Summary
HLA-DP antigens present peptides to CD4+ T cells and play an important role in parasitic infections and autoimmune diseases, yet their influence on HIV-1 susceptibility has not been well studied. Here, we report several HLA-DP genotypes associated with HIV-1 susceptibility in Kenyan sex workers. Among these, one common genotype stands out. DPA1*010301 (frequency=60.4%) was associated with HIV-resistance (P=0.033, odds ratio=1.585, 95% confidence interval=1.036-2.425) and slower seroconversion (P=0.001, log rank=0.595, 95% confidence interval=0.433-0.817). The discovery of common HLA-DP antigens contributing to HIV-1 immunity may help overcome difficulties encountered with highly polymorphic HLA antigens.
Keywords: HIV-1, Sex Workers, HLA-DPA1, HLA-DPB1, Disease Association, Disease Resistance, DNA Sequence Analysis
Heterogeneity in susceptibility to HIV-1 has been described in several cohorts [1-3]. This is exemplified in a subset of highly exposed but uninfected women enrolled in the Pumwani Female Sex Worker cohort [1], which was established in 1985 to prospectively study sexually transmitted infections, and continues to enroll members with biannual follow-up. In the cohort overall seroprevalence is approximately 70% and most HIV-negative women seroconverted within 3 years of enrolment. However, a subgroup appears to be relatively resistant to HIV-1, they remain HIV-seronegative and PCR negative for more than 3 years while continuing active sex work [1]. Several factors have been shown to contribute to this HIV-resistance [4, 5], including class I and II HLA antigens [6-9]. HLA class II DP has a heterodimeric binding cleft comprised of DPA1 and DPB1 [10], and has a key role in presenting antigens to CD4+ T cells [11]. The influence of HLA-DP has not been examined in the context of HIV-1 infection, despite its importance in other diseases [12-16]. To further understand the protective immune response in the HIV-resistant sex workers, we studied the associations of HLA-DP with resistance/susceptibility to HIV-1.
All resistant women in this study enrolled before 2000 (average follow-up: 9.6 ± 4.3 years). HIV-positive women were considered susceptible. HIV-negative women with shorter follow-up were classified as negative and were not included in the comparisons between resistant and positive women. Due to limited DNA, some women could not be typed for both DPA1 and DPB1. DPA1 was genotyped for 114 resistant, 157 HIV-negative and 703 HIV-positive individuals; DPB1 was genotyped for 111 resistant, 221 HIV-negative and 762 HIV-positive individuals; while both DPA1 and DPB1 were genotyped for 111 resistant, 152 HIV-negative and 681 HIV-positive individuals. Ethics committees at the University of Manitoba and University of Nairobi approved this study. Informed consent was obtained from all women enrolled.
Exon 2 of DPA1 and DPB1 was amplified using gene-specific primers [17, 18]. DPA1 and DPB1 gene fragments were sequenced using the BigDye™ Cycle Sequencing Kit (Applied Biosystems, Foster City, CA) [19, 20] and ABI 3100 Prism Genetic Analyzer (Applied Biosystems, Foster City, CA), and genotyped using Codon Express™ [21]. Allele, genotype/haplotype frequencies, and Hardy-Weinberg calculations were estimated using Python for populations-32-0.6.0 (PyPop) [22]. Genotyping results were analysed with biological data using SPSS 15.0. Cross-sectional analysis was performed to identify associations of HLA-DP genotypes with HIV-1 susceptibility, using χ2, Fisher's exact test [23] and crosstabs analysis(odds ratio (OR), 95% confidence interval(95% CI)), were used to determine the relationship between binary outcomes and explanatory variables. Einot and Gabriel adjusted P-values were calculated using modified syntax written by David Nichols of SPSS. Significant cross-sectional associations were included in binary logistic regression using a forward Wald method. To examine the role of HLA-DP in seroconversion, Kaplan-Meier and multivariate Cox regression analyses (backward Wald) were performed. Taking patient enrollment and samples being typed into account, a weighted parameter was generated using logistic regression, to adjust for crosstabs and binary logistic regression.
Several common HLA-DP alleles were identified. Out of the 17 HLA-DP alleles identified, four major alleles accounted for >90%: DPA1*010301 (38.04%), DPA1*0301 (20.07%), DPA1*020101 (17.66%) and DPA1*020202 (16.32%). Three alleles out of 51 identified accounted for >55% of DPB1 alleles: DPB1*010101 (24.36%), DPB1*0402 (19.70%) and DPB1*020102 (13.07%). Allele frequencies did not deviate significantly from Hardy-Weinberg equilibrium. DPA1*020202-DPB1*010101 (14.081%), DPA1*0301-DPB1*0402 (13.631%) and DPA1*010301-DPB1*020102 (11.841%), accounted for nearly 40% of the haplotypes. These frequencies were comparable to other populations around the world [24].
Cross-sectional analysis identified associations of HLA-DP genotypes with HIV-resistance. DPA1*010301 (P=0.033, odds ratio (OR)=1.585, 95% Confidence Interval (CI)=1.036-2.425) and DPB1*3001 (P=0.007, OR=3.274, 95% CI=1.451-7.384), were associated with HIV-resistance. DPA1*010301 homozygotes were enriched in the HIV-1 resistant group. Thirty DPA1-DPB1 haplotypes with five or more copies were examined. DPA1*010301-DPB1*3001 (P=0.022, OR=3.481, 95% CI=1.260-9.611) and DPA1*0301-DPB1*5501 (P=0.019, OR=3.207, 95% CI=1.265-8.132) were associated with resistance to HIV-1 infection. Low-frequency haplotypes with <5 copies in this population were grouped and analyzed together for rare haplotype associations, but no significant associations were found. All observed associations remained significant after adjustment for multiple comparisons. Further analysis using the weighted parameter also confirmed the observed associations.
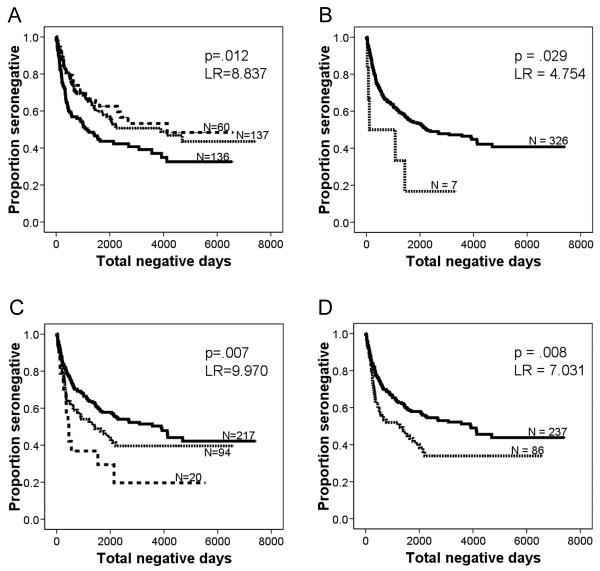
Kaplan Meier analysis showed that individuals with DPA1*010301, the common genotype associated with HIV-resistance, were significantly less likely to seroconvert (P=0.012, log rank (LR)=8.837) (Fig. 1A), but DPA1*010301 homozygosity did not further reduce the risk of seroconversion. In addition, the genotypes and haplotypes associated with HIV-resistance had a trend towards slower seroconversion. Genotypes associated with increased rate of seroconversion included DPA1*0302 (P=0.029, LR=4.754) (Fig. 1B), DPB1*0402 homozygotes (P=0.007, LR=9.970) (Fig. 1C), DPA1*0301-DPB1*0402 (P=0.008, LR=7.031) (Fig. 1D), DPB1*010101/1801 (P=0.022, LR=5.211), and DPA1*020101 homozygotes (P=0.015, LR=5.960). Their corresponding allele frequencies were also enriched in HIV-positive women.
Figure 1. HLA-DP genotypes and haplotypes associated with an altered risk of seroconversion.
A. Women with one copy of DPA1*010301 (dotted line) seroconverted less rapidly than women without this genotype (solid line). Women with two copies of DPA1*010301 (broken line) fared even better, with a decreased rate of seroconversion compared to those with one copy or no copies.
B. Women with DPA1*0302 (dotted line) seroconverted more rapidly than women without this genotype (solid line).
C. Women with one copy of DPB1*0402 (dotted line) seroconverted more rapidly than women without this genotype (solid line). Women with two copies of DPB1*0402 (broken line) fared even worse, with increase in rate of seroconversion compared to those with one copy or no copies.
D. Women with DPA1*0301-DPB1*0402 (dotted line) seroconverted more rapidly than women without this genotype (solid line).
Binary logistic regression showed that the associations of DPA1*010301 and DPB1*3001 genotypes with HIV-resistance were independent of HIV-resistant HLA genotypes previously described, such as DQB1*050301, DQB1*0603, DQB1*0609, DRB*1102, DRB1*01 and the class I supertype A2/6802 [7-9]. The association of DPA1*010301 with slower seroconversion was also independent of DRB1*01 and DRB1*1102 genotypes [8] by Cox regression. Among the HLA-DP genotypes and haplotypes associated with increased rate of seroconversion, only the DPA1* 020101 homozygote became non-significant when analyzed with DRB1*1503-DRB5*010101 haplotype, DRB1*030201 genotype [8], and A*2301 [7], suggesting that they could be linked.
Analysis of protein sequences encoded by exon 2 of DP alleles between genotypes of HIV-resistance/susceptibility revealed variability. At residue 11, DPA1*010301 encoded alanine, while DPA1*0302 encoded methionine. Variability was also observed at residues 8, 9, 11, 55, 56, 69 and 84-87 between DPB1*3001 and DPB1*0402. This suggests that this variability could have functional characteristics and future studies should determine their role in differential antigen presentation.
The DPA1*010301 genotype is possessed by 60.4% of individuals in the cohort. As the most common HLA-DPA1 genotype, its association with HIV-resistance and delayed seroconversion is striking. This suggests that DP molecules play a role that is different from the rare allele advantage observed for HLA class I genes in HIV-1 disease progression [25]. Vaccine strategies based on mechanisms of protection offered by a common HLA class II allele may have broader applications than those associated with class I antigens. Heterozygosity in the HLA class I region is thought to confer an advantage in HIV-1 disease progression, possibly due to the greater ability to present a larger variety of viral epitopes [6, 26]. It has been suggested that this advantage may also extend to HLA class II alleles [27]. Indeed, homozygotes of susceptible alleles, such as DPA1*020101 and DPB1*0402 rapidly seroconverted. While DPA1*010301 was associated with HIV-resistance and slower seroconversion, homozygosity for DPA1*010301 did not provide additional protection for either HIV-resistance or seroconversion. The advantage of two protective alleles may have been neutralized by a disadvantage of homozygosity.
This study is the first to show associations of HLA-DP molecules with resistance/susceptibility to HIV-1 infection. HLA-DP is clearly an important factor in the immune response to HIV-1, and further study of the mechanism of protection is warranted. These findings provide further understanding of the mechanisms of underlying protective immunity to HIV-1, and might ultimately contribute to the design of an effective vaccine.
Acknowledgments
R. Hardie wrote and edited the final drafts of the manuscript, performed experiments and data analysis. E. Knight and B. Bruneau conducted experiments and data analysis, and contributed to writing and editing the paper. C. Semeniuk and K. Gill contributed to experiments and data analysis. N. Nagelkerke edited the paper and consulted on statistical analysis. J. Kimani and C. Wachihi helped with editing, maintaining the cohort and collected biological data. E. Ngugi helped with editing and contributed to establishment of the cohort. M. Luo designed the study, performed data analysis, and edited the paper. F. A. Plummer established and maintained the cohort, designed the study, edited the paper, and secured funding.
This study was supported by a grant from the Bill and Melinda Gates Foundation and the Canadian Institutes of Health Research through the Grand Challenges in Global Health Initiative. The NIH also provided funding. This work would not be possible without the participation of the women of the Pumwani Sex Worker cohort as well as the dedicated staff who work with the cohort. Thomas Bielawny provided technical assistance. Dr. Francis A. Plummer is a Tier 1 CIHR Canada Research Chair.
Financial support: This study was funded by a grant from the Bill and Melinda Gates Foundation and the Canadian Institutes of Health Research (HOP-43135) through the Grand Challenges in Global Health Initiative. This study was also supported through a grant from the NIH (R01 AI56980). Dr. Francis A. Plummer is a Tier I CIHR Canada Research Chair.
Footnotes
The authors have no affiliations, commercial or otherwise, that may pose a conflict of interest.
Information presented at: International AIDS Conference, Toronto, ON, Canada, 13-18 August 2006
International Centre for Infectious Diseases Research and Innovation Retreat, Winnipeg, MB, Canada, 28–29 October 2005, 23–25 October 2006
Public Health Agency of Canada Conference, Winnipeg, MB, Canada, 20–21 March 2006, 12–13 March 2007
Canadian Association for HIV Research Conference, Toronto, ON, Canada, 26–29 April 2007
No author's affiliation or address has changed since the completion of this study.
References
- 1.Fowke KR, Nagelkerke NJ, Kimani J, Simonsen JN, Anzala AO, Bwayo JJ, et al. Resistance to HIV-1 infection among persistently seronegative prostitutes in Nairobi, Kenya. Lancet. 1996;348:1347–1351. doi: 10.1016/S0140-6736(95)12269-2. [DOI] [PubMed] [Google Scholar]
- 2.Promadej N, Costello C, Wernett MM, Kulkarni PS, Robison VA, Nelson KE, et al. Broad human immunodeficiency virus (HIV)-specific T cell responses to conserved HIV proteins in HIV-seronegative women highly exposed to a single HIV-infected partner. J Infect Dis. 2003;187:1053–1063. doi: 10.1086/368127. [DOI] [PubMed] [Google Scholar]
- 3.Yang C, Boone L, Nguyen TX, Rudolph D, Limpakarnjanarat K, Mastro TD, et al. theta-Defensin pseudogenes in HIV-1-exposed, persistently seronegative female sex-workers from Thailand. Infect Genet Evol. 2005;5:11–15. doi: 10.1016/j.meegid.2004.05.006. [DOI] [PubMed] [Google Scholar]
- 4.Alimonti JB, Koesters SA, Kimani J, Matu L, Wachihi C, Plummer FA, et al. CD4+ T cell responses in HIV-exposed seronegative women are qualitatively distinct from those in HIV-infected women. J Infect Dis. 2005;191:20–24. doi: 10.1086/425998. [DOI] [PubMed] [Google Scholar]
- 5.Rowland-Jones S, Sutton J, Ariyoshi K, Dong T, Gotch F, McAdam S, et al. HIV-specific cytotoxic T-cells in HIV-exposed but uninfected Gambian women. Nat Med. 1995;1:59–64. doi: 10.1038/nm0195-59. [DOI] [PubMed] [Google Scholar]
- 6.Carrington M, O'Brien SJ. The influence of HLA genotype on AIDS. Annu Rev Med. 2003;54:535–551. doi: 10.1146/annurev.med.54.101601.152346. [DOI] [PubMed] [Google Scholar]
- 7.MacDonald KS, Fowke KR, Kimani J, Dunand VA, Nagelkerke NJ, Ball TB, et al. Influence of HLA supertypes on susceptibility and resistance to human immunodeficiency virus type 1 infection. J Infect Dis. 2000;181:1581–1589. doi: 10.1086/315472. [DOI] [PubMed] [Google Scholar]
- 8.Lacap P, Huntington J, Luo M, Nagelkerke NJD, Bielawny T, Kimani J, et al. Associations of human leukocyte antigen DRB with resistance or susceptibility to HIV-1 infection in the Pumwani Sex Worker Cohort. AIDS. 2008 doi: 10.1097/QAD.0b013e3282ffb3db. [DOI] [PubMed] [Google Scholar]
- 9.Hardie RA, Luo M, Bruneau B, Knight E, Nagelkerke NJ, Kimani J, et al. Human leukocyte antigen-DQ alleles and haplotypes and their associations with resistance and susceptibility to HIV-1 infection. AIDS. 2008;22:807–816. doi: 10.1097/QAD.0b013e3282f51b71. [DOI] [PubMed] [Google Scholar]
- 10.McFarland BJ, Beeson C. Binding interactions between peptides and proteins of the class II major histocompatibility complex. Med Res Rev. 2002;22:168–203. doi: 10.1002/med.10006. [DOI] [PubMed] [Google Scholar]
- 11.Germain RN. MHC-dependent antigen processing and peptide presentation: providing ligands for T lymphocyte activation. Cell. 1994;76:287–299. doi: 10.1016/0092-8674(94)90336-0. [DOI] [PubMed] [Google Scholar]
- 12.Noble JA, Valdes AM, Thomson G, Erlich HA. The HLA class II locus DPB1 can influence susceptibility to type 1 diabetes. Diabetes. 2000;49:121–125. doi: 10.2337/diabetes.49.1.121. [DOI] [PubMed] [Google Scholar]
- 13.Gilchrist FC, Bunn C, Foley PJ, Lympany PA, Black CM, Welsh KI, et al. Class II HLA associations with autoantibodies in scleroderma: a highly significant role for HLA-DP. Genes Immun. 2001;2:76–81. doi: 10.1038/sj.gene.6363734. [DOI] [PubMed] [Google Scholar]
- 14.May J, Kremsner PG, Milovanovic D, Schnittger L, Loliger CC, Bienzle U, et al. HLA-DP control of human Schistosoma haematobium infection. Am J Trop Med Hyg. 1998;59:302–306. doi: 10.4269/ajtmh.1998.59.302. [DOI] [PubMed] [Google Scholar]
- 15.Meyer CG, Schnittger L, May J. Met-11 of HLA class II DP alpha 1 first domain associated with onchocerciasis. Exp Clin Immunogenet. 1996;13:12–19. [PubMed] [Google Scholar]
- 16.May J, Meyer CG, Kun JF, Lell B, Luckner D, Dippmann AK, et al. HLA class II factors associated with Plasmodium falciparum merozoite surface antigen allele families. J Infect Dis. 1999;179:1042–1045. doi: 10.1086/314661. [DOI] [PubMed] [Google Scholar]
- 17.Rozemuller E, van der Zwan A, Tilanus MGJ. Sequence-based typing for HLA-DPA1, strategy for ABI Sequencing Equipment. In: Tilanus MJG, editor. IHWG Technical Manual, genomic analysis of the human MHC. Chapter 13 Seattle, Washington: IHWG Press; 2002. [Google Scholar]
- 18.Hoshino S, Kimura A, Fukuda Y, Dohi K, Sasazuki T. Polymerase chain reaction--single-strand conformation polymorphism analysis of polymorphism in DPA1 and DPB1 genes: a simple, economical, and rapid method for histocompatibility testing. Hum Immunol. 1992;33:98–107. doi: 10.1016/0198-8859(92)90059-v. [DOI] [PubMed] [Google Scholar]
- 19.Luo M, Bamforth J, Gill K, Cohen C, Brunham RC, Plummer FA. High-resolution sequence-based DPA1 typing identified two novel DPA1 alleles, DPA1*010303 and DPA1*0303, from a Kenyan population. Tissue Antigens. 2005;65:120–122. doi: 10.1111/j.1399-0039.2005.00339.x. [DOI] [PubMed] [Google Scholar]
- 20.Luo M, Ramdahin S, Iqbal S, Pan Y, Jacobson K, Narayansingh MJ, et al. High resolution sequence-based DPB1 typing identified two novel DPB1 alleles, DPB1*9401 and DPB1*9501, from a Kenyan population. Tissue Antigens. 2003;62:182–184. doi: 10.1034/j.1399-0039.2003.00066.x. [DOI] [PubMed] [Google Scholar]
- 21.Luo M, Blanchard J, Pan Y, Brunham K, Brunham RC. High-resolution sequence typing of HLA-DQA1 and -DQB1 exon 2 DNA with taxonomy-based sequence analysis (TBSA) allele assignment. Tissue Antigens. 1999;54:69–82. doi: 10.1034/j.1399-0039.1999.540108.x. [DOI] [PubMed] [Google Scholar]
- 22.Lancaster A, Nelson MP, Meyer D, Thomson G, Single RM. PyPop: a software framework for population genomics: analyzing large-scale multi-locus genotype data. Pac Symp Biocomput. 2003:514–525. [PMC free article] [PubMed] [Google Scholar]
- 23.Hutchon JR. Calculator for confidence intervals of odds ratio in an unmatched case control study For example groups of cases and controls studied to assess a treatment or exposure to a suspected causal factor. 2008. [Google Scholar]
- 24.National Center for Biotechnology Information. dbMHC - Anthropology. Vol. 2007 2007. [Google Scholar]
- 25.Trachtenberg E, Korber B, Sollars C, Kepler TB, Hraber PT, Hayes E, et al. Advantage of rare HLA supertype in HIV disease progression. Nat Med. 2003;9:928–935. doi: 10.1038/nm893. [DOI] [PubMed] [Google Scholar]
- 26.Carrington M, Nelson GW, Martin MP, Kissner T, Vlahov D, Goedert JJ, et al. HLA and HIV-1: heterozygote advantage and B*35-Cw*04 disadvantage. Science. 1999;283:1748–1752. doi: 10.1126/science.283.5408.1748. [DOI] [PubMed] [Google Scholar]
- 27.Zijenah LS, Hartogensis WE, Katzenstein DA, Tobaiwa O, Mutswangwa J, Mason PR, et al. Association of high HIV-1 RNA levels and homozygosity at HLA class II DRB1 in adults coinfected with Mycobacterium tuberculosis in Harare, Zimbabwe. Hum Immunol. 2002;63:1026–1032. doi: 10.1016/s0198-8859(02)00684-5. [DOI] [PubMed] [Google Scholar]