Abstract
For the initiation of DNA replication, dsDNA is unwound by helicases. Primases then recognize specific sequences on the template DNA strands and synthesize complementary oligonucleotide primers that are elongated by DNA polymerases in leading- and lagging-strand mode. The bacterial plasmid RSF1010 provides a model for the initiation of DNA replication, because it encodes the smallest known primase RepB′ (35.9 kDa), features only 1 single-stranded primase initiation site on each strand (ssiA and ssiB, each 40 nt long with 5′- and 3′-terminal 6 and 13 single-stranded nucleotides, respectively, and nucleotides 7–27 forming a hairpin), and is replicated exclusively in leading strand mode. We present the crystal structure of full-length dumbbell-shaped RepB′ consisting of an N-terminal catalytic domain separated by a long α-helix and tether from the C-terminal helix-bundle domain and the structure of the catalytic domain in a specific complex with the 6 5′-terminal single-stranded nucleotides and the C7–G27 base pair of ssiA, its single-stranded 3′-terminus being deleted. The catalytic domains of RepB′ and the archaeal/eukaryotic family of Pri-type primases share a common fold with conserved catalytic amino acids, but RepB′ lacks the zinc-binding motif typical of the Pri-type primases. According to complementation studies the catalytic domain shows primase activity only in the presence of the helix-bundle domain. Primases that are highly homologous to RepB′ are encoded by broad-host-range IncQ and IncQ-like plasmids that share primase initiation sites ssiA and ssiB and high sequence identity with RSF1010.
Keywords: Pri-type, primase DNA complex, single-strand initiator DNA, crystal structure, oriV
At the initiation of DNA replication, the double helix is unwound by helicases, and then primases synthesize complementary oligonucleotide primers on the single-stranded template DNA. These primers are extended by DNA polymerase, continuously at the leading strand and discontinuously at the oppositely-oriented lagging strand, where Okazaki fragments are synthesized. Primase activity depends on specific ssDNA priming sequences that are periodically required for the initiation of the Okazaki fragments on the lagging strand (that are connected by ligase) but only 1 primer is used for the leading strand (1).
The broad-host-range IncQ plasmid RSF1010 is maintained in >30 Gram-negative and 2 Gram-positive bacteria (2, 3). The replication of IncQ and IncQ-like plasmids and their conjugative transfer was studied in Escherichia coli and in vitro. RSF1010 encodes helicase RepA, primase RepB′, and replication initiator protein RepC (4, 5) (Fig. S1A). Additionally, the replication of RSF1010 requires bacterial host replication proteins such as DNA polymerase III and single-stranded binding protein.
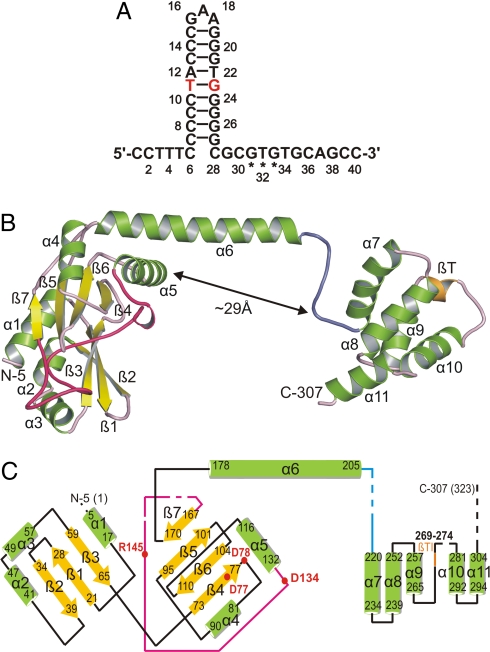
Primase RepB′ is activated on 2 specific, 40-nt-long single-stranded segments termed initiators A and B (ssiA and ssiB) with highly similar nucleotide sequences (6, 7). They are arranged on opposite DNA strands as a palindrome in the origin of vegetative replication (oriV) and most likely form hairpins after strand separation by helicase RepA (8, 9) (Fig. 1A and Fig. S1 B and C). RepB′ recognizes ssiA and ssiB independently of each other (6, 7) and synthesizes primers (10) that are elongated by DNA polymerase III. Replication of RSF1010 depends on the ssi sequences that occur only once on each plasmid strand, suggesting that RSF1010 (11, 12) is replicated in the leading-strand mode in opposite directions along single-stranded template DNA (10, 13, 14) (Fig. S1A).
Fig. 1.
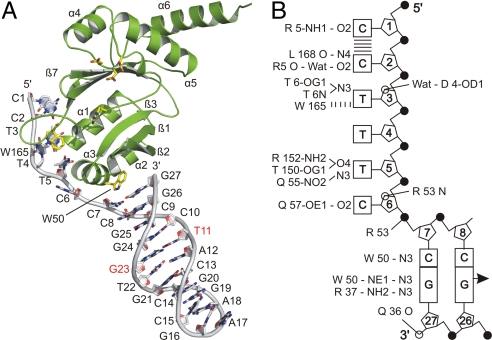
DNA recognition site, secondary and tertiary structure of RepB′. (A) Sequence and hairpin structure of ssiA. The 3 asterisks below the sequence indicate the G31T32G33 priming site (14). Hairpin base pairs of ssiA are as observed in the structure (Fig. 6); the G23T11 wobble is colored red. (B) Crystal structure of RepB′. In the ribbon diagram of RepB′, α-helices are green, β-strands are yellow, β-turn type I (βT) between α9 and α10 is orange, loops are pink except for the long loop connecting α5, β7 (red) and the tether connecting α6, α7 (blue). N and C termini, α-helices, and ß-strands are labeled. A segment of the long loop (red) is located at the bottom of the active site and carries catalytic Arg-145. (C) Topographical scheme of RepB′. Colors and labeling of α-helices and β-sheets are as in B, and their N- and C-terminal residues are indicated by numbers. Parts of the structure that could not be modeled because of disorder are indicated by dashed lines or interruptions (residues 1–4, 154, 159–161, 208–216, 276–278, and 308–323). Conserved catalytic residues are shown as red dots and labeled with 1-letter code. The relative positioning of N- and C-terminal domains of RepB′ are probably influenced by crystal packing.
Crystal structures show that primases belong to 2 classes. One comprises the multidomain DnaG-type primases from bacteria (15, 16) and bacteriophage T7 (17). The other contains heterodimeric Pri-type primases from archaeal bacteria consisting of a small catalytic (PriS) and a large subunit (PriL). Crystal structures are known for PriS (18–20) and the N-terminal domain of PriL (20, 21). Pri-type primases show sequence homology with the large and small subunits of the eukaryotic heterotetrameric Polα-primase and are referred to as archaeal/eukaryotic primases.
Although structurally unrelated, Pri- and DnaG-type primases use the 2-metal-ion mechanism for catalysis of primer synthesis (16, 18, 22) and are zinc dependent (23–25). DnaG-type primase structures exhibit zinc-ribbon domains (15, 26, 27) that are tethered loosely to their catalytic domains and regulate initiation of primer synthesis and length (26–29). Pri-type primases feature zinc-knuckle or zinc-stem structures inserted in their PriS-catalytic domain that were suggested to enforce the interaction with the ssDNA template, but were shown not to be essential for primase activity, in contrast to the zinc ribbon domain (18, 20). As shown by in vitro experiments, primer synthesis of Pri-type primases depends on the PriS-zinc binding structures and PriL (20). Isolated PriS can convey low-level primase activity independently from PriL, but efficient primer synthesis requires the PriS–PriL complex (30).
Structural data showing how primases recognize DNA are limited to the crystal structure of the catalytic domain of primase DnaG from E. coli bound unspecifically to ssDNA (31). Structures of complexes between Pri-type primases and template DNA are lacking. The crystal structure of the nonhomologous end-joining (NHEJ) polymerase domain of ligase LigD from Mycobacterium tuberculosis (NHEJ pol) was determined in a complex with DNA (32). NHEJ polymerase domain lacks a zinc-binding structure, but its scaffold resembles the PriS-catalytic domains (33).
Here, we present the crystal structure of a zinc-independent primase, the full-length RepB′ at 2.0-Å resolution, and that of its catalytic domain in a specific complex with the ssiA DNA fragment comprising nucleotides 1–27 at 2.7-Å resolution. We show by mutagenesis that RepB′ features the same catalytic residues as Pri-type primases and suggest a model for primer synthesis.
Results
Structure Determination of Full-Length RepB′.
The crystal structure of RepB′ was determined by multiple wavelength dispersion (MAD) (see Methods and Table S1). The electron density allowed the modeling of 89% of the amino acid sequence; the remaining 11% (see the legend to Fig. 1C) include residues 308–323 of the C terminus. As outlined in SI Text, this segment is supposed to promote the translocation of mobilization protein MobA (consisting of a relaxase with C-terminally fused RepB′) by the Dot/Icm type IV secretion systems (T4SS).
RepB′ Consists of 2 Distinct Domains.
The dumbbell shape of RepB′ is divided into a large N-terminal domain (residues 1–205), composed of 2 antiparallel β-sheets (βA and βB) flanked by 6 α-helices and a small C-terminal domain (residues 220–323) comprising a bundle of 5 α-helices (Fig. 1 B and C). The 2 domains are ≈29 Å apart and connected by α-helix α6 (residues 178–205) and a 14-aa-long flexible tether (residues 206–219). Helix α6 and the tether are unique to RepB′ and not found in structures of Pri-type primases except for the DnaG-type primase of bacteriophage T7 whose catalytic and zinc ribbon domains are separated by a 26-aa-long tether (26).
RepB′ Shares a Related Fold and Active-Site Residues with Archaeal Primases.
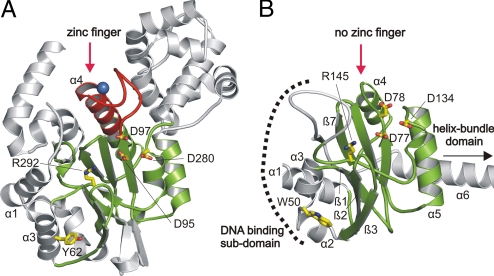
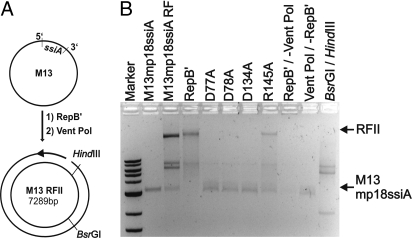
Structure comparisons using the DALI server (34) show that the N-terminal domain of RepB′ fits best with the catalytic domains of the PriS of archaeal primases from Pyrococcus furiosus (Pfu; Fig. 2) (18) and Pho from Pyrococcus horikoshii (19) (Z-score 9.9 for Pfu and Pho, rmsd 2.8 Å for 148 Cα-positions), although RepB′ and archaeal primases share only very low sequence homology. In the core of the large domain of RepB′, the β-sheets βA and βB are normal to each other and form a cleft that opens toward the C-terminal domain (Fig. S2A). Superpositions of the N-terminal domain of RepB′ with the PriS-catalytic domains of archaeal primases Pfu, Pho, Sso from Sulfolobus solfataricus (20) (Z-score 7.8, rmsd 3.0 Å for 140 Cα-positions) and with the polymerase domain of DNA ligase LigD from Pseudomonas aeroginosa (33) (Z-score 9.0, rmsd 3.2 Å for 156 Cα-positions) identify the structurally conserved amino acids Asp-77, Asp-78, Asp-134, and Arg-145 of RepB′ in this cleft (Fig. 2B, Table S2, and Fig. S2B). To test the catalytic influence of these amino acids, we generated point mutations and measured the primase activities of the variants by using circular ssM13 DNA into which ssiA was inserted (Fig. 3A). Compared with WT RepB′ the primase activity was abolished by mutation of Asp-77 or Asp-78 or Asp-134 to alanine and severely reduced by mutation of Arg-145 to alanine (Fig. 3B). RepB′ was inactive on the complementary strand of ssiA (Fig. S2C).
Fig. 2.
Comparison of catalytic centers of Pfu primase (A) and RepB′ (B). Superposition of RepB′ on Pfu (Protein Data Bank ID code 1G71) yields a Z-score of 9.9 and an rmsd of 2.8 Å for 148 Cα positions. The catalytic cores of RepB′ and Pfu primase are shown in the same orientation and scale. Structure elements similar in both molecules are in green, those that differ are in gray and red. (A) The zinc knuckle motif of Pfu primase is located at the N terminus of helix α4 (red); the bound Zn2+ is indicated by the blue sphere. (B) The corresponding helix α4 of RepB′ is truncated (red arrow). The ssiA DNA binding subdomain of RepB′ (α1–α3, β1–β3, and β7; see Fig. 6B) is indicated by a dashed black line. The proposed catalytic amino acids of both primases and DNA-binding Tyr-62 (Pfu) and Trp-50 (RepB′) are shown as yellow stick models (carbon yellow, oxygen red, nitrogen blue) and enlarged for better visibility.
Fig. 3.
Complementary strand synthesis initiated by RepB′. (A) Principle of primase assay. A complete reaction mixture contains circular ssM13 DNA carrying ssiA, primase RepB′, Vent-DNA polymerase, and dNTPs. RepB′ synthesizes a primer (black arrow) complementary to the ssiA recognition sequence that is extended by Vent-DNA polymerase. The primase activity is indicated by the ability to initiate dsDNA synthesis on the recombinant ssM13 DNA carrying ssiA. Newly-synthesized M13 dsDNA contains a single-strand nick and is called replicative form II (RFII, RF), contrasting to replicative form I (RFI) that corresponds to circularly-closed supercoiled M13dsDNA. The restriction sites Hind III (6,281 bp) and BsrGI (1,021 bp) are labeled on M13 RFII. (B) Primase activity of RepB′ variants. If not otherwise denoted all reaction mixtures contain dNTPs, ssM13mp18ssiA (circular ssM13 DNA carrying ssiA; mp18, Max-Planck strain 18) and Vent polymerase. Reaction mixtures contained either RepB′ or variants D77A or D78A or D134A and ssM13mp18ssiA. For controls only either ssM13mp18ssiA or M13mp18ssiA RF (replicative forms of M13mp18ssiA isolated from infected E. coli cells) is in the reaction mixture. Vent Pol/-RepB′, reaction in the absence of RepB′ and in the presence of Vent polymerase. RepB′/-Vent Pol, Vent-DNA-polymerase absent and RepB′ present. BsrGI and HindIII, product analysis of a complete primase reaction with specific dsDNA restriction enzymes BsrGI and HindIII resulted in 2 DNA fragments of the expected sizes, 2.0 and 5.2 kb. Marker (2-log-DNA-ladder, NEB): 10, 8, 6, 5 (4, 3, double band), 2, 1.5, 1.2 (kB).
RepB′ Lacks a Zinc Binding Motif.
The segment forming the zinc-knuckle structure on helix α4 of the catalytic domain of Pfu primase is truncated in RepB′ (Fig. 2). Unlike the DnaG- and Pri-type primase structures, RepB′ does not contain a bound zinc ion, neither on the catalytic nor on the helix-bundle domain. To verify the absence of zinc in RepB′, the primase activity was tested in the presence of 0.001–10 mM ZnCl2 or Zn2+ chelator N,N,N′,N′-tetrakis-(2-pyridylmethyl)-ethylenediamine but no difference was found.
The Helix-Bundle Domain Conveys Primase Activity.
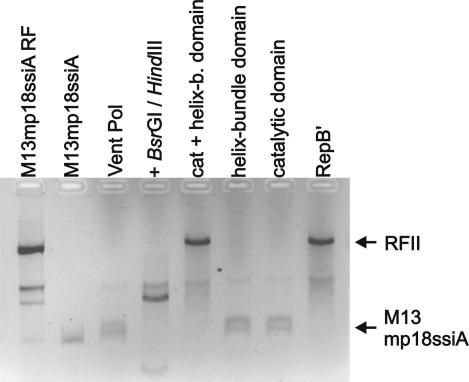
To assess whether the helix-bundle domain of RepB′ influences the initiation of primer synthesis, we carried out primase assays in the presence of either the isolated catalytic or the helix-bundle domain and in the presence of both isolated domains (Fig. 4). The catalytic or the helix-bundle domains were unable to promote independently the initiation of DNA synthesis on circular single-stranded M13-DNA templates carrying ssiA (Fig. 3A). However, using catalytic and helix-bundle domains together, we found that the primase activity was restored. Compared with WT RepB′, full primase activity was only reached with an equimolar ratio of the catalytic and helix-bundle domains. Decreasing the concentration of one domain (50 nM to 1 nM) relative to the other domain (50 nM) reduced the primase activity (Fig. 4B and Fig. S2E).
Fig. 4.
Complementation assay is shown for the catalytic and helix-bundle domain (see the legend for Fig. 3B). Cat + helix-b. domain, isolated catalytic and helix bundle domains in molar ratio of 1:1. Catalytic domain, primase assay in the presence of the catalytic domain. Helix-bundle domain, primase assay with helix-bundle domain. Controls: product analysis by BsrGI and HindIII digestion. Vent Pol, reaction mixture without catalytic and helix-bundle domain, but with Vent polymerase. RepB′, reaction in presence of RepB′. M13mp18ssiA RF isolated from E. coli and ssM13mp18ssiA.
To test whether the helix-bundle domain of the mutant proteins can also be active in trans, we measured the primase activity of full-length RepB′ variants D77A, D78A, D134A, and R145A supplemented with the WT catalytic domain of RepB′ (Fig. S2F). Primase activity of the inactive variants of RepB′ was fully recovered by addition of the catalytic domain and that of the weakly active RepB′ R145A was significantly increased.
The Helix-Bundle Domain Binds to ssiA DNA.
A structural search with the DALI server using the helix-bundle domain showed no similarity to other proteins, suggesting that this domain exhibits a unique fold. However, the antiparallel helices α8 and α9 and the oblique DNA-recognition helix α10 share structural similarity to dsDNA-binding homeodomains (Fig. S3A). Except for helix α7 of RepB′, 4 α-helices of the helix-bundle domain superimpose with the transposase dsDNA-binding domain of phage Mu (Z-score 3.3 and rmsd 2.9 Å for 58 Cα-positions; Fig. S3B). In this and other superpositions with related dsDNA-binding domains, helix α10 of RepB′ coincides with (proven or potential) DNA-recognition helices, which insert into the large groove of dsDNA.
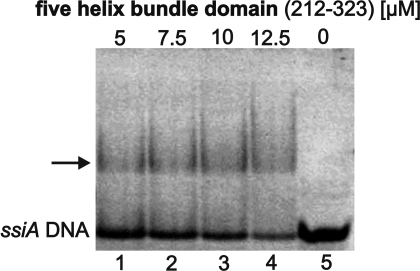
Indeed, EMSA confirmed complex formation between ssiA DNA and the helix-bundle domain (residues 212–323; Fig. 5). Via analytical ultracentrifugation the dissociation constant of this complex was determined to be 27.0 μM (Table S3), and that formed with ssiA DNA lacking the single-stranded 3′-tail [ssiA(3′Δ13)] was similar, 25.8 μM (Table S3), indicating that the 3′ tail of ssiA does not contribute significantly to complex formation.
Fig. 5.
EMSA of ssiA DNA with the helix-bundle domain of RepB′. The shifted protein/DNA complexes are marked by an arrow; the lower band is ssiA DNA. Lanes 1–4, ssiA DNA with addition of 5–12.5 μM helix-bundle domain; lane 5, only ssiA DNA.
Crystal Structure of the Catalytic Domain in Complex with ssiA(3′Δ13) DNA.
Attempts to cocrystallize RepB′ with ssiA DNA or the fragments described in Methods failed, because RepB′ was specifically cleaved after Glu-207. The catalytic domain could only be cocrystallized with ssiA(3′Δ13), not with intact ssiA; for structure determination see Methods (Table S4). Residues 1–3, 156–162, and 199–212 could not be modeled because of disorder, whereas those residues interacting with ssiA(3′Δ13) are well defined in the electron density (Fig. S4A).
The Catalytic Domain Recognizes the Complete ssiA 5′ Tail and the First Base Pair of the Hairpin.
In the complex of the catalytic domain with ssiA(3′Δ13) the single-stranded 5′ tail and the first base pair (C7–G27) of the hairpin are recognized specifically because of 17 favorable interactions with 13 aa, of which 11 form hydrogen bonds with the bases and 3 with the sugar-phosphate backbone, 2 being mediated by water (Fig. 6). In the single-stranded 5′ tail of ssiA(3′Δ13), base stacking is only found for C1 on C2 (Fig. S4 A and B) as Trp-165 stacks with T3 and displaces T4, thereby abolishing stacking of T3 with C2 or T4. C6 does not stack with neighboring bases, because helix α3 inserts between T5 and the first C7–G27 base pair of the hairpin, pushing C6 away from T5 (Fig. 6A).
Fig. 6.
Architecture of the DNA–primase complex. (A) Crystal structure of the catalytic domain of RepB′ in complex with ssiA(3′Δ13). Catalytic domain in ribbon representation is green, and ssiA(3′Δ13) is shown as a gray stick model with deoxyribose-phosphate backbones as smooth lines. Nucleotides are numbered and labeled in 1-letter code. The DNA double helix features a G23T11 wobble (red letters; see also Fig. S4C). (B) Topographical scheme of hydrogen bonding and stacking interactions between the catalytic domain and ssiA(3′Δ13). Bases are squares/rectangles, deoxyriboses are 5-membered rings (labeled with sequential numbers), and phosphates are black-filled circles, O4′ is open circles. Direct and water (Wat)-mediated hydrogen bonds formed between amino acids and ssiA(3′Δ13) are indicated by solid lines; amino acids are in 3-letter code. When hydrogen bonds involve peptide bonds, only the residue number and peptide atoms are given. Parallel lines ( ) indicate stacking of bases C1 on C2 and base T3 on W165.
) indicate stacking of bases C1 on C2 and base T3 on W165.
The Single-Stranded 5′ Tail of the Recognition Sequence Is Crucial for Binding of ssiA to RepB′.
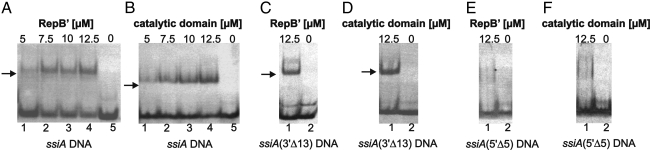
The structure of the catalytic domain in complex with ssiA(3′Δ13) DNA agrees with EMSA studies (Fig. 7) showing that the 5′ tail of ssiA or of ssiA(3′Δ13) DNA is crucial for complex formation with RepB′ and the catalytic domain, respectively. The dissociation constants as determined by ultracentrifugation for the complexes formed between the catalytic domain of RepB′ and ssiA (2.01 μM) are a factor of 2 lower than for ssiA(3′Δ13) DNA (5.22 μM), suggesting that at least some of the single-stranded 3′-terminal nucleotides contribute when binding to the catalytic domain (Table S3). However, binding of RepB′ to ssiA DNA is abolished when the 5′tail of ssiA is truncated by 5 nt (ssiA(5′Δ5); Fig. 7 E and F).
Fig. 7.
EMSA shows that the single-stranded 5′ tail of ssiA DNA is essential for binding to RepB′. Shifted bands indicating protein/DNA complexes marked by arrows. (A) Lanes 1–4, ssiA DNA with addition of 5–12.5 μM RepB′; lane 5, only ssiA DNA. (B) The same as A with RepB′ replaced by the catalytic domain; lane 5 only ssiA DNA. (C) Lane 1, ssiA(3′Δ13) with addition of 12.5 μM RepB′; lane 2 only ssiA(3′Δ13). (D) Lane 1, ssiA(3′Δ13) with addition of 12.5 μM catalytic domain; lane 2, only ssiA(3′Δ13). (E) Lane 1, ssiA DNA without single-stranded 5′ tail [ssiA(5′Δ5)] with the addition of 12.5 μM RepB′; lane 2, only ssiA(5′Δ5). (F) Lane 1, ssiA(5′Δ5) with the addition of 12.5 μM catalytic domain; lane 2, only ssiA(5′Δ5).
The ssiA DNA Hairpin Contains a G–T Wobble.
The double helical hairpin adopts the B-form and features a G–T wobble (T11-G23; Fig. S4C) that contradicts the proposed hairpin structure with looped-out thymidine T11 (10) (Fig. S1B). The electron density shows that the nucleotide sequence in the double helix is shifted by 1 nt such that a regular Watson–Crick double helix is formed (Figs. 1 A and 6 A). Consequently, the single-stranded 5′-tail consists of 6 instead of 5 nt. Nucleotides 16–18 form the loop of the hairpin and are stacked on base pair C15–G19.
The Catalytic Domain Contains a Specific DNA Binding and a Catalytic Subdomain.
Based on the structure of the complex formed by the catalytic domain and ssiA(3′Δ13), we divide the catalytic domain into 2 subdomains (Fig. 2B): one (helices α1–α3, β-strands β1–β3 and β7) recognizing ssiA(3′Δ13), and the other (α4–α6, β4–β6, and loop 133–166) harboring the catalytically-active amino acids.
Structure comparisons of PriS catalytic domains between RepB′, Pfu, Pho, Sso, and NHEJ polymerase domain show that the specific DNA-binding subdomain of RepB′ diverges from the PriS catalytic domain fold of Pfu and Pho (Fig. 2). Moreover, a steric clash would be caused by the single-stranded 5′ tail of ssiA(3′Δ13) and an additional helix α1 that is conserved between the structures of primases Pfu (Fig. 2A), Pho, Sso, and the NHEJ polymerease domain, but not found in RepB′. Among the structures compared above, helix α3 and the β-strands β1–β3 and β7 of the RepB′–DNA-binding subdomain are invariant. Helix α3 of RepB′ features the structurally-conserved Trp-50 that recognizes base pair C7–G27 of ssiA(3′Δ13) (Fig. 6A). Trp-50 corresponds to Tyr-62 (Pfu, Pho; Fig. 2), Tyr 63 (Sso), and Trp-101 (NHEJ polymerase domain) that are solvent-exposed except for Trp-101 of the NHEJ polymerase domain, which does not participate in DNA binding, in contrast to Trp-50 of RepB′.
RepB′ Is the Prototype of a Distinct Class of Primases with Exclusive Leading Strand Replication Mode Encoded by Broad-Host-Range Plasmids.
Although sharing related catalytic domain folds with structurally and functionally well-conserved residues Asp-77, Asp-78, Asp-134, and Arg-145 (Fig. S2B), the amino acid sequences of RepB′ and Pri-type primases Sso, Pfu, and Pho align very poorly. In addition, a zinc-binding motif is absent in RepB′, and the conserved 100DXD102 motif (RepB′ numbering) found in archaeal primases is replaced in RepB′ by 77DD78. The EGYATA motif found in several plasmid-encoded primases (35) does not occur in RepB′.
By contrast, the amino acid sequence of RepB′ aligns far better with another group of primases encoded by IncQ and IncQ-like broad-host-range plasmids, whose 3D structures are not yet known, the identity being >38% and the homology >63%. These even increase to 70% identity and 93% homology if one of the sequences, Q9AMP4 PXF5823, is deleted (Fig. S5A). Catalytic and DNA-binding amino acids found in RepB′ are conserved among these primases. Because these plasmids also contain ssiA and ssiB DNA segments that share high sequence identity with those of plasmid RSF1010 and are able to form hairpins (Figs. S1 B and C and S5), RepB′ is apparently the prototype of a distinct class of primases (36) with high sequence homology and exclusive leading-strand replication mode.
Discussion
Because it is replicated exclusively in leading-strand mode, plasmid RSF1010 provides a paradigmatic priming system. On each of the 2 primase recognition sites, ssiA and ssiB, RepB′ synthesizes independently 2 primers that are elongated by host DNA polymerase III.
The catalytic domains of RepB′ and the PriS primases share a conserved mixed α/β polymerase fold harboring the catalytic amino acids (Fig. S2B). RepB′ and Pri-type primases exhibit structurally different helix-bundle domains (18, 19) (Fig. 2). The function of the Pri-type helix-bundle domains is unknown. However, the wide separation of catalytic and helix-bundle domains in RepB′ is not found in any of the PriS structures (Figs. 1B and 2). Complementation studies show that the helix-bundle domain of RepB′ is essential for primase activity (Fig. 4 and Fig. S2 D and E) and suggest that catalytic and helix-bundle domains form a specific complex on ssiA. EMSA confirms that the isolated helix-bundle domain binds to ssiA (Fig. 5 and Table S3), but the binding site of the helix-bundle domain remains elusive. In contrast to archaeal primases, RepB′ lacks a zinc-binding motif that supports ssDNA template binding (Fig. 2) but features the DNA-binding subdomain on the catalytic domain that affects recognition of the 5′-terminal single-stranded tail of ssiA, as also shown by EMSA studies (Fig. 7) and may substitute for the missing zinc structure as the ssDNA binding motif (see below). None of the ssiA binding amino acids of RepB′ are conserved in archaeal primase structures except for solvent-exposed Trp-50 that corresponds to Tyr-62 (Pfu, Pho; Fig. 2), Tyr-63 (Sso), and Trp-101 (NHEJ polymerase domain).
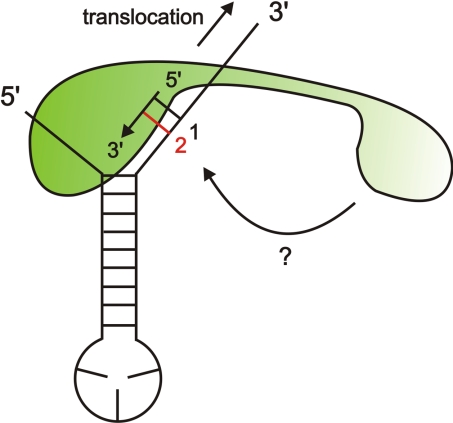
In the crystal structure of the RepB′-ssiA (3′Δ13) complex (Fig. 6A), the 3′ terminus (G27) of ssiA(3′Δ13) is closer to the catalytic residues of RepB′ than the 5′ tail, suggesting that an extended 3′ tail of ssiA/ssiB would bind along the active center and serve as a DNA template for primer synthesis. This finding supports studies by Lin and Meyer (10) who proposed that primer synthesis is initiated within the G31T32G33 priming site on the 3′ tail (* in Fig. 1A). Because the primer is synthesized complementary to the template DNA, the template DNA must be translocated relative to RepB′ (Fig. 8). Because the 5′ tail of ssiA(3′Δ13) is trapped by the ssDNA-binding subdomain of RepB′, we suggest that only the 3′ tail is translocated during primer synthesis. Translocation of the 3′ tail is possible, because the 3′ priming site of ssiA is dispensable for binding to RepB′ (Fig. 7 C and D). Unwinding of the hairpin becomes essential when the primer exceeds the length of 6 nt, including the proposed G31T32G33 priming site (10).
Fig. 8.
Model for primer synthesis and translocation of ssiA. RepB′ (green) is bound to the intact ssiA DNA segment. The 5′ tail and the first base pair of the hairpin are specifically recognized and trapped by the catalytic domain. The 3′ tail is bound along the active center of the catalytic domain and serves as DNA template. The primer is synthesized complementary to the DNA template in the 5′ → 3′ direction, and the 3′ tail is translocated in the opposite direction (arrow near 3′ end). The first formed base pair of the template-primer hybrid is shown in black and labeled 1, and the additional base pair formed by the second incoming nucleotide is denoted red and labeled 2. Possible binding of the helix-bundle domain to the ssiA priming site is indicated by a bent arrow marked ?.
The helix-bundle domain was shown by EMSA and analytical ultracentrifugation to bind to ssiA and ssiA(3′Δ13) (Fig. 5 and Table S3), and complementation assays indicate that it is essential for primase activity (Fig. 4 and Fig. S2D). To initiate primer synthesis at the catalytic domain, the helix-bundle domain would have to move (Fig. 8) from its position shown in the RepB′ crystal structure (Fig. 1B). This could be achieved by structural changes of the flexible tether, similar to the zinc-ribbon domain of DnaG-type primases that is linked by a 26-residue-long tether to its catalytic domain.
RepB′ is the prototype of a distinct family of primases encoded by IncQ and IncQ-like plasmids (Fig. S5A) that show substantial amino acid sequence identity and share very low sequence homology with Pri-type primases (37). Compared with Pri-type primases, the RepB′ family features structural differences regarding the primase architecture, the ssDNA template recognition, and the requirements for primase activity. RepB′ and the known primases of the IncQ family are encoded by plasmids that feature only 1 copy of ssiA and ssiB on each strand, suggesting that they replicate exclusively in the leading-strand mode as does plasmid RSF1010. The unusually complex ssi recognition sites (Fig. 1A and Fig. S1C) are found only on IncQ and IncQ-like plasmids, but not on bacterial or archaeal genomes.
Although RSF1010 has been exclusively found in eubacteria, RepB′ shares with the archaeal Pri-type primases the structural organization of the active site (Fig. 2) and conserved catalytic amino acids (Fig. S2B). Interestingly, it was reported that RepB′ is still active after heating to 80 °C for 30 min (38). The remarkable thermal stability of RepB′ can now be explained by its structural relationship to the thermophilic archaeal primases Pfu, Pho, and Sso and rises questions of the ancestry not only of RepB′, but also of plasmid RSF1010.
Methods
Expression and Purification of RepB′ and Its N- and C-Terminal Domains.
We followed published procedures (14) with modifications (see SI Text).
Purification of the Complex Formed by Catalytic Domain and ssiA(3′Δ13) DNA.
The complex formed by mixing equimolar amounts of purified catalytic domain (SI Text) and ssiA(3′Δ13) DNA (TIB MOLBIOL) was incubated for 30 min at 37 °C, purified by gel filtration on a Superdex75 column (GE Healthcare) using 10 mM Tris·HCl (pH 7.5), 300 mM NaCl, and 0.01 mM EDTA and concentrated to 10 mg/mL for crystallization.
RepB′: Crystallization, Data Collection, and Processing.
Hanging drop vapor diffusion at 18 °C yielded rod-like crystals that were soaked with OsO3 × 2 pyridine to afford a heavy atom derivative. Difference Patterson methods located 3 osmium sites in the crystal asymmetric unit. After MAD phasing, the electron density map was interpreted and the model was refined until convergence (see SI Text).
Catalytic Domain/ssiA (3′Δ13) Complex: Crystallization, Data Collection, and Processing.
Under hanging drop vapor diffusion, rod-like crystals grew from a solution of the purified catalytic domain/ssiA(3′Δ13) DNA. The phase problem was solved by molecular replacement (39) using the structure of the catalytic domain (residues 4–205) as search model (see SI Text).
EMSAs with RepB′, Catalytic, and Helix-Bundle Domains.
For EMSA procedures see SI Text. Three different ssDNA molecules were used in EMSA (nucleotides in the hairpin in bold). The hairpin as proposed earlier (9) (Fig. S1B) is underlined: ssiA DNA, intact (5′-CCTTTCCCCCTACCCGAAGGGTGGGGGCGCGTGTGCAGCC-3′); ssiA(3′Δ13), truncated 3′ tail (5′-CCTTTCCCCCTACCCGAAGGGTGGGGG-3′); ssiA(5′Δ5), truncated 5′ tail (5′-CCCCCTACCCGAAGGGTGGGGGCGCGTGTGCAGCC-3′).
Structure Searches.
Structure searches using the DALI server (34) were performed separately with the N- and C-terminal domains of RepB′, residues 5–205 and 220–307. All figures were prepared by using PYMOL (40). For further details, see SI Text.
Analytical Ultracentrifugation.
Analytical ultracentrifugation was performed as described (41). Protein and DNA were dissolved in buffer containing 100 mM MgCl2, 50 mM KCl, and 20 mM Hepes, pH 8.0. Extinction coefficients ε (l/mol × cm) used for these studies were calculated by using ProtParam (42) from the expasy-server (see SI Text).
Production of Recombinant M13.
The sequences ssiA and ssiB were molecularly cloned separately in the replicative form of phages M13mp18 and M13mp19 (mp18, mp19, Max-Planck strain 18, 19) carrying oppositely-oriented multicloning sites. For the production and purification of the recombinant M13 phages published protocols were followed (43). Four constructs were made: M13mp18ssiA and M13mp18ssiB carrying the matrix ssDNAs of ssiA and ssiB; M13mp19ssiA and M13mp19ssiB carrying the complementary ssiA and ssiB ssDNA strands.
Generation and Purification of the Mutant Proteins of RepB′.
To introduce point mutations in the repB′ gene, the QuikChange mutagenesis kit II (Stratagene) was used following the supplied protocol, affording the variants of RepB′ D77A, D78A, D134A, and R145A. All mutant proteins were purified to homogeneity as described for WT RepB′.
Primase Assay.
The reaction mixture (20 μL) contained 50 nM RepB′ or one of its variants, 30 ng of recombinant M13 DNA (see above), 0.5 unit of Vent-DNA polymerase (NEB), and 375 μM dATP, dCTP, dGTP, and dTTP. The primase assay was performed in NEB buffer 2 [50 mM NaCl, 10 mM Tris·HCl (pH 7.9), 10 mM MgCl2, 1 mM DTT] and incubated at 37 °C and 72 °C for 10 min. The incubation time was 20 min for reactions with RepB′ variants. To analyze products, the M13 RF II (replicative form II) was cleaved with restriction enzymes BsrGI and HindIII for 15 min at 37 °C. The reactions were stopped by addition of SDS to a final concentration of 0.8%. The reaction mixtures were loaded onto a 1% agarose-gel with 1× Tris-acetate-EDTA (TAE) buffer. Electrophoresis was performed at 9 V/cm for 170 min at room temperature, and DNA was stained with ethidium bromide (Figs. 3 and 4 and Fig. S2D).
Supplementary Material
Acknowledgments.
We thank the staffs at the protein crystallography beamline ID 14.2 of the European Synchrotron Radiation Facility/Grenoble and the protein structure factory beamline of Freie Universität Berlin at Berliner Elektronenspeicherring-Gesellschaft für Synchrotronstrahlung/Berlin for help; Dr. Ron Clarke for critically reading the manuscript; Dr. Joachim Behlke for analytical ultracentrifugation; Dr. Werner Schröder for N-terminal sequencing; Drs. Chris Weise and Peter Franke for MALDI-TOF analysis of RepB′; Claudia Alings for technical assistance; and Drs. Eugene Koonin, Thomas Spreter, and Wilhelm Weihofen for suggestions. This work was supported by the Deutsche Forschungsgemeinschaft.
Footnotes
The authors declare no conflict of interest.
Data deposition: The atomic coordinates have been deposited in the Protein Data Bank, www.pdb.org (PDB ID codes 2P96 and 3C1W).
This article contains supporting information online at www.pnas.org/cgi/content/full/0902910106/DCSupplemental.
References
- 1.Frick DN, Richardson CC. DNA primases. Annu Rev Biochem. 2001;70:39–80. doi: 10.1146/annurev.biochem.70.1.39. [DOI] [PubMed] [Google Scholar]
- 2.Rawlings DE, Tietze E. Comparative biology of IncQ and IncQ-like plasmids. Microbiol Mol Biol Rev. 2001;65:481–496. doi: 10.1128/MMBR.65.4.481-496.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Frey J, Bagdasarian M. In: Promiscuous Plasmids of Gram-Negative Bacteria. Thomas CM, editor. London: Academic; 1989. pp. 79–93. [Google Scholar]
- 4.Scherzinger E, et al. Replication of the broad host range plasmid RSF1010: Requirement for three plasmid-encoded proteins. Proc Natl Acad Sci USA. 1984;81:654–658. doi: 10.1073/pnas.81.3.654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Scherzinger E, Haring V, Lurz R, Otto S. Plasmid RSF1010 DNA replication in vitro promoted by purified RSF1010 RepA, RepB, and RepC proteins. Nucleic Acids Res. 1991;19:1203–1211. doi: 10.1093/nar/19.6.1203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Honda Y, et al. Functional division and reconstruction of a plasmid replication origin: Molecular dissection of the oriV of the broad-host-range plasmid RSF1010. Proc Natl Acad Sci USA. 1991;88:179–183. doi: 10.1073/pnas.88.1.179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Honda Y, et al. DnaG-dependent priming signals can substitute for the two essential DNA initiation signals in oriV of the broad host-range plasmid RSF1010. Nucleic Acids Res. 1992;20:1733–1737. doi: 10.1093/nar/20.7.1733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Miao DM, et al. A base-paired hairpin structure essential for the functional priming signal for DNA replication of the broad host range plasmid RSF1010. Nucleic Acids Res. 1993;21:4900–4903. doi: 10.1093/nar/21.21.4900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Honda Y, et al. Mutational analysis of the specific priming signal essential for DNA replication of the broad host-range plasmid RSF1010. FEBS Lett. 1993;324:67–70. doi: 10.1016/0014-5793(93)81534-7. [DOI] [PubMed] [Google Scholar]
- 10.Lin LS, Meyer RJ. DNA synthesis is initiated at two positions within the origin of replication of plasmid R1162. Nucleic Acids Res. 1987;15:8319–8331. doi: 10.1093/nar/15.20.8319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Scholz P, et al. Complete nucleotide sequence and gene organization of the broad-host-range plasmid RSF1010. Gene. 1989;75:271–288. doi: 10.1016/0378-1119(89)90273-4. [DOI] [PubMed] [Google Scholar]
- 12.Lin LS, Meyer RJ. Nucleotide sequence and functional properties of DNA encoding incompatibility in the broad host-range plasmid R1162. Mol Gen Genet. 1984;194:423–431. doi: 10.1007/BF00425554. [DOI] [PubMed] [Google Scholar]
- 13.Tanaka K, et al. Functional difference between the two oppositely oriented priming signals essential for the initiation of the broad host-range plasmid RSF1010 DNA replication. Nucleic Acids Res. 1994;22:767–772. doi: 10.1093/nar/22.5.767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Haring V, Scherzinger E. In: Promiscuous Plasmids of Gram-Negative Bacteria. Thomas CM, editor. London: Academic; 1989. pp. 95–124. [Google Scholar]
- 15.Pan H, Wigley DB. Structure of the zinc-binding domain of Bacillus stearothermophilus DNA primase. Structure (London) 2000;8:231–239. doi: 10.1016/s0969-2126(00)00101-5. [DOI] [PubMed] [Google Scholar]
- 16.Keck JL, Roche DD, Lynch AS, Berger JM. Structure of the RNA polymerase domain of E. coli primase. Science. 2000;287:2482–2486. doi: 10.1126/science.287.5462.2482. [DOI] [PubMed] [Google Scholar]
- 17.Toth EA, et al. The crystal structure of the bifunctional primase-helicase of bacteriophage T7. Mol Cell. 2003;12:1113–1123. doi: 10.1016/s1097-2765(03)00442-8. [DOI] [PubMed] [Google Scholar]
- 18.Augustin MA, Huber R, Kaiser JT. Crystal structure of a DNA-dependent RNA polymerase (DNA primase) Nat Struct Biol. 2001;8:57–61. doi: 10.1038/83060. [DOI] [PubMed] [Google Scholar]
- 19.Ito N, et al. Crystal structure of the Pyrococcus horikoshii DNA primase-UTP complex: Implications for the mechanism of primer synthesis. Genes Cells. 2003;8:913–923. doi: 10.1111/j.1365-2443.2003.00693.x. [DOI] [PubMed] [Google Scholar]
- 20.Lao-Sirieix SH, et al. Structure of the heterodimeric core primase. Nat Struct Mol Biol. 2005;12:1137–1144. doi: 10.1038/nsmb1013. [DOI] [PubMed] [Google Scholar]
- 21.Ito N, Matsui I, Matsui E. Molecular basis for the subunit assembly of the primase from an archaeon Pyrococcus horikoshii. FEBS J. 2007;274:1340–1351. doi: 10.1111/j.1742-4658.2007.05690.x. [DOI] [PubMed] [Google Scholar]
- 22.Steitz TA. A mechanism for all polymerases. Nature. 1998;391:231–232. doi: 10.1038/34542. [DOI] [PubMed] [Google Scholar]
- 23.Bernstein JA, Richardson CC. A 7-kDa region of the bacteriophage T7 gene 4 protein is required for primase but not for helicase activity. Proc Natl Acad Sci USA. 1988;85:396–400. doi: 10.1073/pnas.85.2.396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Powers L, Griep MA. Escherichia coli primase zinc is sensitive to substrate and cofactor binding. Biochemistry. 1999;38:7413–7420. doi: 10.1021/bi983059f. [DOI] [PubMed] [Google Scholar]
- 25.Kusakabe T, Hine AV, Hyberts SG, Richardson CC. The Cys4 zinc finger of bacteriophage T7 primase in sequence-specific single-stranded DNA recognition. Proc Natl Acad Sci USA. 1999;96:4295–4300. doi: 10.1073/pnas.96.8.4295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kato M, et al. Modular architecture of the bacteriophage T7 primase couples RNA primer synthesis to DNA synthesis. Mol Cell. 2003;11:1349–1360. doi: 10.1016/s1097-2765(03)00195-3. [DOI] [PubMed] [Google Scholar]
- 27.Corn JE, Pease PJ, Hura GL, Berger JM. Cross-talk between primase subunits can act to regulate primer synthesis in trans. Mol Cell. 2005;20:391–401. doi: 10.1016/j.molcel.2005.09.004. [DOI] [PubMed] [Google Scholar]
- 28.Kusakabe T, Richardson CC. The role of the zinc motif in sequence recognition by DNA primases. J Biol Chem. 1996;271:19563–19570. doi: 10.1074/jbc.271.32.19563. [DOI] [PubMed] [Google Scholar]
- 29.Qimron U, Lee SJ, Hamdan SM, Richardson CC. Primer initiation and extension by T7 DNA primase. EMBO J. 2006;25:2199–2208. doi: 10.1038/sj.emboj.7601112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Matsui E, et al. Distinct domain functions regulating de novo DNA synthesis of thermostable DNA primase from hyperthermophile Pyrococcus horikoshii. Biochemistry. 2003;42:14968–14976. doi: 10.1021/bi035556o. [DOI] [PubMed] [Google Scholar]
- 31.Corn JE, Pelton JG, Berger JM. Identification of a DNA primase template tracking site redefines the geometry of primer synthesis. Nat Struct Mol Biol. 2008;15:163–169. doi: 10.1038/nsmb.1373. [DOI] [PubMed] [Google Scholar]
- 32.Brissett NC, et al. Structure of a NHEJ polymerase-mediated DNA synaptic complex. Science. 2007;318:456–459. doi: 10.1126/science.1145112. [DOI] [PubMed] [Google Scholar]
- 33.Zhu H, et al. Atomic structure and nonhomologous end-joining function of the polymerase component of bacterial DNA ligase D. Proc Natl Acad Sci USA. 2006;103:1711–1716. doi: 10.1073/pnas.0509083103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Holm L, Sander C. Touring protein fold space with Dali/FSSP. Nucleic Acids Res. 1998;26:316–319. doi: 10.1093/nar/26.1.316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Strack B, Lessl M, Calendar R, Lanka E. A common sequence motif, -E-G-Y-A-T-A-, identified within the primase domains of plasmid-encoded I- and P-type DNA primases and the α protein of the Escherichia coli satellite phage P4. J Biol Chem. 1992;267:13062–13072. [PubMed] [Google Scholar]
- 36.Leipe DD, Aravind L, Koonin EV. Did DNA replication evolve twice independently? Nucleic Acids Res. 1999;27:3389–3401. doi: 10.1093/nar/27.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Iyer LM, Koonin EV, Leipe DD, Aravind L. Origin and evolution of the archaeo-eukaryotic primase superfamily and related palm-domain proteins: Structural insights and new members. Nucleic Acids Res. 2005;33:3875–3896. doi: 10.1093/nar/gki702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Haring V. PhD thesis. Berlin: Univ of Berlin; 1986. The replication proteins of the plasmid RSF1010: Overproduction, purification and characterization of the plasmid encoded enzymes. [Google Scholar]
- 39.Storoni LC, McCoy AJ, Read RJ. Likelihood-enhanced fast rotation functions. Acta Crystallogr D. 2004;60:432–438. doi: 10.1107/S0907444903028956. [DOI] [PubMed] [Google Scholar]
- 40.DeLano WL. PyMol. San Carlos, CA: DeLano Scientific; 2002. [Google Scholar]
- 41.Behlke J, Ristau O, Schönfeld HJ. Nucleotide-dependent complex formation between the Escherichia coli chaperonins GroEL and GroES studied under equilibrium conditions. Biochemistry. 1997;36:5149–5156. doi: 10.1021/bi962755h. [DOI] [PubMed] [Google Scholar]
- 42.Wilkins MR, et al. Protein identification and analysis tools in the ExPASy server. Methods Mol Biol. 1999;112:531–552. doi: 10.1385/1-59259-584-7:531. [DOI] [PubMed] [Google Scholar]
- 43.Scherzinger E, et al. Bacteriophage-T7-induced DNA-priming protein. A novel enzyme involved in DNA replication. Eur J Biochem. 1977;72:543–558. doi: 10.1111/j.1432-1033.1977.tb11278.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.