Abstract
Histone deacetylase inhibitors (HDACi) represent a new group of drugs currently being tested in a wide variety of clinical applications. They are especially effective in preclinical models of cancer where they show antiproliferative action in many different types of cancer cells. Recently, the first HDACi was approved for the treatment of cutaneous T cell lymphomas. Most HDACi currently in clinical development act by unspecifically interfering with the enzymatic activity of all class I HDACs (HDAC1, 2, 3, and 8), and it is widely believed that the development of isoform-specific HDACi could lead to better therapeutic efficacy. The contribution of the individual class I HDACs to different disease states, however, has so far not been fully elucidated. Here, we use a genetic approach to dissect the involvement of the different class I HDACs in tumor cells. We show that deletion of a single HDAC is not sufficient to induce cell death, but that HDAC1 and 2 play redundant and essential roles in tumor cell survival. Their deletion leads to nuclear bridging, nuclear fragmentation, and mitotic catastrophe, mirroring the effects of HDACi on cancer cells. These findings suggest that pharmacological inhibition of HDAC1 and 2 may be sufficient for anticancer activity, providing an experimental framework for the development of isoform-specific HDAC inhibitors.
Keywords: cancer, mitotic catastrophe, HDAC inhibitor, acetylation, tumorigenesis
Histone deacetylases (HDACs) comprise an ancient family of enzymes that play crucial roles in numerous biological processes (1). There are 11 different HDAC isoforms encoded in mammalian genomes, which can be classified in four different families (class I, IIa, IIb, and IV). They function by removing acetyl groups from proteins and can act on histones and on nonhistone proteins, thereby counteracting the activity of their antagonists, the histone acetyltransferases (HATs) (2). Deacetylation of histones generally leads to repression of gene expression caused by compaction of chromatin, which in turn leads to reduced accessibility of transcription factors and other regulatory molecules (1). The dynamic interplay of acetylation and deacetylation serves as a key regulatory mechanism governing the control of gene expression, differentiation, and development (1). It is thus somewhat surprising that drugs interfering with this basic molecular mechanism [i.e., HDAC inhibitors (HDACi)] are well tolerated in humans (3) and show therapeutic benefit in a wide variety of disease models (1).
In humans, the HDACi suberoylanilide hydroxamic acid (SAHA) has been approved by the FDA for the treatment of cutaneous T cell lymphoma in patients who have not responded to other therapies (4, 5). Well over 100 clinical trials have been performed or are currently ongoing to test the efficacy of HDACi as mono- or combination therapy (6), with most of these testing HDACi as anticancer agents. The majority of current HDACi inhibit HDACs unspecifically, for example the IC50 values for SAHA lie between 30 and 50 nM for Hdac1, Hdac3, and Hdac6 (7). Although the involvement of class I HDACs in cancer biology has been well documented in recent years, the contribution of the specific HDAC isoforms has remained controversial (3, 8–26).
As recent analyses of class I HDAC knockout mice have revealed highly specific functions for individual HDAC isoforms in development and the control of gene expression programs (27–31), we hypothesized that a distinct subset of the class I HDACs might be responsible for the antiproliferative action of HDAC inhibitors on cancer cells. Using a genetic ex vivo model, we show that HDAC1 and 2 play redundant and essential roles in tumor cell survival.
Results
Generation of Tumor Cell Lines with Conditional Alleles for Class I Hdacs.
To circumvent the lack of well characterized isoform-specific HDACi (3) and the problem of possible off-target effects when using knockdown methods (32), we used a genetic approach to delete class I HDAC isoforms specifically in a well-defined tumor model (33, 34). We generated conditional alleles of Hdac1 (28), Hdac2 (28), Hdac3 (27), and Hdac8 by flanking exons encoding crucial domains of the enzymes with loxP sites. After removal of neomycin resistance cassettes with Flpe-expressing mice (35), the alleles were bred to homozygosity, and primary cell lines were established from the outgrowth of tail biopsies. Primary cells were immortalized after the first passage by infection with a pBabe-derived retrovirus encoding simian virus 40 (SV40) large T antigen and consecutively transformed by infecting with a pBabe-derived virus encoding H-Ras V12G (Fig. 1A). The derived cell lines form subcutaneous tumors (see below) and are susceptible to HDACi at concentrations described for other tumor cell lines [nanomolar range for trichostatin A (TSA) and micromolar range for SAHA] (Fig. 1 B and C). As expected, treatment of these cells with TSA leads to hyperacetylation of nuclear, and to a lesser degree, cytoplasmic proteins as detected by immunofluorescent staining using a pan acetyllysine antibody (Fig. 1D).
Fig. 1.
Generation of tumor cell lines with conditional class I Hdac alleles. (A) Tail biopsies were taken from mice of the indicated genotypes and primary fibroblasts obtained by outgrowth. Primary cells were immortalized by using retrovirally encoded SV40 large T antigen and subsequently transformed with retrovirus encoding H-Ras V12G. (B and C) Cells were treated with increasing concentrations of the HDACi TSA and SAHA. Cell density was visualized by using crystal violet stain after 1 week. (Scale bars: 200 μm.) (D) Staining of cells for acetylated lysine after treatment with 1 μM TSA for 24 h. [Scale bars: 100 μm (Upper); 20 μm (Lower).] ac-K, acetyllysine.
Cre-Mediated Deletion of Single Class I Hdac Alleles.
We initially used a transgenically encoded tamoxifen-inducible Cre (36) to delete the conditional alleles but observed significant leakiness of Cre activity during the transformation of the cell lines. To circumvent the leakiness, we used adenovirus-mediated Cre delivery for deletion of the floxed alleles. Efficient deletion of the loxP-flanked exons was demonstrated by PCR with genomic DNA extracted 72 h after deletion (Fig. 2B), and previous studies have shown that 72 h after knockdown of HDACs almost no protein is detectable by Western blotting (11). We then tested the cells for viability and proliferation by seeding them at low density (≈1/20 confluence) and visualized them by crystal violet staining after 1 week. Surprisingly, loss of any single HDAC isoform was well tolerated with neither cell morphology nor proliferation rates showing major changes after single HDAC deletion (Fig. 2A).
Fig. 2.
Deletion of single HDAC alleles in tumor cells. (A) Cells of the indicated genotypes were incubated with either GFP- or Cre-expressing adenovirus. Cells were stained 1 week after Hdac inactivation with crystal violet. (Scale bars: 200 μm.) (B) Genotyping of Cre-treated tumor cells by genomic PCR using primers binding in the deleted region of each HDAC gene.
Redundancy of Hdac1 and 2 in Tumor Cells.
We previously demonstrated a genetic redundancy between Hdac1 and Hdac2 in mouse development, such that deletion of three alleles in a given organ system (i.e., heart, brain, lung, endothelial cells, smooth muscle cells) was well tolerated if only one copy of either Hdac1 or Hdac2 was left intact (28). We therefore generated a tumor cell line with double conditional alleles for Hdac1 and Hdac2 (Hdac1flox/flox Hdac2flox/flox). When both Hdac1 and 2 were deleted, a profound cell viability and proliferation phenotype was observed (Fig. 3 A and B). Cell numbers started to decline 3 days after Hdac1/2 deletion, and 6 days after Hdac inactivation almost no viable cells could be observed (Fig. 3B). Strikingly, many of the Hdac1/2-null cells showed a multinuclear morphology when visualized by crystal violet stain (Fig. 3C). The presence of multiple nuclei was verified by visualizing the DNA directly with Hoechst stain (Fig. 3D).
Fig. 3.
Deletion of Hdac1 and Hdac2 leads to cell death and mitotic catastrophe. (A) Hdac1flox/flox Hdac2flox/flox cells were incubated with either GFP- or Cre-expressing adenovirus. Cells were stained 1 week after Hdac inactivation with crystal violet. (Scale bars: 200 μm.) (B) Time course analysis and quantification of reduced proliferation and cell death in Hdac1/2 double-null cells. (C) Multinuclear morphology of surviving Hdac1/2 double-null cells. Cell stain is crystal violet. (Scale bar: 40 μm.) (D) Multiple nuclei in an Hdac1/2 double-null cell visualized by Hoechst stain. Red counterstain is phalloidin. (Scale bar: 20 μm.) (E) Nuclear bridging in Hdac1/2 double-null cells. Note the DNA filament stretching between the two cells (arrows). (Scale bar: 20 μm.) (F) Nuclear fragmentation in Hdac1/2 double-null cells. Note the multiple micronuclei surrounding the main nucleus. [Scale bar: 40 μm (Left and Center); 10 μm (Right).]
Mitotic Catastrophe in Hdac1- and 2-Deficient Tumor Cells.
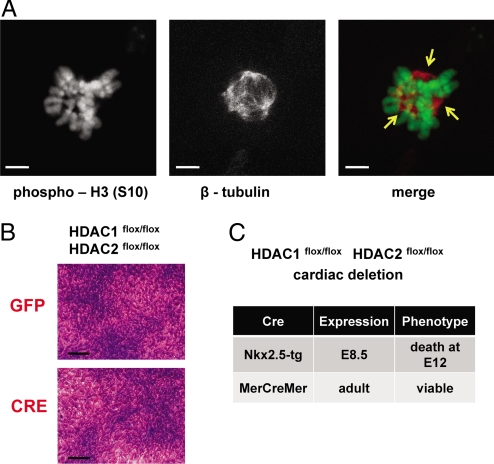
Hoechst staining revealed additional phenotypes in Hdac1/2-null cells such as nuclear bridging (Fig. 3E) and nuclear fragmentation (Fig. 3F). These nuclear phenotypes are generally associated with mitotic catastrophe in tumor cells (37), a phenotype that is common in HDACi-treated cancer cells and that has been associated with a defective spindle checkpoint (38–43). We observed multiple spindle poles and an aberrant spindle apparatus in Hdac1/2-deficient cells, indicating slippage of the spindle assembly checkpoint (Fig. 4A). These observations suggest that a defective mitotic checkpoint leading to mitotic catastrophe is a major mechanism of tumor cell death due to Hdac1/2 deletion. A testable prediction of this hypothesis is that nonproliferating cells would tolerate the deletion of Hdac1/2. We therefore deleted Hdac1 and Hdac2 in immortalized fibroblasts that were quiescent because of contact inhibition. Deletion of Hdac1/2 was well tolerated in these nonproliferating cells (Fig. 4B). Nonproliferating primary cells such as confluent fibroblasts and primary calvarial osteoblasts also tolerated deletion of Hdac1/2, whereas proliferating primary cells showed signs of mitotic catastrophe after several rounds of mitoses.
Fig. 4.
Spindle abnormalities in Hdac1/2 double-null cells leads to cell death in proliferating cells. (A) Multiple spindle poles (yellow arrows) and an aberrant spindle apparatus in Hdac1/2-deficient cells indicating slippage of the spindle assembly checkpoint. Cells were stained with antibodies against phosphorylated histone H3S10 and β-tubulin. (Scale bars: 5 μm.) (B) Lethal phenotype of Hdac1/2 deletion depends on cell proliferation. Hdac1/2 were deleted in contact inhibited, quiescent fibroblasts. Cells were stained with crystal violet after 1 week. (Scale bars: 100 μm.) (C) Cardiac deletion of Hdac1/2 in embryogenesis is lethal, whereas deletion in postmitotic adult cardiomyocytes is well tolerated.
Hdac1 and 2 Are Dispensable for Postmitotic Cells but Are Required for Tumor Growth in Vivo.
We next tested whether deletion of Hdac1 and 2 would be tolerated by postmitotic cells in vivo. We used a tamoxifen-inducible Cre under the control of the α-myosin heavy chain promoter (44) to delete Hdac1 and 2 in vivo specifically in cardiomyocytes. Deletion of Hdac1 and 2 from the adult heart was well tolerated with no obvious phenotype, whereas embryonic deletion with Nkx2.5-Cre, when cardiomyocytes are still proliferating, was invariably lethal (Fig. 4C).
Finally, we tested whether deletion of Hdac1 and Hdac2 would be sufficient to inhibit the growth of tumor cells in vivo. We incubated 105 Hdac1/2 double-conditional tumor cells with either GFP or Cre encoding adenovirus for 8 h. After extensive washing the cells were injected s.c. into nude mice and the tumor size measured every 5 days. GFP-treated cells invariably formed visible tumors in the first 10 days. In contrast, the deletion of Hdac1/2 led to a complete block of tumor growth (Fig. 5 A and B).
Fig. 5.
Inhibition of tumor cell growth in vivo. (A) Hdac1flox/flox Hdac2flox/flox cells were incubated for 8 h with either GFP- or Cre-expressing virus and injected s.c. into nude mice. Tumor growth is shown 3 weeks after injection. (B) Time course analysis of tumor growth after s.c. injection of 1 × 105 Hdac1/2 conditional cells treated with either Ad-GFP or Ad-Cre. Eight animals were used in each group.
Discussion
The results of this study indicate that Hdac1 and Hdac2 play redundant and essential roles in tumor cell survival. This suggests that Hdac1 and Hdac2 mediate a significant subset of the antiproliferative actions of current HDACi.
Because Hdac1 and Hdac2 are the most closely related of the HDAC superfamily (45), inhibitors specifically targeting these isoforms, with little or no inhibition of the remaining HDACs, could potentially be identified (46, 47). Although current pan-HDACi are relatively well tolerated, they have been associated with numerous side effects such as cardiac arrhythmia, bone marrow depression, clotting disorders, diarrhea, fatigue, and electrolyte disturbances in phase I and II studies (48). It is expected that compounds that specifically inhibit the subset of enzymatic isoforms required to obtain a desired clinical outcome will substantially broaden the therapeutic window of HDACi (49).
Numerous studies have analyzed the role of different HDAC isoforms in cancer cell biology, but the results have often been conflicting (8–26, 31, 50–53), and evidence from genetic mouse models is missing for most of these associations. Recent exceptions are the establishment of an Hdac1-null ES cell line (31), a conditional Hdac3 3T3 cell line (54) and the development of Hdac6-deficient cell lines (55, 56). Loss of Hdac1 in ES cells leads to the dysregulation of a specific set of genes without any overt mitotic defects (31). Interestingly, the loss of Hdac1 is invariably associated with an up-regulation of Hdac2 in this system (28, 30, 31), suggesting that Hdac2 might compensate for the loss of HDAC activity. This is in line with the observed redundancy between Hdac1 and Hdac2 in later embryogenesis (28).
Loss of Hdac3 leads to defective DNA double-strand break repair in vitro and to a mild reduction of phosphorylated histone H3Ser10; however, these cells do not show overt mitotic defects (54). Inhibition of Hdac3 in vivo appears to be undesirable because knockout studies in mice have shown that Hdac3 is a crucial regulator of metabolic homeostasis (27, 29). Hdac6-null cell lines, however, show diminished growth in soft agar, and Hdac6-knockout mice display a reduction of carcinogen-induced skin tumors (55). Pharmacological inhibition of Hdac6 synergizes with the tumor-suppressive activity of proteasome inhibitors in vitro (57, 58), consistent with its role as the main cytoplasmic deacetylase (2). This is in contrast to the mainly nuclear localization of class I Hdacs and the nuclear phenotype observed in Hdac1/2-deleted cells (Fig. 4A) and might suggest additive or synergistic effects of Hdac1/2 and Hdac6 inhibition. However, antitumor efficacy of specific Hdac6 inhibitors has not been demonstrated conclusively in vivo so far.
In summary, we provide evidence that pharmacological targeting of HDAC1 and HDAC2 might be desirable in the development of specific HDACi for anticancer applications. Although loss of Hdac1 and Hdac2 in vivo is lethal when occurring early in embryonic development (28), deletion in adult, postmitotic cells is well tolerated (Fig. 4C). This is consistent with the viability of quiescent Hdac1/2-deleted cells (Fig. 4B) and provides an explanation for the selective HDACi sensitivity of fast cycling cells such as tumor cells.
Methods
Materials.
Tamoxifen and TSA were obtained from Sigma and SAHA from Cayman Chemicals. Anti-acetylated lysine, phospho-H3 (S10), and tubulin antibodies were from Cell Signaling.
Generation of Cell Lines.
Cell lines were derived from tail biopsies of Hdac mutant mice. The construction of conditional alleles for Hdac1, Hdac2, and Hdac3 has been described (27, 28). A conditional allele for Hdac8 was constructed by flanking exon 4 with loxP sites using standard techniques. Mice used in this study were on a mixed genetic background (SV129/C57BL6/CD1). Tail biopsies (1 cm) were taken from 6- to 8-week-old mice, washed with ethanol, minced, placed in a 6-well dish, and carefully overlayed with tissue culture medium (DMEM, 10% FBS, antibiotic-antimycotic mix; Invitrogen). After outgrowth of primary fibroblasts, cells were infected at the first passage with a retrovirus encoding SV40 large T antigen. Cells were passaged 10 times with a splitting ratio of 1:10 and then were infected with a retrovirus encoding H-Ras V12G. Two days after the second infection, cells were selected with Puromycin (1 μg/mL) and Zeocin (200 μg/mL). Primary osteoblasts were generated from neonatal calvaria by using standard protocols as described in ref. 59.
Viruses.
Retrovirus constructs were generated by using the pBabe/pCL-Eco system, with standard methods. Briefly, HEK 293T cells were transfected in 10-cm dishes by using 6 μg of pBabe and 6 μg of pCL-Eco with FuGENE (Roche). Medium was changed 24 h later and virus harvested at 48 and 72 h. Viral supernatant was filtered, supplemented with Polybrene (Sigma) at 1 μg/mL, and frozen at − 80 °C. The plasmids pCL-Eco (Addgene 12371), pBabe-puro SV40 LT (Addgene 13970), pBabe-zeo large T genomic (Addgene 1778) and pBabe-puro Ras V12 (Addgene 1768) were obtained from Addgene. pBabe-zeo Ras V12 was generated by subcloning H-Ras V12G from pBabe-puro Ras V12 into pBabe-zeo large T genomic via BamHI and SalI. Adenovirus constructs Ad5CMVCre, Ad5CMVntlacZ, Ad5CMVCre-eGFP, and Ad5CMVeGFP were obtained from the University of Iowa Gene Transfer Vectore Core and used at a multiplicity of infection of 103.
Gene Deletion and Proliferation Assays.
Conditional alleles were deleted by using adenoviral Cre delivery. Cells were trypsinized and counted, and then 5 × 105 cells were seeded in a 6-well dish and incubated with control or Cre-expressing adenovirus overnight. The next day (considered day 0 of deletion) medium was changed. The following day, 1 × 105 cells were seeded in a 10-cm dish and used for proliferation assays. For qualitative analysis, cells were stained with a 1% crystal violet solution. Quantitative analysis was performed by trypsinizing, counting, and reseeding of cells in triplicate. Cell numbers were determined in a Coulter counter. Genotyping of the Hdac1–3 cell lines was performed as described for the mice from which they were generated (27, 28). Genotyping for Hdac8 cell lines was done by using standard PCR protocols for 32 cycles at 58 °C annealing temperature by using primers spanning the 5′ loxP site 8A: TCAGCCTTGGATATGCTAGCC and 8B: TTGCCAGAGTAGACCTAAGTGCT.
Immunofluorescence.
Immunofluorescence analysis was performed by using standard protocols and visualized with a confocal microscope.
Tumor Growth Study.
For in vivo tumor formation, 5 × 105 cells were incubated with control or Cre-expressing virus for 8 h. Cells were washed, and 1 × 105 cells were injected s.c. in the flanks of athymic nude mice (Hsd:Athymic Nude-nu; Harlan). Eight animals were used in each group. Tumor size was calculated by using the formula π/6 × [(L + W)/2]3. All animal experiments were conducted according to procedures approved by the Institutional Animal Care and Use Committee at the University of Texas Southwestern Medical Center.
Tamoxifen Treatment.
Tamoxifen was solubilized in 5% ethanol/95% sesame oil at 100 mg/mL. One hundred microliters of this solution was administered by gavage at 5 consecutive days.
Statistical Analysis.
Results are expressed as means ± SD. Differences between groups were tested for statistical significance by using the unpaired 2-tailed Student t test with Welch correction. P values of <0.05 were considered significant.
Acknowledgments.
We thank Scott Hiebert and Christian Seiser for their insights and comments. Work in the laboratory of E.N.O. was supported by grants from the National Institutes of Health, the Donald W. Reynolds Foundation for Clinical Cardiovascular Research, the Robert A. Welch Foundation, and the Sandler Foundation for Asthma Research. M.H was supported by Deutsche Forschungsgemeinschaft Grant HA 3335/2-1.
Footnotes
The authors declare no conflict of interest.
References
- 1.Haberland M, Montgomery RL, Olson EN. The many roles of histone deacetylases in development and physiology: Implications for disease and therapy. Nat Rev Genet. 2009;10:32–42. doi: 10.1038/nrg2485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Yang XJ, Seto E. The Rpd3/Hda1 family of lysine deacetylases: From bacteria and yeast to mice and men. Nat Rev Mol Cell Biol. 2008;9:206–218. doi: 10.1038/nrm2346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bolden JE, Peart MJ, Johnstone RW. Anticancer activities of histone deacetylase inhibitors. Nat Rev Drug Discov. 2006;5:769–784. doi: 10.1038/nrd2133. [DOI] [PubMed] [Google Scholar]
- 4.Mann BS, Johnson JR, Cohen MH, Justice R, Pazdur R. FDA approval summary: Vorinostat for treatment of advanced primary cutaneous T cell lymphoma. Oncologist. 2007;12:1247–1252. doi: 10.1634/theoncologist.12-10-1247. [DOI] [PubMed] [Google Scholar]
- 5.Olsen EA, et al. Phase IIb multicenter trial of vorinostat in patients with persistent, progressive, or treatment refractory cutaneous T cell lymphoma. J Clin Oncol. 2007;25:3109–3115. doi: 10.1200/JCO.2006.10.2434. [DOI] [PubMed] [Google Scholar]
- 6.Dokmanovic M, Clarke C, Marks PA. Histone deacetylase inhibitors: Overview and perspectives. Mol Cancer Res. 2007;5:981–989. doi: 10.1158/1541-7786.MCR-07-0324. [DOI] [PubMed] [Google Scholar]
- 7.Lee AY, et al. Quantitative analysis of histone deacetylase-1 selective histone modifications by differential mass spectrometry. J Proteome Res. 2008;7:5177–5186. doi: 10.1021/pr800510p. [DOI] [PubMed] [Google Scholar]
- 8.Ropero S, et al. A truncating mutation of HDAC2 in human cancers confers resistance to histone deacetylase inhibition. Nat Genet. 2006;38:566–569. doi: 10.1038/ng1773. [DOI] [PubMed] [Google Scholar]
- 9.Zhu P, et al. Induction of HDAC2 expression upon loss of APC in colorectal tumorigenesis. Cancer Cell. 2004;5:455–463. doi: 10.1016/s1535-6108(04)00114-x. [DOI] [PubMed] [Google Scholar]
- 10.Oehme I, et al. Histone deacetylase 8 in neuroblastoma tumorigenesis. Clin Cancer Res. 2009;15:91–99. doi: 10.1158/1078-0432.CCR-08-0684. [DOI] [PubMed] [Google Scholar]
- 11.Senese S, et al. Role for histone deacetylase 1 in human tumor cell proliferation. Mol Cell Biol. 2007;27:4784–4795. doi: 10.1128/MCB.00494-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Choi JH, et al. Expression profile of histone deacetylase 1 in gastric cancer tissues. Jpn J Cancer Res. 2001;92:1300–1304. doi: 10.1111/j.1349-7006.2001.tb02153.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Weichert W, et al. Association of patterns of class I histone deacetylase expression with patient prognosis in gastric cancer: A retrospective analysis. Lancet Oncol. 2008;9:139–148. doi: 10.1016/S1470-2045(08)70004-4. [DOI] [PubMed] [Google Scholar]
- 14.Weichert W, et al. Histone deacetylases 1, 2, and 3 are highly expressed in prostate cancer and HDAC2 expression is associated with shorter PSA relapse time after radical prostatectomy. Br J Cancer. 2008;98:604–610. doi: 10.1038/sj.bjc.6604199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Weichert W, et al. Class I histone deacetylase expression has independent prognostic impact in human colorectal cancer: Specific role of class I histone deacetylases in vitro and in vivo. Clin Cancer Res. 2008;14:1669–1677. doi: 10.1158/1078-0432.CCR-07-0990. [DOI] [PubMed] [Google Scholar]
- 16.Miyake K, et al. Expression of hypoxia-inducible factor-1α, histone deacetylase 1, and metastasis-associated protein 1 in pancreatic carcinoma: Correlation with poor prognosis with possible regulation. Pancreas. 2008;36:e1–9. doi: 10.1097/MPA.0b013e31815f2c2a. [DOI] [PubMed] [Google Scholar]
- 17.Halkidou K, et al. Up-regulation and nuclear recruitment of HDAC1 in hormone refractory prostate cancer. Prostate. 2004;59:177–189. doi: 10.1002/pros.20022. [DOI] [PubMed] [Google Scholar]
- 18.Rikimaru T, et al. Clinical significance of histone deacetylase 1 expression in patients with hepatocellular carcinoma. Oncology. 2007;72:69–74. doi: 10.1159/000111106. [DOI] [PubMed] [Google Scholar]
- 19.Sasaki H, et al. Histone deacetylase 1 mRNA expression in lung cancer. Lung Cancer. 2004;46:171–178. doi: 10.1016/j.lungcan.2004.03.021. [DOI] [PubMed] [Google Scholar]
- 20.Krusche CA, et al. Histone deacetylase-1 and -3 protein expression in human breast cancer: A tissue microarray analysis. Breast Cancer Res Treat. 2005;90:15–23. doi: 10.1007/s10549-004-1668-2. [DOI] [PubMed] [Google Scholar]
- 21.Zhang Z, et al. Quantitation of HDAC1 mRNA expression in invasive carcinoma of the breast. Breast Cancer Res Treat. 2005;94:11–16. doi: 10.1007/s10549-005-6001-1. [DOI] [PubMed] [Google Scholar]
- 22.Keshelava N, et al. Histone deacetylase 1 gene expression and sensitization of multidrug-resistant neuroblastoma cell lines to cytotoxic agents by depsipeptide. J Natl Cancer Inst. 2007;99:1107–1119. doi: 10.1093/jnci/djm044. [DOI] [PubMed] [Google Scholar]
- 23.Inoue S, Mai A, Dyer MJ, Cohen GM. Inhibition of histone deacetylase class I but not class II is critical for the sensitization of leukemic cells to tumor necrosis factor-related apoptosis-inducing ligand-induced apoptosis. Cancer Res. 2006;66:6785–6792. doi: 10.1158/0008-5472.CAN-05-4563. [DOI] [PubMed] [Google Scholar]
- 24.Huang BH, et al. Inhibition of histone deacetylase 2 increases apoptosis and p21Cip1/WAF1 expression, independent of histone deacetylase 1. Cell Death Differ. 2005;12:395–404. doi: 10.1038/sj.cdd.4401567. [DOI] [PubMed] [Google Scholar]
- 25.Zimmermann S, et al. Reduced body size and decreased intestinal tumor rates in HDAC2-mutant mice. Cancer Res. 2007;67:9047–9054. doi: 10.1158/0008-5472.CAN-07-0312. [DOI] [PubMed] [Google Scholar]
- 26.Balasubramanian S, et al. A novel histone deacetylase 8 (HDAC8)-specific inhibitor PCI-34051 induces apoptosis in T cell lymphomas. Leukemia. 2008;22:1026–1034. doi: 10.1038/leu.2008.9. [DOI] [PubMed] [Google Scholar]
- 27.Montgomery RL, et al. Maintenance of cardiac energy metabolism by histone deacetylase 3 in mice. J Clin Invest. 2008;118:3588–3597. doi: 10.1172/JCI35847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Montgomery RL, et al. Histone deacetylases 1 and 2 redundantly regulate cardiac morphogenesis, growth, and contractility. Genes Dev. 2007;21:1790–1802. doi: 10.1101/gad.1563807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Knutson SK, et al. Liver-specific deletion of histone deacetylase 3 disrupts metabolic transcriptional networks. EMBO J. 2008;27:1017–1028. doi: 10.1038/emboj.2008.51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lagger G, et al. Essential function of histone deacetylase 1 in proliferation control and CDK inhibitor repression. EMBO J. 2002;21:2672–2681. doi: 10.1093/emboj/21.11.2672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zupkovitz G, et al. Negative and positive regulation of gene expression by mouse histone deacetylase 1. Mol Cell Biol. 2006;26:7913–7928. doi: 10.1128/MCB.01220-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hochegger H, Takeda S, Hunt T. Cyclin-dependent kinases and cell-cycle transitions: Does one fit all? Nat Rev Mol Cell Biol. 2008;9:910–916. doi: 10.1038/nrm2510. [DOI] [PubMed] [Google Scholar]
- 33.Hahn WC, et al. Creation of human tumour cells with defined genetic elements. Nature. 1999;400:464–468. doi: 10.1038/22780. [DOI] [PubMed] [Google Scholar]
- 34.Land H, Parada LF, Weinberg RA. Tumorigenic conversion of primary embryo fibroblasts requires at least two cooperating oncogenes. Nature. 1983;304:596–602. doi: 10.1038/304596a0. [DOI] [PubMed] [Google Scholar]
- 35.Rodriguez CI, et al. Tumorigenic conversion of primary embryo fibroblasts requires at least two cooperating oncogenes. Nat Genet. 2000;25:139–140. doi: 10.1038/304596a0. [DOI] [PubMed] [Google Scholar]
- 36.Hayashi S, McMahon AP. Efficient recombination in diverse tissues by a tamoxifen-inducible form of Cre: A tool for temporally regulated gene activation/inactivation in the mouse. Dev Biol. 2002;244:305–318. doi: 10.1006/dbio.2002.0597. [DOI] [PubMed] [Google Scholar]
- 37.Okada H, Mak TW. Pathways of apoptotic and non-apoptotic death in tumour cells. Nat Rev Cancer. 2004;4:592–603. doi: 10.1038/nrc1412. [DOI] [PubMed] [Google Scholar]
- 38.Magnaghi-Jaulin L, Eot-Houllier G, Fulcrand G, Jaulin C. Histone deacetylase inhibitors induce premature sister chromatid separation and override the mitotic spindle assembly checkpoint. Cancer Res. 2007;67:6360–6367. doi: 10.1158/0008-5472.CAN-06-3012. [DOI] [PubMed] [Google Scholar]
- 39.Cimini D, Mattiuzzo M, Torosantucci L, Degrassi F. Histone hyperacetylation in mitosis prevents sister chromatid separation and produces chromosome segregation defects. Mol Biol Cell. 2003;14:3821–3833. doi: 10.1091/mbc.E03-01-0860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Qiu L, et al. Histone deacetylase inhibitors trigger a G2 checkpoint in normal cells that is defective in tumor cells. Mol Biol Cell. 2000;11:2069–2083. doi: 10.1091/mbc.11.6.2069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Dowling M, et al. Mitotic spindle checkpoint inactivation by trichostatin a defines a mechanism for increasing cancer cell killing by microtubule-disrupting agents. Cancer Biol Ther. 2005;4:197–206. [PubMed] [Google Scholar]
- 42.Xu WS, Parmigiani RB, Marks PA. Mitotic spindle checkpoint inactivation by trichostatin A defines a mechanism for increasing cancer cell killing by microtubule-disrupting agents. Oncogene. 2007;26:5541–5552. [Google Scholar]
- 43.Xu WS, Perez G, Ngo L, Gui CY, Marks PA. Induction of polyploidy by histone deacetylase inhibitor: A pathway for antitumor effects. Cancer Res. 2005;65:7832–7839. doi: 10.1158/0008-5472.CAN-04-4608. [DOI] [PubMed] [Google Scholar]
- 44.Sohal DS, et al. Temporally regulated and tissue-specific gene manipulations in the adult and embryonic heart using a tamoxifen-inducible Cre protein. Circ Res. 2001;89:20–25. doi: 10.1161/hh1301.092687. [DOI] [PubMed] [Google Scholar]
- 45.Gregoretti IV, Lee YM, Goodson HV. Molecular evolution of the histone deacetylase family: Functional implications of phylogenetic analysis. J Mol Biol. 2004;338:17–31. doi: 10.1016/j.jmb.2004.02.006. [DOI] [PubMed] [Google Scholar]
- 46.Khan N, et al. Molecular evolution of the histone deacetylase family: Functional implications of phylogenetic analysis. Biochem J. 2008;409:581–589. [Google Scholar]
- 47.Bieliauskas AV, Pflum MK. Isoform-selective histone deacetylase inhibitors. Chem Soc Rev. 2008;37:1402–1413. doi: 10.1039/b703830p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bruserud Ø, Stapnes C, Ersvaer E, Gjertsen BT, Ryningen A. Histone deacetylase inhibitors in cancer treatment: A review of the clinical toxicity and the modulation of gene expression in cancer cell. Curr Pharm Biotechnol. 2007;8:388–400. doi: 10.2174/138920107783018417. [DOI] [PubMed] [Google Scholar]
- 49.Moradei O, Maroun CR, Paquin I, Vaisburg A. Histone deacetylase inhibitors: Latest developments, trends and prospects. Curr Med Chem Anticancer Agents. 2005;5:529–560. doi: 10.2174/1568011054866946. [DOI] [PubMed] [Google Scholar]
- 50.Ropero S, et al. Transforming pathways unleashed by a HDAC2 mutation in human cancer. Oncogene. 2008;27:4008–4012. doi: 10.1038/onc.2008.31. [DOI] [PubMed] [Google Scholar]
- 51.Halkidou K, Cook S, Leung HY, Neal DE, Robson CN. Nuclear accumulation of histone deacetylase 4 (HDAC4) coincides with the loss of androgen sensitivity in hormone refractory cancer of the prostate. Eur Urol. 2004;45:382–389. doi: 10.1016/j.eururo.2003.10.005. author reply 389. [DOI] [PubMed] [Google Scholar]
- 52.Krusche CA, et al. Class I histone deacetylase expression in the human cyclic endometrium and endometrial adenocarcinomas. Hum Reprod. 2007;22:2956–2966. doi: 10.1093/humrep/dem241. [DOI] [PubMed] [Google Scholar]
- 53.Zhang Z, et al. Class I histone deacetylase expression in the human cyclic endometrium and endometrial adenocarcinomas. Clin Cancer Res. 2004;10:6962–6968. [Google Scholar]
- 54.Bhaskara S, et al. Deletion of histone deacetylase 3 reveals critical roles in S phase progression and DNA damage control. Mol Cell. 2008;30:61–72. doi: 10.1016/j.molcel.2008.02.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Lee YS, et al. The cytoplasmic deacetylase HDAC6 is required for efficient oncogenic tumorigenesis. Cancer Res. 2008;68:7561–7569. doi: 10.1158/0008-5472.CAN-08-0188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zhang Y, et al. Mice lacking histone deacetylase 6 have hyperacetylated tubulin but are viable and develop normally. Mol Cell Biol. 2008;28:1688–1701. doi: 10.1128/MCB.01154-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Hideshima T, et al. Small-molecule inhibition of proteasome and aggresome function induces synergistic antitumor activity in multiple myeloma. Proc Natl Acad Sci USA. 2005;102:8567–8572. doi: 10.1073/pnas.0503221102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Bazzaro M, et al. Ubiquitin proteasome system stress underlies synergistic killing of ovarian cancer cells by bortezomib and a novel HDAC6 inhibitor. Clin Cancer Res. 2008;14:7340–7347. doi: 10.1158/1078-0432.CCR-08-0642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Schmidt K, et al. The high mobility group transcription factor Sox8 is a negative regulator of osteoblast differentiation. J Cell Biol. 2005;168:899–910. doi: 10.1083/jcb.200408013. [DOI] [PMC free article] [PubMed] [Google Scholar]