Abstract
Many human genes are associated with dispersed arrays of transcriptional enhancers that regulate their expression in time and space. Studies in invertebrate model systems have suggested that these elements could function as discrete and independent regulatory units, but the in vivo combinatorial properties of vertebrate enhancers remain poorly understood. To explore the modularity and regulatory autonomy of human developmental enhancers, we experimentally concatenated up to four enhancers from different genes and used a transgenic mouse assay to compare the in vivo activity of these compound elements with that of the single modules. In all of the six different combinations of elements tested, the reporter gene activity patterns were additive without signs of interference between the individual modules, indicating that regulatory specificity was maintained despite the presence of closely-positioned heterologous enhancers. Even in cases where two elements drove expression in close anatomical proximity, such as within neighboring subregions of the developing limb bud, the compound patterns did not show signs of cross-inhibition between individual elements or novel expression sites. These data indicate that human developmental enhancers are highly modular and functionally autonomous and suggest that genomic enhancer shuffling may have contributed to the evolution of complex gene expression patterns in vertebrates.
Keywords: enhancer, cis-regulatory, combinatorial, evolution
Introduction
The regulation of many human genes is controlled by multiple discrete enhancer sequences with different tissue specificities (e.g. references [1; 2; 3; 4; 5; 6]). Such enhancers activate gene expression independent of their orientation [7] and are commonly scattered across large noncoding intervals [8; 9], in some extreme cases functioning >1 Mb from their gene promoter target [10; 11]. While progress towards genome-wide annotation of developmental enhancers has been made by coupling comparative genomic and chromatin immunoprecipitation-sequencing approaches to experimental studies in mice and fish [12; 13; 14; 15], the functional and evolutionary significance of their dispersed arrangement remains unclear. Structural modularity of enhancer architecture may facilitate evolutionary fine tuning of distinct aspects of expression patterns [16; 17], but observations in invertebrate models [18; 19] have also raised the possibility that intergenic translocation of preformed enhancer modules may have contributed to the evolution of complex gene expression patterns in vertebrates. However, an open question remains whether this proposed mechanism of regulatory evolution is feasible since it requires that enhancers accurately retain their individual activities when placed into a new genomic context.
Results
Heterologous Pairs of Enhancers Have Additive Regulatory Properties
To explore the prevalence of possible positive or negative interactions among human developmental enhancers, we recombined enhancer modules from different, functionally unrelated genes and studied their regulatory in vivo properties during embryonic development in transgenic mice. We selected for this purpose six in vivo validated enhancers [12; 13]; E1–E6, Fig. 1a; Suppl. Table 1). When individually coupled to a minimal heat shock protein 68 promoter linked to a LacZ reporter gene (Fig. 1b; [13; 20]), each of these elements drove reproducible tissue-specific expression in transgenic mouse embryos. Representative patterns observed with these single enhancers are shown in Fig. 1c. Of note, the enhancers were selected for analysis based on their expression patterns which are easily distinguished at the resolution of whole-mount staining, yet also include features that are located in close spatial proximity. In particular, activity patterns comprise different subregions of the developing forebrain (E2 and E6), the midbrain (E5 and E6), the hindbrain (E5 and E6), and three different subregions of the developing limb (E1, E3, and E4). A detailed overview of the strong reproducibility of these features in independent transgenic embryos, as well as additional reproducible features that were observed for the individual patterns, is provided in Suppl. Table 2. All elements are located on different human chromosomes and thereby are expected to regulate independent gene(s), with the exception of E1 and E2 which are within introns of different genes on chromosome 1 but are located more than 5Mb apart from each other. All elements are located at least 30kb away from the closest known promoter, with 4 elements found within genes (E1, E2, E3 and E6) and the remaining 2 elements found between genes (E4 and E5).
Figure 1. Spatial additivity of tissue-specific enhancers fused from different genes.
a) genomic environment of six conserved enhancers used in this study. A 50kb genomic interval bracketing each enhancer is shown, including intron/exon structure of overlapping genes (black) and conservation in 17 vertebrates (color shaded boxes; [26]). All enhancers included an ultraconserved core region (Suppl. Table 1; [27]). b) Single and compound enhancer constructs for in vivo testing. c) Enhancer activity of single elements at mouse embryonic day 11.5. d)-i) In vivo activity of heterologous compound enhancers. d) E1+E2, e) E2+E5, f) E3+E4, g) E1+E5, h) E5+E6, i) E1+E2+E5+E6. Only one representative embryo is shown for each single and compound pattern, see Suppl. Table 2 for reproducibility across independent transgenic animals.
These enhancers are experimentally defined by their ability to drive reporter gene expression in transgenic mouse embryos, but (as for most human developmental enhancers) it is unknown whether additional cis-regulatory cues such as general or tissue-specific repressor/silencer activities are also embedded in these elements that might affect the activity of other regulatory elements in their vicinity. To determine the combinatorial properties of these human enhancers, we generated five constructs containing pair-wise tandem fusions of the heterologous enhancers described above (Fig. 1b). For each construct, we obtained multipletransgenic embryos at e11.5 (representing 8 to 15 independent transgenic integration events, see Suppl. Table 2). In each of the cases studied we observed reproducible patterns that were a direct superimpositionof the two individual patterns (Fig. 1d–h). For instance, as one representative example, a construct containing E2 (forebrain) coupled to E5 (medial-dorsal and lateral cell populations of the midbrain and ventral hindbrain) targets reporter gene expression to the same respective subregions of the fore-, mid- and hindbrain as observed for E2 and E5 alone (Fig. 1c,e). Using the presence of reproducible staining in individual anatomical structures as a qualitative measure, we observed no disruption of enhancer activities in the tandem fusion constructs in comparison to each enhancer element alone (Suppl. Table 2). Conversely, the concatenation of heterologous enhancers did not result in any reproducible staining in additional anatomical structures or domains outside of those observed for the individual enhancer constructs. These data indicate that in all instances tested, the two enhancers retained their specificity independent of each other despite the artificial coupling of enhancer modules that regulate different genes.
Complex but Predictable Regulatory Output from a Heterologous Multi-Enhancer Array
To test whether more complex combinations of enhancer modules would result in positive or negative interactions, we concatenated four different enhancers from unrelated genes and tested the activity of this compound construct at embryonic day 11.5. Similar to the additive results observed in the combination of two discrete enhancers, independent transgenic embryos for the compound 4-mer construct had highly reproducible patterns that included all the major features of the individual patterns, while not introducing additional reproducibly stained structures (Fig. 1i, Suppl. Table 2). Thus, even when multiple distant-acting enhancers were combined in close proximity in a single construct, the individual enhancer units retained their distinct spatial specificities at this time-point.
Both Spatial and Temporal Regulatory Specificity are Preserved
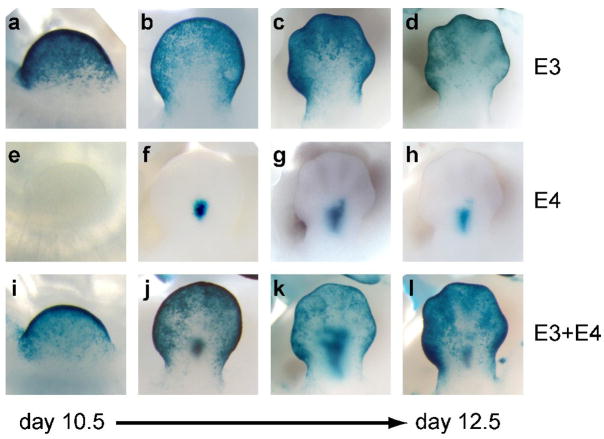
In addition to spatial properties, we also examined temporal aspects of expression driven by concatenated enhancers. We selected for this purpose construct E3+E4, containing a pair of enhancers that drove expression in the developing limb (Fig. 1f), where morphological changes enable precise developmental stage matching of independently generated transgenic embryos. When tested individually at stages ranging from e10.5 to e12.5, enhancer E3 alone targets expression to the apical ectodermal ridge (AER) and the surface ectoderm of the limb bud (Fig. 2a–d). In contrast, E4 alone does not drive limb staining at e10.5, but targets expression to a sharply restricted central cell population in the limb at e11.5 that continues throughout e12.5 (Fig. 2e–h). In order to compare the developmental progression of expression driven by constructs E3, E4, and E3+E4, we collected multiple transgenic embryos at e10.5, e11.5 and e12.5 for each construct. We found that the compound construct E3+E4 drove AER expression at all time-points examined, whereas the medial expression domain was first observed at e11.5 as with E4 alone, indicating that the developmental onset of expression driven by E4 is not affected by the presence of E3 in its immediate proximity. Taken together, these results indicate that the functional independence of human developmental enhancers and the absence of obvious regulatory interference among them could allow the generation of complex spatiotemporal expression patterns through modular intergenic recombination of enhancers.
Figure 2. Temporal and spatial additivity of individual enhancer activities within the developing limb.
a)-l) dorsal surface view of forelimb buds of individual embryos transgenic for E3 (a-d), E4 (e–h) and the compound enhancer E3+E4 (i–l). For each construct, embryos were collected at e10.5 (a,e,i), e11.5 (b,f,j), and e12.5 (c,d,g,h,k,l) and representative limbs were stage-matched based on morphology. The transgenic status of the embryo shown in e) was confirmed by genotyping.
Discussion
The high degree of regulatory autonomy observed in this study suggests that functional independence and spatiotemporal additivity are common features of human distant-acting enhancer modules. Only a limited number of permutations of a small subset of human enhancers were tested in the present study, therefore it remains to be determined whether these properties are a universal feature applying to all developmental enhancers. Moreover, the LacZ-based approach used in this study provides direct evidence for qualitative additivity of patterns, yet we cannot exclude the possibility that subtle quantitative differences in enhancer activities between single and compound constructs were not detectable. These potential limitations notwithstanding, our results indicate that emerging collections of human and other vertebrate enhancers [12; 13; 14; 15] provide a toolbox enabling the design of regulatory composites driving customized, complex in vivo expression patterns in a predictable manner due to the additive nature of the individual components.
These observations also have potential evolutionary implications, as it has been proposed that duplication of regulatory elements into new genomic locations may have contributed to the emergence of complex gene expression patterns [17; 21; 22]. Bona fide examples of intergenic enhancer shuffling in the human genome remain to be identified, but recent comparative genomic evidence suggests that exaptation of transposable genome elements occurred on a pervasive scale [23]. While some of these mobile elements gave rise to functional enhancers [24; 25], it remains uncertain whether such transposon-derived elements typically arose de novo or if partially preformed cis-regulatory functions were already embedded at the time of their translocation.
The remarkable modular additivity of spatial and temporal enhancer activities indicates that functional distant-acting enhancers, if translocated into new genomic environments, have the potential to transfer aspects of expression patterns between genes without disrupting the function of pre-existing enhancers. We expect that the combinatorial properties of human enhancers demonstrated through our experiments will provide confidence and a conceptual basis for constructing increasingly complex arrays of enhancers by homologous recombination in mice, which is expected to provide further insights into enhancer shuffling as a potential mechanism of vertebrate genome evolution.
Materials and Methods
Enhancer reporter constructs
All enhancer sequences were PCR amplified from human genomic DNA (Clontech) using the primers listed in Suppl. Table 1. PCR fragments were cloned into the pENTR plasmid (Invitrogen), transferred into an Hsp68-LacZ reporter vector containing a Gateway cassette using LR recombination (Invitrogen; [13; 20]) and sequence validated.
Compound enhancers
To generate compound enhancers, inserts from individual constructs were subcloned by standard molecular cloning techniques. Sequence and orientation of enhancers in the final constructs are indicated in Suppl. Table 1. Residual multiple cloning site fragments of up to 48bp residing between enhancers are also listed in Suppl. Table 1.
Transgenic mice
Transgenic mouse embryos were generated by pronuclear injection in accordance with protocols reviewed and approved by the Lawrence Berkeley National Laboratory Animal Welfare and Research Committee. Zygotes at 0.5dpc for pronuclear injection were collected from FVB strain donor females (Charles River) and, after injection, transferred into pseudopregnant CD-1 strain recipient females (Charles River). Embryos were collected and stained for LacZ activity as previously described [8].
Assessment of reporter gene expression
Only anatomical structures in which reporter gene expression was present in at least three embryos resulting from independent transgene integration events were considered reproducible. Reproducibilities for all patterns observed with individual and compound enhancers are listed in Suppl. Table 2.
Acknowledgments
The authors wish to thank Shyam Prabhakar, Rotem Sorek, Marcelo Nobrega, James Noonan and Nadav Ahituv for critical comments on the manuscript; Keith Lewis, Amy Holt, Ingrid Plajzer-Frick, Sengthavy Phouanenavong, and Sumita Bhardwaj for technical support. L.A.P./E.M.R. were supported by the Berkeley-PGA, under the Programs for Genomic Applications, funded by National Heart, Lung, & Blood Institute, and L.A.P. by the National Human Genome Research Institute. Research was performed under Department of Energy Contract DE-AC02-05CH11231, University of California, E.O. Lawrence Berkeley National Laboratory. A.V. was supported by an American Heart Association postdoctoral fellowship.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Simonet WS, Bucay N, Pitas RE, Lauer SJ, Taylor JM. Multiple tissue-specific elements control the apolipoprotein E/C-I gene locus in transgenic mice. J Biol Chem. 1991;266:8651–4. [PubMed] [Google Scholar]
- 2.Schwartz RJ, Olson EN. Building the heart piece by piece: modularity of cis-elements regulating Nkx2-5 transcription. Development. 1999;126:4187–92. doi: 10.1242/dev.126.19.4187. [DOI] [PubMed] [Google Scholar]
- 3.Burch JB. Regulation of GATA gene expression during vertebrate development. Semin Cell Dev Biol. 2005;16:71–81. doi: 10.1016/j.semcdb.2004.10.002. [DOI] [PubMed] [Google Scholar]
- 4.Abbasi AA, Paparidis Z, Malik S, Goode DK, Callaway H, Elgar G, Grzeschik KH. Human GLI3 Intragenic Conserved Non-Coding Sequences Are Tissue-Specific Enhancers. PLoS ONE. 2007;2:e366. doi: 10.1371/journal.pone.0000366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bender MA, Roach JN, Halow J, Close J, Alami R, Bouhassira EE, Groudine M, Fiering SN. Targeted deletion of 5′HS1 and 5′HS4 of the beta-globin locus control region reveals additive activity of the DNaseI hypersensitive sites. Blood. 2001;98:2022–7. doi: 10.1182/blood.v98.7.2022. [DOI] [PubMed] [Google Scholar]
- 6.Landry JR, Bonadies N, Kinston S, Knezevic K, Wilson NK, Oram SH, Janes M, Piltz S, Hammett M, Carter J, Hamilton T, Donaldson IJ, Lacaud G, Frampton J, Follows G, Kouskoff V, Gottgens B. Expression of the leukaemia oncogene Lmo2 is controlled by an array of tissue specific elements dispersed over 100kb and bound by Tal1/Lmo2, Ets and Gata factors. Blood. 2009 doi: 10.1182/blood-2008-11-187757. Epub ahead of print . [DOI] [PubMed] [Google Scholar]
- 7.Banerji J, Rusconi S, Schaffner W. Expression of a beta-globin gene is enhanced by remote SV40 DNA sequences. Cell. 1981;27:299–308. doi: 10.1016/0092-8674(81)90413-x. [DOI] [PubMed] [Google Scholar]
- 8.Nobrega MA, Ovcharenko I, Afzal V, Rubin EM. Scanning human gene deserts for long-range enhancers. Science. 2003;302:413. doi: 10.1126/science.1088328. [DOI] [PubMed] [Google Scholar]
- 9.de la Calle-Mustienes E, Feijoo CG, Manzanares M, Tena JJ, Rodriguez-Seguel E, Letizia A, Allende ML, Gomez-Skarmeta JL. A functional survey of the enhancer activity of conserved non-coding sequences from vertebrate Iroquois cluster gene deserts. Genome Res. 2005;15:1061–72. doi: 10.1101/gr.4004805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lettice LA, Heaney SJ, Purdie LA, Li L, de Beer P, Oostra BA, Goode D, Elgar G, Hill RE, de Graaff E. A long-range Shh enhancer regulates expression in the developing limb and fin and is associated with preaxial polydactyly. Hum Mol Genet. 2003;12:1725–35. doi: 10.1093/hmg/ddg180. [DOI] [PubMed] [Google Scholar]
- 11.Sagai T, Hosoya M, Mizushina Y, Tamura M, Shiroishi T. Elimination of a long-range cis-regulatory module causes complete loss of limb-specific Shh expression and truncation of the mouse limb. Development. 2005;132:797–803. doi: 10.1242/dev.01613. [DOI] [PubMed] [Google Scholar]
- 12.Visel A, Prabhakar S, Akiyama JA, Shoukry M, Lewis KD, Holt A, Plajzer-Frick I, Afzal V, Rubin EM, Pennacchio LA. Ultraconservation identifies a small subset of extremely constrained developmental enhancers. Nat Genet. 2008;40:158–60. doi: 10.1038/ng.2007.55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pennacchio LA, Ahituv N, Moses AM, Prabhakar S, Nobrega MA, Shoukry M, Minovitsky S, Dubchak I, Holt A, Lewis KD, Plajzer-Frick I, Akiyama J, De Val S, Afzal V, Black BL, Couronne O, Eisen MB, Visel A, Rubin EM. In vivo enhancer analysis of human conserved non-coding sequences. Nature. 2006;444:499–502. doi: 10.1038/nature05295. [DOI] [PubMed] [Google Scholar]
- 14.Woolfe A, Goodson M, Goode DK, Snell P, McEwen GK, Vavouri T, Smith SF, North P, Callaway H, Kelly K, Walter K, Abnizova I, Gilks W, Edwards YJ, Cooke JE, Elgar G. Highly conserved non-coding sequences are associated with vertebrate development. PLoS Biol. 2005;3:e7. doi: 10.1371/journal.pbio.0030007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Visel A, Blow M, Li Z, Zhang T, Akiyama J, Holt A, Plajzer-Frick I, Shoukry M, Wright C, Chen F, Afzal V, Ren B, Rubin E, Pennacchio L. ChIP-seq Accurately Predicts Tissue-Specific Activity of Enhancers. Nature. 2009;457:854–8. doi: 10.1038/nature07730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Davidson EH. The Regulatory Genome: Gene Regulatory Networks In Development And Evolution. Academic Press; Burlington, MA: 2006. [Google Scholar]
- 17.Carroll SB, Grenier J, Weatherbee S. From DNA to Diversity: Molecular Genetics and the Evolution of Animal Design. Wiley-Blackwell; Oxford: 2005. [Google Scholar]
- 18.Kirchhamer CV, Bogarad LD, Davidson EH. Developmental expression of synthetic cis-regulatory systems composed of spatial control elements from two different genes. Proc Natl Acad Sci U S A. 1996;93:13849–54. doi: 10.1073/pnas.93.24.13849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gray S, Szymanski P, Levine M. Short-range repression permits multiple enhancers to function autonomously within a complex promoter. Genes Dev. 1994;8:1829–38. doi: 10.1101/gad.8.15.1829. [DOI] [PubMed] [Google Scholar]
- 20.Kothary R, Clapoff S, Darling S, Perry MD, Moran LA, Rossant J. Inducible expression of an hsp68-lacZ hybrid gene in transgenic mice. Development. 1989;105:707–14. doi: 10.1242/dev.105.4.707. [DOI] [PubMed] [Google Scholar]
- 21.Britten RJ, Davidson EH. Repetitive and non-repetitive DNA sequences and a speculation on the origins of evolutionary novelty. Q Rev Biol. 1971;46:111–38. doi: 10.1086/406830. [DOI] [PubMed] [Google Scholar]
- 22.Kermekchiev M, Pettersson M, Matthias P, Schaffner W. Every enhancer works with every promoter for all the combinations tested: could new regulatory pathways evolve by enhancer shuffling? Gene Expr. 1991;1:71–81. [PMC free article] [PubMed] [Google Scholar]
- 23.Lowe CB, Bejerano G, Haussler D. Thousands of human mobile element fragments undergo strong purifying selection near developmental genes. Proc Natl Acad Sci USA. 2007;104:8005–10. doi: 10.1073/pnas.0611223104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Santangelo AM, de Souza FS, Franchini LF, Bumaschny VF, Low MJ, Rubinstein M. Ancient exaptation of a CORE-SINE retroposon into a highly conserved mammalian neuronal enhancer of the proopiomelanocortin gene. PLoS Genet. 2007;3:1813–26. doi: 10.1371/journal.pgen.0030166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bejerano G, Lowe CB, Ahituv N, King B, Siepel A, Salama SR, Rubin EM, Kent WJ, Haussler D. A distal enhancer and an ultraconserved exon are derived from a novel retroposon. Nature. 2006;441:87–90. doi: 10.1038/nature04696. [DOI] [PubMed] [Google Scholar]
- 26.Siepel A, Bejerano G, Pedersen JS, Hinrichs AS, Hou M, Rosenbloom K, Clawson H, Spieth J, Hillier LW, Richards S, Weinstock GM, Wilson RK, Gibbs RA, Kent WJ, Miller W, Haussler D. Evolutionarily conserved elements in vertebrate, insect, worm, and yeast genomes. Genome Res. 2005;15:1034–50. doi: 10.1101/gr.3715005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bejerano G, Pheasant M, Makunin I, Stephen S, Kent WJ, Mattick JS, Haussler D. Ultraconserved elements in the human genome. Science. 2004;304:1321–5. doi: 10.1126/science.1098119. [DOI] [PubMed] [Google Scholar]