Abstract
IFN-stimulated gene 56 (ISG56) is one of the first identified proteins induced by viruses and type I IFNs. In this study, we identified ISG56 as a virus-induced protein associated with MITA, an adapter protein involved in virus-triggered induction of type I IFNs. Overexpression of ISG56 inhibited Sendai virus-triggered activation of IRF3, NF-κB, and the IFN-β promoter, whereas knockdown of ISG56 had opposite effects. Consistently, overexpression of ISG56 reversed cytoplasmic poly(I:C)-induced inhibition of vesicular stomatitis virus (VSV) replication, whereas knockdown of ISG56 inhibited VSV replication. Competitive coimmunoprecipitation experiments indicated that ISG56 disrupted the interactions between MITA and VISA or TBK1, two components in the virus-triggered IFN signaling pathways. These results suggest that ISG56 is a mediator of negative-feedback regulation of virus-triggered induction of type I IFNs and cellular antiviral responses.
Keywords: interferon, MITA, negative-feedback regulation, IRF3, NF-κB
The innate immune system is the first line of host defense against viral infection. The host antiviral responses are initiated by the recognition of viral components by host pathogen recognition receptors (PRRs), which lead to the induction of type I IFNs consisting of IFN-α and IFN-β family cytokines (1–3). Type I IFNs further induces downstream proteins, which cause suppression of viral replication, clearance of virus-infected cells, and facilitation of adaptive immune response (1–3).
Among PRRs, certain Toll-like receptors (TLRs) and RIG-I-like helicases (RLHs) are known to detect viral infection. For example, TLR3 is a transmembrane receptor that recognizes viral dsRNA released by infected cells (4). Engagement of TLR3 by dsRNA triggers TRIF-mediated signaling pathways, leading to activation of the transcription factors IRF3 and NF-κB (5–7). Unlike TLR3, which acts as a transmembrane receptor, the RLH family members RIG-I and MDA5 are cytoplasmic viral RNA sensors (8, 9). Both RIG-I and MDA5 are RNA helicase proteins that contain 2 CARD modules at their N terminus and a DexD/H-box RNA helicase domain at their C terminus. The helicase domains of RIG-I and MDA5 serve as intracellular viral RNA receptors, whereas the CARD modules are responsible for transmitting signals to downstream CARD-containing adaptor VISA (also known as IPS-1, MAVS, and Cardif) (10–13). VISA is a mitochondrial membrane protein that is associated with another mitochondrial membrane adapter protein MITA/STING. It has been demonstrated that MITA recruits TBK1 and IRF3 to the VISA-associated complex in a viral infection-dependent process (14, 15). In this complex, IRF3 is phosphorylated by TBK1, leading to its dimerization and translocation into the nucleus. VISA is also associated with TRAF6, which activates NF-κB through IKK (10). The activated IRF3 and NF-κB collaboratively induce transcription of type I IFN genes (1–3).
Type I IFNs are undetectable under physiological conditions but rapidly induced after viral infection in most cell types. Type I IFNs activate the JAK-STAT signal transduction pathways, leading to transcriptional induction of >300 IFN-stimulated genes (ISGs) (16). Numerous ISGs, such as ISG15, the GTPase Mx1, RNaseL, and PKR, have been shown to function as antiviral effectors. Mice with mutations or deficiencies in these proteins or the key components of the pathways triggered by these proteins have increased susceptibility to viral infection (17). ISG56 (IFIT1) and ISG54 (IFIT2) are two of the first identified ISGs belonging to the same family (18, 19). All members in this family contain multiple tetratricopeptide (TPR) motifs that are known to mediate protein–protein interactions through scaffolds formed among tandem TPR repeats (20). ISG56 and ISG54 are induced in response to type I IFNs, dsRNAs, and viruses (21, 22). Although ISG56 and ISG54 are among the first identified ISGs, little is known about their functions. It has been reported that ISG56 and ISG54 interact with the translation initiation factor eIF-3, leading to the inhibition of translation initiation and protein synthesis (22). ISG56 has also been implicated in antiviral actions of IFNs against hepatitis C virus, West Nile virus, and lymphocytic choriomeningitis virus (23, 24). A recent study demonstrates that ISG56 inhibits human papillomavirus DNA replication by binding to the viral protein E1 (25).
Although type I IFNs are critically involved in host defense against viral infection, production of these cytokines have to be properly regulated to prevent excessive harmful immune responses. Various molecules, including A20, DUBA, RNF125, Pin1, RBCK1, NLRX1, SIKE, DAK, and LGP2, have been shown to regulate induction of type I IFNs by targeting distinct components of the virus-triggered signaling pathways (26–28). In this report, we found that ISG56 was associated with the adapter protein MITA and disrupted the interaction of MITA with VISA or TBK1, leading to the inhibition of virus-induced IRF3 activation, IFN-β expression, and cellular antiviral responses. Our findings revealed a negative-feedback regulatory mechanism of cellular antiviral responses mediated by ISG56.
Results
Identification of ISG56 and ISG54 as MITA-Interacting Proteins.
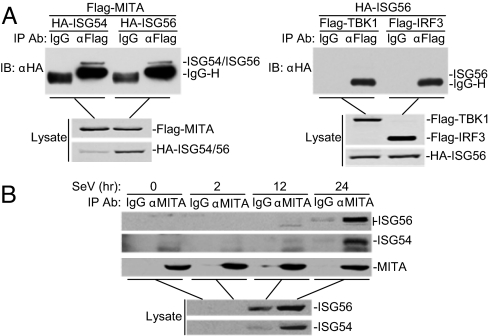
Recently, we and others have identified MITA as a critical adapter protein in virus-triggered IRF3 activation, type I IFN production and cellular antiviral responses (14, 15). To further investigate how the functions of MITA in the virus-triggered signaling pathways are regulated, we attempted to unambiguously identify MITA-associated proteins through a biochemical purification approach. We transfected 293 cells with Flag-tagged MITA alone or in combination with HA-tagged TBK1. The MITA-associated complexes were purified by anti-Flag affinity columns and eluted with Flag peptides. A shotgun mass spectrum analysis demonstrated that ISG56 and ISG54 were two proteins specifically associated with Flag-MITA/TBK1 but not with Flag-MITA alone (Fig. S1). To determine whether ISG56 and ISG54 are associated with MITA or TBK1, we performed transient transfection and coimmunoprecipitation experiments. The results indicated that ISG56 and ISG54 interacted with MITA but not TBK1 or IRF3 (Fig. 1A). Previously, we have shown that overexpression of MITA only weakly activates IRF3, whereas TBK1 or TBK1 plus MITA strongly activates IRF3 (14). It is possible that ISGs were strongly induced in the Flag-MITA/TBK1-transfected cells but only weakly induced in the Flag-MITA-alone-transected cells, which may account for the observation that ISG56 and ISG54 were identified in the Flag-MITA/TBK1 but not Flag-MITA sample.
Fig. 1.
ISG56 and ISG54 interact with MITA. (A) ISG56 and ISG54 interact with MITA but not TBK1 or IRF3. 293 cells (2 × 106) were transfected with the indicated plasmids (5 μg each). Coimmunoprecipitation and immunoblot analysis were performed with the indicated antibodies (Upper). Expression of the transfected proteins was analyzed by immunoblots with anti-HA and anti-Flag (Lower). (B) Endogenous ISG56 and ISG54 are associated with MITA. 293 cells (2 × 107) were infected with SeV for the indicated times or left uninfected. Immunoprecipitation and immunoblot analysis were performed with the indicated antibodies.
To determine whether endogenous ISG56 and ISG54 are associated with MITA, we induced expression of ISG56 and ISG54 by Sendai virus (SeV) infection for 2, 12, and 24 h, and then performed coimmunoprecipitation experiments. The results suggest that endogenous ISG56 and ISG54 induced by viral infection are associated with endogenous MITA (Fig. 1B). Interestingly, we found that ISG56 was phosphorylated at 24 h after SeV infection. The phosphorylation was confirmed by phosphatase treatment. The significance of the phosphorylation is unknown at this time.
Overexpression of ISG56 Inhibits Virus-Triggered Activation of ISRE and the IFN-β Promoter.
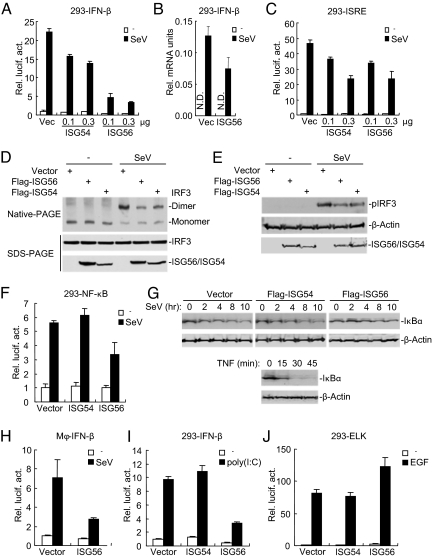
Because ISG56 and ISG54 are associated with MITA, we determined whether ISG56 and ISG54 regulate virus-triggered signaling. In reporter assays, overexpression of ISG56 strongly inhibited SeV-induced activation of the IFN-β promoter in a dose-dependent manner in 293 cells, whereas overexpression of ISG54 had minor inhibitory effect (Fig. 2A). Real-time PCR experiments also indicated that ISG56 could inhibit SeV-induced expression of IFN-β mRNA (Fig. 2B).
Fig. 2.
ISG56 inhibits virus-induced signaling. (A) ISG56 and ISG54 inhibit SeV-induced activation of the IFN-β promoter in a dose-dependent manner. 293 cells (1 × 105) were transfected with an IFN-β promoter reporter (0.1 μg) and increased amount of ISG56 or ISG54 expression plasmids. Eighteen hours after transfection, cells were left uninfected or infected with SeV for 12 h before luciferase assays were performed. (B) ISG56 inhibits SeV-induced expression of IFN-β mRNA. 293 cells (2 × 105) were transfected control or ISG56 expression plasmid (1 μg). Eighteen hours after transfection, cells were left uninfected or infected with SeV for 12 h before real-time PCRs were performed. (C) ISG56 and ISG54 inhibit SeV-induced ISRE activation in a dose-dependent manner. Experiments were performed similarly as in A except that an ISRE reporter was used. (D and E) ISG56 and ISG54 inhibit SeV-induced dimerization (D) and phosphorylation (E) of IRF3. 293 cells were transfected with the indicated plasmids for 24 h. Cells were then left uninfected or infected with SeV for 8 h (D) or 6 h (E) before native or SDS/PAGE and immunoblots with the indicated antibodies were performed. (F) Effects of ISG56 and ISG54 on SeV-induced activation of NF-κB. The experiments were similarly performed as in A. (G) Effects of ISG56 and ISG54 on SeV-induced degradation of IκBα. 293 cells (2 × 105) were transfected with the indicated expression plasmids (1 μg each). Eighteen hours after transfection, cells were left untreated or infected with SeV for the indicated times before immunoblot analysis was performed. Shown in the lower panels is TNF-induced IκBα degradation. (H) ISG56 inhibits SeV-induced activation of the IFN-β promoter in human primary macrophages. The experiments were similarly performed as in A except that the cells were transfected with the Nucleofactor method. (I) Effects of ISG56 and ISG54 on cytoplasmic poly(I:C)-induced activation of the IFN-β promoter. 293 cells (1 × 105) were transfected with the indicated plasmids (0.5 μg each) for 18 h. Cells were then further transfected with poly(I:C) (1 μg) or left untransfected for 24 h before luciferase assays were performed. (J) Effects of ISG56 and ISG54 on EGF-induced activation of Elk. 293 cells (1 × 105) were transfected with a pFR-luc reporter (0.2 μg) and an Elk reporter (0.02 μg) together with the indicated expression plasmids (0.5 μg each). Eighteen hours after transfection, cells were treated with EGF (50 ng/ml) for 12 h before luciferase assays were performed.
It has been demonstrated that induction of type I IFNs requires coordinated and cooperative action of the transcription factors IRF3 and NF-κB (1–3). In reporter assays, both ISG56 and ISG54 inhibited SeV-triggered activation of ISRE, a conserved enhancer motif recognized by activated IRF3 (Fig. 2C). ISG56 and ISG54 also inhibited SeV-induced IRF3 dimerization (Fig. 2D) and phosphorylation (Fig. 2E), which are hallmarks of IRF3 activation. In reporter assays, ISG56 inhibited SeV-induced NF-κB activation, whereas ISG54 had no inhibitory effect (Fig. 2F). Consistently, ISG56, but not ISG54, inhibited SeV-induced IκBα degradation (Fig. 2G). The effect of ISG56 on virus-induced IFN signaling is not cell type specific because ISG56 also inhibited SeV-induced IFN-β promoter activation in human blood monocyte-derived macrophages (Fig. 2H), HeLa and A549 cells (Fig. S2). Furthermore, ISG56 and ISG54 had different effects on cytoplasmic poly(I:C)-induced IFN-β promoter activation. Overexpression of ISG56 inhibited cytoplasmic poly(I:C)-induced activation of the IFN-β promoter, whereas ISG54 had no inhibitory effect (Fig. 2I). In similar experiments, both ISG56 and ISG54 had no inhibitory role on EGF-induced activation of the Elk transcription factor (Fig. 2J). Taken together, these data suggest that ISG56 negatively regulates virus-triggered activation of IRF3, NF-κB, and the IFN-β promoter, whereas ISG54 has minor effect.
Knockdown of ISG56 Potentiates Virus-Induced Activation of ISRE, NF-κB, and the IFN-β Promoter.
To determine whether ISG56 and ISG54 are involved in the regulation of virus-triggered type I IFN induction under physiological conditions, we examined the effects of knockdown of these two proteins on virus-triggered signaling. We constructed multiple RNAi plasmids for ISG56 and ISG54, respectively (Fig. 3A). Reporter assays indicated that knockdown of ISG56 potentiated SeV-induced ISRE activation, whereas knockdown of ISG54 had little effect (Fig. 3B). The potentiation of ISG56 RNAi plasmids on SeV-induced ISRE activation was correlated with their abilities to down-regulate ISG56 expression (Fig. 3 A and B). We selected ISG56-RNAi-1 and ISG54-RNAi-1 plasmids for additional experiments described below. These RNAi plasmids could also knockdown SeV-induced expression of endogenous ISG56 and ISG54, respectively (Fig. 3A). In reporter assays, knockdown of ISG56 potentiated SeV-triggered activation of NF-κB (Fig. 3C) and the IFN-β promoter (Fig. 3D) in 293 cells, whereas knockdown of ISG54 had little or no effects (Fig. 3 C and D). RT-PCR experiments confirmed that knockdown of ISG56 could potentiate virus-induced expression of endogenous IFN-β, Rantes, and ISG15 (Fig. 3E). Interestingly, knockdown of ISG54 caused elevated expression of ISG56, whereas knockdown of ISG56 caused elevated expression of ISG54 (Fig. 3E). These results suggest that ISG54 and ISG56 can compensate for decreased expression of the other protein. However, this may not be responsible for the observation that knockdown of ISG54 had minor effect on SeV-induced IFN-β promoter activation because double knockdown of ISG54 and ISG56 did not markedly enhance the potentiation effect on SeV-induced IFN-β promoter activation compared with knockdown of ISG56 alone (Fig. 3D). Knockdown of ISG56 also potentiated SeV-induced IFN-β promoter activation in human primary macrophages (Fig. 3F) and dendritic cells (DCs) (Fig. 3G), as well as HeLa and A549 cells (Fig. S3). Consistently, knockdown of ISG56 potentiated cytoplasmic poly(I:C)-induced activation of the IFN-β promoter, whereas knockdown of ISG54 had a lesser effect (Fig. 3H). These data suggest that endogenous ISG56 negatively regulated SeV-induced activation of IFN-β pathway.
Fig. 3.
Effects of RNAi-mediated knockdown of ISG56 and ISG54 on virus-induced signaling. (A) Effects of ISG56 and ISG54 RNAi plasmids on expression of ISG56 and ISG54. In the upper panels, 293 cells (2 × 105) were transfected with the indicated expression (0.5 μg each) and RNAi plasmids (1 μg each) for 24 h before immunoblot analysis was performed with anti-Flag. In the lower two panels, 293 cells (2 × 105) were transfected with the indicated RNAi plasmids (1 μg each) for 24 h, and then infected with SeV for 10 h before immunoblot analysis was performed with the indicated antibodies. (B) Effects of ISG56 and ISG54 RNAi on SeV-induced ISRE activation. 293 cells (1 × 105) were transfected with the indicated RNAi plasmids (0.5 μg each) for 24 h, and then infected with SeV or left uninfected for 12 h before reporter assays were performed. (C) Effects of ISG56 and ISG54 RNAi on SeV-induced NF-κB activation. The experiments were similarly performed as in B except that NF-κB reporter plasmid was used. (D) Effects of ISG56 and ISG54 RNAi on SeV-induced activation of the IFN-β promoter. 293 cells (1 × 105) were transfected with the indicated amount of RNAi plasmids. An empty vector was added to ensure that each transfection received the same amount of total DNA. The reporter assays were similarly performed as in B. (E) Effects of ISG56 and ISG54 RNAi on SeV-induced expression of downstream genes. 293 cells (2 × 105) were transfected with the indicated RNAi plasmid (1 μg each). Twenty-four hours after transfection, cells were left uninfected or infected with SeV for 12 h before RT-PCR was performed. (F and G) Effects of ISG56 RNAi on SeV-induced activation of the IFN-β promoter in human primary macrophages (F) or DCs (G). The experiments were similarly performed as in B except the cells were transfected with the Nucleofactor method. (H) Effects of ISG56 and ISG54 RNAi on cytoplasmic poly(I:C)-induced activation of the IFN-β promoter. 293 cells (1 × 105) were transfected with the indicated RNAi plasmids (0.5 μg each). Twenty-four hours later, cells were further transfected with poly(I:C) (1 μg) or left untransfected and luciferase assays were performed 24 h later.
ISG56 Negatively Regulates Cellular Antiviral Responses.
Because ISG56 negatively regulates virus- and cytoplasmic poly(I:C)-triggered IRF3 activation and IFN-β induction, we investigated whether ISG56 plays a role in cellular antiviral response. In plaque assays, overexpression of ISG56 mildly enhanced vesicular stomatitis virus (VSV) replication and significantly reversed cytoplasmic poly(I:C)-mediated inhibition of VSV replication (Fig. 4A). Conversely, knockdown of ISG56 significantly inhibited VSV replication and further enhanced the inhibitory effect triggered by cytoplasmic poly(I:C) (Fig. 4B). In these experiments, the effects of ISG54 are minimal (Fig. 4 A and B). These data suggest that ISG56 negatively regulates cellular antiviral responses.
Fig. 4.
Roles of ISG56 and ISG54 in cellular antiviral response. (A) Overexpression of ISG56 increases VSV replication. 293 cells (1 × 105) were transfected with the indicated expression plasmids (0.5 μg each). Eighteen hours later, cells were further transfected with poly(I:C) (1 μg) or left untransfected. Twenty-four hours after transfection, cells were infected with VSV [multiplicity of infection (moi), 0.1], and the supernatants were harvested at 24 h postinfection. Supernatants were analyzed for VSV production with standard plaque assays. Graphs show mean ± SD (n = 3). (B) Knockdown of ISG56 inhibits VSV replication. Plaque assays were performed as in A except that a control or ISG56/54 RNAi plasmid (no. 1) (1 μg) was transfected. Graphs show mean ± SD (n = 3).
ISG56 Disrupts the Interaction of MITA with VISA or TBK1.
Because ISG56 interacted with MITA and negatively regulated virus-triggered IRF3 activation and IFN-β induction, we investigated whether ISG56 acts through disruption of MITA-associated complexes. We have previously demonstrated that MITA is associated with VISA and acts as a scaffold protein to recruit TBK1 and IRF3 to the VISA complex (14). We wondered whether ISG56 inhibits virus-triggered signaling through disrupting the interaction of MITA with either its upstream protein VISA or its downstream protein TBK1. To test this, we performed competitive coimmunoprecipitation experiments. The results indicated that ISG56 disrupted the MITA–VISA and MITA–TBK1 interactions dose-dependently (Fig. 5A). Interestingly, ISG54 also disrupted these interactions (Fig. S4), consistent with its ability to inhibit SeV-induced ISRE and IFN-β promoter activation (Fig. 2 A and C). In similar experiments, ISG56 did not disrupt the interactions between VISA and MDA5, an interaction important for cytoplasmic poly(I:C)-triggered signaling, or between TRIF and TBK1, an interaction involved in TLR3 signaling (5) (Fig. 5B). These data suggest that ISG56 specifically disrupts the interactions of MITA with VISA and TBK1.
Fig. 5.
ISG56 disrupts the interaction of MITA with VISA or TBK1. (A) ISG56 disrupts the association of MITA with VISA or TBK1 in a dose-dependent manner. 293 cells (2 × 106) were transfected with the indicated plasmids (5 μg each) and an increase amount of ISG56 expression plasmids. Coimmunoprecipitation and immunoblot analysis were performed as in Fig. 1A. (B) ISG56 does not disrupt MDA5–VISA or TRIF–TBK1 interactions. 293 cells were transfected with the indicated plasmids and the coimmunoprecipitation and immunoblot analysis were performed as in Fig. 1A. (C) Kinetics of MITA–VISA and MITA–TBK1 associations after viral infection. 293 cells were left uninfected or infected with SeV for the indicated times. Immunoprecipitation and immunoblot analysis were performed with the indicated antibodies.
To determine whether endogenous association between MITA and VISA or TBK1 is disrupted after induction of ISG56, we performed endogenous coimmunoprecipitation experiments at various time points after viral infection. Consistent with our previous studies (18), we found that MITA interacted with VISA in uninfected cells. However, MITA interacted with TBK1 at 2 h after SeV infection. The MITA–VISA and MITA–TBK1 associations were diminished at 12-h time point and undetectable at 24-h time point (Fig. 5). The kinetics of disassociation between these proteins is correlated with the expression of endogenous ISG56 and its association with MITA (Figs. 1B, 5C). These results support the conclusion that induction of ISG56 disrupts the MITA–VISA and MITA–TBK1 interactions.
Discussion
ISG56 and ISG54 are two of the first identified proteins that are induced by type I IFNs, dsRNAs, and viruses (19, 22). In this study, we identified ISG56 and ISG54 as two proteins associated with MITA, a critical adapter protein involved in virus-triggered induction of type I IFNs (14, 15). Overexpression of ISG56 inhibited SeV-triggered activation of IRF3, NF-κB, and the IFN-β promoter, whereas knockdown of ISG56 had an opposite effect. Consistently, overexpression of ISG56 reversed cytoplasmic poly(I:C)-induced inhibition of VSV replication, whereas knockdown of ISG56 inhibited VSV replication. These results suggest that ISG56 negatively regulates virus-triggered IFN-β induction and cellular antiviral responses. Previously, it has been established that ISG56 is induced by viral infection and type I IFNs (21). Taken together, these studies suggest that ISG56 is a mediator of negative-feedback regulation of cellular antiviral responses.
Interestingly, although ISG54 is homologous to ISG56 and also disrupted MITA–VISA and MITA–TBK1 interactions in mammalian overexpression systems, either overexpression or knockdown of ISG54 had only minor effects on SeV-induced activation of the IFN-β promoter as well as cellular antiviral responses. Previously, it has been demonstrated that ISG56 mRNA expression was much stronger and longer than ISG54 in response to viruses such as influenza virus and herpes simplex virus type 1 (29, 30). It has also been noted that ISG56 mRNA is the most abundant IFN-induced mRNAs in a gene array analysis (16). These observations suggest that ISG56 is a major mediator of virus-triggered negative-feedback regulation of type I IFNs and cellular antiviral responses, whereas ISG54 plays much lesser role in these processes.
In coimmunoprecipitation experiments, ISG56 interacted with MITA but not TBK1 or IRF3. Competitive coimmunoprecipitation experiments indicated that ISG56 disrupted the MITA–VISA and MITA–TBK1 interactions in a dose-dependent manner. In similar experiments, ISG56 did not disrupt the MDA5–VISA and TRIF–TBK1 interactions. Based on these results, we propose that ISG56 negatively regulates virus-triggered signaling and cellular antiviral responses through specific disruption of the VISA–MITA–TBK1 complex by steric hindrance.
To prevent harmful effects resulting from spontaneous production in uninfected cells or overproduction of type I IFNs during an acute infection, host cells have developed distinct strategies to control excessive antiviral innate immune responses. At least two distinct mechanisms have been identified. One of the mechanisms involves the ubiquitin–proteosome system. Several members of the E3 ubiquitin ligase family, such as A20, RNF125, Pin1, RBCK1, RNF5, and Ro52, target the key components of the virus-induced type I IFN signaling pathways for degradation (26–28, 31, 32). Second, some inhibitory proteins, such as DAK, SIKE, and NLRX1, are constitutively expressed and physically associated with key components of virus-induced type I IFN signaling pathways to sequester them in inactive forms (26, 27, 33). Viral infection leads to release of these inhibitors, resulting in activation of the signaling components. In this study, we identified a distinct mechanism on regulation of virus-triggered induction of type I IFNs and cellular antiviral response: ISG56 is strongly induced by viral infection and then down-regulates virus-triggered induction of type I IFNs and cellular antiviral responses by disrupting the VISA–MITA–TBK1 complex.
Previous reports suggest that ISG56 acts as a suppressor of viral replication and protein translation. In light of these and our current findings, it is possible that ISG56 has multiple functions and is an important integrator of inhibition of viral replication and control of excessive antiviral responses. It is also possible that the functions of ISG56 are temporally regulated during viral infection. Although more studies are needed to understand the delicate regulatory mechanisms of ISG56 in viral replication and antiviral responses, our findings reveal a previously undescribed role for ISG56 in regulating cellular antiviral responses.
Methods
Reagents.
Flag peptide (Sigma), poly(I:C) (Invitrogen), EGF (R&D Systems); fetal bovine serum (ExCell); mouse monoclonal antibodies against Flag, HA, and β-actin (Sigma); and rabbit polyclonal antibody against IRF3 (Santa Cruz Biotechnology) were purchased from the indicated manufacturers. SeV, VSV, and rabbit and mouse anti-MITA antibodies were described in ref. 14. Mouse anti-ISG56 and ISG54 antiserum were raised against recombinant human ISG56 and ISG54.
Constructs.
NF-κB, ISRE, and the IFN-β promoter luciferase reporter plasmids, mammalian expression plasmids for HA- or Flag-tagged RIG-I, MDA5, VISA, TBK1, TRIF, and IRF3 were described in refs. 10, 14, and 33. Mammalian expression plasmids for human HA- or Flag-tagged ISG56 and ISG54 were constructed by standard molecular biology techniques.
Protein Purification and Mass Spectrometry Analysis.
293 cells (5 × 107) were transfected with Flag-MITA alone or in combination with HA-TBK1. The transfected cells were lysed, and the lysates were subjected to anti-Flag affinity purification. The anti-Flag associated proteins were eluted with Flag peptides. The eluted proteins were digested by trypsin in solution. The tryptic peptides were analyzed by HPLC-ESI/MS/MS with a Thermo Finnigan LTQ adapted for nanospray ionization. The tandem spectra were searched against Homo sapiens National Center for Biotechnology Information reference database using the SEQUEST. Results was filtered by Xcorr +1 > 1.9, +2 > 2.2, +3 > 3.5, sp > 500, Deltcn > 0.1, Rsp <= 5.
Transfection, Reporter Assays, Coimmunoprecipitation, Blot Analysis and RT-PCR, Generation and Transfection of Human Primary Macrophages and DCs.
These experiments were performed as described in refs. 10, 14, and 32.
RNAi Experiments.
Double-strand oligonucleotides corresponding to the target sequences were cloned into the pSuper.retro RNAi plasmid (Oligoengine). The target sequences for human ISG56 cDNA are as follows: no. 1, GGATAAAGCTCTTGAGTTA; no. 2, CAGAAAAGCTGAAGAGAAT; no. 3, GACAAGGTGGAGAACATTT; and no. 4, CTACAAATTGGAAGGAAAT. The target sequences for human ISG54 cDNA are as follows: no. 1, CGTAAAGCTGAAGAGTTAA; no. 2, GAGAGAAGTTAGTTGAAGA; no. 3, CCAGAAATCAAGGGAGAAA; and no. 4, CAAATTGGGTGCTGCTATA.
Supplementary Material
Acknowledgments.
We thank other members of the H.-B.S. laboratory for discussions and help with techniques. This work was supported by Chinese 973 Program Grant 2006CB504301, National Natural Science Foundation of China Grants 30630019 and 30700431, Chinese 863 Program Grant 2006AA02A306, and Chinese Science and Technology Key Project Grant 2008ZX10002-014.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0900818106/DCSupplemental.
References
- 1.Akira S, Uematsu S, Takeuchi O. Pathogen recognition and innate immunity. Cell. 2006;124:783–801. doi: 10.1016/j.cell.2006.02.015. [DOI] [PubMed] [Google Scholar]
- 2.Hiscott J. Convergence of the NF-kappaB and IRF pathways in the regulation of the innate antiviral response. Cytokine Growth Factor Rev. 2007;18:483–490. doi: 10.1016/j.cytogfr.2007.06.002. [DOI] [PubMed] [Google Scholar]
- 3.Honda K, Takaoka A, Taniguchi T. Type I interferon [corrected] gene induction by the interferon regulatory factor family of transcription factors. Immunity. 2006;25:349–360. doi: 10.1016/j.immuni.2006.08.009. [DOI] [PubMed] [Google Scholar]
- 4.Alexopoulou L, Holt AC, Medzhitov R, Flavell RA. Recognition of double-stranded RNA and activation of NF-kappaB by Toll-like receptor 3. Nature. 2001;413:732–738. doi: 10.1038/35099560. [DOI] [PubMed] [Google Scholar]
- 5.Han KJ, et al. Mechanisms of the TRIF-induced interferon-stimulated response element and NF-kappaB activation and apoptosis pathways. J Biol Chem. 2004;279:15652–15661. doi: 10.1074/jbc.M311629200. [DOI] [PubMed] [Google Scholar]
- 6.Oshiumi H, Matsumoto M, Funami K, Akazawa T, Seya T. TICAM-1, an adaptor molecule that participates in Toll-like receptor 3-mediated interferon-beta induction. Nat Immunol. 2003;4:161–167. doi: 10.1038/ni886. [DOI] [PubMed] [Google Scholar]
- 7.Yamamoto M, et al. Cutting edge: A novel Toll/IL-1 receptor domain-containing adapter that preferentially activates the IFN-beta promoter in the Toll-like receptor signaling. J Immunol. 2002;169:6668–6672. doi: 10.4049/jimmunol.169.12.6668. [DOI] [PubMed] [Google Scholar]
- 8.Andrejeva J, et al. The V proteins of paramyxoviruses bind the IFN-inducible RNA helicase, mda-5, and inhibit its activation of the IFN-beta promoter. Proc Natl Acad Sci USA. 2004;101:17264–17269. doi: 10.1073/pnas.0407639101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yoneyama M, et al. The RNA helicase RIG-I has an essential function in double-stranded RNA-induced innate antiviral responses. Nat Immunol. 2004;5:730–737. doi: 10.1038/ni1087. [DOI] [PubMed] [Google Scholar]
- 10.Xu LG, et al. VISA is an adapter protein required for virus-triggered IFN-beta signaling. Mol Cell. 2005;19:727–740. doi: 10.1016/j.molcel.2005.08.014. [DOI] [PubMed] [Google Scholar]
- 11.Kawai T, et al. IPS-1, an adaptor triggering RIG-I- and Mda5-mediated type I interferon induction. Nat Immunol. 2005;6:981–988. doi: 10.1038/ni1243. [DOI] [PubMed] [Google Scholar]
- 12.Seth RB, Sun L, Ea CK, Chen ZJ. Identification and characterization of MAVS, a mitochondrial antiviral signaling protein that activates NF-kappaB and IRF 3. Cell. 2005;122:669–682. doi: 10.1016/j.cell.2005.08.012. [DOI] [PubMed] [Google Scholar]
- 13.Meylan E, et al. Cardif is an adaptor protein in the RIG-I antiviral pathway and is targeted by hepatitis C virus. Nature. 2005;437:1167–1172. doi: 10.1038/nature04193. [DOI] [PubMed] [Google Scholar]
- 14.Zhong B, et al. The adaptor protein MITA links virus-sensing receptors to IRF3 transcription factor activation. Immunity. 2008;29:538–550. doi: 10.1016/j.immuni.2008.09.003. [DOI] [PubMed] [Google Scholar]
- 15.Ishikawa H, Barber GN. STING is an endoplasmic reticulum adaptor that facilitates innate immune signalling. Nature. 2008;455:674–678. doi: 10.1038/nature07317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Der SD, Zhou A, Williams BR, Silverman RH. Identification of genes differentially regulated by interferon alpha, beta, or gamma using oligonucleotide arrays. Proc Natl Acad Sci USA. 1998;95:15623–15628. doi: 10.1073/pnas.95.26.15623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sadler AJ, Williams BR. Interferon-inducible antiviral effectors. Nat Rev Immunol. 2008;8:559–568. doi: 10.1038/nri2314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wathelet M, et al. Molecular cloning, full-length sequence and preliminary characterization of a 56-kDa protein induced by human interferons. Eur J Biochem. 1986;155:11–17. doi: 10.1111/j.1432-1033.1986.tb09452.x. [DOI] [PubMed] [Google Scholar]
- 19.Levy D, Larner A, Chaudhuri A, Babiss LE, Darnell JE., Jr Interferon-stimulated transcription: Isolation of an inducible gene and identification of its regulatory region. Proc Natl Acad Sci USA. 1986;83:8929–8933. doi: 10.1073/pnas.83.23.8929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lamb JR, Tugendreich S, Hieter P. Tetratrico peptide repeat interactions: To TPR or not to TPR? Trends Biochem Sci. 1995;20:257–259. doi: 10.1016/s0968-0004(00)89037-4. [DOI] [PubMed] [Google Scholar]
- 21.Guo J, Peters KL, Sen GC. Induction of the human protein P56 by interferon, double-stranded RNA, or virus infection. Virology. 2000;267:209–219. doi: 10.1006/viro.1999.0135. [DOI] [PubMed] [Google Scholar]
- 22.Terenzi F, Hui DJ, Merrick WC, Sen GC. Distinct induction patterns and functions of two closely related interferon-inducible human genes, ISG54 and ISG56. J Biol Chem. 2006;281:34064–34071. doi: 10.1074/jbc.M605771200. [DOI] [PubMed] [Google Scholar]
- 23.Wang C, et al. Alpha interferon induces distinct translational control programs to suppress hepatitis C virus RNA replication. J Virol. 2003;77:3898–3912. doi: 10.1128/JVI.77.7.3898-3912.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wacher C, et al. Coordinated regulation and widespread cellular expression of interferon-stimulated genes (ISG) ISG-49, ISG-54, and ISG-56 in the central nervous system after infection with distinct viruses. J Virol. 2007;81:860–871. doi: 10.1128/JVI.01167-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Terenzi F, Saikia P, Sen GC. Interferon-inducible protein, P56, inhibits HPV DNA replication by binding to the viral protein E1. EMBO J. 2008;27:3311–3321. doi: 10.1038/emboj.2008.241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Komuro A, Bamming D, Horvath CM. Negative regulation of cytoplasmic RNA-mediated antiviral signaling. Cytokine. 2008;43:350–358. doi: 10.1016/j.cyto.2008.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Takeuchi O, Akira S. Innate immunity to virus infection. Immunol Rev. 2009;227:75–86. doi: 10.1111/j.1600-065X.2008.00737.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhang M, et al. Negative feedback regulation of cellular antiviral signaling by RBCK1-mediated degradation of IRF3. Cell Res. 2008;18:1096–1104. doi: 10.1038/cr.2008.277. [DOI] [PubMed] [Google Scholar]
- 29.Baas T, et al. Integrated molecular signature of disease: Analysis of influenza virus-infected macaques through functional genomics and proteomics. J Virol. 2006;80:10813–10828. doi: 10.1128/JVI.00851-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pasieka TJ, et al. Functional genomic analysis of herpes simplex virus type 1 counteraction of the host innate response. J Virol. 2006;80:7600–7612. doi: 10.1128/JVI.00333-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Higgs R, et al. The E3 ubiquitin ligase Ro52 negatively regulates IFN-beta production post-pathogen recognition by polyubiquitin-mediated degradation of IRF3. J Immunol. 2008;181:1780–1786. doi: 10.4049/jimmunol.181.3.1780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhong B, et al. The ubiquitin ligase RNF5 regulates antiviral responses by mediating degradation of the adpater protein MITA. Immunity. 2009;30:1–11. doi: 10.1016/j.immuni.2009.01.008. [DOI] [PubMed] [Google Scholar]
- 33.Diao F, et al. Negative regulation of MDA5- but not RIG-I-mediated innate antiviral signaling by the dihydroxyacetone kinase. Proc Natl Acad Sci USA. 2007;104:11706–11711. doi: 10.1073/pnas.0700544104. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.