Abstract
Adult BMD, an important risk factor for fracture, is the result of genetic and environmental interactions. A quantitative trait locus (QTL) for the phenotype of volumetric BMD (vBMD), named Bmd8, was found on mid-distal chromosome (Chr) 6 in mice. This region is homologous to human Chr 3p25. The B6.C3H-6T (6T) congenic mouse was previously created to study this QTL. Using block haplotyping of the 6T congenic region, expression analysis in the mouse, and examination of nonsynonymous SNPs, peroxisome proliferator activated receptor γ (Pparg) was determined to be the most likely candidate gene for the Bmd8 QTL of the 630 genes located in the congenic region. Furthermore, in the C3H/HeJ (C3H) strain, which is the donor strain for the 6T congenic, several polymorphisms were found in the Pparg gene. On challenge with a high-fat diet, we found that the 6T mouse has a lower areal BMD (aBMD) and volume fraction of trabecular bone (BV/TV%) of the distal femur compared with B6 mice. Interactions between SNPs in the PPARG gene and dietary fat for the phenotype of BMD were examined in the Framingham Offspring Cohort. This analysis showed that there was a similar interaction of the PPARG gene and diet (fat intake) on aBMD in both men and women. These findings suggest that dietary fat has a significant influence on BMD that is dependent on the alleles present for the PPARG gene.
Key words: skeletal genetics, animal models, candidate gene, environmental interaction, human population
INTRODUCTION
Multiple studies have shown that between 55% and 85% of the variance in peak bone mass is determined by heritable factors.(1) Whereas numerous quantitative trait loci (QTLs) have been identified for the phenotype of peak bone mass in both mice and humans, the identification of the underlying genes has remained elusive.(1–3) Environmental factors such as diet and exercise contribute to peak bone mass and are likely to complicate candidate gene analysis.
We previously identified a QTL for the phenotype of peak volumetric BMD (vBMD) on mid-distal mouse chromosome (Chr) 6 in a cross between the C57BL/6J (B6) and C3H/HeJ (C3H) inbred strains of mice.(4) This QTL was named Bmd8 and was found to be coincident in genetic location for a QTL for the phenotype of serum IGF-1, named Igf1sl1, that also had been identified in the B6×C3H cross.(5) The B6.C3H-6T (6T) congenic strain of mice was developed for the purpose of gaining insight into the genetics and biology underlying both phenotypes. The congenic strain was made by introgressing a region of Chr 6 from C3H onto a B6 background by 10 generations of selective backcrossing, followed by several generations of intercrossing. The 6T mice are homozygous for B6 alleles for the entire genome except for the region between the genetic markers D6Mit93 and D6Mit150, where this congenic is homozygous for the C3H alleles.(6) Female 6T mice have lower femoral and vertebral vBMD, a smaller mid-diaphyseal periosteal circumference, slightly shorter femurs, and lower serum IGF-1 levels than the B6 background strain. In addition, female 6T mice also exhibit a significant decrease in whole body areal BMD (aBMD) and in trabecular bone volume fraction (BV/TV%) of the distal femur and lumbar vertebrae.(6) This latter decrease in BV/TV% is coincident with an increase in marrow adipocytes.(6,7) One primary genetic mapping study and two meta-analyses studies have identified a QTL for aBMD in humans with a peak LOD score at 3p25, the region in humans that is homologous with the distal portion of our 6T congenic region.(8–10)
Dietary factors have long been known to influence BMD. Several studies have examined the association between types of dietary fat and BMD in humans, but the results are conflicting. For example, saturated fat intake was found to be negatively associated with aBMD in men and women in the NHANES III cohort study,(11) yet Brownbill and Ilich(12) did not find any association between saturated fat intake and aBMD in a study of white postmenopausal women. Two studies examining γ-linolenic acid and eicosapentaenoic acid intake in postmenopausal women also yielded conflicting results.(13,14) One possible explanation for these conflicting results might relate to a possible interaction between dietary intake of fat and genetic factors.
In this study, we used a mouse model, the 6T congenic, to tease out a complicated gene (Pparg) by environment (dietary fat) interaction that impacts on aBMD. We further verified this association in a human cohort. By showing a PPARG gene by dietary fat interaction in both mice and humans, these results highlight the role of genetic variation in the skeletal response to the environment and potentially explain why previous studies examining the association between PPARG and aBMD were conflicting.
MATERIALS AND METHODS
Animal husbandry
All studies and procedures involving mice were approved by the Institutional ACUC of The Jackson Laboratory, and all mice were obtained from the Jackson Laboratory (Bar Harbor, ME, USA). Mice were maintained in groups of 2–3 in polycarbonate boxes (130 cm2) on bedding of sterilized white pine shavings under conditions of 14h light; 10h darkness, with free access to acidified water (pH 2.5 with HCl to retard bacterial growth) that contains 0.4 mg/ml of vitamin K (menadione Na bisulfite). All diets used in these studies were provided ad libitum.
Assessment of the effect of dietary fat
Diets used to examine the effects of fat on aBMD were obtained from Research Diets (Cat. D12489B, D12266B, and D12492). B6 and 6T female mice were weaned at 22 days of age onto one of three diets (11%, 32%, and 60% kJ from fat, respectively). Body weight and whole body aBMD data were collected at 16 wk of age. The Comprehensive Cage Animal Monitoring System (Columbus Instruments) was used to simultaneously determine calorimetric parameters and food and water consumption as previously described.(15) The mice were killed at 16 wk of age, and the femurs were collected and fixed in 70% ethanol. BV/TV% of the distal femur was determined by μCT analysis and marrow adiposity was assessed by static histomorphometry.
Genotyping
B6.C3H-6T mice were genotyped for the purpose of further defining the C3H-like congenic region. Genotyping of the mice, including the preparation of the DNA and PCR reaction conditions, has been previously described.(16) Genetic markers used to define the ends of the congenic have been previously published and primer sequences are freely available at http://www.informatics.jax.org/.
Microarray
The microarray, including sample preparation and methodology used for expression analysis, has been described in detail elsewhere.(16) The raw data from this microarray has been deposited in NCBI's Gene Expression Omnibus (GEO, http://www.ncbi.nlm.nih.gov/geo/), and these data are available through GEO Series accession number GSE5959. In short, differential gene expression data were determined using the MOE430v2.0* GeneChip arrays (Affymetrix) on liver collected from fasted 8-wk-old B6 and 6T female mice fed a standard chow diet (LabDiets, 16% fat by kilojoules, 6% fat by weight, Cat. 5K52).
Haplotype analysis
For haplotype analysis, BMD was measured in 16-wk-old female mice from the following mouse stocks: C57BL/6J (B6), C57BL/6J-Chr6A/NaJ, C57BL/6J-Chr6PWD/Ph/ForeJ, and 6T. A total of 4160 SNPs from the Broad Institute SNP database (http://www.broad.mit.edu/) located within the 6T congenic region were used for high-density SNP-block-haplotype mapping. Using the methodology of Park et al., we located haplotype blocks where A/J and B6 shared common alleles yet had different alleles than C3H and PWD/PhJ (PWD).(17) A block was considered to begin and end one base pair distal and one base pair proximal to the first SNP on either side of the block were the allele for that SNP did not meet the criteria of (B6 = A/J) ≠ (C3H = PWD). Any gene with a portion of the coding region found between these two flanking SNP was considered to be within the block.
Sequencing of Pparg in mouse
The coding region, promoters and 3′ UTR of Pparg were sequenced in both B6 and C3H. Cycle sequencing of DNA templates was performed using Applied Biosystems' BigDye Terminator v3.1 cycle sequencing kit. Purified reactions were run on Applied Biosystems 3730xl DNA Analyzer using POP 7 polymer. Raw data were analyzed using Applied Biosystems DNA Sequencing Analysis Software, Version 5.2.
Body composition, BMD, and trabecular bone architecture
Body composition, total body aBMD, lean mass, and fat mass were measured using the PIXImus DXA (GE-Lunar) as previously described.(6) Isolated femur lengths were measured with digital calipers (Stoelting), and distal femurs were scanned using a desktop μCT imaging system (μCT40; Scanco Medical) to measure trabecular bone volume fraction and microarchitecture in the secondary spongiosa of the distal femur as previously described.(18)
Histomorphometry
Femurs were dissected, fixed in 70% ethanol, dehydrated, and embedded undecalcified in methyl methacrylate. Longitudinal sections, 5 μm thick, were cut on a Microm microtome (Richards-Allan Scientific) and stained with toluidine blue, pH 6.4. Because this was fixed tissue, the lipid content of the adipocyte was stripped, leaving “holes” that could be counted. The number of adipocytes per area was determined using an OsteoMeasure morphometry system (Osteometrics).
Statistical analysis in mouse studies
Data are expressed as mean ± SE in tables and figures. Statistical evaluation of bone parameters and body composition was conducted using JMP version 6 software (SAS). To account for differences in body size between strains, a stepwise analysis of covariance (ANCOVA) approach was used for DXA and μCT data using body weight and femur length as covariates. Nonsignificant covariates and interactions were removed in a stepwise fashion until the final model was obtained.
Framingham Offspring study cohort
The Framingham Offspring cohort is comprised of 2616 adult offspring of couples from the Original Framingham cohort (51%), 898 adult offspring with one parent in the Original Cohort at greater risk of cardiovascular disease (17.5%), 34 stepchildren (<1%), and 1576 spouses of these individuals (30.8%). Nearly all (96.4%) of the Offspring Cohort are whites, with origins in Eastern and Western Europe. After their initial evaluation, these individuals have undergone repeat examinations approximately every 4 yr and participated in the Framingham Osteoporosis study between 1996 and 2001, as described elsewhere.(19)
Informed consent was obtained from participants of each cohort before entry into the study, through a protocol approved by the Boston University Institutional Review Board for Human Subjects Research and the Hebrew Rehabilitation Center Institutional Review Board. Participants in this study are a subset of unrelated individuals from the Framingham Offspring Cohort who provided blood samples for DNA, had aBMD measurements of the hip and spine, and had dietary intake data gathered.(19,20) A total of 1792 of these (867 men and 925 women) also had genotyping done on 13 SNPs that were used for this study. These subjects were not selected on the basis of any trait, but only on the basis that they were biologically unrelated with available DNA.
Phenotyping measurements
The participants underwent bone densitometry of the femoral neck by DXA using a Lunar DPX-L instrument to measure aBMD (g/cm2) in 1996–2001. The CV in normal subjects for the DPX-L at the femoral neck was 1.7%.(21) Dietary fat intake information was collected from a food frequency questionnaire that had previously been validated in both men and women.(22–25)
SNP selection, DNA extraction, and genotyping
Initially common genetic variations in the region spanning PPARG on Chr 3 (Entrez GeneID: 5468) were surveyed, and a dense SNP map in the Center d'Etude du Polymorphisme Humain (CEPH) panel was constructed. This led to the development of working assays for 55 polymorphic SNPs. Again in the CEPH population, tag SNPs selection software TagSNPs (http://www-rcf.usc.edu/∼stram) was used to choose 11 tag SNPs that were genotyped in Framingham participants along with two common mutations associated with diabetes and cardiac disease (Pro12Ala or rs1801282) and with serum leptin concentrations(26) and obesity (His477His or rs3856806).(27) These SNP are listed in Table 1. The TagSNPs selection algorithm is based on optimizing the squared correlation between estimates of the number of copies of a particular haplotype h and the true number of copies of haplotype h (Rh 2) carried by a subject, averaging over all possible genotype data under an assumption of Hardy-Weinberg equilibrium.(28)
Table 1.
SNPs in PPARG Genotyped in This Study in the Framingham Offspring Cohort
| SNP number | SNP rs number | Location | Position* | Alleles | Minor allele frequency |
| 1 | rs2028760 | 5′ UTR | 12347882 | G>A | 0.25 |
| 2 | rs1801282 | 5′ UTR | 12368125 | C>G | 0.11 |
| 3 | G531a24 | 5′ UTR | 12370645 | G>A | 0.12 |
| 4 | rs1805192 | Exon 1 | 12396238 | C>G | 0.11 |
| 5 | rs1151996 | Intron 3 | 12420807 | T>G | 0.37 |
| 6 | rs1151999 | Intron 3 | 12422153 | A/C | 0.47 |
| 7 | rs709150 | Intron 4 | 12426337 | C>G | 0.47 |
| 8 | rs1175544 | Intron 5 | 12442044 | C>T | 0.32 |
| 9 | rs1152002 | Intron 5 | 12446871 | G>A | 0.48 |
| 10 | rs3856806 | Exon 6 | 12450557 | G>A | 0.13 |
| 11 | rs1152004 | Intergenic | 12458104 | T>C | 0.20 |
| 12 | rs1175381 | Intergenic | 12460844 | T>C | 0.04 |
| 13 | rs1186464 | Intergenic | 12462511 | A>G | 0.11 |
* NCBI 36 assembly of the human genome, November 2005.
All SNP genotyping, except Pro12Ala and His477His, was performed as part of the NHLBI Program in Genomic Applications (PGA; http://cardiogenomics.med.harvard.edu/home). PGA genomic DNA was extracted from peripheral white blood cells (WBCs), or EB-transformed B-lymphocytic cells. Genotyping was performed using the Sequenom MassArray platform with products of different masses obtained for each allele of each SNP resolved by mass spectrometry (MALDI-TOF). Automated allele-calling algorithms were used to process the data. Genotyping of two SNPs, Pro12Ala and His477His, was performed using the TaqMan assay (Perkin-Elmer). Products were amplified using 0.9 μM each of the forward and the reverse primers, 30 ng DNA, 5.0 mM MgCl2, and 1× Taqman Universal PCR Master Mix containing AmpliTaq Gold DNA Polymerase in a 27-μl reaction volume. After an initial step of 2 min at 50°C and 10 min at 95°C to activate the AmpliTaq Gold, the products were amplified using 40 cycles of 15 s at 95°C and 1 min at 60°C. A total of 0.2 μM of each of the sequence-specific probes was used in the allele discrimination assay, and allele detection and genotype calling were performed using the ABI 7900 and the Sequence Detection System software (Perkin-Elmer). Blind duplicates and negative controls were introduced as part of the quality control procedures.
Statistical analysis of data from the Framingham Offspring cohort
Means ± SDs for continuous variables or proportions for categorical variables were computed for all study variables separately for men and women. For all SNPs, Hardy-Weinberg equilibrium was evaluated using a 1 degree of freedom χ2 statistic; genotype frequencies did not deviate from Hardy-Weinberg equilibrium expectations. We used three genotype groups (2 df test) for all SNPs, except for rs1175381. For rs1175381 (minor allele frequency < 5%), we combined the heterozygote with the minor allele homozygotes to compare with the common homozygote groups.
To test for interaction effects between the PPARG SNP genotypes and percent of energy intake from total fat on femoral neck aBMD, we performed ANCOVA techniques using Proc GLM in SAS (SAS Institute). Femoral neck aBMD was used as a dependent variable; percent energy intake from total fat, indicator variables for individual genotypes, and interaction terms between indicator variables of the individual SNP genotypes and percent energy intake from total fat as primary independent variables of interest; age, BMI, height, diabetes, total energy intake, and estrogen status in women were used as covariates. For estrogen status, women were classified into two categories: (1) premenopausal or postmenopausal on estrogen or (2) postmenopausal not on estrogen, where menopause was defined as having no menstrual period for at least 1 yr. Percent of energy intake from total fat was evaluated both as a continuous and dichotomous variable (dichotomized at the median value of 27%). Analyses were performed for men and women separately.
RESULTS
Identification of candidate genes for Bmd8
Block haplotyping for the purpose of identifying candidate genes underlying a QTL is most powerful when as many different strains as possible can be added to the analysis. To include a strain in the analysis, one must first know if the alleles present in a given genomic interval from that strain influence the phenotype of interest compared with a baseline strain. For this reason, BMD was assessed in two consomic strains: the C57BL/6J-Chr6A/NaJ strain that carries A/J alleles for all of Chr 6 on an otherwise B6 background and the C57BL/6J-Chr6PWD/Ph/ForeJ strain that carries PWD/PhJ (PWD) alleles for all of Chr 6 on an otherwise B6 background. When comparing C57BL/6J-Chr6A/NaJ to the B6 strain, no differences in BMD were found, whereas the C57BL/6J-Chr6PWD/Ph/ForeJ strain was found to have significantly higher BMD (data not shown). From these data, it can be concluded that there are no A/J-like alleles on Chr 6 that affect BMD, whereas alleles present on Chr 6 from PWD do change BMD.
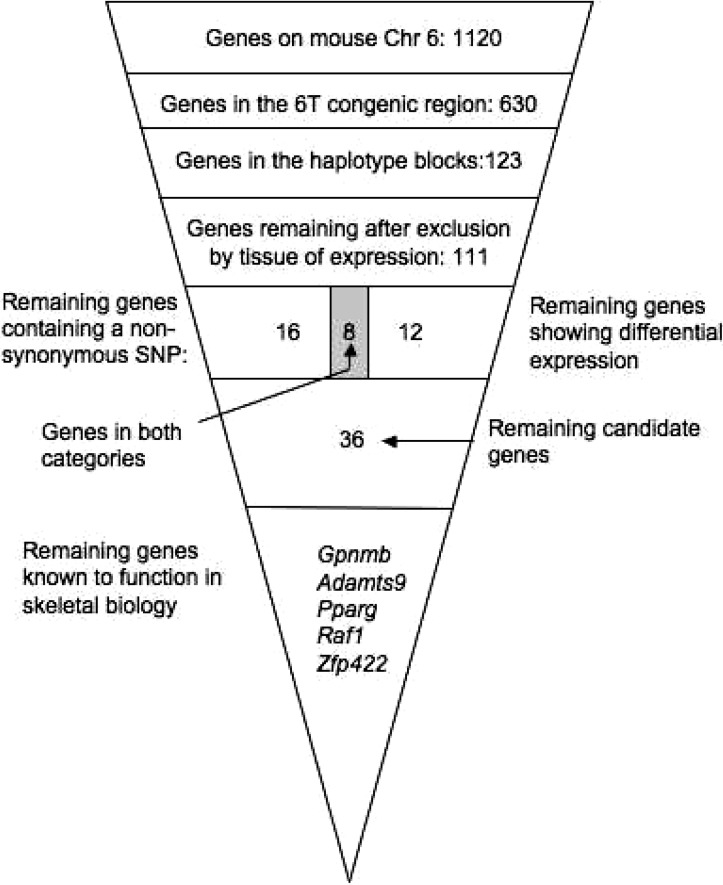
Further genotyping of the 6T congenic at the N10 generation determined that the C3H-like donated region actually extended as far proximal as D6Mit274, but not as far as D6Mit207. At the distal end, the congenic region extended as far as D6Mit216, but not as far distal as D6Mit134. We had previously determined that the 6T congenic has lower vBMD than age- and sex-matched B6 mice.(6) Using this information, block haplotyping was performed for the 6T congenic region using the method of Park et al.(17) by examining the alleles carried at each of 4160 SNPs for the C3H, B6, A/J, and PWD strains. We found 15 haplotype blocks where B6 and A/J carried the same alleles, yet C3H and PWD did not. A total of 123 genes were found to be completely contained within, or have coding region that overlapped with a haplotype block and were therefore considered potential candidate genes for skeletal phenotypes. Of these 123 genes, 12 genes were eliminated as candidate genes because of expression only in tissues unrelated to our phenotypes (i.e., retina) or because of expression being restricted to specific developmental time points (http://symatlas.gnf.org/SymAtlas/). For example, the gene H1foo (MGI:2176207) was found to be located in the last haplotype block, but expression of this gene is restricted to the oocyte and fertilized egg, thus eliminating this gene as a potential candidate. This left 111 potential candidate genes, as shown in Fig. 1.
FIG. 1.
Systematic identification of the most likely candidate gene for the mouse QTL Bmd8. In generating the 6T congenic strain, 43.6% of the genes on the sixth Chr could be eliminated as candidates for Bmd8. Using block haplotyping, the number of candidates was further reduced to 123 genes. Examination of tissue of expression eliminated another 12 genes, resulting in 111 candidate genes. Of these genes, 16 were found to contain a nonsynonymous SNPs, 12 were found to be differentially expressed in the 6T mouse, and an additional 8 were found to be differentially expressed and contain a nonsynonymous SNP, leaving 36 genes. For 5 of these 36 genes, literature references existed describing a role in bone biology.
IGF-1 is a critical growth factor for several tissues, including bone. Because 6T mice exhibit markedly reduced hepatic expression of IGF-I compared with B6 and C3H,(6) we examined differential gene expression in the livers of B6 and 6T by microarray. Expression analysis showed that 20 of the remaining 111 candidate genes had differential expression when comparing B6-6T. Examination of all known SNPs identified 24 genes with a nonsynonymous SNP. When these two lists of genes were compared, it was found that eight of these genes had both a nonsynonymous SNP and expression differences. As shown in Fig. 1, this left us with 36 candidate genes (20 genes with just differential expression, 24 genes with just a nonsynonymous SNP, and 8 genes having both). These 36 candidate genes are listed in Table 2.
Table 2.
Short List of 36 Candidate Genes for Bmd8
| Gene symbol | Fold change for expression in liver | Non-synonymous SNPs | Role in bone biology |
| Gpnumb (MGI:1934765) | rs13478745 | Expressed in osteoblasts(29) | |
| Smyd1 (MGI:104790) | −1.508677 | ||
| Rmnd5a (MGI: 1915727) | −1.527938 | ||
| Jmjd1a (MGI: 98847) | −1.201164 | ||
| Mrpl35 (MGI: 1913473) | −1.377875 | ||
| Immt (MGI: 1923864) | rs30691060 | ||
| Ptcd3 (MGI: 1917206) | rs30690719, rs30687220, rs30688074, rs30690874, rs30267257, rs30697183 | ||
| Rpo1–4 (MGI: 1096397) | −1.423717 | rs30703724, rs30707379, rs30707379, rs30707379 | |
| St3gal5 (MGI: 1339963) | −1.611648 | ||
| Vamp5 (MGI: 1858622) | −1.45543 | ||
| Vamp8 (MGI: 1336882) | rs30078477 | ||
| Mat2a (MGI: 2443731) | −1.905166 | ||
| Rbed1 (MGI: 2445168) | −1.330913 | rs37332129 | |
| Retsat (MGI: 1914692) | −1.528719 | rs30809096 | |
| Tgoln1 (MGI: 105080) | −2.15785 | rs38907202 | |
| Dnahc6 (MGI: 107744) | rs13478818, rs30716987 | ||
| Gpr175 (MGI: 1345190) | −1.533345 | ||
| Adamts9 (MGI: 1916320) | rs30983269, rs30980342 | Chondrocytes(30) | |
| Magi1 (MGI: 1203522) | −1.584799 | rs38831890 | |
| Slc25a26 (MGI: 1914832) | rs36315133 | ||
| Ttll3 (MGI: 2141418) | rs38208806 | ||
| Syn2 (MGI:103020) | rs31483470 | ||
| Pparg (MGI:97747) | −1.587832 | Mesenchymal stem cell differentiation(56) | |
| Raf1 (MGI:97847) | rs31487978, rs31488644 | Chondrocytes(31) | |
| Tmem40 (MGI: 2137870) | rs31495179, rs31498799, rs31498257 | ||
| Cand2 (MGI: 1914338) | rs31503553, rs31504157, rs31504159 | ||
| BC060267 (MGI: 2681834) | −1.329998 | rs31503452, rs31500907 | |
| Mbd4 (MGI: 1333850) | −1.391686 | rs31502440, rs31503102, rs31501809, rs31503026 | |
| D6Wsu116e (MGI: 106463) | rs13460813 | ||
| Anubll (MGI: 1914742) | −1.607684 | rs31539045 | |
| March8 (MGI: 1919029) | −1.483522 | ||
| Olfr212 (MGI: 3030046) | rs31551252, rs31551918, rs31553514 | ||
| Olfr215 (MGI: 3030049) | rs36378907, rs3626396 | ||
| Zfp422 (MGI: 1914505) | −1.359005 | Tooth Eruption(32) | |
| Rassf4 (MGI: 2386853) | −1.33672 | ||
| C230095G01Rik (MGI: 2442707) | rs36770584 |
A careful search of the literature was performed for these 36 genes, and 5 were found to have a previously described role in bone biology. These genes included Gpnmb, Adamts9, Pparg, Raf1, and Zfp422.(29–33) The hepatic gene expression signature in 6T was also remarkable for a consistent pattern of significant upregulation of several genes essential for adipocyte differentiation (i.e., Lpl [lipoprotein lipase], Fasn [fatty acid synthase], Cd36 [CD36 antigen], Fabp4 [fatty acid binding protein 4, i.e., AP2], Lpin1 [Lipin 1], and Srebf1 [sterol regulatory element binding factor 1], known target genes for Pparg). Last, we had previously found the expression of Pparg was altered in cultured calvarial osteoblasts from 6T congenic mice.(7) We therefore chose to focus on Pparg as our most likely candidate gene.
Sequencing the Pparg gene in C3H/HeJ
The promoters, coding region, and 3′ UTR of the Pparg gene in both B6 and C3H were sequenced. A total of 27 polymorphisms were found when comparing mice of the C3H strain to B6 mice. Six of these polymorphisms were in the coding region, but all were found to be synonymous. The SNP rs8254779, a nonsynonymous coding region SNP, has been previously reported to be polymorphic between B6 and C3H (http://www.jax.org/phenome). This polymorphic difference was not found in our mice and therefore was not included in Table 2. The C3H-like Pparg sequence has been submitted to GenBank and can be retrieved using the following accession numbers: EF062476, EF062477, EF062478, and EF062479.
Effects of dietary fat on body composition and bone in mice
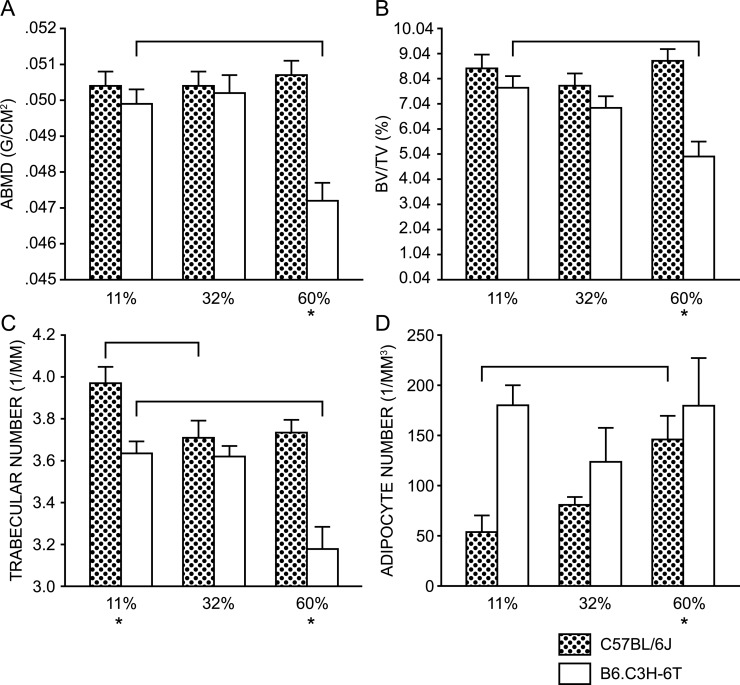
Dietary fat is a potent activator of PPARG.(34,35) Therefore, we sought to determine the effect of dietary fat on bone mass and body composition in both B6 and 6T female mice. Total body aBMD did not differ in B6 mice across the three diets but was markedly reduced in 6T at the highest fat intake (Fig. 2A). Trabecular bone volume fraction (BV/TV%) of the distal femur showed a similar strain pattern (Fig. 2B), and the decline in BV/TV% was principally caused by a reduction in trabecular number (Fig. 2C) and not trabecular thickness (data not shown). Additionally, marrow fat increased with fat feeding in B6 mice but not 6T mice (Fig. 2D). Food intake and metabolic parameters such as oxygen consumption and CO2 production were measured. Whereas the kilojoule density of the three diets is different, the kilojoules consumed per mouse per day were not contingent on strain or diet (data not shown), indicating that the effects observed were as a consequence of the diet constituents and not the number of kilojoules consumed. The respiratory quotient displayed the appropriate changes with regard to fat intake and diurnal changes in both strains, and no significant differences were noted between the strains (data not shown).
FIG. 2.
Effects of dietary fat on bone and body composition in the B6 versus 6T strains. Brackets above the graphs denote statistically significant differences (p < 0.05) compared with the group fed the 11% fat diet. Comparisons were done within a strain by ANOVA. *Statistical significance when comparing the two strains fed the same diet, as determined by Student's t-test (p < 0.05). Stipled bars denote B6 and white bar denote 6T. BV/TV% and trabecular number were assessed by μCT. B6 and 6T mice reacted very differently with regards to aBMD (A) and BV/TV% (B) when fed diets containing differing %KJ from fat. Body weight was used as a covariate for aBMD. Neither femoral length nor body weight was found to be an appropriate covariate in the fit model for either BV/TV% or trabecular number. The decrease in BV/TV% associated with the increase in dietary fat seen in 6T was primarily caused by a decrease in trabecular number (C), but no change in trabecular thickness was observed (data not shown). There was an increase in the absolute number of adipocytes in the distal femur of B6 fed the higher %KJ from fat diets, whereas there was no change observed in 6T (D).
Interaction between PPARG alleles and dietary fat in the Framingham Offspring cohort
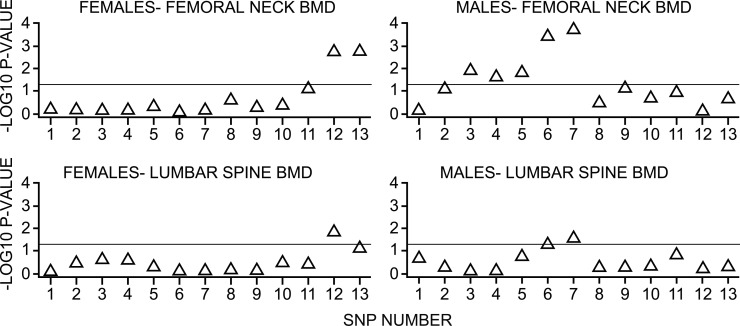
To evaluate these findings in humans, a subset of unrelated men (n = 867) and women (n = 925) from the Framingham Offspring cohort was studied (Table 3). Participants were genotyped for the 13 SNPs in the PPARG gene listed in Table 1. Each of the 13 SNPs was examined for sex-specific interaction with percent of energy intake from total fat on aBMD of the femoral neck. Figure 3 shows the p values for interaction between 13 SNPs across the PPARG gene and dietary fat intake above versus below the median. In men, at the femoral neck, there were significant interactions between SNPs 3, 4, 5, 6, and 7 and fat intake (Fig. 3, right). In women, at the femoral neck, there were significant interactions between SNPs 12 and 13 and fat intake (Fig. 3, left). Similar results were observed for aBMD of the trochanter and the lumbar spine (data not shown).
Table 3.
Characteristics of Subjects Participating in the Framingham Offspring Cohort Osteoporosis Study
| Characteristics | All (N = 1792) | Males (N = 867) | Females (N = 925) |
| Age (yr) | 61.3 ± 9.1 | 62.2 ± 9.1 | 60.5 ± 9.1 |
| BMI (kg/m2) | 28.1 ± 5.1 | 28.7 ± 4.6 | 27.5 ± 5.5 |
| Height (in) | 66.1 ± 3.7 | 68.8 ± 2.7 | 63.5 ± 2.5 |
| Estrogen status positive* | 439 (47.5%) | — | 439 (47.5%) |
| Diabetes [N (%)]† | 185 (10.3%) | 113 (13.0%) | 72 (7.8%) |
| Total caloric intake | 1856.35 ± 577.22 | 1979.15 ± 593.99 | 1741.85 ± 536.69 |
| Percent caloric intake from total fat (median) | 27.20 ± 5.61 (27.31) | 27.40 ± 5.61 (27.49) | 27.01 ± 5.60 (27.04) |
| Femoral neck BMD | 0.93 ± 0.15 | 0.98 ± 0.14 | 0.88 ± 0.14 |
Values are mean ± SD, unless otherwise specified.
* Estrogen positive includes premenopausal women and postmenopausal women taking estrogen.
† Diabetes defined as fasting glucose ≥140 or on medication for diabetes.
FIG. 3.
p values for interaction between fat intake and PPARG SNPs. p values for interaction between 13 SNPs across the PPARG gene and dietary fat intake (%KJ in the diet obtained from fat) above vs. below median. The y-axis provides the –log10 of the p values such that values above the line are <0.05. The top two panels present data from the femoral neck, whereas the bottom two panels present data for the lumbar spine.
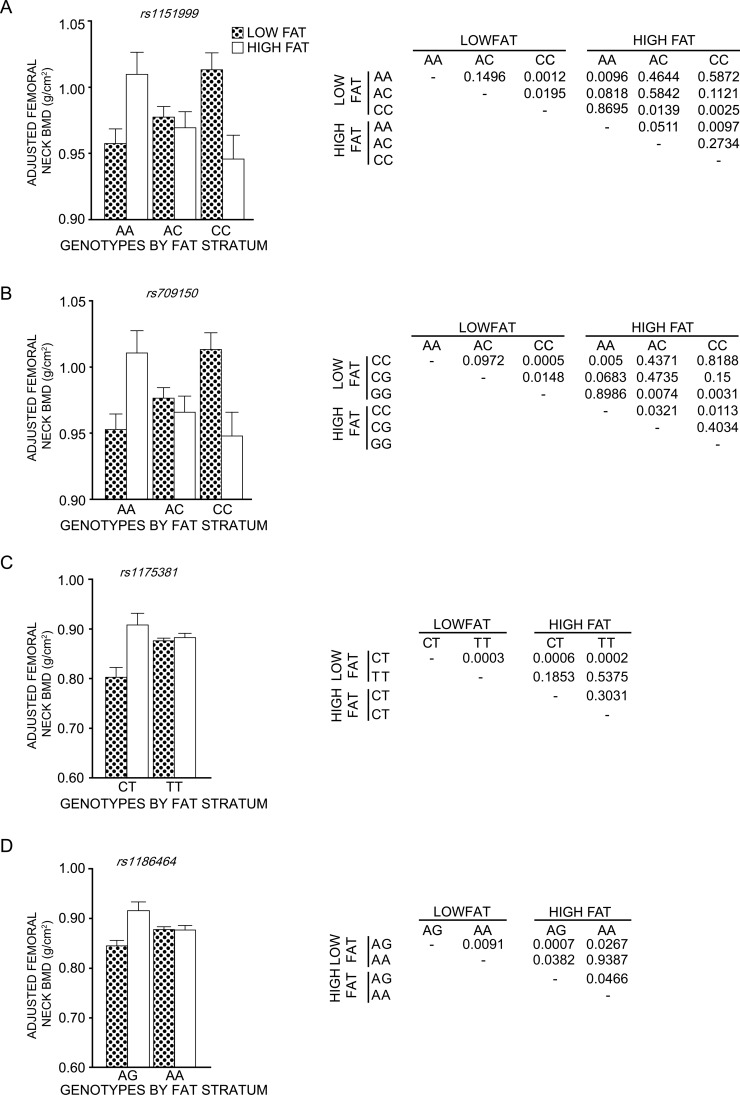
As shown in Fig. 4, the effect of dietary fat was clearly dependent on the alleles at a given SNP. For example, compared with men on a low-fat diet, men on a high-fat diet had lower BMD of the femoral neck when homozygous for the C allele for SNP 6 (Fig. 4A) but a higher BMD of the femoral neck when homozygous for the A allele at this same SNP. An identical pattern was observed for SNP 7 in men (data not shown). For SNP13, in women (Fig. 4C), the AA genotype yielded lower BMD with the high-fat intake than did the AG or GG combined group. Conversely, the AA genotype group yielded higher BMD with low-fat intake, indicating that fat intake affected the two genotypes differentially. A similar observation was made for SNP 12, wherein the TT genotype in women yielded higher aBMD with low-fat intake (Fig. 4B).
FIG. 4.
Diet by SNP allele interactions in the Framingham Offspring cohort. A significant SNP allele by percent energy derived from dietary fat interaction was observed for SNP 6 (A, rs1151999) and SNP 7 (B, rs709150) in men and for SNP 12 (C, rs1175381) and SNP 13 (D, rs1186464) in women. SNPs 6 and 7 are in strong linkage disequilibrium (LD), with a D′ of 0.996 and an r 2 of 0.976, and as a consequence, yielded virtually the same result. In addition, a D′ of 1 and an r 2 of 0.338 were noted for SNPs 12 and 13. The p values for the interaction between a given SNP and energy intact (%KJ from total fat) were as follows: SNP 6, p = 0.0004; SNP7, p = 0.0002; SNP12, p = 0.002; SNP13, p = 0.002). This interaction is visually presented in for each SNP in the graph. The results to the right of each graph shows the p values obtained from pairwise comparisons by fat stratum and genotypes.
DISCUSSION
There are 1130 known or predicted genes on the sixth chromosome in mice, with 630 of these genes located within the 6T congenic region. Using block haplotyping, expression profiling, and by examining the existence of the coding region SNPs, we eliminated all but 36 of these gene as candidates for our BMD and serum IGF-1 QTLs. By examination of the published data that exist for these 36 genes, we were able to determine that 5 of these genes had a role in bone biology: Gpnmb, Adamts9, Pparg, Raf1, and Zfp422.(29–33) Gpnmb, also known as osteoactivin, is expressed in osteoblasts and may play a role in BMP2 signaling.(29) Adamts9 expression has been found in chondrocytes,(30) as has expression of Raf1.(31) The expression of Zfp422 has been found in craniofacial bones, and it is thought to play a role in tooth eruption.(32) The fifth gene was Pparg.
The PPARG protein is the third member of the PPAR family of nuclear receptors. PPARG forms a heterodimer with the retinoid X receptor α (RXRa), and this heterodimer binds to DNA to induce gene transcription.(36) This nuclear receptor is essential for adipocyte differentiation, as has been shown in two separate in vivo mouse models.(37,38) PPARG has been shown to play a key role in the maturation of marrow mesenchymal stem cells (MSCs) into either adipocytes or osteoblasts.(39) The haploinsufficient Pparg mouse (Ppargtm1Tka) has been shown to have high cancellous bone volume and high BMD.(33) Activation of PPARG in Swiss-Webster mice and in older male B6 mice with the exogenous ligand rosiglitazone results in increases in marrow adiposity that are coincident with bone loss.(40,41)
One of the most striking phenotypes of our 6T congenic mouse was the increase in marrow adiposity.(7) Because PPARG protein has a known role in the differentiation of MSC into adipocytes(42) and overexpression of Pparg has been shown to suppress IGF-1 secretion in UAMS-γ33 cells,(43) we focused on the Pparg gene as the most likely candidate gene for our QTLs. Because dietary fat is a known ligand for PPARG,(34,35) we challenged our congenic mouse with a high-fat diet and found that increased dietary fat intake was associated with decreased aBMD and BV/TV% in the 6T mouse. No effect on bone was observed in B6 mice, suggesting a strong diet by gene interaction for this phenotype. We directly translated these findings to humans by examining this interaction of dietary fat by PPARG allele in the Framingham Osteoporosis study. We found that there was indeed a strong interaction between SNPs in the PPARG gene and dietary fat in both men and women for the phenotypes of aBMD of the trochanter, femoral neck, and lumbar spine.
Pparg is likely not the only gene that contributes to the final phenotype of the 6T congenic strain, nor is this gene likely the only gene that contributes to the Bmd8 and Igfsl1 QTLs. Increasing evidence has suggested that a QTL may be a cluster of genes that together contribute to a phenotype.(44,45) In this study, five genes were found by genetic and expression analysis to be possible candidate genes for the QTLs of Bmd8 and Igfsl1. Interestingly, three of these genes, Pparg, Raf1, and Zfp422, all are located within the same haplotype block. Future studies will investigate the gene networks that are involved in the phenotypes of this congenic mouse.
Another gene found within the same haplotype block as Pparg is the Alox5 gene. This gene was recognized as a potential candidate gene for a BMD QTL identified in a cross between the B6 and DBA/2J inbred mouse strains.(46) The protein product of this gene, 5-lipoxygenase, functions in the synthesis of leukotrienes from arachidonic acid, and arachidonic acid is a known endogenous ligand for PPARG.(47) The DBA/2J strain contains a nonsynonymous SNP that converts residue 646 from the B6-like valine to an isoleucine (rs30121304),(48) and this polymorphism has been reported to render the 5-lipoxygenase enzyme nonfunctional.(49) Unlike the DBA/2J strain, the C3H/HeJ strain is B6-like for this SNP. This suggests that, whereas these two QTLs influence the same pathway, the affected genes are different. One recent study has examined SNPs in the ALOX5 gene for association with BMD in a large cohort of both men and women. No association was found in either sex, but environmental factors were not examined in that analysis.(50)
The identification of candidate genes and proof that a candidate gene is responsible for the phenotype associated with a given QTL has been the subject of many papers and reviews. The “White Paper” written by 80 members of the international Complex Trait Consortium has thoroughly addressed this issue.(51) It has generally been concluded that several lines of evidence are required as proof for a given candidate gene, including: coding or regulatory region polymorphisms, appropriate gene function, functional studies in an in vivo model, and proof of principle in another species, preferably humans.(51) In this study, we believe that we have met many of these burdens of proof. More specifically, we found several polymorphisms in the promoter regions of the Pparg gene in the C3H donor strain for our 6T congenic. The PPARG protein has a documented role in bone biology,(33) in MSC differentiation and trafficking,(42,52) and in the regulation of IGF-1.(43) In an in vivo animal model, we showed a strong gene by diet interaction for the phenotype of aBMD. In our human study cohort, we were able to show a similar interaction between dietary fat intake and PPARG genotypes in association with aBMD.
There were several potential limitations of our approach. First, the methodology used to find our candidate gene in the mouse was restricted by the reliability of the databases and sources used. For example, genes containing nonsynonymous SNPs were included as candidate genes. It is possible that our selection of candidate genes was not complete, because nonsynonymous SNPs in the C3H strain were not represented in the available databases. In addition, genes were included or excluded based on the presence or absence of expression differences at the transcript level. Because transcription does not necessarily reflect translation, genes could have been inappropriately excluded. Notwithstanding these limitations, the focus of this paper was not to find every gene that could contribute to our QTL but rather to generate a manageable list of genes that should be explored further in our human study cohort.
In summary, we established a nutrient by gene interaction in mice and humans, indicating that a high-fat diet may be detrimental or beneficial to bone mass depending on the presence of specific allelic variants in the PPARG gene. Three studies have examined PPARG as a candidate gene for BMD in humans. In the first two studies, the rs3856806 SNP in exon 6 was investigated. Ogawa et al.(53) found a positive association with the T allele for this SNP and BMD, yet Rhee et al.(54) found no association. More recently, Casado-Diaz et al.(55) found no association between BMD and alleles of the PPARG SNP, rs10865710. Our findings of strong gene by diet interaction may partially explain these conflicting/negative results, because diet was not considered as a co-factor in these three studies. Our findings emphasize the need to understand the effects of diet on PPARG activation on the skeleton.
ACKNOWLEDGMENTS
The authors thank Jesse Hammer for assistance in the preparation of this manuscript. This work was funded by the following: NIH Grants AR43433, AR054604, AG030910-01, AR/AG41398, AR050066, DK042424, DK045227, DK073267, and HL54776 and National Heart, Lung and Blood Institute's Framingham Heart Study (Contract N01-HC-25195). Genotyping in the Framingham Study was supported by the Program in Genomic Applications (CardioGenomics Project U01 HL 66582). In addition, this study was supported by the following contracts: 53-K06-5-10 from NIH and 58-1950-9-001 from the U.S. Department of Agriculture Research Service.
Footnotes
The authors state that they have no conflicts of interest.
REFERENCES
- 1.Ralston S, de Crombrugghe B. Genetic regulation of bone mass and susceptibility to osteoporosis. Genes Dev. 2006;20:2492–2506. doi: 10.1101/gad.1449506. [DOI] [PubMed] [Google Scholar]
- 2.Williams F, Spector T. Recent advances in the genetics of osteoporosis. J Musculoskelet Neuronal Interact. 2006;6:27–35. [PubMed] [Google Scholar]
- 3.Huang QY, Recker RR, Deng HW. Searching for osteoporosis genes in the post-genome era: Progress and challenges. Osteoporos Int. 2003;14:701–715. doi: 10.1007/s00198-003-1445-9. [DOI] [PubMed] [Google Scholar]
- 4.Beamer WG, Shultz KL, Donahue LR, Churchill G, Sen S, Wergedal JR, Baylink DJ, Rosen CJ. Quantitative trait loci for femoral and lumbar vertebral bone mineral density in C57BL/6J and C3H/HeJ inbred strains of mice. J Bone Miner Res. 2001;16:1195–1206. doi: 10.1359/jbmr.2001.16.7.1195. [DOI] [PubMed] [Google Scholar]
- 5.Rosen CJ, Churchill G, Donahue LR, Shultz K, Burgess JK, Powell DR, Ackert C, Beamer WG. Mapping quantitative trait loci for serum insulin-like growth factor-I levels in mice. Bone. 2000;27:521–528. doi: 10.1016/s8756-3282(00)00354-9. [DOI] [PubMed] [Google Scholar]
- 6.Bouxsein ML, Rosen CJ, Turner CH, Ackert C, Shultz K, Donahue LR, Churchill G, Adamo ML, Powell DR, Turner RT, Muller R, Beamer WG. Generation of a new congenic mouse strain to test the relationship among serum insulin-like growth factor I, bone mineral density and skeletal morphology in vivo. J Bone Miner Res. 2002;17:570–579. doi: 10.1359/jbmr.2002.17.4.570. [DOI] [PubMed] [Google Scholar]
- 7.Rosen CJ, Ackert-Bicknell C, Adamo ML, Shultz K, Rubin J, Donahue LR, Horton L, Delahunty KM, Beamer WG, Sipos J, Clemmons D, Nelson T, Bouxsein ML, Horowitz M. Congenic mice with low serum IGF-1 have increased body fat, reduced bone mineral density, and an altered osteoblast differentiation program. Bone. 2004;35:1046–1058. doi: 10.1016/j.bone.2004.07.008. [DOI] [PubMed] [Google Scholar]
- 8.Deng HW, Xu FH, Huang QY, Shen H, Deng H, Conway T, Liu YJ, Lui YZ, Li JL, Zhang HT, Davies KM, Recker RR. A whole-genome linkage scan suggests several genomic regions potentially containing quantitative trait loci for osteoporosis. J Clin Endocrinol Metab. 2002;87:5151–5159. doi: 10.1210/jc.2002-020474. [DOI] [PubMed] [Google Scholar]
- 9.Lee YH, Rho YH, Choi SJ, Ji JD, Song GG. Meta-analysis of genome-wide linkage studies for bone mineral density. J Hum Genet. 2006;51:480–486. doi: 10.1007/s10038-006-0390-9. [DOI] [PubMed] [Google Scholar]
- 10.Ioannidis J, Ng M, Sham P, Zintzaras E, Lewis C, Deng H, Econs M, Karasik D, Devoto M, Kammerer C, Spector T, Andrew T, Cupples L, Foroud T, Kiel D, Koller D, Langdahl B, Mitchell B, Peacock M, Recker R, Shen H, Sol-Church K, Spotila L, Uitterlinden A, Wilson S, Kung A, Ralston S. Meta-analysis of genome wide scans provides evidence for gender and site specific regulation of bone mass. J Bone Miner Res. 2007;22:173–183. doi: 10.1359/jbmr.060806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Corwin R, Hartmen T, Maczuga S, Graubard B. Dietary saturated fat is inversely associated with bone density in humans: Analysis of NHANES III. J Nutr. 2006;136:159–165. doi: 10.1093/jn/136.1.159. [DOI] [PubMed] [Google Scholar]
- 12.Brownbill R, Ilich J. Lipid profile and bone paradox: Higher serum lipids are asociated with higher bone mineral density in postmenopausal women. J Womens Health. 2006;15:261–270. doi: 10.1089/jwh.2006.15.261. [DOI] [PubMed] [Google Scholar]
- 13.Kruger M, Coetzer H, de Winter R, Gericke G, van Papendorp D. Calcium, gamma-linolenic acid and eicosapentaenoic acid supplementation in senile osteoporosis. Aging Clin Exp Res. 1998;10:385–394. doi: 10.1007/BF03339885. [DOI] [PubMed] [Google Scholar]
- 14.Bassey E, Littlewood J, Rothwell M, Pye D. Lack of effect of supplementation with essential fatty acids on bone mineral density in healthy pre- and postmenopausal women: Two randomized control trials of Efacal v. calcium alone. Br J Nutr. 2000;83:629–635. doi: 10.1017/s0007114500000805. [DOI] [PubMed] [Google Scholar]
- 15.Svenson K, Bogue M, Peters L. Identifying new mouse models of cardiovascular disease: A review of high-throughput screens of mutagenized and inbred strains. J Appl Physiol. 2003;94:1650–1659. doi: 10.1152/japplphysiol.01029.2003. [DOI] [PubMed] [Google Scholar]
- 16.Ackert-Bicknell C, Salisbury J, Horowitz M, Demambro V, Horton L, Shultz KL, Lecka-Czernik B, Rosen CJ. A chromosomal inversion within a Quantitative Trait Locus has a major effect on adipogenesis and osteoblastogenesis. Ann NY Acad Sci. 2008;1116:291–305. doi: 10.1196/annals.1402.010. [DOI] [PubMed] [Google Scholar]
- 17.Park Y, Clifford R, Buetow K, Hunter K. Multiple cross and inbred strain haplotype mapping of complex-trait candidate genes. Genome Res. 2003;13:118–121. doi: 10.1101/gr.786403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Delahunty KM, Shultz KL, Gronowicz G, Koczon-Jaremko B, Adamo ML, Horton L, Lorenzon J, Donahue LR, Ackert-Bicknell C, Kream B, Beamer WG, Rosen CJ. Congenic mice provide in vivo evidence fr a genetic locus that modulates serum IGF-1 and bone acquisition. Endocrinology. 2006;147:3915–3923. doi: 10.1210/en.2006-0277. [DOI] [PubMed] [Google Scholar]
- 19.Ferrari S, Karasik D, Liu J, Karamohamed S, Herbert A, Cupples LA, Kiel DP. Interactions of interleukin-6 promoter polymorphisms with dietary factors and their associations with bone mass in men and in women from the Framingham Osteoporosis Study. J Bone Miner Res. 2004;19:552–559. doi: 10.1359/JBMR.040103. [DOI] [PubMed] [Google Scholar]
- 20.Karasik D, Myers R, Cupples LA, Hannan MT, Gagnon DR, Herbert A, Kiel DP. Genome screen for quantitative trait loci contributing to normal variation in bone mineral density: The Framingham Study. J Bone Miner Res. 2002;17:1718–1727. doi: 10.1359/jbmr.2002.17.9.1718. [DOI] [PubMed] [Google Scholar]
- 21.Hannan MT, Tucker KL, Dawson-Hughes B, Cupples LA, Felson D, Kiel DP. Effect of dietary protein on bone loss in elderly men and women: The Framingham Osteoporosis Study. J Bone Miner Res. 2000;15:2504–2512. doi: 10.1359/jbmr.2000.15.12.2504. [DOI] [PubMed] [Google Scholar]
- 22.London S, Sacks F, Caesar J, Stampfer M, Siguel E, Willett W. Fatty acid composition of subcutaneous adipose tissue and diet in postmenopausal US women. Am J Clin Nutr. 1991;54:340–345. doi: 10.1093/ajcn/54.2.340. [DOI] [PubMed] [Google Scholar]
- 23.Garland M, Sacks F, Colditz G, Rimm E, Sampson L, Willett W, Hunter D. The relationship between dietary intake and adipose tissue composition of selected fatty acids in US women. Am J Clin Nutr. 1998;67:25–30. doi: 10.1093/ajcn/67.1.25. [DOI] [PubMed] [Google Scholar]
- 24.Hunter D, Rimm E, Sacks F, Stampfer M, Colditz G, Litin L, Willett W. Comparison measures of fatty acid intake by subcutaneous fat aspirate, food frequency questionnaire, and diet records in a free-living population of US men. Am J Epidemiol. 1992;135:418–427. doi: 10.1093/oxfordjournals.aje.a116302. [DOI] [PubMed] [Google Scholar]
- 25.Willett W, Stampfer M, Chu N, Spiegelman D, Holmes M, Rimm E. Assessment of questionnaire validity for measuring total fat intake using plasma lipid levels as criteria. Am J Epidemiol. 2001;154:1107–1112. doi: 10.1093/aje/154.12.1107. [DOI] [PubMed] [Google Scholar]
- 26.Meirhaeghe A, Fajas L, Helbecque N, Cottel D, Lebel P, Dallongeville J, Deeb S, Auwerx J, Amouyel P. A genetic polymorphism of the peroxisome proliferator-activated receptor gamma gene influences plasma leptin levels in obese humans. Hum Mol Genet. 1998;7:435–440. doi: 10.1093/hmg/7.3.435. [DOI] [PubMed] [Google Scholar]
- 27.Knouff C, Auwerx J. Peroxisome proliferator-activated receptor-gamma calls for activation in moderation: Lessons from genetics and pharmacology. Endocr Rev. 2004;25:899–918. doi: 10.1210/er.2003-0036. [DOI] [PubMed] [Google Scholar]
- 28.Stram D, Haiman C, Hirschhorn J, Altshuler D, Kolonel L, Henderson B, Pike M. Choosing haplotype-tagging SNPS based on unphased genotype data using preliminary sample of unrelated subjects with an example from the Multiethnic Cohort Study. Hum Hered. 2003;51:27–36. doi: 10.1159/000071807. [DOI] [PubMed] [Google Scholar]
- 29.Abdelmagid S, Barbe M, Arango-Hisijara I, Owen T, Popoff S, Safadi F. Osteoactivin acts as downstream mediator of BMP-2 effects on osteoblast function. J Cell Physiol. 2007;210:26–37. doi: 10.1002/jcp.20841. [DOI] [PubMed] [Google Scholar]
- 30.Davidson R, Waters J, Kevorkian L, Darrah C, Cooper A, Donell S, Clark I. Expression profiling of metalloproteinases and their inhibitors in synovium and cartilage. Arthritis Res Ther. 2006;8:R124. doi: 10.1186/ar2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kaneko Y, Tanzawa H, Sato K. The proto-oncogene C-raf-1 is highly expressed only in the hypertrophic zone of the growth plate. Calcif Tissue Int. 1994;54:426–430. doi: 10.1007/BF00305531. [DOI] [PubMed] [Google Scholar]
- 32.Gao Y, Kobayashi H, Ganss B. The human KROX-26/ZNF22 gene is expressed at sites of tooth formation and maps to the locus for permanent tooth agenesis (He-Zhao deficiency) J Dent Res. 2003;82:1002–1007. doi: 10.1177/154405910308201213. [DOI] [PubMed] [Google Scholar]
- 33.Akune T, Ohba S, Kamekura S, Yamaguchi M, Chung U-i, Kubota N, Terauchi Y, Harada Y, Azuma Y, Nakamura K, Kadowaki T, Kawaguchi H. PPARgamma insufficiency enhances osteogenesis through osteoblast formation from bone marrow progenitors. J Clin Invest. 2004;113:846–855. doi: 10.1172/JCI19900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Inoue M, Ohtake T, Motomura W, Takahashi N, Hosoki Y, Miyoshi S, Suzuki Y, Saito H, Kohgo Y, Okumura T. Increased expression of PPARgamma in high fat diet-induced liver steatosis in mice. Biochem Biophys Res Commun. 2005;336:215–222. doi: 10.1016/j.bbrc.2005.08.070. [DOI] [PubMed] [Google Scholar]
- 35.Yu S, Matsusue K, Kashireddy P, Cao W-Q, Yeldandi V, Yeldandi A, Rao M, Gonzalez FJ, Reddy JK. Adipocyte-specific gene expression and adipogenic steatosis in the mouse liver due to Peroxisome Proliferator-activated Receptor gamma1 (PPARgamma1) overexpression. J Biol Chem. 2003;278:498–505. doi: 10.1074/jbc.M210062200. [DOI] [PubMed] [Google Scholar]
- 36.Spiegelman BM. PPAR-gamma: Adipogenic regulator and thiazolidinedione receptor. Diabetes. 1998;47:507–514. doi: 10.2337/diabetes.47.4.507. [DOI] [PubMed] [Google Scholar]
- 37.Barak Y, Nelson MC, Ong ES, Jones YZ, Ruiz-Lozano P, Chien KR, Koder A, Evans RM. PPAR gamma is required for placental, cardiac, and adipose tissue development. Mol Cell. 1999;4:585–595. doi: 10.1016/s1097-2765(00)80209-9. [DOI] [PubMed] [Google Scholar]
- 38.Rosen E, Sarraf P, Troy AE, Bradwin G, Moore K, Milstone DS, Speigelman BM, Mortensen RM. PPAR gamma is required for the differentiation of adipose tissue in vivo and in vitro. Mol Cell. 1999;4:611–617. doi: 10.1016/s1097-2765(00)80211-7. [DOI] [PubMed] [Google Scholar]
- 39.Lecka-Czernik B, Suva L. Resolving the two “bony” faces of PPAR-gamma. PPAR Res. 2008;2006:27489. doi: 10.1155/PPAR/2006/27489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Afshan A, Weinstein RS, Stewart S, Parfitt AM, Manolagas SC, Jilka RL. Rosiglitazone causes bone loss in mice suppressing osteoblast differentiation and bone formation. Endocrinology. 2005;146:1226–1235. doi: 10.1210/en.2004-0735. [DOI] [PubMed] [Google Scholar]
- 41.Lazarenko O, Rzonca S, Hogue W, Swain F, Suva L, Lecka-Czernik B. Rosiglitazone induces decreases in bone mass and strength that are reminiscent of aged bone. Endocrinology. 2007;148:2669–2680. doi: 10.1210/en.2006-1587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lecka-Czernik B, Moerman EJ, Grant DF, Lehmann JM, Manolagas SC, Jilka RL. Divergent effects of selective peroxisome proliferator-activated receptor-gamma2 ligands on adipocyte versus osteoblast differentiation. Endocrinology. 2002;143:2376–2384. doi: 10.1210/endo.143.6.8834. [DOI] [PubMed] [Google Scholar]
- 43.Lecka-Czernik B, Ackert-Bicknell C, Adamo ML, Marmolejos V, Churchill GA, Shockley K, Reid I, Grey A, Rosen CJ. Activation of peroxisome proliferator-activated receptor gamma (PPARgamma) by rosiglitazone suppresses components of the IGF regulatory system in vitro and in vivo. Endocrinology. 2007;148:903–911. doi: 10.1210/en.2006-1121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Griffiths M, Remmers E. Genetic analysis of collagen-induced arthritis in rats: A polygenic model for rheumatoid arthritis predicts a common framework of cross-species inflammatory/autoimmune disease loci. Immunol Rev. 2001;184:172–183. doi: 10.1034/j.1600-065x.2001.1840116.x. [DOI] [PubMed] [Google Scholar]
- 45.de Haan G, Bystrykh L, Weersing E, Dontje B, Geiger H, Ivanova N, Lemischka I, Vellenga E, Van Zant G. A genetic and genomic analysis identifies a cluster of genes associated with hematopoietic cell turnover. Blood. 2002;100:2056–2062. doi: 10.1182/blood-2002-03-0808. [DOI] [PubMed] [Google Scholar]
- 46.Mehrabian M, Allayee H, Stockton J, Lum P, Drake T, Castellani L, Suh M, Armour C, Edwards S, Lamb J, Lusis A, Schadt E. Integrating genotypic and expression data in a segregating mouse population to identify 5-lipoxygenase as a susceptibility gene for obesity and bone traits. Nat Genet. 2005;37:1224–1233. doi: 10.1038/ng1619. [DOI] [PubMed] [Google Scholar]
- 47.Berger J, Moller D. The mechanisms of action of PPARs. Annu Rev Med. 2002;53:409–435. doi: 10.1146/annurev.med.53.082901.104018. [DOI] [PubMed] [Google Scholar]
- 48.Lyons M. Arachidonate 5-lipoxygenase variants in atherosclerosis, obesity, and bone traits. Circ Res. 2006;98:e66. doi: 10.1161/01.RES.0000219674.81938.63. [DOI] [PubMed] [Google Scholar]
- 49.Kuhn H, Anton M, Gerth C, Habenicht A. Amino acid differences in the deduced 5-lipoxygenase sequence of CAST atherosclerosis-resistance mice confer impaired activity when introduced into the human ortholog. Arterioscler Thromb Vasc Biol. 2003;23:1072–1076. doi: 10.1161/01.ATV.0000074167.01184.48. [DOI] [PubMed] [Google Scholar]
- 50.Foroud T, Ichikawa S, Koller DL, Lai D, Curry L, Edenberg H, Hui SL, Peacock M, Econs MJ. Association studies of ALOX5 and bone mineral density in healthy adults. Osteoporos Int. 2008;19:637–643. doi: 10.1007/s00198-007-0484-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Consortium Mot CT. The nature and identification of quantitative trait loci: A community's review. Nat Rev Genet. 2003;4:911–916. doi: 10.1038/nrg1206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Crossno J, Jr, Majka S, Grazia T, Gill R, Klemm D. Rosiglitazone promotes development of a novel adipocyte population from bone marrow-derived circulating progenitor cells. J Clin Invest. 2006;116:3220–3228. doi: 10.1172/JCI28510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ogawa S, Urano T, Hosoi T, Miyao M, Hoshino S, Fujita M, Shiraki M, Orimo H, Ouchi Y, Inoue S. Association of bone mineral density with a polymorphism of the peroxisome proliferator-activated receptor gamma expression in osteoblasts. Biochem Biophys Res Commun. 1999;260:122–126. doi: 10.1006/bbrc.1999.0896. [DOI] [PubMed] [Google Scholar]
- 54.Rhee E-J, Oh K-W, Lee W-Y, Kim S-Y, Oh E-S, Baek K-H, Kang M-I, Kim S-W. The effects of C161→T polymorphism in exon 6 of peroxisome proliferator-activated receptor-gamma gene on bone mineral metabolism and serum osteoprotegerin levels in healthy middle-aged women. Am J Obstet Gynecol. 2005;192:1087–1093. doi: 10.1016/j.ajog.2004.09.033. [DOI] [PubMed] [Google Scholar]
- 55.Casado-Diaz A, Cuenca-Acevedo R, Quesada JM, Dorado G. Individual single tube genotyping and DNA pooling by allele-specific PCR to uncover associations of polymorphisms with complex diseases. Clin Chim Acta. 2007;376:155–162. doi: 10.1016/j.cca.2006.08.014. [DOI] [PubMed] [Google Scholar]
- 56.Lecka-Czernik B, Gubrij I, Moerman EJ, Kajkenova O, Lipschitz DA, Manolagas SC, Jilka RL. Inhibition of Osf2/Cbfa1 expression and terminal osteoblast differentiation by PPARgamma2. J Cell Biochem. 1999;74:357–371. [PubMed] [Google Scholar]