gly 27, which are the most protected regions of the fibril (Fig. 4C). Our data thus reinforce the crucial role of residues comprising the RT loop and the DT in the native state in the formation of amyloid fibrils.
On the Mechanism of Aggregation.
Several mechanisms have been proposed to explain amyloid fibril formation (24); among them, the nucleated conformational conversion (NCC) mechanism (25) is supported by a range of experimental and theoretical observations (26–28). In the NCC mechanism, a group of monomers initially present in solution coalesce to form amorphous oligomers. These oligomers subsequently undergo a reorganization process and eventually give rise to organized oligomers and fibrils rich in β-sheet structure. Up to now, a detailed empirical description of such a process at the molecular level has remained elusive because of the challenge in describing the early stages of aggregation of polypeptide chains by experiment, primarily because of the difficulties in detecting and characterizing the small, structurally heterogenous, and transient species that are involved.
When the pulse-labeling HD exchange approach is used to probe the aggregation behavior of the PI3-SH3 domain under AM conditions, only the initial, Smon-Uagg, and final, Fagg, states are detected (Fig. 4A). Explanations for the lack of detection of Oagg species include the possibility that they are present at very low populations, or that their protected regions are highly dynamic, resulting in fluctuations that are large enough to give rise to exchange during the labeling pulse. The latter possibility could be rationalized by the fact that intermolecular interactions are less favorable at the lower ionic strength that exists at pH 2.0 than at pH 1.5.
Application of the same pulse-labeling experiment under PF conditions reveals that species detected early in the aggregation process, referred to throughout this article as Dagg, are disordered on the basis of the low degree of amide hydrogen exchange protection measured by MS and NMR methods (Fig. 5B). The intermediate species, referred to as Oagg, however, are found to adopt highly ordered conformations, particularly from residues Tyr 12 to Ile 22, a conclusion based on the very low proton occupancies of these residues (Fig. 5C).
The fact that under PF conditions an oligomer-specific antibody, thought to be conformation specific, recognizes some or all of the intermediate species, but not their precursors or the subsequent fibrils (Fig. 3C), further reinforces the conclusion that a conformational change has occurred before their formation. This antibody is likely to recognize conformational characteristics of the intermediate species, perhaps because of the exposure of hydrophobic side chains (28, 29) or the adoption of a different conformational form (30). The most protected residues in the Oagg species are also the most protected ones in the Fagg species, implying that these intermediates are likely to be species that progress to fibril formation. Under PF conditions, a variety of species, including transient ones, has been detected and indeed trapped. Their exchange data are consistent, at least, with the NCC mechanism because aggregation occurs through initially formed disordered species, Dagg, that subsequently evolve and assemble into ordered aggregates, Oagg, that eventually lead to amyloid fibrils, Fagg.
Conclusions
The results presented here have shown that a pulse-labeling HD exchange strategy is a powerful means of gaining detailed kinetic, mechanistic, and structural insights into the nature and interconversion of different species populated during the aggregation of an SH3 domain into amyloid fibrils. Similar approaches should be applicable to other peptide and protein systems including those associated with important medical disorders. In addition, besides examining the aggregates themselves, the effect of other molecular species on the aggregating system can be investigated suggesting that extension of this approach would have significant biological and pharmaceutical importance.
Materials and Methods
Protein Expression.
Unlabeled PI3-SH3, uniformly 15N, and 13C, 15N labeled PI3-SH3 were obtained as described in ref. 16.
Pulse-Labeling HD Exchange Experiments.
PI3-SH3 aggregation was initiated by incubating freshly dissolved protein in D2O (Euriso-top) at a concentration of 0.4 mM, at either pH* 1.6 or pH* 1.1, corresponding to pH 2.0 and 1.5, at 34 °C. For both aggregation buffers, the pH*, the reading on the pH meter when measuring D2O solutions, was adjusted by using HCI. Duplicate aliquots of 150 μL were withdrawn after different Δtagg intervals. Aliquots of the solutions were taken at Δtagg ranging from 0 to 21 days. After the variable aggregation period, Δtagg, in the D2O buffer, the buffer was exchanged to give an H2O-based buffer by means of a fixed labeling time, Δtlabel. Two different means of buffer exchange were used: dialysis and a 1/15 dilution. For each of them, Δtlabel was determined experimentally and adjusted to be the minimum time required for exposed deuterons to exchange with solvent. HD exchange is slower at pH 2.0 than at pH 1.5, so appropriate dialysis and dilution controls were performed at pH 2.0. At this pH, the optimal Δtlabel was found to be 2 h for dialysis and 30 min for dilution (Fig. S5). The same Δtlabel times were used at pH 2.0 and at pH 1.5. Dialysis was performed for 2 h with a 100 MWCO membrane (Spectra/Por) and H2O (Riedel-de-Haën) adjusted to pH 1.5 or pH 2.0 with HCl (Riedel-de-Haën). After the labeling pulse by means of dialysis, from each 150-μL aliquot a 20-μL aliquot was taken for ESI-MS analysis and a 100-μL aliquot for NMR analysis, both of which were immediately freeze-dried to quench exchange. When the labeling pulse was applied by means of dilution, from each 150-μL aliquot, 20-μL and 100-μL aliquots were divided for ESI-MS and NMR analysis, respectively. These aliquots were diluted 1/15 with H2O that was adjusted to pH 1.5 or 2.0 with HCl. The dilution buffer at pH 1.5 was adjusted with formic acid (Riedel-de-Haën) because the use of HCl gives rise to strong adduct peaks in the MS. Controls to determine that formic acid was not affecting the nature of the species under investigation were carried out (Fig. S6 and Fig. S7). After dilution, samples were immediately freeze-dried to quench exchange. Further considerations on the pulse-labeling experiments are given in the online SI Materials and Methods.
HD Exchange Analyzed by ESI-MS and NMR.
Lyophilized samples were transferred into a solution of 95% DMSO-d6/5% D2O at pH* 4.6 that solubilizes the various aggregates into monomers and preserves the deuterium content of the protein molecules (9). The solubilized samples were then analyzed by ESI-MS (8, 16) and NMR (9, 10, 16). Exchange profile for Fagg species detected under AM conditions were obtained by deconvoluting the NMR data obtained at a given Δtagg (for 10, 11, 12, 13, 17, and 19 days) by using the population distributions obtained by ESI-MS at that Δtagg and the NMR data of samples prepared after Δtagg equal to 8 days as corresponding to the NMR exchange profile for the Smon-Uagg species. Exchange profiles for Dagg, Oagg, and Fagg species detected under PF conditions were obtained by multi-linear regression analyses combining the NMR data obtained for samples prepared at a given Δtagg (equal to 0, 2, 6, 10, 13, and 15 days) with the population distributions obtained by ESI-MS at that Δtagg by using the program GNUMERIC. Further details on the ESI-MS and NMR analysis are provided in the online SI Material and Methods.
Morphological Characterization of Amyloid Fibril Formation.
EM and AFM analysis were carried out with samples withdrawn from the D2O solution at different Δtagg times. EM analysis was carried out as reported (16). AFM imaging was performed with a commercial multimode atomic force microscope controlled by a Nanoscope IV electronics (Digital Instruments). Ten microliters of the sample were allowed to adsorb for ≈5–10 min at room temperature on freshly cleaved, highly ordered pyrolytic graphite (Nt-MDT Co.), and finally imaged in milliQ water and in tapping mode operation.
Oligomer-Specific Antibody.
Three-microliter aliquots of PI3-SH3 samples withdrawn from the D2O solution under AM and PF conditions were applied to a nitrocellulose membrane at different Δtagg. The membrane was then immunodetected with the oligomer-specific antibody, A11 (Biosource) (14), following manufacturers procedures. Immunoreactive bands were detected with the ECL Plus system (Amersham Biosciences).
Supplementary Material
Acknowledgments.
This work was supported by Program Grants from Fundació La Caixa, Fundació La Marató de TV3 (to N.C. and E.G.), and Ministerio de Ciencia e Innovación-Fondo Europeo de Desarrollo Regional BIO2008-00799 (to E.G.), by the Royal Society and the Walters-Kundert Trust (C.V.R.), and by the Wellcome Trust and the Leverhulme Trust (C.M.D.). M.Z. received support from the Eurpean Union 3D repertoire, and M.A. received a Ministerio de Educación y Ciencia predoctoral Formación de Personal Universitario Fellowship.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0812227106/DCSupplemental.
References
- 1.Selkoe DJ. Folding proteins in fatal ways. Nature. 2003;426:900–904. doi: 10.1038/nature02264. [DOI] [PubMed] [Google Scholar]
- 2.Chiti F, Dobson CM. Protein misfolding, functional amyloid, and human disease. Annu Rev Biochem. 2006;75:333–366. doi: 10.1146/annurev.biochem.75.101304.123901. [DOI] [PubMed] [Google Scholar]
- 3.Rochet JC, Lansbury PT., Jr Amyloid fibrillogenesis: Themes and variations. Curr Opin Struct Biol. 2000;10:60–68. doi: 10.1016/s0959-440x(99)00049-4. [DOI] [PubMed] [Google Scholar]
- 4.Cleary JP, et al. Natural oligomers of the amyloid-beta protein specifically disrupt cognitive function. Nat Neurosci. 2005;8:79–84. doi: 10.1038/nn1372. [DOI] [PubMed] [Google Scholar]
- 5.Haass C, Selkoe DJ. Soluble protein oligomers in neurodegeneration: Lessons from the Alzheimer's amyloid beta-peptide. Nat Rev Mol Cell Biol. 2007;8:101–112. doi: 10.1038/nrm2101. [DOI] [PubMed] [Google Scholar]
- 6.Petkova AT, et al. Self-propagating, molecular-level polymorphism in Alzheimer's beta-amyloid fibrils. Science. 2005;307:262–265. doi: 10.1126/science.1105850. [DOI] [PubMed] [Google Scholar]
- 7.Chen M, Margittai M, Chen J, Langen R. Investigation of alpha-synuclein fibril structure by site-directed spin labeling. J Biol Chem. 2007;282:24970–24979. doi: 10.1074/jbc.M700368200. [DOI] [PubMed] [Google Scholar]
- 8.Kheterpal I, Zhou S, Cook KD, Wetzel R. Abeta amyloid fibrils possess a core structure highly resistant to hydrogen exchange. Proc Natl Acad Sci USA. 2000;97:13597–13601. doi: 10.1073/pnas.250288897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hoshino M, et al. Mapping the core of the beta(2)-microglobulin amyloid fibril by H/D exchange. Nat Struct Biol. 2002;9:332–336. doi: 10.1038/nsb792. [DOI] [PubMed] [Google Scholar]
- 10.Luhrs T, et al. 3D structure of Alzheimer's amyloid-beta(1–42) fibrils. Proc Natl Acad Sci USA. 2005;102:17342–17347. doi: 10.1073/pnas.0506723102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bitan G, Teplow DB. Rapid photochemical cross-linking—A new tool for studies of metastable, amyloidogenic protein assemblies. Acc Chem Res. 2004;37:357–364. doi: 10.1021/ar000214l. [DOI] [PubMed] [Google Scholar]
- 12.Bernstein SL, et al. Amyloid beta-protein: Monomer structure and early aggregation states of Abeta42 and its Pro19 alloform. J Am Chem Soc. 2005;127:2075–2084. doi: 10.1021/ja044531p. [DOI] [PubMed] [Google Scholar]
- 13.Nettleton EJ, et al. Characterization of the oligomeric states of insulin in self-assembly and amyloid fibril formation by mass spectrometry. Biophys J. 2000;79:1053–1065. doi: 10.1016/S0006-3495(00)76359-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kayed R, et al. Common structure of soluble amyloid oligomers implies common mechanism of pathogenesis. Science. 2003;300:486–489. doi: 10.1126/science.1079469. [DOI] [PubMed] [Google Scholar]
- 15.Kheterpal I, et al. Abeta protofibrils possess a stable core structure resistant to hydrogen exchange. Biochemistry. 2003;42:14092–14098. doi: 10.1021/bi0357816. [DOI] [PubMed] [Google Scholar]
- 16.Carulla N, et al. Molecular recycling within amyloid fibrils. Nature. 2005;436:554–558. doi: 10.1038/nature03986. [DOI] [PubMed] [Google Scholar]
- 17.Guijarro JI, et al. Amyloid fibril formation by an SH3 domain. Proc Natl Acad Sci USA. 1998;95:4224–4228. doi: 10.1073/pnas.95.8.4224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zurdo J, et al. Dependence on solution conditions of aggregation and amyloid formation by an SH3 domain. J Mol Biol. 2001;311:325–340. doi: 10.1006/jmbi.2001.4858. [DOI] [PubMed] [Google Scholar]
- 19.Bucciantini M, et al. Inherent toxicity of aggregates implies a common mechanism for protein misfolding diseases. Nature. 2002;416:507–511. doi: 10.1038/416507a. [DOI] [PubMed] [Google Scholar]
- 20.Miranker A, Robinson CV, Radford SE, Dobson CM. Investigation of protein folding by mass spectrometry. FASEB J. 1996;10:93–101. doi: 10.1096/fasebj.10.1.8566553. [DOI] [PubMed] [Google Scholar]
- 21.Bouchard M, et al. Formation of insulin amyloid fibrils followed by FTIR simultaneously with CD and electron microscopy. Protein Sci. 2000;9:1960–1967. doi: 10.1110/ps.9.10.1960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ventura S, et al. Short amino acid stretches can mediate amyloid formation in globular proteins: The Src homology 3 (SH3) case. Proc Natl Acad Sci USA. 2004;101:7258–7263. doi: 10.1073/pnas.0308249101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ding F, et al. Molecular dynamics simulation of the SH3 domain aggregation suggests a generic amyloidogenesis mechanism. J Mol Biol. 2002;324:851–857. doi: 10.1016/s0022-2836(02)01112-9. [DOI] [PubMed] [Google Scholar]
- 24.Gsponer J, Vendruscolo M. Theoretical approaches to protein aggregation. Protein Pept Lett. 2006;13:287–293. doi: 10.2174/092986606775338407. [DOI] [PubMed] [Google Scholar]
- 25.Serio TR, et al. Nucleated conformational conversion and the replication of conformational information by a prion determinant. Science. 2000;289:1317–1321. doi: 10.1126/science.289.5483.1317. [DOI] [PubMed] [Google Scholar]
- 26.Petty SA, Decatur SM. Experimental evidence for the reorganization of beta-strands within aggregates of the Abeta(16–22) peptide. J Am Chem Soc. 2005;127:13488–13489. doi: 10.1021/ja054663y. [DOI] [PubMed] [Google Scholar]
- 27.Bader R, et al. Probing the mechanism of amyloidogenesis through a tandem repeat of the PI3-SH3 domain suggests a generic model for protein aggregation and fibril formation. J Mol Biol. 2006;356:189–208. doi: 10.1016/j.jmb.2005.11.034. [DOI] [PubMed] [Google Scholar]
- 28.Cheon M, et al. Structural reorganisation and potential toxicity of oligomeric species formed during the assembly of amyloid fibrils. PLoS Comput Biol. 2007;3:1727–1738. doi: 10.1371/journal.pcbi.0030173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Orte A, et al. Direct characterization of amyloidogenic oligomers by single-molecule fluorescence. Proc Natl Acad Sci USA. 2008;105:14424–14429. doi: 10.1073/pnas.0803086105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Daggett V. Alpha-sheet: The toxic conformer in amyloid diseases? Acc Chem Res. 2006;39:594–602. doi: 10.1021/ar0500719. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.