Abstract
Genetic variation influences gene expression, and this can be efficiently mapped to specific genomic regions and variants. We used gene expression profiling of EBV-transformed lymphoblastoid cell lines of all 270 individuals of the HapMap consortium to elucidate the detailed features of genetic variation underlying gene expression variation. We find gene expression levels to be heritable and differentiation between populations in agreement with earlier small-scale studies. A detailed association analysis of over 2.2 million common SNPs per population (5% frequency HapMap) with gene expression identified at least 1348 genes with association signals in cis and at least 180 in trans. Replication in at least one independent population was achieved for 37% of cis- signals and 15% of trans- signals, respectively. Our results strongly support an abundance of cis- regulatory variation in the human genome. Detection of trans- effects is limited but suggests that regulatory variation may be the key primary effect contributing to phenotypic variation in humans. Finally, we explore a variety of methodologies that improve the current state of analysis of gene expression variation.
Understanding the molecular basis of human phenotypic variation is a key goal of human genetics, encompassing disease susceptibility, variable response to drugs and ultimately treatment and public health. Over the past decades studies have described and analyzed the genetic basis of human phenotypic variation ranging from whole organism phenotypes such as height 1, to molecular level phenotypes such as lipid levels 2,3. Previous studies have also investigated the effects of nucleotide variation in specific genes or genomic regions on complex and monogenic diseases. Recently, there has been an explosion of genome-wide studies examining the genetic basis of complex diseases by exploring the effects of genetic variation such as single nucleotide polymorphisms (SNPs) 4-7 and copy number variants (CNVs) 8-10 some of which are clearly in non-coding regions of the genome 4-7,11. Technological advances have now made genome-wide association studies a reasonable and affordable approach to the study of complex phenotypes 12.
While association methodologies can identify genomic regions harbouring the genetic variant(s) underlying disease and other phenotypes, on their own they provide very little insight into which is the functional variant and / or mechanism. There is a need for methodologies that allow both the interpretation of functional consequences of variants and the description of functionally important variants 13,14. Gene expression (i.e. transcription) level is a quantitative phenotype that is directly linked to DNA variation and can be affected by polymorphisms in cis regulatory regions 15-19 or exonic variants altering transcript stability or splicing 20. In addition, gene expression (or mRNA levels) can be measured accurately and consistently in tissues and cell lines in humans. Many studies have described the genetic basis of transcriptional variation and have convincingly demonstrated that it is a heritable trait 16,17,21. The highly-dense, recently-released phase II HapMap 22,23 now allows for the first time a fine-scale analysis of the genomic location and properties of the variants associated with gene expression. Furthermore, we can advance current knowledge by investigating the genetic basis of gene expression variation within and between populations as well as its implications and relationship to genome function.
In this study we used transcriptional profiling of the 270 individuals of the four HapMap populations and their genotypes of nearly 4 million SNPs described by the HapMap Consortium 22,23 to elucidate some of the key features of the genetics of gene expression. We provide new biological insights in terms of variable gene expression and the fine-scale genomic properties of cis- and trans-acting regulatory variants. We also present new methodological implementations that uncover shared functional genetic effects between populations, as well as describing the degree and characteristics of population differentiation with respect to genetic effects. Our analysis describes extensive replication of signals in multiple populations and provides the first comprehensive exploration of biological signals underlying regulatory associations
Results
Data generation and biological properties of the data
We quantified gene expression in the 270 individuals genotyped in the International HapMap Project with Illumina's human whole-genome expression (WG-6 version 1) arrays which contain 47,294 probes in 4 technical replicates as described in 19. The population samples include 30 Caucasian trios of Northern and Western European origin (CEU), 45 unrelated Chinese individuals from Beijing University (CHB), 45 unrelated Japanese individuals from Tokyo (JPT), and 30 Yoruba trios from Ibadan, Nigeria (YRI). Expression signal values were log2 transformed and normalized using quantile normalization across the four replicates of an individual followed by median normalization across all 270 individuals to allow comparison of expression values across populations 24,25.
We estimated the median and variance of each of the 47,294 probe types for each population, and analyzed the distribution of variance and median values of normalized values by Gene Ontology (GO) categories 26 after summarizing them in GO-slim categories 27. Specific GO-slim categories such as “chaperone regulatory activity” showed an excess of high variance of gene expression, while genes with extracellular function showed low levels of variation. “Chaperone regulatory activity” genes and “translational regulatory activity” genes had highest median expression levels across all populations. The latter category was mainly driven by very high levels of expression of ribosomal proteins.
We estimated heritability of expression phenotypes independently in the CEU and YRI trios by performing midparent-offspring regressions. Of the 47,294 analyzed probes,4,829 and 6,482 (10% and 13%, respectively) demonstrate heritability greater than 0.2 in CEU and YRI, respectively, with an overlap of 958 genes, while 154 CEU genes and 217 YRI had heritability higher than 0.5 with an overlap of just 9 genes. This suggests that even with only 30 trios per population we can detect a substantial number of heritable expression phenotypes, through the low heritability estimates indicate substantial additional non-genetic sources of variation in gene expression.
We tested for population differences in gene expression levels as recent studies have shown it to be considerable 16,28. To avoid potential age and batch effects occurring at the establishment of the cell lines, we used the trios to establish critical values of differentiation. This approach (see Supplementary Methods) results in a difference in median log2 values of 0.2 (16% difference in median expression levels). Using this threshold, we tested all pairs of populations, after pooling CHB and JPT into one population (ASN) and calculated the number of genes exceeding this threshold. In total, 5,359 genes exceeded the threshold in one or more of the three population pairs (Figure S1), with most of them different in only a single population pair as expected. If the number of genes expressed in lymphoblastoid cell lines (LCLs) is about 50% of the total (about 12,000 genes) then we estimate that the fraction of genes with significant gene expression differences between any two populations is between 17% and 29%. However, one needs to be cautious with such a result since we observed that the CEU population was the most divergent, while we expected it to cluster closer to ASN. This is likely due to the much older age of the CEU cell lines relative to the most recently established YRI and ASN cell lines.
For subsequent analyses, we defined a reduced set of 14,456 probes (13,643 distinct genes) selected on criteria of variance and population differentiation (see Supplementary Methods). This is smaller than our previously described set of 14925 probes (14072 genes) 19, because we removed probes with either multiple mapping positions (370) or with SNPs within them that generated false associations (99). All association analyses were performed with the 14,456 probe set corresponding to 13,643 genes.
Comparison of expression measurements between genome-wide and custom expression arrays
To maximize the power to detect genetic effects, it is important to establish that expression measurements are robust to experimental variables. We previously described expression data for 60 CEU cell lines (all of the CEU HapMap parents) generated with Illumina's low-density (~700 genes) custom arrays 18. A total of 539 probes on this custom array have identical sequence as probes on the genome-wide array, which allows direct comparison of expression signals across experiments. For probes where there was variable signal intensity across individuals, there is highly significant correlation between signals from the two experiments (Figure S2). Note that the RNA used in each experiment for a given individual was extracted from a different cell line batch. The RNA labelling, hybridization and normalization were also done independently. The high degree of correlation illustrates that the transcript level measurements are stable despite differential growth and treatment of cell lines and samples.
Cis-associations of gene expression with SNPs
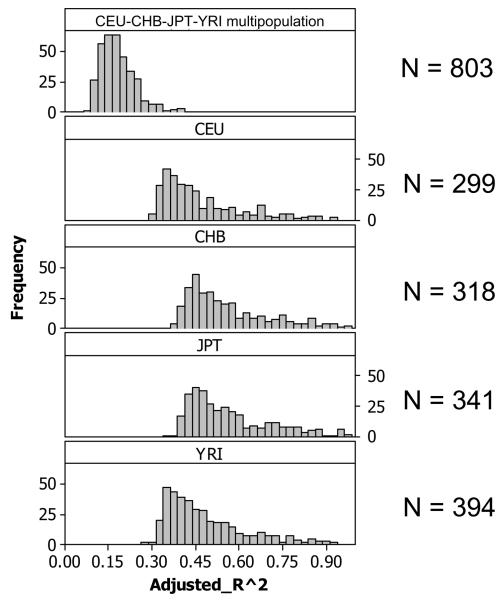
We selected HapMap phase II SNPs with minor allele frequency greater than 5% from each of the four populations (CEU, CHB, JPT, and YRI; approximately 2.2 million SNPs per population). We tested for association between SNP variation and expression variation for each of 13,643 genes independently in each population, considering unrelated individuals only. For each population, we employed a linear regression model. To analyze those SNPs potentially acting in cis, we tested only those SNPs located within 1Mb upstream or downstream of the expression probe midpoint (“candidate region approach” 18). For large genes ( > 500 Kb), we also used the transcription start site (TSS) as the center of the 2Mb window, discovering only one additional association relative to those discussed below. To determine significance of the regression p-values, we performed 10,000 permutations of the data independently for each gene (and population) and analyzed in depth those associations significant at the 0.001 permutation threshold (see Supplementary Methods). At this level of significance, we expect approximately 14 genes to have at least one significant association by chance, and we detect 299, 318, 341 and 394 for CEU, CHB, JPT and YRI, respectively, with a false discovery rate (FDR) of 4-5% per population (Table 1 and Table S1). In total, there is a non-redundant set of 831 genes exhibiting a significant cis- association in at least one population, 310 genes in at least two populations, and 62 in all four. As expected, due to the small sample size, the detected genetic effects are large, with R2 values ranging from 0.27 to close to 1. A total of 209 out of 299 CEU-significant genes and 247 out of 394 YRI-significant genes had heritability estimates above 0.2. Of the set of 831 genes with significant cis associations, 431 had heritability estimates above 0.2. Heritability overall correlated reasonably well with cis-association significance (Figure S4) but the heritability estimates have large variance and are to be taken with caution on a per gene basis. When evaluated in the context of GO-slim terms, we detected a significant deficit of genes involved in “cellular processes” among the set of genes with significant cis- associations. This is not surprising since segregating genetic variation affecting such basic cell functions would be expected to be detrimental.
Table 1.
Cis-associations detected with Linear Regression and Spearman rank Correlation.
| 0.001 | |||||
|---|---|---|---|---|---|
| linear regression |
Spearman | overlap | |||
| significant genes |
significant genes |
LR & Spearman |
% LR |
% Spear |
|
| CEU | 299 | 293 | 242 | 0.81 | 0.83 |
| CHB | 318 | 274 | 238 | 0.75 | 0.87 |
| JPT | 341 | 326 | 264 | 0.77 | 0.81 |
| YRI | 394 | 363 | 302 | 0.77 | 0.83 |
|
Non-
redundant |
831 | 783 | 642 | ||
| 4 pops | 62 | 57 | 46 | ||
| >= 2 pops | 310 | 283 | 239 | ||
| 0.01 | |||||
|
linear regression |
Spearman | overlap | |||
|
significant genes |
significant genes |
LR & Spearman |
% LR
|
% Spear
|
|
| CEU | 606 | 591 | 450 | 0.74 | 0.76 |
| CHB | 634 | 598 | 460 | 0.73 | 0.77 |
| JPT | 679 | 667 | 521 | 0.77 | 0.78 |
| YRI | 742 | 730 | 564 | 0.76 | 0.77 |
|
Non-
redundant |
1746 | 1737 | 1282 | ||
| 4 pops | 114 | 99 | 87 | ||
| >= 2 pops | 533 | 513 | 423 | ||
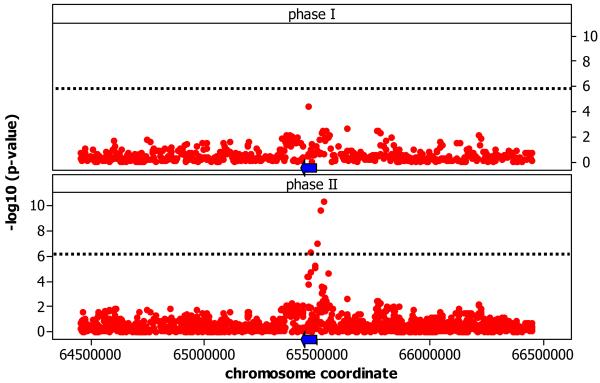
When we compare the sets of genes exhibiting cis- associations identified using the phase I HapMap 19 to those identified using the phase II HapMap data, we observe that the phase I HapMap captured 79%-87% (depending on the population) of the genes detected with phase II. Only in YRI is there is a substantial gain in numbers of cis- associated genes (Figure S5 and Figure 1A). This is due to the fact that in the other three populations, linkage disequilibrium (LD) decays much more slowly, so that instead of capturing the vast majority of common haplotype diversity, in the YRI the phase II captures additional functional genetic variation relative to phase I HapMap.
Figure 1.
Associations of SNPs with gene expression of SPRED2 on chromosome 2. Panels contrast the results obtained using phase I HapMap SNPs and phase II HapMap SNPs. Coordinates are in NCBI Build 35. Blue arrows represent the location (not to scale) and direction of transcription of the associated gene.
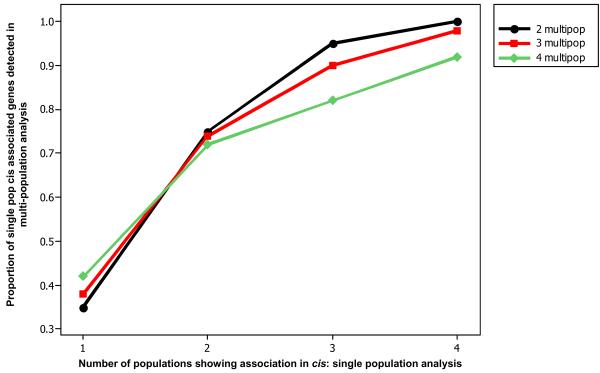
It would be desirable to be able to use the full sample of all 210 unrelated individuals to detect genetic effects on gene expression that are common but of smaller magnitude than those detected within each individual population. However, one cannot simply pool the samples without appropriate corrections, since population differentiation will generate spurious associations. Conditional permutations allow us to reveal the relevant associations while masking inflated associations 29,30. We repeated the association analysis after pooling unrelated individuals of: i) all four populations, ii) a subset of three (CEU-CHB-JPT) populations, and iii) two (CHB-JPT) populations. The rationale for the choice of population combinations was to pool those sets of populations that are more closely-related. To correct for inflation of the p-values, we performed conditional permutations, such that expression values from an individual of a given population were only assigned to another individual of the same population. This corrects for the p-value inflation since p-values from permuted datasets are also inflated. A total of 803, 735 and 651 genes were detected as significant for the 4-population, 3-population and 2-population pools, corresponding to 1083 distinct genes and FDR at 1-2%. The overlap between the multi-population and the single population analysis (Figure 2A) demonstrates that this methodology captures the vast majority of population shared associations that were detected in the single population analysis – as expected - but also captures a large number of additional cis effects (example in Figure 2B). Most of the effects detected in the multi-population analysis are smaller (R2 = 0.08-0.41) than those detected in single populations (Figure 2C), which is a direct function of the increase in the analyzed sample size.
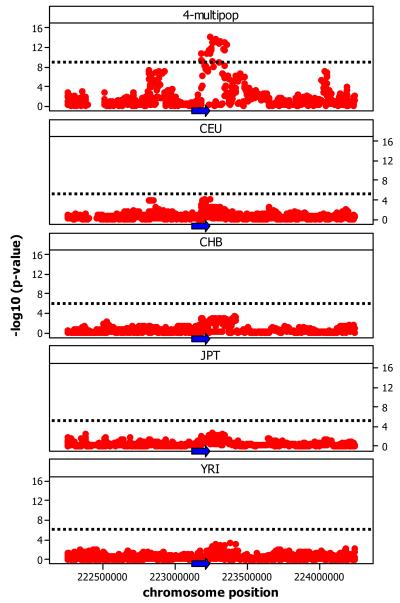
Figure 2.
Comparison of detected cis associations between single and multi-population analysis. (A) Numbers of genes with significant cis- associations as uncovered by single and multi-population analysis and proportion overlap of associations across the two methodologies. (B) Associations of SNPs of the phase II HapMap with gene expression of SGPP2 on chromosome 2. Coordinates are in NCBI Build 35. Panels show results of 4-population multi-population analysis, and individual population analysis for CEU, CHB, JPT, and YRI. Blue arrows represent the location (not to scale) and direction of transcription of the associated gene. In this case the SNP was not rare in any of the populations (MAF was between 0.08 and 0.44) but the effect was small (R2 = 0.25 and slope = 0.25) so it could only be detected when we pooled the populations increasing the sample size.
(C) Comparison of the adjusted R2 values (proportion of the variance in expression explained by the linear relationship between genotype and phenotype) of cis significant associations obtained from single and multi-population analysis (0.001 permutation threshold).
Most previous studies have used a linear regression (LR) model to associate SNP genotypes with gene expression. We employed an alternative non-parametric method to evaluate the sensitivity of our results to the statistical methodology used. We used Spearman rank correlation (SRC) to perform the same cis- analysis described above. We detected 293, 274, 326 and 363 cis- associations for CEU, CHB, JPT and YRI, corresponding to 783 distinct genes and FDR of 4-5%. Of those, 283 genes were detected in at least 2 populations and 57 in all four. The overlap of SRC with LR was between 77% and 86% of the genes, depending on the population (Table 1). We conclude that SRC performs at an equivalent level as LR.
Allelic effects between populations
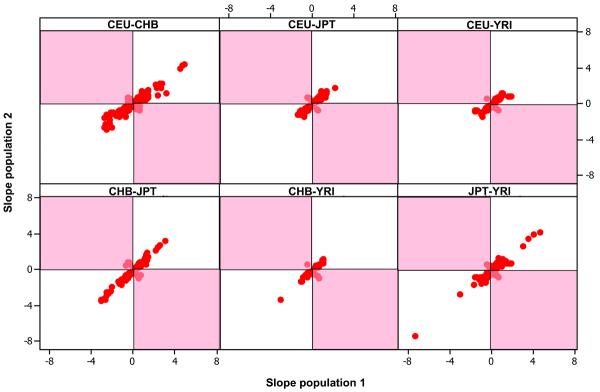
We have reported that a large fraction of genes exhibiting cis- associations at the 0.001 permutation threshold are shared (about 37%) in at least two populations (Table 2 and 19). This comparison refers to the across-population association of the same gene and in most cases the same SNPs. The gold standard for association replication requires the same SNP to be associated with the same phenotype, and the allelic effects to be in the same direction across multiple independent populations. We compared the allelic directions of SNP-gene associations shared in all pairs of populations. In 95-97% of the shared associations, the direction of the allelic effect was the same across populations (Figure 3), and the discordant 3-5% is of the same order as the FDR. This further corroborates that the associations we observed represent real genetic effects on gene expression.
Table 2.
Cis- associations in single populations and multi-population subsets
| 0.001 | |||||||
|---|---|---|---|---|---|---|---|
| 1 | 2 | 3 | 4 | ||||
| linear regression |
CEU-CHB-JPT- YRI |
CEU-CHB- JPT |
CHB-JPT | overlap | overlap | overlap | |
| significant genes |
multipop | multipop | multipop | 1&2 | 1&3 | 1&4 | |
| CEU | 299 | 845 | 750 | 851 | 207 | 215 | 180 |
| CHB | 318 | 845 | 750 | 851 | 233 | 255 | 276 |
| JPT | 341 | 845 | 750 | 851 | 247 | 261 | 287 |
| YRI | 394 | 845 | 750 | 851 | 219 | 185 | 174 |
|
Non-
redundant |
831 | ||||||
| 4 pops | 62 | ||||||
| >= 2 pops | 310 | ||||||
| 0.01 | |||||||
| 1 | 2 | 3 | 4 | ||||
|
linear regression |
CEU-CHB-JPT-
YRI |
CEU-CHB-
JPT |
CHB-
JPT |
overlap | overlap | overlap | |
|
significant genes |
multipop | multipop | multipop | 1&2 | 1&3 | 1&4 | |
| CEU | 606 | 1211 | 1157 | 1071 | 358 | 374 | 312 |
| CHB | 634 | 1211 | 1157 | 1071 | 378 | 407 | 430 |
| JPT | 679 | 1211 | 1157 | 1071 | 419 | 436 | 473 |
| YRI | 742 | 1211 | 1157 | 1071 | 373 | 340 | 312 |
|
Non-
redundant |
1746 | ||||||
| 4 pops | 114 | ||||||
| >= 2 pops | 533 | ||||||
Figure 3.
Comparison of the direction of shared SNP-gene allelic effects across all pairs of populations (0.001 permutation threshold). White panels indicate effects in the same direction.
We also investigated whether the extent to which those associations not shared between populations could be attributed to differential allele frequencies across populations as recently reported 16. For each pair of populations we split the associated SNPs (0.001 permutation threshold) into 3 categories: i) SNPs significant for the same gene in both populations (SNP-shared associations); ii) gene associations in both populations but with different SNPs (Gene-shared associated SNPs); iii) population-specific associations (unshared associations). For these 3 categories of SNPs, we computed the difference in expected heterozygosity (2pq) in the same direction (e.g. Hetpopulation1-Hetpopulation2) and compared the distributions of the differences among the three categories. As expected, median difference in heterozygosity was the lowest for SNP-shared associations, with gene-shared associated SNPs exhibiting the second lowest difference (Figure S4). Our results are consistent with the Spielman et al. 16 observation. One small caveat is that because of small sample sizes, there could be slight fluctuations in allele frequencies simply due to sampling variance that may affect detection of associations above a certain threshold.
Associations with respect to genome annotation and evolutionary conservation
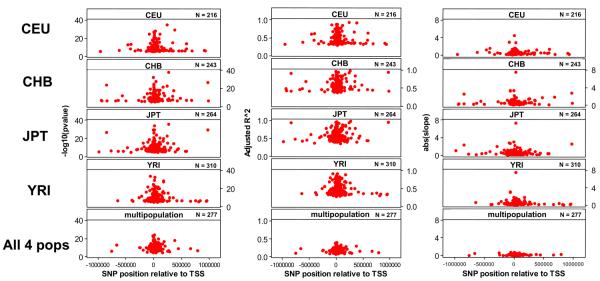
The high SNP density of the HapMap makes it possible that some of the SNPs interrogated are actually the causal variants (it is estimated that 30-50% of SNPs with MAF > 5% are represented in the HapMap 22,23), which means that evaluation of the genomic annotation where associated SNPs are found may be informative. For each of the 4 populations, we mapped the most significant SNP for each of the genes with significant cis- associations from the single population analysis relative to the transcription start site (TSS) of genes with annotated 5′UTRs. We found a strong signal for the SNPs to be located very close to the TSS (Figure 4), with no discernable trend toward 3′ or 5′. This symmetrical trend was also evident in the recent analysis of the ENCODE consortium 31. When we considered the most significant SNP of the 341 additional genes detected in the 4-population multi-population analysis, the signal was even tighter around the TSS. Three of the associated SNPs (rs10998076, rs869736, rs1010167) have been previously shown in promoter transfection assays to have a direct effect on transcriptional activation 32 in kidney and brain cell lines.
Figure 4.
Statistical significance, adjusted R2 of the association (proportion of the expression variance explained by the linear relationship between genotype and phenotype), and absolute value of the slope of the linear regression, as a function of distance from the transcription start site, of the most significantly associated SNP per gene in each of the 4 populations (order CEU, CHB, JPT and YRI) and the pooled sample of all 4 populations.
Next we mapped the location of the most significant SNP of the 831 cis- associated genes from the single population analysis with respect to gene promoters, coding sequences, and conserved non-coding sequences. We observed significant excess of associated SNPs (relative to those tested) in promoters and coding sequences (Fisher's Exact test; P = 8.94 × 10−24, P = 4.52 × 10−12, respectively), and under-representation in conserved non-coding sequences (CNCs; P= 0.00358). The first two signals are quite expected and partly confounded since most of the causal variants are found in the genic regions, so SNPs in LD with causal variants may also map to coding sequences. Not surprisingly, we observed enrichment of associated SNPs in regions that align in multiple mammals and as far as fish or chicken (Fisher's exact test, P = 10−5. The apparent contradiction between enrichment in conserved nucleotides and deficit in CNCs is probably due to the fact that the majority of the signal for conserved nucleotide comes from exonic sequences that are close to the TSS where the associations are mainly found. The deficit of associated SNPs in CNC sequences is somewhat surprising because previous studies have suggested that they are selectively constrained 31,33,34 and in some cases have been shown to play a role in gene regulation 35-37. It is possible that the skew toward large effect sizes also skews the distribution of causal regulatory variants.
Trans-associations of gene expression with SNPs
The availability of whole genome expression and SNP data allows the elucidation of genetic effects acting in trans. The number of 2.2 million SNPs per population is large (MAF > 0.05) so testing all SNPs against all genes becomes computationally and statistically challenging (correcting for millions of tests). We took a “candidate variant approach” by testing only putatively functional SNPs. The goal of the analysis is not to compare the numbers of cis vs. trans effects, which is an irrelevant question in the genome-wide context especially given differential power in detection. The goal is to assess the relative contribution of primary molecular variants in trans. Towards this end, we selected four categories of SNPs to analyze for trans effects: i) SNPs with functional effects on gene expression in cis (as determined above in the single population analyses), ii) non-synonymous SNPs (Ensembl v41 annotation); iii) SNPs influencing splicing (Ensembl v41 annotation) iv) and SNPs within microRNAs (as annotated in miRBase). These correspond to approximately 25,000 SNPs (MAF > 0.05) per population (see Table 3 for counts in each category).
Table 3.
Number and source category of SNPs used in the trans analysis
| CEU |
CHB |
JPT |
YRI |
|
|---|---|---|---|---|
| cis- associated (rSNPs) | 13221 | 13133 | 13191 | 13375 |
| Non-synonymous | 9904 | 9383 | 9378 | 10727 |
| Splicing | 1756 | 1585 | 1594 | 1950 |
| miRNA | 34 | 34 | 32 | 37 |
| Non-redundanta | 24635 | 23854 | 23907 | 25797 |
Note: all SNPs are > 0.05 frequency in the respective population.
For each population, we employed a linear regression model as described above to test for association between SNP variation and expression variation. We confined the trans analysis to those SNP-gene combinations where the genomic distance between probe midpoint and SNP was greater than 1Mb (or where probe and SNP were on different chromosomes). Significance was evaluated through 10,000 permutations as described above. We identified 43, 37, 38, 23 genes in CEU, CHB, JPT and YRI, respectively, with significant trans- associations (0.001 permutation threshold). In total, 108 genes show significant trans- association in at least one population (16 genes or 15% show a significant trans- association in at least 2 populations and 5 in all four populations). We also performed analysis in pooled populations as described above and detected 44, 52 and 39 genes for the 4-, 3- and 2-population pools respectively. Overall there seems to be low power to detect trans effects in these cell lines and sample sizes (Table 4), as indicated by the low number of discovered genes and consequently the high FDR.
Table 4.
Trans- associations in single and multi-population analysis
| 0.001 | |||||||
|---|---|---|---|---|---|---|---|
| 1 | 2 | 3 | 4 | ||||
| linear regression | CEU-CHB-JPT- YRI |
CEU-CHB- JPT |
CHB- JPT |
overlap | overlap | overlap | |
| significant genes |
multipop | multipop | multipop | 1&2 | 1&3 | 1&4 | |
| CEU | 43 | 44 | 52 | 39 | 9 | 12 | 12 |
| CHB | 37 | 44 | 52 | 39 | 10 | 14 | 15 |
| JPT | 38 | 44 | 52 | 39 | 10 | 14 | 16 |
| YRI | 23 | 44 | 52 | 39 | 7 | 7 | 7 |
|
Non-
redundant |
108 | ||||||
| 4 pops | 5 | ||||||
| >= 2 pops | 16 | ||||||
| 0.01 | |||||||
| 1 | 2 | 3 | 4 | ||||
| linear regression |
CEU-CHB-JPT-
YRI |
CEU-CHB-
JPT |
CHB-
JPT |
overlap | overlap | overlap | |
|
significant genes |
multipop | multipop | multipop | 1&2 | 1&3 | 1&4 | |
| CEU | 247 | 171 | 329 | 208 | 14 | 28 | 18 |
| CHB | 210 | 171 | 329 | 208 | 11 | 22 | 20 |
| JPT | 196 | 171 | 329 | 208 | 15 | 20 | 20 |
| YRI | 159 | 171 | 329 | 208 | 15 | 17 | 15 |
|
Non-
redundant |
756 | ||||||
| 4 pops | 9 | ||||||
| >= 2 pops | 32 | ||||||
At the 0.001 threshold, the majority of trans associations are caused by SNPs from the first category, i.e. those with cis-regulatory effects, showing 3- to 6-fold enrichment relative to the total SNPs tested (Fisher's exact test p-values 10−10-10−24, depending on the population except the YRI population, where it is not significant). Some SNPs were significantly associated with expression of multiple genes (up to 6 genes for a single SNP). The numbers of SNPs that are associated with more than 1 gene are: CEU = 29 SNPs, CHB = 13, JPT = 7, YRI = 4. A total of 8 genes had a trans association on the same chromosome (distance > 1 Mb) with distances ranging from 1003413 bps (potential cis effect) to 187659746 bps. If gene expression perturbations are similar in underlying genetic effects as whole organism perturbations and disease, then this last result suggests that the majority of the common phenotypic variation in humans is driven by variants in regulatory elements, rather than variants in protein-coding sequences, providing some potential answers to the long standing question of the relative contribution of regulatory and coding causal variants to complex phenotypes.
Discussion
We performed a comprehensive analysis of genetic effects on gene expression variation in human lymphoblastoid cell lines, presenting evidence for cis regulatory effects of 1348 genes and their biological properties by adopting a “candidate region approach”. The limited power of our analysis means that we detect only a subset of the existing functional regulatory effects in these populations. In addition, as we have only interrogated a single cell type, variation manifested only in other cell types is not represented here. These two facts argue for an abundance of cis regulatory variants segregating in human populations, some of which may be responsible for higher-order phenotypic variation and susceptibility to disease.
Our analysis goes beyond the mere detection of cis regulatory effects. We have performed to our knowledge, the most comprehensive analysis to date of the properties of the cis association signals, and we have systematically described characteristics of the expression data. Together these analyses provide us with confidence in the detected signals. In addition, we have demonstrated that the detected association signals replicate very well across populations, even though the populations are quite divergent and the sample sizes are small. We also detect the effect of population differentiation on gene expression. In this respect, we confirm what has been previously documented in smaller scale studies 16,28; Among-population allele frequency differences exist and provide a framework for the study of phenotypic differences among populations.
We have provided new methodological insights into the analysis of gene expression variation. By employing pooling of divergent populations and conditional permutation schemes, we increased the sensitivity of our analysis, detecting smaller regulatory effects shared across populations. One can imagine a more sophisticated conditional permutation scheme that would permit pooling of any set of populations for which the population identities or relatedness metrics are known. We have also employed a non-parametric test, namely Spearman rank correlation, and demonstrated that it has enough power to be used in such studies. In addition, SRC has some advantages over linear regression due to the fact that, contrary to the linear regression where outliers can have a large impact on the p-values, SRC is not sensitive to them and therefore the nominal p-values can be used directly in methods that estimate FDR (example given in Figure S6).
The evolutionary and annotation properties of cis regulatory associations are very relevant since the density of the phase II HapMap allows for a fine-scale analysis of the association signal. The vast majority of detected cis regulatory effects map very close to the TSS and are enriched in regions of high sequence conservation. This information provides a useful framework to search for cis regulatory variants in the human genome and suggests that most of the large effect variants are in the genic and immediate intergenic regions. The association data will become available at the Ensembl web site in the October 2007 as Distributed Annotation System (DAS) tracks to enable browsing and downloading.
Finally, we have attempted to analyze effects in trans by adopting a “candidate variants approach” assigning prior relevance to those SNPs already known to be associated with cis regulation, protein sequence variation (amino acid or splicing variation), or miRNA structure, and this approach made correction by permutation feasible. There were fewer genes exhibiting significant trans effects than exhibiting cis effects. This is a function of the fact that trans effects are often more indirect and therefore weaker, so our sample size does not provide us with enough power in conjunction with the much larger number of tests we have to correct for. In general, the detection of trans effects in humans has been less successful than in yeast 38,39. This may be because the yeast cell comprises the entire organism, so study of the biological interactions in a yeast cell has the potential to detect all of the interactions, while the human cell is just a small part of the organism so many of the intercellular effects mediating trans effects cannot be discovered. Finally, we have provided evidence that among a set of potential variants that could have effects in trans, we observe a large enrichment in the contribution of cis regulatory variants, which may suggest that cis regulatory variation explains much of the complex phenotypic variation in humans, at least at the molecular level.
We have described the most comprehensive analysis to date of gene expression variation in human populations, and provide a detailed characterization of the genetic as well as the positional effects in the genome. This detailed analysis provides a robust and useful framework for the future analysis of gene expression variation in large cohorts with larger sample sizes but lower SNP densities and potentially multiple cell types. It will also greatly facilitate the interpretation and follow up of disease association studies by allowing the dissection of biological effects in regions that carry strong statistical signals of association. This and future studies will lead to a detailed map of functional variation in the human genome that will complement functional and variation studies towards the complete understanding of phenotypic variation in human populations.
Methods
RNA preparation
Total RNA was extracted from lymphoblastoid cell lines of the 270 individuals of the HapMap (22; Coriell, Camden, New Jersey, United States). Two, one-quarter scale Message Amp II reactions (Ambion, Austin, Texas, United States) were performed for each RNA extraction using 200 ng of total RNA as previously described 18. 1.5 μg of the cRNA was hybridized to an array 19.
Gene expression quantification
To assay transcript levels in the cell lines, we used Illumina's commercial whole genome expression array, Sentrix Human-6 Expression BeadChip version 1 (Illumina, San Diego, California, United States) 40.
Post-experimental raw data normalization
Background-corrected values for a single bead type are subsequently summarized by Illumina software and output to the user as a set of 47,294 intensity values for each individual hybridization 25. To combine data from our multiple replicate hybridizations, raw data were read using the beadarray R package 24 and then normalized on a log scale using a quantile normalization method 41 across replicates of a single individual, followed by a median normalization method across all 270 individuals.
Association analyses
Of the 47,294 probes for which we collected expression data, we initially selected a set of 14,925 probes to analyze as described in Stranger et al. 19. We subsequently discarded from our analyses any probe that mapped to more than one Ensembl gene (Ensembl version 42) or that had an associated SNP underlying the probe sequence. This resulted in a set of 14,456 probes that were analyzed in the association analyses, corresponding to 13,643 unique autosomal genes.
Association and multiple-test correction (individual populations)
For each of the selected probes interrogating expression and for each SNP, we fit a linear regression model as previously described 18,19. We also performed Spearman Rank Correlation. Both of these analyses were applied to each population separately, including the unrelated individuals only.
For the cis- association, we limited the analysis to those probes and SNPs (MAF > 5%) where the distance from probe genomic midpoint to SNP genomic location was less than or equal to 1Mb.
For the trans- association, we selected a subset of phase II HapMap SNPs that have a higher probability of being functional than randomly selected SNPs of the genome. We selected SNPs of four categories: i) All SNPs with significant cis- associations, ii) All nsSNPs (rs numbers from Ensembl v41, genotypes extracted from HapMap v21), iii) All splice SNPs (rs numbers from Ensembl v41, genotypes extracted from HapMap v21), and iv) microRNA SNPs (as annotated in miRBase; genotypes from HapMap v21). Together these categories comprised a set of approximately 29,000 SNPs with MAF > 5% in each of the four populations.
An association to a gene expression phenotype was considered significant if the p-value from the analysis of the observed data (nominal p-value) was lower than the threshold of the 0.001 tail of the distribution of the minimal p-values (among all comparisons for a given gene) from 10,000 permutations of the expression phenotypes 42,43.
Association and multiple-test correction (multiple population panels)
With the aim of increasing the power of our cis- association analysis, data were combined (normalized expression values and SNP genotypes) for unrelated individuals of multiple populations to comprise three different multiple population analysis panels: 1) CEU-CHB-JPT-YRI, 2) CEU-CHB-JPT, and 3) CHB-JPT. The cis- association was performed separately for each of these panels using linear regression as described above, only considering those SNPs located less than 1Mb away from the probe midpoint. Conditional permutations were performed to assess significance of the nominal p-values 29,30.
Accession Numbers
The expression data reported in this paper have been previously deposited in the Gene Expression Omnibus (GEO) (http://www.ncbi.nlm.nih.gov/geo) database (Series Accession Number GSE6536 19).
Supplementary Material
Acknowledgements
We thank the HapMap consortium for data availability, Michael Smith for assistance with software development and Mark Gibbs, Jill Orwick and Chad Gerringer for technical support. Funding was provided by the Wellcome Trust to ETD, PD, NIH ENDGAME to ETD and ST, CRUK to ST and MRC to MD. ST is a Royal Society Wolfson Research Merit Award holder.
References
- 1.Ferrari SL, et al. Polymorphisms in the low-density lipoprotein receptor-related protein 5 (LRP5) gene are associated with variation in vertebral bone mass, vertebral bone size, and stature in whites. Am J Hum Genet. 2004;74:866–75. doi: 10.1086/420771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bansal A, et al. Association testing by DNA pooling: an effective initial screen. Proc Natl Acad Sci U S A. 2002;99:16871–4. doi: 10.1073/pnas.262671399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mahley RW, Huang Y. Apolipoprotein E: from atherosclerosis to Alzheimer's disease and beyond. Curr Opin Lipidol. 1999;10:207–17. doi: 10.1097/00041433-199906000-00003. [DOI] [PubMed] [Google Scholar]
- 4.Kleinjan DA, van Heyningen V. Long-range control of gene expression: emerging mechanisms and disruption in disease. Am J Hum Genet. 2005;76:8–32. doi: 10.1086/426833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Valentonyte R, et al. Sarcoidosis is associated with a truncating splice site mutation in BTNL2. Nat Genet. 2005;37:357–64. doi: 10.1038/ng1519. [DOI] [PubMed] [Google Scholar]
- 6.Saxena R, et al. Genome-Wide Association Analysis Identifies Loci for Type 2 Diabetes and Triglyceride Levels. Science. 2007 doi: 10.1126/science.1142358. [DOI] [PubMed] [Google Scholar]
- 7.Zeggini E, et al. Replication of Genome-Wide Association Signals in U.K. Samples Reveals Risk Loci for Type 2 Diabetes. Science. 2007 doi: 10.1126/science.1142364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fanciulli M, et al. FCGR3B copy number variation is associated with susceptibility to systemic, but not organ-specific, autoimmunity. Nat Genet. 2007 doi: 10.1038/ng2046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Stankiewicz P, Lupski JR. Genome architecture, rearrangements and genomic disorders. Trends Genet. 2002;18:74–82. doi: 10.1016/s0168-9525(02)02592-1. [DOI] [PubMed] [Google Scholar]
- 10.Merla G, et al. Submicroscopic deletion in patients with williams-beuren syndrome influences expression levels of the nonhemizygous flanking genes. Am J Hum Genet. 2006;79:332–41. doi: 10.1086/506371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Li M, et al. CFH haplotypes without the Y402H coding variant show strong association with susceptibility to age-related macular degeneration. Nat Genet. 2006;38:1049–54. doi: 10.1038/ng1871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hirschhorn JN, Daly MJ. Genome-wide association studies for common diseases and complex traits. Nat Rev Genet. 2005;6:95–108. doi: 10.1038/nrg1521. [DOI] [PubMed] [Google Scholar]
- 13.Stranger BE, Dermitzakis ET. The genetics of regulatory variation in the human genome. Hum Genomics. 2005;2:126–31. doi: 10.1186/1479-7364-2-2-126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Stranger BE, Dermitzakis ET. From DNA to RNA to disease and back: the ‘central dogma’ of regulatory disease variation. Hum Genomics. 2006;2:383–90. doi: 10.1186/1479-7364-2-6-383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Doss S, Schadt EE, Drake TA, Lusis AJ. Cis-acting expression quantitative trait loci in mice. Genome Res. 2005;15:681–91. doi: 10.1101/gr.3216905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cheung VG, et al. Mapping determinants of human gene expression by regional and genome-wide association. Nature. 2005;437:1365–9. doi: 10.1038/nature04244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Schadt EE, et al. Genetics of gene expression surveyed in maize, mouse and man. Nature. 2003;422:297–302. doi: 10.1038/nature01434. [DOI] [PubMed] [Google Scholar]
- 18.Stranger BE, et al. Genome-wide associations of gene expression variation in humans. PLoS Genet. 2005;1:e78. doi: 10.1371/journal.pgen.0010078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Stranger BE, et al. Relative impact of nucleotide and copy number variation on gene expression phenotypes. Science. 2007;315:848–53. doi: 10.1126/science.1136678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Knight JC. Regulatory polymorphisms underlying complex disease traits. J Mol Med. 2005;83:97–109. doi: 10.1007/s00109-004-0603-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Monks SA, et al. Genetic inheritance of gene expression in human cell lines. Am J Hum Genet. 2004;75:1094–105. doi: 10.1086/426461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.International HapMap Consortium A haplotype map of the human genome. Nature. 2005;437:1299–320. doi: 10.1038/nature04226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.International HapMap Consortium The phase II haplotype map of the human genome. Nature. 2007 in review. [Google Scholar]
- 24.Dunning MJ, Smith DR, Thorne NP, Tavare S. beadarray: An R Package to analyse llumina BeadArrays. R News. 2006;6:17. [Google Scholar]
- 25.Dunning MJ, Thorne NP, Camilier I, Smith ML, Tavare S. Quality control and low-level statistical analysis of Illumina BeadArrays. Revstat. 2006;4:1–30. [Google Scholar]
- 26.Ashburner M, et al. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat Genet. 2000;25:25–9. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Camon E, Barrell D, Lee V, Dimmer E, Apweiler R. The Gene Ontology Annotation (GOA) Database--an integrated resource of GO annotations to the UniProt Knowledgebase. In Silico Biol. 2004;4:5–6. [PubMed] [Google Scholar]
- 28.Storey JD, Akey JM, Kruglyak L. Multiple locus linkage analysis of genomewide expression in yeast. PLoS Biol. 2005;3:e267. doi: 10.1371/journal.pbio.0030267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lee SI, Pe'er D, Dudley AM, Church GM, Koller D. Identifying regulatory mechanisms using individual variation reveals key role for chromatin modification. Proc Natl Acad Sci U S A. 2006;103:14062–7. doi: 10.1073/pnas.0601852103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Koren M, et al. ATM haplotypes and breast cancer risk in Jewish high-risk women. Br J Cancer. 2006;94:1537–43. doi: 10.1038/sj.bjc.6603062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Birney E, et al. Identification and analysis of functional elements in 1% of the human genome by the ENCODE pilot project. Nature. 2007;447:799–816. doi: 10.1038/nature05874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hoogendoorn B, et al. Functional analysis of polymorphisms in the promoter regions of genes on 22q11. Hum Mutat. 2004;24:35–42. doi: 10.1002/humu.20061. [DOI] [PubMed] [Google Scholar]
- 33.Dermitzakis ET, et al. Evolutionary discrimination of mammalian conserved non-genic sequences (CNGs) Science. 2003;302:1033–5. doi: 10.1126/science.1087047. [DOI] [PubMed] [Google Scholar]
- 34.Drake JA, et al. Conserved noncoding sequences are selectively constrained and not mutation cold spots. Nat Genet. 2006;38:223–7. doi: 10.1038/ng1710. [DOI] [PubMed] [Google Scholar]
- 35.Bejerano G, et al. Ultraconserved elements in the human genome. Science. 2004;304:1321–5. doi: 10.1126/science.1098119. [DOI] [PubMed] [Google Scholar]
- 36.Abbasi AA, et al. Human GLI3 Intragenic Conserved Non-Coding Sequences Are Tissue-Specific Enhancers. PLoS ONE. 2007;2:e366. doi: 10.1371/journal.pone.0000366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Woolfe A, et al. Highly conserved non-coding sequences are associated with vertebrate development. PLoS Biol. 2005;3:e7. doi: 10.1371/journal.pbio.0030007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Brem RB, Kruglyak L. The landscape of genetic complexity across 5,700 gene expression traits in yeast. Proc Natl Acad Sci U S A. 2005;102:1572–7. doi: 10.1073/pnas.0408709102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yvert G, et al. Trans-acting regulatory variation in Saccharomyces cerevisiae and the role of transcription factors. Nat Genet. 2003;35:57–64. doi: 10.1038/ng1222. [DOI] [PubMed] [Google Scholar]
- 40.Kuhn K, et al. A novel, high-performance random array platform for quantitative gene expression profiling. Genome Res. 2004;14:2347–56. doi: 10.1101/gr.2739104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bolstad BM, Irizarry RA, Astrand M, Speed TP. A comparison of normalization methods for high density oligonucleotide array data based on variance and bias. Bioinformatics. 2003;19:185–93. doi: 10.1093/bioinformatics/19.2.185. [DOI] [PubMed] [Google Scholar]
- 42.Churchill GA, Doerge RW. Empirical threshold values for quantitative trait mapping. Genetics. 1994;138:963–71. doi: 10.1093/genetics/138.3.963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Doerge RW, Churchill GA. Permutation tests for multiple loci affecting a quantitative character. Genetics. 1996;142:285–94. doi: 10.1093/genetics/142.1.285. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.