Abstract
Because particular inbred strains of experimental animals are informative for only a subset of the genes underlying variability in BMD, we undertook a genome screen to identify quantitative trait loci (QTLs) in 828 F2 progeny (405 males and 423 females) derived from the Copenhagen 2331 (COP) and dark agouti (DA) strains of rats. This screen was performed to complement our study in female Fischer 344 (F344) and Lewis (LEW) rats and to further delineate the factors underlying the complex genetic architecture of BMD in the rat model. Microsatellite genotyping was performed using markers at an average density of 20 cM. BMD was measured by pQCT and DXA. These data were analyzed in the R/qtl software to detect QTLs acting in both sexes as well as those having sex-specific effects. A QTL was detected in both sexes on chromosome 18 for midfemur volumetric BMD (vBMD; genome-wide, p < 0.01). On distal chromosome 1, a QTL was found for femur and vertebral aBMD as well as distal femur vBMD, and this QTL appears distinct from the proximal chromosome 1 QTL impacting BMD in our F344/LEW cross. Additional aBMD and vBMD QTLs and several sex-specific QTLs were also detected. These included a male-specific QTL (p < 0.01) on chromosome 8 and a female-specific QTL on chromosomes 7 and 14 (p < 0.01). Few of the QTLs identified showed overlap with the significant QTLs from the F344/LEW cross. These results confirm that the genetic influence on BMD in the rat model is quite complex and would seem to be influenced by a number of different genes, some of which have sex-specific effects.
Key words: quantitative trait loci, BMD, inbred rats, synteny
INTRODUCTION
Osteoporotic fractures at the hip and spine represent a major public health problem in developed countries.(1) Reduced BMD at one or more skeletal sites is the cardinal feature of osteoporosis. BMD in humans and mammalian models follows a pattern of increase during growth and puberty, followed by a steady state after skeletal maturity (peak BMD) and then decline in later life. In humans, peak BMD is the primary determinant of osteoporotic fracture risk among older individuals, with high peak BMD levels providing protection against osteoporosis later on in life. Peak BMD is quite variable among individuals in the normal human population(2) and largely caused by heritable factors. However, major gene effects underlying this complex phenotype have not yet been identified.(3)
We previously reported results from a microsatellite genome screen for peak BMD phenotypes in the laboratory rat (Rattus norvegicus), using the inbred Lewis (LEW) and Fischer 344 (F344) strains to generate a population of F2 females.(4) Highly significant quantitative trait loci (QTLs) for BMD in this cross were detected at several chromosomal positions in the rat genome, with LOD scores of 7 or higher obtained for hip or spine density phenotypes on rat chromosomes 1, 2, 8, 10, and 19. The chromosomal regions detected in the F344/LEW cross provide a resource for comparison with human QTLs and candidate gene studies of BMD.
Because of the breeding designs used to create inbred strains, however, a given pair of animal strains will be polymorphic and thus genetically informative only for a subset of the markers available for genome screens in the rat model. For this reason, we have undertaken a second F2 cross in a different pair of inbred rat strains that differ for key BMD phenotypes. The strains selected for this second cross were the Copenhagen (COP) and dark agouti (DA) inbred rats. We performed areal BMD (aBMD) measurement by DXA and volumetric BMD (vBMD) measurement by pQCT in large samples of both male and female F2 offspring. Accordingly, in addition to the female BMD QTLs detected in the prior study, we have the ability to detect QTLs that influence BMD in one or both sexes. This design also provides an opportunity to replicate female BMD QTLs from the F344/LEW cross in genomic regions where both the COP/DA and F344/LEW crosses are informative.
MATERIALS AND METHODS
Animal breeding
Reciprocal mating of 12 breeding pairs of COP rats with DA rats was performed to first create an F1 population, and the 190 F1 rats were intercrossed to create 828 F2 offspring (405 males and 423 females). The rats were allowed to grow to 26 wk of age, thereby attaining peak BMD, before they were killed. Rat identities were recorded on data chips implanted subcutaneously and were verified using a scanner from Biomedic Data System (Seaford, DE, USA). The rats were housed at Indiana University's Laboratory Animal Resource Center (LARC), two rats per cage, and provided standard rat chow (NIH-31 Mouse/Rat diet 7017; Harlan Teklad, Madison, WI, USA) and water ad libitum. After death, rat left femora and L3–L5 vertebrae were dissected out and stripped of muscle and transferred to 70% ethyl alcohol at 4°C for densitometry analyses. The excised spleens were immediately stored in liquid nitrogen before transferring to −80°C. The procedures performed throughout the experiment followed the guidelines of the Indiana University Animal Care and Use Committee (IACUC).
Phenotypic measurements
BMD in humans is typically measured clinically using DXA, which produces an aBMD value. In this study, we used CT for measurement of true vBMD, as well as DXA density measurement for direct comparison with the many human studies on which it has been used. To maintain maximal clinical relevance, we continue to focus on the same skeletal sites as in our previous report: whole femur and lumbar (L3–L5) spine for the aBMD measures, and midfemur, distal femur, and lumbar vertebrae for the vBMD measurements by CT.
pQCT
The left femurs were placed in plastic tubes filled with 70% ethyl alcohol and centered in the gantry of a Norland Stratec XCT Research SA+pQCT (Stratec Electronics, Pforzheim, Germany). Single slice measurements of 0.26 mm thickness and a voxel size of 0.07 mm were taken perpendicularly through the midfemoral shaft, distal femur, and the L5 vertebral body. For each slice, the X-ray source was rotated through 180° of projection. Total vBMD (mg/cm3) was measured using the XCT Research SA Plus, software version 5.40. This is the BMC divided by the total volume of the bone cross-section, including the marrow. The accuracy and repeatability of the pQCT measurements was confirmed using a method described previously.(5)
DXA
The femur and L3–L5 lumbar vertebrae were scanned using a fan-beam Hologic QDR 4500A DXA machine (Hologic, Bedford, MA, USA) equipped with Hologic version 11.2:3 software. The machine was calibrated daily with an anthropomorphic spine phantom, as described previously. Repeatability of the aBMD measures was assessed as described previously for pQCT.(4)
DNA isolation and microsatellite marker genotyping
Genomic DNA was isolated from the individual rat spleen using the Puregene DNA isolation kit (Gentra Systems, Minneapolis, MN, USA) according to the manufacturer's protocol. Genotyping for each animal was accomplished using PCR with microsatellite markers (Research Genetics, Birmingham, AL, USA) previously shown to be polymorphic for COP and DA rats. The entire genome-wide screen (chromosomes 1–20, X) included 93 markers at an average interval of 20 cM were analyzed using automated fluorescent microsatellite analysis. PCR products were sized on an ABI 3100 Genetic Analyzer (PE, Applied Biosystem, Foster City, CA, USA) by use of the Genotyper program, version 3.6. All genotyping data were confirmed by two independent readers. Chromosomal positions, marker order, and map positions were obtained from the Rat Genome Database (RGD) website (http://rgd.mcw.edu/). Genotypic data in the F2 animals for each marker were tested to ensure the expected mendelian ratios.
Quantitative genetic analysis
A total of 93 markers were genotyped in the 828 F2 progeny. Marker maps were generated using MAPMARKER/EXP with genotype data from all F2 animals.(6) Marker order and distances were compared with previously published RGD maps. This marker map was used by the program R/qtl, and genome-wide linkage analyses were performed using the EM algorithm, with body weight included as a covariate for all QTL analyses.(7) The following phenotypes were used as quantitative traits in the QTL screen: femur aBMD, vertebral aBMD, midfemur vBMD, distal femur vBMD, and vertebral vBMD. Heritability of each phenotypic measure was estimated as unity minus the ratio of the pooled variance in the parental animals (COP and DA) to the phenotypic variance in the F2 progeny. ANOVA was performed using the most significant marker in each QTL region to further characterize significant genotypic group differences.
Permutation tests were performed to obtain appropriate genome-wide significance levels for the linkage results.(8) Because the phenotypes are correlated, the set of phenotypes for each rat was kept together and randomly reassigned (permuted) to another rat in the sample. This phenotypic reassignment was performed for each of the 828 F2 animals to generate a permutated sample. This process was repeated to generate 5000 permutated datasets, and genome-wide linkage analysis was performed in each of the 5000 permutated datasets. All phenotypes measured for each rat were included in the permutation approach. Thus, genome-wide significance thresholds appropriate for each phenotype were obtained. The maximum logarithm of the odds (LOD) scores for linkage for each phenotype was recorded in each permutated dataset. In this manner, the LOD significance thresholds for the 95th and 99th percentile of the maximum genome-wide LOD scores across all phenotypes were found to be 3.5 and 4.3, respectively.
Sex specificity for each statistically significant QTL identified was evaluated using the method described by Solberg et al.(9) This method involves calculating the likelihood of the genotype and phenotype data under two models. The first (full) model contains effects for the QTL, sex, body weight, and a QTL × sex interaction term, whereas the second (reduced) model contains terms for QTL, sex, and body weight; thus, the models differ only in the QTL × sex interaction effect. At the position of each QTL achieving genome-wide significance in the primary screen (LOD > 4.3), a traditional likelihood ratio test (1-df χ2) was performed using the full and reduced models. This represents a test for sex specificity of the QTL for the phenotype and chromosomal position considered.
RESULTS
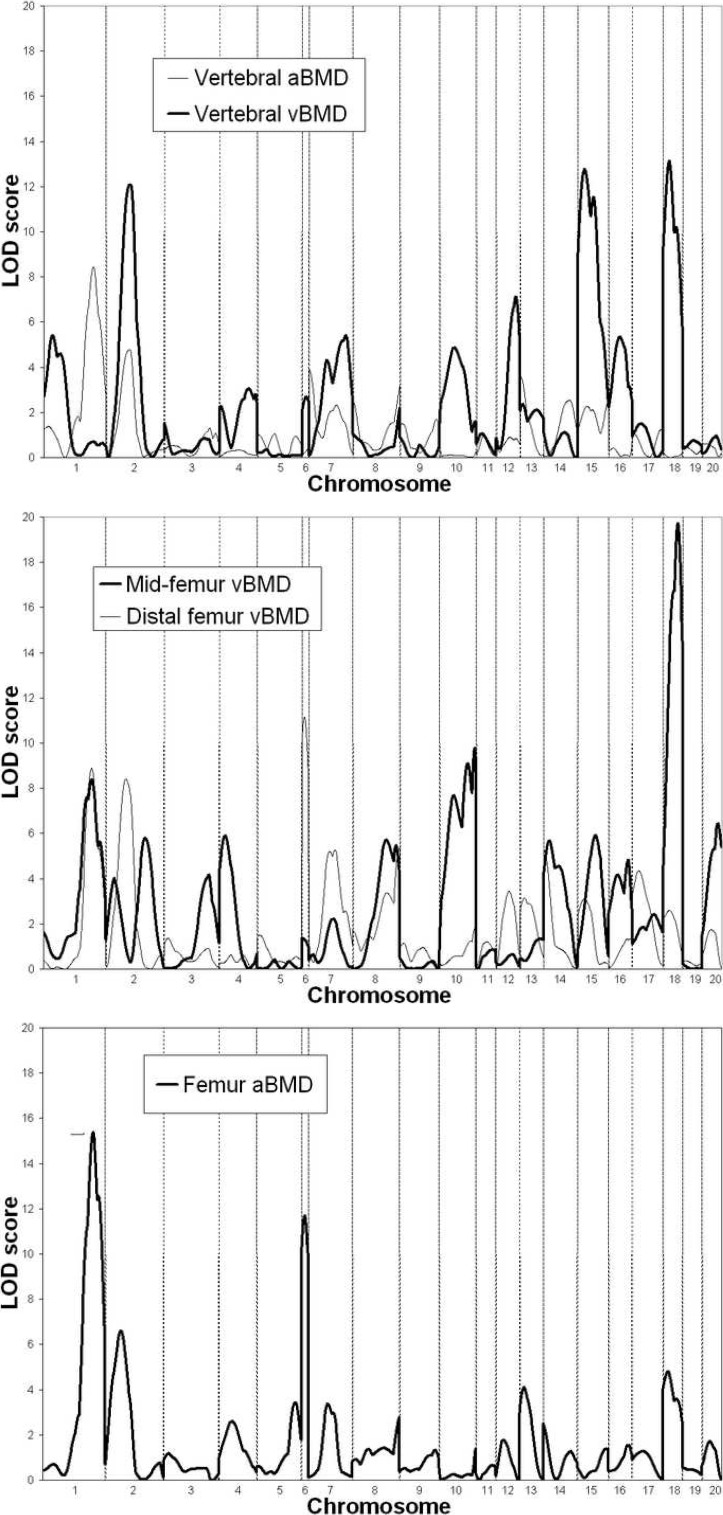
Summary statistics for each of the BMD phenotypes measured in the study are presented in Table 1, along with the estimate of heritability for each measure. Genome screen results for the five BMD phenotypes are shown in Fig. 1 and summarized in Table 2. The QTLs reported explained a total proportion of phenotypic variation ranging from 7.4% for vertebral aBMD to 35.1% for midfemur vBMD. The linkage result with the highest level of statistical significance observed in the analysis of the COP/DA cross was for midfemur vBMD on proximal rat chromosome 18, with a LOD score of 19.7. Both the male and female subgroups showed strong evidence of linkage to this chromosomal region (LOD = 10.3 and 11.3, respectively). The chromosome 18 QTL also exerted a strong pleiotropic effect on other BMD phenotypes in the COP/DA F2 animals. A LOD score of 13.1 was observed at proximal rat chromosome 18 for vertebral vBMD, and a LOD score of 4.8 was obtained in the same chromosomal region for femur aBMD. Notably, a QTL in this region for spine or femur BMD was not detected in our prior study of 595 female F2 animals derived from the F344 and LEW rat strains.
Table 1.
BMD Properties of the Copenhagen and Dark Agouti Rat Strains Used in the Experimental Cross
|
Copenhagen |
Dark agouti |
Heritability | |||
| Mean | SD | Mean | SD | ||
| Vertebral aBMD | 0.24 | 0.01 | 0.23 | 0.01 | 0.79 |
| Midfemur vBMD | 1033.57 | 14.79 | 881.93 | 22.80 | 0.71 |
| Distal femur vBMD | 732.44 | 28.07 | 702.41 | 22.36 | 0.91 |
| Femur aBMD | 0.25 | 0.01 | 0.23 | 0.01 | 0.58 |
| Vertebral vBMD | 720.28 | 24.44 | 711.95 | 21.80 | 0.69 |
FIG. 1.
Genome screen LOD plots for the five BMD phenotypes measured in the COP/DA rat F2 experiment. Chromosome boundaries are denoted by vertical dashed lines.
Table 2.
Summary of Genome Screen LOD Scores for BMD Phenotypes
| Chromosome | Phenotype |
LOD (position in cM)
|
Human synteny and phenotype | ||
| All | Male | Female | |||
| 1 (tel) | Femur aBMD | 15.4 (113) | 9.7 (109) | 7.7 (118) | 6q25–27, Spine and trochanter BMD(13,20) |
| Distal femur vBMD | 8.9 (108) | 5.6 (102) | 5.0 (117) | 9p24, Wrist BMD(21) | |
| Vertebral aBMD | 8.4 (111) | 6.2 (104) | 9q21, Wrist bone size(22) | ||
| Mid-femur vBMD | 8.4 (109) | 8.0 (108)* | 10q26, Hip BMD(11,15) | ||
| 1 (cen) | Vertebral vBMD | 5.4 (33) | 11q12–13, Spine BMD(23) | ||
| 2 (cen) | Vertebral vBMD | 12.1 (38) | 8.5 (39) | 3q24–26, Pelvic axis length(24,25) | |
| Distal femur vBMD | 8.4 (34) | 4.6 (35) | 3q24–26, Femur head, shaft width(24,25) | ||
| Femur aBMD | 6.6 (28) | 13q14, Trochanter BMD(13) | |||
| Vertebral aBMD | 4.8 (38) | ||||
| 2 (tel) | Mid-femur vBMD | 5.8 (77) | 4.8 (78) | ||
| 4 | Mid-femur vBMD | 5.9 (34) | 4.8 (30) | ||
| 6 | Femur aBMD | 11.7 (11) | 4.4 (11) | 6.8 (15) | 2p21–25, Forearm and hip BMD(12,26) |
| Distal femur vBMD | 11.2 (9) | 6.5 (8) | 4.7 (10) | 7p21, Spine BMD(20) | |
| 14q21, Spine BMD(14) | |||||
| 14q31–32, Spine and trochanter BMD(14,15) | |||||
| 7 | Vertebral vBMD | 5.4 (71) | 4.6 (72) | 8q24, Ward's BMD(14) | |
| Distal femur vBMD | 5.3 (50) | 9.6 (57)* | 22q13, Spine BMD(15) | ||
| 8 (cen) | Mid-femur vBMD | 5.7 (53) | 4.7 (50) | ||
| 8 (tel) | Distal femur vBMD | 5.4 (82) | |||
| Distal femur vBMD | 5.5 (48) | ||||
| 10 | Mid-femur vBMD | 9.8 (69) | 5.5 (69) | 16p13, Spine BMD(16) | |
| Vertebral vBMD | 4.9 (34) | 17p11–12, Hip BMD(11) | |||
| 17q21–23, Wrist bone size(22) | |||||
| 12 | Vertebral vBMD | 7.1 (46) | 12q24, Forearm and spine BMD(11,13) | ||
| 14 | Mid-femur vBMD | 5.7 (11) | 4.6 (9) | ||
| Distal femur vBMD | 5.3 (2) | ||||
| 15 | Vertebral vBMD | 12.8 (17) | 10.5 (17) | 13q14, Trochanter BMD(13) | |
| Mid-femur vBMD | 5.9 (35) | 13q21, Hip BMD(13) | |||
| 14q11, Hip bone size(27) | |||||
| 16 | Vertebral vBMD | 5.3 (19) | 4q32–35, Hip BMD(12) | ||
| 18 | Mid-femur vBMD | 19.7 (28) | 10.3 (24) | 11.3 (31) | |
| Vertebral vBMD | 13.1 (14) | 9.7 (13) | |||
| Femur aBMD | 4.8 (12) | ||||
| 20 | Mid-femur vBMD | 6.4 (32) | |||
* Sex-specific QTL effect (p < 0.01). Other QTLs shown were not sex specific at the α = 0.05 level.
Strong evidence of a QTL for femur aBMD was detected on distal rat chromosome 1 (LOD = 15.4). This QTL also showed pleiotropic effects on other phenotypes, because vertebral aBMD, midfemur vBMD, and distal femur vBMD resulted in LOD scores on distal chromosome 1 between 8 and 9. This QTL detected in the COP/DA F2 offspring seems to be distinct from the proximal chromosome 1 QTL found in our F344/LEW F2 cross. The COP/DA F2 cross, however, did show evidence of linkage for proximal chromosome 1 for the vertebral vBMD phenotype (LOD = 5.4), near where other BMD phenotypes were found to link in the F344/LEW experiment.
Several statistically significant QTLs in the COP/DA F2 offspring were also found underlying phenotypic variation in vertebral vBMD as measured by pQCT,. These QTLs included a region of rat midchromosome 15 (LOD = 12.8), midchromosome 2 (LOD = 12.1), and midchromosome 12 (LOD = 7.1). Among these chromosomal regions, only the QTL on chromosome 2 appeared in the F344/LEW cross and only for femur BMD phenotypes in that case. The evidence for the COP/DA QTL on chromosome 15 was stronger in females than in males (LOD = 10.5 versus 4.1 in males), although this does not represent evidence of sex specificity of the QTL by the method of Solberg et al. (p = 0.13(9)). Interestingly, however, we did not observe evidence of linkage to this region in the large sample of female F344/LEW F2 rats previously studied for any vBMD or aBMD phenotype.(5)
A novel pleiotropic QTL was detected on proximal rat chromosome 6 in the COP/DA F2 animals. This QTL impacts variability in both femur aBMD (LOD = 11.7) and distal femur vBMD (LOD = 11.2). The chromosome 6 QTL achieves genome-wide significance in each sex separately for both the aBMD and vBMD phenotypes.
In addition to the major QTL for midfemur vBMD on chromosome 18 reported above, we also detected QTLs underlying variability in this phenotype on rat chromosomes 10 (LOD = 9.8) and 20 (LOD = 6.4). The chromosome 10 QTL also exerts a pleiotropic effect on the vertebral vBMD phenotype (LOD = 4.9). This region also seems to provide evidence for a common QTL having an effect on BMD in both the COP/DA and F344/LEW crosses. The same vBMD phenotypes and chromosomal regions were detected with LOD scores >6.0 in the female F344/LEW F2 offspring we reported previously.
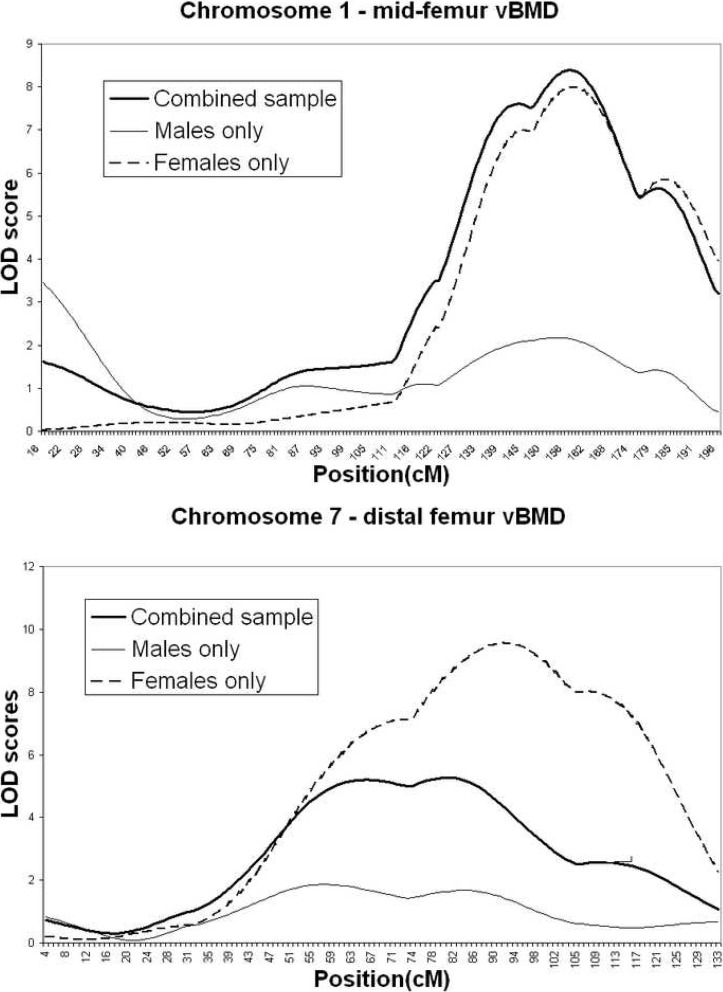
The majority of the QTL findings seem to be supported by both the male and female rat subgroups. However, a small number of the findings seem to be driven mainly by the male or female F2 subgroups. For two of the QTLs, this trend reaches statistical significance according to the method of Solberg et al.(9) The QTL for midfemur vBMD on distal chromosome 1 is female specific (sex-specific p = 0.006; Fig. 2). The QTL for distal femur vBMD on chromosome 7 is female specific as well (p = 0.004). Several other QTLs showed trends toward male- or female-specific effects, but none of these reached significance (p > 0.05).
FIG. 2.
Chromosome LOD plots for QTLs determined to be sex specific at the α = 0.05 level according to the method of Solberg et al.(9)
Table 3 provides information regarding the magnitude and direction of the effect of the QTLs detected in the COP/DA cross. Many of the QTLs, including those for aBMD on chromosomes 1 and 2, showed an increase in mean BMD as the number of alleles from the dark agouti (DA) grandparent increases. However, there are also multiple QTLs where the allele from the Copenhagen grandparent (COP) is associated with increased BMD; many of the latter QTLs were detected for the midfemur vBMD phenotype, such as those on chromosomes 2, 4, 8, and 10. This shows the complexity of the genetics of BMD variability in the rat model, as discussed below.
Table 3.
Genotypic Mean Values (aBMD in g/cm2, vBMD in mg/cm3, area in mm2) Adjusted by Body Weight for Phenotypes With Evidence of Linkage to Chromosomes 1, 2, 4, 6, 7, 8, 10, 12, 14, 15, 16, 18, and 20
| Chr | Marker | Phenotype | c/c | Genotype c/d | d/d | ANOVA p value |
| 1 | D1Rat169 | Femur aBMD | 0.1640 ± 0.0007 | 0.1678 ± 0.0004 | 0.1710 ± 0.0006 | <0.0001* |
| D1Rat69 | Distal femur vBMD | 657 ± 4.1 | 681 ± 2.8 | 689 ± 3.9 | <0.0001† | |
| D1Rat69 | Vertebral aBMD | 0.1502 ± 0.0010 | 0.1547 ± 0.0007 | 0.1579 ± 0.0010 | <0.0001* | |
| D1Rat69 | Midfemur vBMD | 1004 ± 2.5 | 996 ± 1.7 | 983 ± 2.4 | <0.0001* | |
| D1Rat261 | Vertebral vBMD | 721 ± 2.7 | 722 ± 1.9 | 708 ± 2.6 | <0.0001‡ | |
| 2 | D2Rat280 | Vertebral vBMD | 731 ± 2.6 | 719 ± 1.8 | 704 ± 2.6 | <0.0001* |
| D2Rat280 | Distal femur vBMD | 689 ± 3.9 | 681 ± 2.8 | 657 ± 3.9 | <0.0001‡ | |
| D2Rat198 | Femur aBMD | 0.1693 ± 0.0006 | 0.1681 ± 0.0005 | 0.1651 ± 0.0007 | <0.0001‡ | |
| D2Rat54 | Midfemur vBMD | 984 ± 2.6 | 995 ± 1.7 | 1003 ± 2.5 | <0.0001* | |
| D2Rat280 | Vertebral aBMD | 0.1578 ± 0.0010 | 0.1547 ± 0.0007 | 0.1512 ± 0.0010 | <0.0001* | |
| 4 | D4Rat103 | Midfemur vBMD | 1002 ± 2.7 | 996 ± 1.7 | 986 ± 2.4 | <0.0001‡ |
| 6 | D6Rat46 | Femur aBMD | 0.1647 ± 0.0007 | 0.1675 ± 0.0005 | 0.1710 ± 0.0006 | <0.0001* |
| D6Rat46 | Distal femur vBMD | 658 ± 4.0 | 675 ± 2.8 | 696 ± 3.8 | <0.0001* | |
| 7 | D7Rat78 | Vertebral vBMD | 708 ± 2.6 | 720 ± 1.9 | 727 ± 2.8 | <0.0001† |
| D7Rat23 | Distal femur vBMD | 661 ± 4.1 | 685 ± 2.7 | 675 ± 4.2 | <0.0001† | |
| 8 | D8Rat15 | Midfemur vBMD | 1004 ± 2.6 | 994 ± 1.7 | 987 ± 2.5 | <0.0001* |
| D8Rat2 | Distal femur vBMD | 691 ± 4.2 | 681 ± 3.0 | 664 ± 3.5 | <0.0001‡ | |
| 10 | D10Rat15 | Midfemur vBMD | 1006 ± 2.5 | 995 ± 1.8 | 983 ± 2.4 | <0.0001* |
| D10Rat162 | Vertebral vBMD | 712 ± 2.7 | 717 ± 1.9 | 728 ± 2.8 | <0.0001‡ | |
| 12 | D12Rat30 | Vertebral vBMD | 708 ± 2.6 | 717 ± 1.9 | 728 ± 2.6 | <0.0001* |
| 14 | D14Rat54 | Midfemur vBMD | 1004 ± 2.5 | 992 ± 1.8 | 990 ± 2.4 | <0.0001† |
| D14Rat54 | Distal femur vBMD | 691 ± 4.0 | 678 ± 2.9 | 663 ± 3.9 | <0.0001* | |
| 15 | D15Rat117 | Vertebral vBMD | 704 ± 2.6 | 718 ± 1.9 | 731 ± 2.7 | <0.0001* |
| D15Rat117 | Midfemur vBMD | 1004 ± 2.5 | 994 ± 1.8 | 987 ± 2.5 | <0.0001† | |
| 16 | D16Rat60 | Vertebral vBMD | 727 ± 2.7 | 718 ± 1.9 | 709 ± 2.6 | <0.0001* |
| 18 | D18Rat50 | Midfemur vBMD | 1009 ± 2.2 | 994 ± 1.7 | 979 ± 2.6 | <0.0001* |
| D18Rat50 | Vertebral vBMD | 732 ± 2.5 | 715 ± 1.9 | 708 ± 2.8 | <0.0001† | |
| D18Rat65 | Femur aBMD | 0.1697 ± 0.0006 | 0.1678 ± 0.0005 | 0.1658 ± 0.0006 | <0.0001‡ | |
| 20 | D20Rat29 | Midfemur vBMD | 1004 ± 2.4 | 994 ± 1.8 | 986 ± 2.7 | <0.0001* |
Values are means ± SE.
* Mean phenotypic value for all three pairs of genotypes differs at p = 0.05.
† Mean phenotypic value for d/d vs. c/d rats does not differ at p = 0.05.
‡ Mean phenotypic value for c/c vs. c/d rats does not differ at p = 0.05.
c/c, homozygous for COP/COP alleles; d/d, homozygous for DA/DA alleles; c/d, heterozygous.
DISCUSSION
The sequencing of the rat genome is complete,(10) and a large number of genomic resources are now available for this species (RGD, http://rgd.mcw.edu/). Further genetic studies of the rat will be facilitated by the rapidly growing database of SNPs (currently >40,000) that have proven to be polymorphic among experimental rat strains. Numerous genome scans for BMD phenotypes have been performed in both humans(11–16) and mice,(17–19) and it is relatively straightforward to use genomic information in the rat, mouse, and human to evaluate linkage findings in the three species for possible shared synteny. The rat model also provides the ability to identify genes within these QTL regions through congenic lines and other breeding strategies. Novel biological pathways implicated in BMD variation by these powerful screening strategies in experimental animal crosses can also provide more focused hypotheses for genetic studies of bone density and osteoporosis in humans.
Several of the QTLs identified in our QTL screen are located in rat chromosomal regions that are in fact syntenic to human chromosomal regions containing BMD QTLs that have been reported previously by our group and others (Table 2). These findings exemplify an advantage of analyses in animal models of complex phenotypes such as BMD, namely, that genes that are not detectable in the human linkage screen are able to reach significance in the experimental animal cross, implicating human genomic regions by synteny that otherwise would likely have been passed over. The study of animal models also permits the study of phenotypes that would be impractical to study in human subjects, such as bone strength and structure measures that we have also mapped in the COP/DA cross.(29)
In addition to the novel sex-specific QTLs detected in the COP/DA rat cross, we also found confirmatory evidence for QTLs detected in the F344/LEW rat cross on chromosomes 1, 2, and 15. These three common BMD QTLs were detected along with 13 other QTLs in the COP/DA cross and 9 other QTLs in the F344/LEW F2 females. The number of unique QTLs in each of the two crosses suggests a great degree of complexity underlying the genetic architecture of BMD variability in the rat model. For example, the highly significant QTL for midfemur vBMD that we report here on proximal rat chromosome 18 was not detected in the Fischer/Lewis cross or in linkage studies of human BMD variation. This shows the use of mapping in multiple animal crosses, because not all chromosomal regions will be informative in any one cross.
This genetic complexity is also shown by BMD-increasing alleles derived from either strain (COP or DA), depending on which QTL is being considered, as well as the skeletal site-specific effects of the majority of the QTLs detected in this experimental cross. This study further shows the power of the F2 design in experimental animals to detect chromosomal positions of underlying genetic effects on bone phenotypes. The remaining genetic variability is likely caused by epistatic effects (interaction between two or more loci(28)) and interactions between genes and environmental factors, which will require larger samples, sophisticated study designs, and additional phenotyping to detect.
ACKNOWLEDGMENTS
This work was supported by the National Institutes of Health through the following grants: R01AR047822 (CHT) and P01AG018397 (CHT, DLK, TF, MJE).
Footnotes
The authors state that they have no conflicts of interest.
REFERENCES
- 1.National Institutes of Health. Osteoporosis prevention, diagnosis, and therapy. NIH Consens Statement. 2000;17:1–45. [PubMed] [Google Scholar]
- 2.Marshall D, Johnell O, Wedel H. Meta-analysis of how well measures of bone mineral density predict occurrence of osteoporotic fractures. BMJ. 1996;312:1254–1259. doi: 10.1136/bmj.312.7041.1254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Peacock M, Turner CH, Econs MJ, Foroud T. Genetics of osteoporosis. Endocr Rev. 2002;23:303–326. doi: 10.1210/edrv.23.3.0464. [DOI] [PubMed] [Google Scholar]
- 4.Koller DL, Alam I, Sun Q, Liu L, Fishburn T, Carr LG, Econs MJ, Foroud T, Turner CH. Genome screen for bone mineral density phenotypes in Fisher 344 and Lewis rat strains. Mamm Genome. 2005;16:578–586. doi: 10.1007/s00335-004-2459-0. [DOI] [PubMed] [Google Scholar]
- 5.Alam I, Sun Q, Liu L, Koller DL, Fishburn T, Carr LG, Econs MJ, Foroud T, Turner CH. Whole-genome scan for linkage to bone strength and structure in inbred Fischer 344 and Lewis rats. J Bone Miner Res. 2005;20:1589–1596. doi: 10.1359/JBMR.050512. [DOI] [PubMed] [Google Scholar]
- 6.Lander ES, Green P, Abrahamson J, Barlow A, Daly MJ, Lincoln SE, Newburg L. MAPMAKER: An interactive computer package for constructing primary genetic linkage maps of experimental and natural populations. Genomics. 1987;1:174–181. doi: 10.1016/0888-7543(87)90010-3. [DOI] [PubMed] [Google Scholar]
- 7.Broman KW, Wu H, Sen S, Churchill GA. R/qtl: QTL mapping in experimental crosses. Bioinformatics. 2003;19:889–890. doi: 10.1093/bioinformatics/btg112. [DOI] [PubMed] [Google Scholar]
- 8.Doerge RW, Churchill GA. Permutation tests for multiple loci affecting a quantitative character. Genetics. 1996;142:285–294. doi: 10.1093/genetics/142.1.285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Solberg LC, Baum AE, Ahmadiyeh N, Shimomura K, Li R, Turek FW, Churchill GA, Takahashi JS, Redei EE. Sex- and lineage-specific inheritance of depression-like behavior in the rat. Mamm Genome. 2004;15:648–662. doi: 10.1007/s00335-004-2326-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mullins LJ, Mullins JJ. Insights from the rat genome sequence. Genome Biol. 2004;5:221. doi: 10.1186/gb-2004-5-5-221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Deng HW, Xu FH, Huang QY, Shen H, Deng H, Conway T, Liu YJ, Liu YZ, Li JL, Zhang HT, Davies KM, Recker RR. A whole-genome linkage scan suggests several genomic regions potentially containing quantitative trait Loci for osteoporosis. J Clin Endocrinol Metab. 2002;87:5151–5159. doi: 10.1210/jc.2002-020474. [DOI] [PubMed] [Google Scholar]
- 12.Devoto M, Shimoya K, Caminis J, Ott J, Tenenhouse A, Whyte MP, Sereda L, Hall S, Considine E, Williams CJ, Tromp G, Kuivaniemi H, Ala-Kokko L, Prockop DJ, Spotila LD. First-stage autosomal genome screen in extended pedigrees suggests genes predisposing to low bone mineral density on chromosomes 1p, 2p and 4q. Eur J Hum Genet. 1998;6:151–157. doi: 10.1038/sj.ejhg.5200169. [DOI] [PubMed] [Google Scholar]
- 13.Kammerer CM, Schneider JL, Cole SA, Hixson JE, Samollow PB, O'Connell JR, Perez R, Dyer TD, Almasy L, Blangero J, Bauer RL, Mitchell BD. Quantitative trait loci on chromosomes 2p, 4p, and 13q influence bone mineral density of the forearm and hip in Mexican Americans. J Bone Miner Res. 2003;18:2245–2252. doi: 10.1359/jbmr.2003.18.12.2245. [DOI] [PubMed] [Google Scholar]
- 14.Karasik D, Myers RH, Cupples LA, Hannan MT, Gagnon DR, Herbert A, Kiel DP. Genome screen for quantitative trait loci contributing to normal variation in bone mineral density: The Framingham Study. J Bone Miner Res. 2002;17:1718–1727. doi: 10.1359/jbmr.2002.17.9.1718. [DOI] [PubMed] [Google Scholar]
- 15.Koller DL, Econs MJ, Morin PA, Christian JC, Hui SL, Parry P, Curran ME, Rodriguez LA, Conneally PM, Joslyn G, Peacock M, Johnston CC, Foroud T. Genome screen for QTLs contributing to normal variation in bone mineral density and osteoporosis. J Clin Endocrinol Metab. 2000;85:3116–3120. doi: 10.1210/jcem.85.9.6778. [DOI] [PubMed] [Google Scholar]
- 16.Wilson SG, Reed PW, Bansal A, Chiano M, Lindersson M, Langdown M, Prince RL, Thompson D, Thompson E, Bailey M, Kleyn PW, Sambrook P, Shi MM, Spector TD. Comparison of genome screens for two independent cohorts provides replication of suggestive linkage of bone mineral density to 3p21 and 1p36. Am J Hum Genet. 2003;72:144–155. doi: 10.1086/345819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Beamer WG, Shultz KL, Churchill GA, Frankel WN, Baylink DJ, Rosen CJ, Donahue LR. Quantitative trait loci for bone density in C57BL/6J and CAST/EiJ inbred mice. Mamm Genome. 1999;10:1043–1049. doi: 10.1007/s003359901159. [DOI] [PubMed] [Google Scholar]
- 18.Beamer WG, Shultz KL, Donahue LR, Churchill GA, Sen S, Wergedal JR, Baylink DJ, Rosen CJ. Quantitative trait loci for femoral and lumbar vertebral bone mineral density in C57BL/6J and C3H/HeJ inbred strains of mice. J Bone Miner Res. 2001;16:1195–1206. doi: 10.1359/jbmr.2001.16.7.1195. [DOI] [PubMed] [Google Scholar]
- 19.Klein RF, Mitchell SR, Phillips TJ, Belknap JK, Orwoll ES. Quantitative trait loci affecting peak bone mineral density in mice. J Bone Miner Res. 1998;13:1648–1656. doi: 10.1359/jbmr.1998.13.11.1648. [DOI] [PubMed] [Google Scholar]
- 20.Duncan EL, Brown MA, Sinsheimer J, Bell J, Carr AJ, Wordsworth BP, Wass JA. Suggestive linkage of the parathyroid receptor type 1 to osteoporosis. J Bone Miner Res. 1999;14:1993–1999. doi: 10.1359/jbmr.1999.14.12.1993. [DOI] [PubMed] [Google Scholar]
- 21.Deng HW, Shen H, Xu FH, Deng HY, Conway T, Zhang HT, Recker RR. Tests of linkage and/or association of genes for vitamin D receptor, osteocalcin, and parathyroid hormone with bone mineral density. J Bone Miner Res. 2002;17:678–686. doi: 10.1359/jbmr.2002.17.4.678. [DOI] [PubMed] [Google Scholar]
- 22.Deng HW, Xu FH, Liu YZ, Shen H, Deng H, Huang QY, Liu YJ, Conway T, Li JL, Davies KM, Recker RR. A whole-genome linkage scan suggests several genomic regions potentially containing QTLs underlying the variation of stature. Am J Med Genet. 2002;113:29–39. doi: 10.1002/ajmg.10742. [DOI] [PubMed] [Google Scholar]
- 23.Johnson ML, Gong G, Kimberling W, Recker SM, Kimmel DB, Recker RB. Linkage of a gene causing high bone mass to human chromosome 11 (11q12-13) Am J Hum Genet. 1997;60:1326–1332. doi: 10.1086/515470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Koller DL, Liu G, Econs MJ, Hui SL, Morin PA, Joslyn G, Rodriguez LA, Conneally PM, Christian JC, Johnston CC, Jr, Foroud T, Peacock M. Genome screen for quantitative trait loci underlying normal variation in femoral structure. J Bone Miner Res. 2001;16:985–991. doi: 10.1359/jbmr.2001.16.6.985. [DOI] [PubMed] [Google Scholar]
- 25.Koller DL, White KE, Liu G, Hui SL, Conneally PM, Johnston CC, Econs MJ, Foroud T, Peacock M. Linkage of structure at the proximal femur to chromosomes 3, 7, 8, and 19. J Bone Miner Res. 2003;18:1057–1065. doi: 10.1359/jbmr.2003.18.6.1057. [DOI] [PubMed] [Google Scholar]
- 26.Niu T, Chen C, Cordell H, Yang J, Wang B, Wang Z, Fang Z, Schork NJ, Rosen CJ, Xu X. A genome-wide scan for loci linked to forearm bone mineral density. Hum Genet. 1999;104:226–233. doi: 10.1007/s004390050940. [DOI] [PubMed] [Google Scholar]
- 27.Deng HW, Shen H, Xu FH, Deng H, Conway T, Liu YJ, Liu YZ, Li JL, Huang QY, Davies KM, Recker RR. Several genomic regions potentially containing QTLs for bone size variation were identified in a whole-genome linkage scan. Am J Med Genet. 2003;119:121–131. doi: 10.1002/ajmg.a.20100. [DOI] [PubMed] [Google Scholar]
- 28.Koller DL, Liu L, Alam I, Sun Q, Econs MJ, Foroud T, Turner CH. Epistatic effects contribute to variation in bone density in Fischer 344 x Lewis F2 rats. J Bone Miner Res. 2008;23:41–47. doi: 10.1359/JBMR.071001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sun Q, Alam I, Liu L, Koller DL, Carr LG, Econs MJ, Foroud T, Turner CH. Genetic loci affecting bone structure and strength in inbred COP and DA rats. Bone. 2008;42:547–553. doi: 10.1016/j.bone.2007.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]