Abstract
Adenosine deaminases that act on RNA (ADARs) are RNA-editing enzymes that convert adenosine to inosine within double-stranded RNA. In the 12 years since the discovery of ADARs only a few natural substrates have been identified. These substrates were found by chance, when genomically encoded adenosines were identified as guanosines in cDNAs. To advance our understanding of the biological roles of ADARs, we developed a method for systematically identifying ADAR substrates. In our first application of the method, we identified five additional substrates in Caenorhabditis elegans. Four of those substrates are mRNAs edited in untranslated regions, and one is a noncoding RNA edited throughout its length. The edited regions are predicted to form long hairpin structures, and one of the RNAs encodes POP-1, a protein involved in cell fate decisions.
Adenosine deaminases that act on RNA (ADARs) have been detected in every metazoan examined (reviewed in refs. 1 and 2), and cDNAs encoding various members of the enzyme family have been cloned (3). The product of adenosine deamination, inosine, is found within mRNA in tissue-specific amounts that correlate with the amounts of ADAR in various tissues (4), and the observed levels suggest many mRNAs are acted on by ADARs. Yet, in the 12 years since the discovery of ADARs (5–8), only a few natural substrates have been identified (reviewed in ref. 2). These substrates have been found entirely by chance, when genomically encoded adenosines were identified as guanosines in cDNAs. Such A to G transitions are diagnostic of A to I conversions, because inosine prefers to pair with cytidine and is changed to a G during cDNA synthesis.
Studies of the serendipitously discovered substrates show that one function of ADARs is to deaminate adenosines within codons. In this way, multiple protein isoforms can be synthesized from a single encoded mRNA. ADARs are involved in producing functionally important isoforms of mammalian serotonin receptors (9), several mammalian glutamate receptor subunits (10–12), and the virally encoded hepatitis delta antigen (13). In addition, A to G transitions have been detected within codons of several other viral and cellular transcripts where function has not yet been verified (reviewed in ref. 2).
Many additional functions of ADARs have been proposed. Conceivably, A to I conversions could affect any process that involves sequence-specific interactions, so effects on RNA processing, stability, and translatability are all possible. Further, because adenosine deamination can alter RNA structure, sequence-independent processes also could be affected. In fact, the intrinsic properties of ADARs suggest their primary and primordial functions remain to be elucidated. For example, ADARs act promiscuously on completely base-paired double-stranded RNA (dsRNA) substrates, deaminating ≈50% of the adenosines in a single molecule. Obviously, such rampant deamination is ill-suited for generating protein isoforms of precise function. In mRNAs where ADARs are known to produce a functionally important codon change, deamination occurs more selectively, apparently because the structures surrounding the editing sites are not completely double-stranded, but interrupted by mismatches, bulges, and loops (reviewed in ref. 2).
Clearly it would be advantageous to have a way to identify inosine-containing RNAs, not only to identify other RNAs where ADARs act to create protein isoforms, but also to identify other ways ADARs modify gene expression. To this end we developed a method that allows the identification of large numbers of ADAR substrates systematically, rather than by chance. In our first application of the method, we identified five additional ADAR substrates, thus increasing the number of known ADAR substrates about 50%. Interestingly, none of the deamination sites are within codons.
METHODS
RNA Isolation.
The guanidinium thiocyanate/phenol method was used to prepare Caenorhabditis elegans RNA from frozen worm (strain N2) pellets harvested from 1 liter of mixed stage liquid cultures. Poly(A)+ RNA was purified through two rounds of selection on oligo(dT)-cellulose (Collaborative Biomedical Products type 3 or type 2). Before the second round, RNA was heated (55°C, 5 min) in DMSO/buffered LiCl (14) to disrupt rRNA-mRNA interactions.
Inosine-Specific Cleavage and Tailing of Cleavage Sites.
Before inosine-specific cleavage of poly(A)+ RNA (Fig. 1A, step 1), 3′ hydroxyls were oxidized with sodium periodate to prevent elongation of the original poly(A) tails during step 2. Reactions contained 10 μl (25 μg) of RNA, 3.3 μl of 0.5 M sodium acetate (pH 5.5), and 3.3 μl of 50 mM sodium periodate (freshly dissolved). After 1 hr in the dark at room temperature, 16.6 μl of 2% ethylene glycol was added and incubation continued for 10 min. The reaction was diluted to 400 μl with water, and RNA was precipitated with ethanol. Inosine-specific cleavage was performed on two 5-μg aliquots of oxidized poly(A)+ RNA as described (15). Two additional aliquots of RNA were treated identically except RNase T1 was omitted from the inosine-specific cleavage reaction. Twenty microliters of polyadenylation reactions (Fig. 1A, step 2) contained 5 μg of RNA, 1× reaction buffer (United States Biochemical), 0.5 mM ATP, 0.05 mM cordycepin triphosphate, and 250 units of poly(A) polymerase (United States Biochemical). After 1 hr at 30°C, 0.5 μl of 10 mg/ml proteinase K (Boehringer Mannheim) was added, and the reaction was incubated for 20 min at 37°C. Samples were extracted twice with phenol-chloroform-isoamyl alcohol (25:24:1), and RNA was precipitated with ethanol.
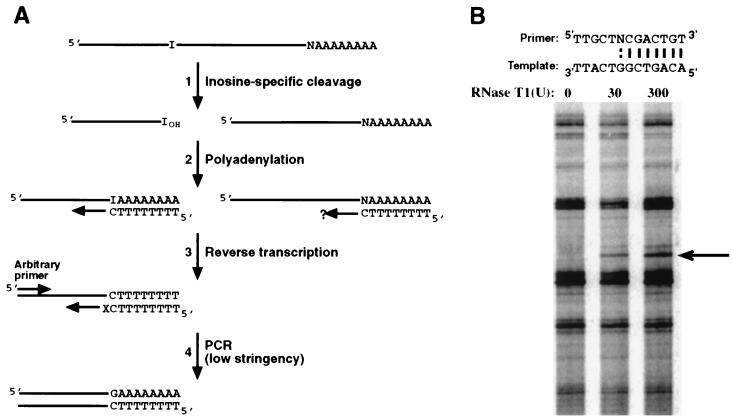
Figure 1.
A protocol for identifying endogenous ADAR substrates. (A) N, nucleotide 5′ of poly(A) tail. X, nucleotide(s) on the 3′ end of the downstream PCR primer (see Methods). Step 1: Poly(A)+ RNA is specifically cleaved 3′ of inosines as described (15). Step 2: A poly(A) tail is added 3′ to the inosine at the cleavage site to create a primer binding site for first-strand cDNA synthesis. Step 3: Because inosine pairs with cytidine, reverse transcription is performed with a T12C primer to enrich for cleaved molecules. The question mark indicates that the primer will extend on uncleaved RNA only if n = G. Step 4: Multiple aliquots of the cDNA are analyzed by using numerous combinations of primers (see Methods for primer design). Importantly, bands that derive from inosine-containing molecules depend on the addition of RNase T1 and are identified by comparison to samples not treated with ribonuclease. (B) Optimization of the method. A 386-nt synthetic RNA (0.5 fmol) containing a single inosine (15) was spiked into 5 μg of total yeast RNA and subjected to the protocol of A by using 0, 30, or 300 units of RNase T1. The complementarity between the upstream arbitrary PCR primer, and its priming site is shown above the gel; N represents a randomized position. For the downstream PCR primer, X=G because the nucleotide on the 5′ side of the inosine in the control RNA was a C. The arrow points to an RNase T1-dependent band whose sequence confirmed that it derived from the synthetic RNA cleaved precisely 3′ to its single inosine.
Differential Display Reverse Transcription–PCR.
First-strand cDNA (Fig. 1A, step 3) was synthesized from each of the four RNA samples. Twenty-microliter reactions contained 5 μg RNA, 1× reaction buffer (BRL), 1 mM DTT, 20 μM dNTPs, 0.5 μM T12C primer, and 200 units of murine leukemia virus reverse transcriptase (BRL). The mixture was heated for 5 min at 65°, 5 min at 37°C, RT was added, and after 1 hr at 37°C, 2.5 μl of 1 M NaOH was added. RNA was hydrolyzed (30 min, 50°C), and the solution was neutralized by adding 2.5 μl of 1 M HCl, and then diluted to 1 ml with water. Arbitrarily primed PCR (Fig. 1A, step 4) was performed on two (plus and minus RNase T1) of the four cDNA samples. Ten-microliter PCRs contained 2 μl of diluted cDNA, 1× PCR buffer II (Perkin–Elmer), 2.5 mM MgCl2, 20 μM dNTPs, 2 μCi [α-33P]-dATP (NEN), 1 μM primers, and 1.25 units of Amplitaq gold (Perkin–Elmer). Each reaction contained one arbitrary 13-mer and one of seven different downstream primers. The sixth nucleotide from the 5′ end of each arbitrary primer was randomized to improve sensitivity (see Fig. 1B). (Arbitrary primers are not random primers but have a fixed sequence that is chosen arbitrarily; ref. 16.) Downstream primers were one of seven different primers of the form GAGACCAGT12CX, where X represents G, A, C, TG, TA, TC, or TT; X = TG, TA, TC, or TT were required because primers with a single 3′ T were inefficiently extended. The first 8 nt at the 5′ end, GAGACCAT, were added to improve efficiency of reamplification of gel-purified PCR products. (The substrate called 52G was identified by using a downstream primer that lacked the eight extra nucleotides; see Results.) Amplification conditions were: 94°C, 9 min (to activate enzyme); 50 cycles of 94°C, 1 min, 40°C, 2 min, 72°C, 1 min; 72°C, 5 min. We performed 420 PCRs (60 arbitrary primers × seven downstream primers) on each of the two cDNA samples (plus and minus RNase T1). Six microliters of each PCR product was added to 4 μl of formamide loading buffer and 5 μl was loaded onto a 6% sequencing gel. Sequencing gels were dried, and PCR products were visualized by autoradiography.
Identification of Candidate ADAR Substrates.
Autoradiograms were examined for bands unique to samples treated with RNase T1 (see Fig. 2). Each PCR that produced an RNase-T1-dependent band was repeated by using the duplicate cDNA samples. Eighty-two bands were reproducibly dependent on RNase T1; these were excised from the gel, reamplified, and cloned (15). Because different PCR products often comigrated, we sequenced three clones for each excised band. When at least two clones were the same, the majority sequence was carried to the next step. A majority sequence was found for 49 of the 82 bands. The 49 sequences were submitted to a blast search to identify genomic sequences. Forty-six of the 49 sequences were found in various databases. blast searches were performed by using National Center for Biotechnology Information (www.ncbi.nlm.nih.gov/BLAST/) and the Sanger Centre (www.sanger.ac.uk/Projects/C_elegans/) web sites. Substrate locations in the C. elegans database are: 1TC, W10C8.2 (N terminus) and W03D8.4 (C terminus); 9A, ZC239.6; 16G, St. Louis unfinished data, chromosome III, Y6D11.contig 147; 52G, F55A4.
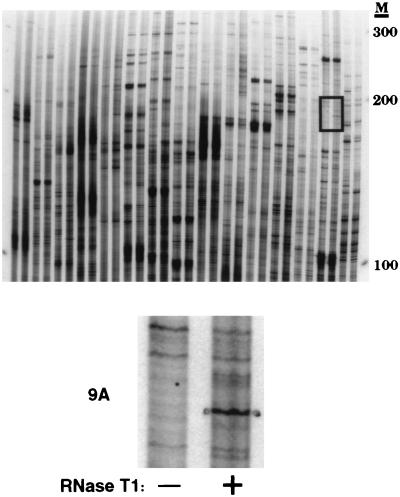
Figure 2.
A differential display gel from the analysis of C. elegans poly(A)+ RNA. A region containing a T1-dependent band (boxed) is enlarged below. Each pair of lanes corresponds to a different primer pair, minus (Left) or plus (Right) RNase T1 (100 units). The dots on each side of the band are pinholes used to mark its position for excision and elution of the DNA. The T1-dependent band was called 9A because it was generated by using upstream arbitrary primer 9 and the GAGACCAGT12CA downstream primer; this nomenclature was used for all T1-dependent bands. M, DNA ladder with numbers in bp.
Limited Primer Extension.
Sequences surrounding editing sites were amplified from cDNA or genomic DNA and used as templates for primer extension. Primers were designed to anneal to the opposite strand a few bases away from the editing site, so that there were no adenosines or guanosines between the primer and site. 5′ End-labeled primers were extended with Sequenase (United States Biochemical) in the presence of either dGTP, dCTP, dTTP, and ddATP or dATP, dCTP, dTTP, and ddGTP. Extension products were separated on a 20% polyacrylamide, 8 M urea gel and quantified by PhosphorImager analysis.
RESULTS
Development of a Method to Identify Inosine-Containing RNAs.
The method we developed to identify additional ADAR substrates is outlined in Fig. 1A. Poly(A)+ RNA was first subjected to a protocol that allows specific cleavage of phosphodiester bonds 3′ to inosines (15). In brief, RNA is treated with glyoxal, followed by digestion with ribonuclease T1 (RNase T1). Although RNase T1 normally cleaves 3′ to both guanosine and inosine, glyoxal reacts specifically with guanosines to preclude T1 cleavage. Cleaved molecules were then identified by using a reverse transcription–PCR strategy based on differential display (ref. 17; see Methods). Each step of the differential display protocol was optimized by monitoring its ability to detect a synthetic RNA containing a single inosine, spiked into the background of yeast RNA to approximate a rare message (Fig. 1B). As another control, we successfully used our differential display strategy to isolate known ADAR substrates (GluR-B and 5HT2C serotonin receptor mRNAs) from rat brain poly(A)+ RNA (data not shown).
Identification of Candidate ADAR Substrates in C. elegans.
By using perfectly duplexed RNA as substrate, ADAR activity has been detected in every metazoan examined, including C. elegans (D.P.M., M. Krause, and B.L.B., unpublished data). The C. elegans genome encodes two apparent ADAR family members (T20H4.4 and H15N14.1), but it is not yet known which, if either, is an active ADAR. We chose to search for new ADAR substrates in C. elegans because its genome sequencing project is essentially complete, which allowed us to easily identify false positives (see below). The procedure of Fig. 1A was performed on two 5-μg samples of C. elegans poly(A)+ RNA that differed only in whether or not they were treated with RNase T1. Because inosine-containing sequences should be amplified only in the presence of RNase T1, candidate ADAR substrates were identified by comparing the two samples. Fig. 2 shows a typical differential display gel that contains an example of an RNase T1-dependent band. PCRs that produced a candidate band were repeated with duplicate cDNA samples. Each reproducibly T1-dependent band was excised from a dried gel, and the DNA was eluted, reamplified, cloned, and sequenced as described (15).
Because guanosines in glyoxalated RNA are not completely resistant to RNase T1, we anticipated that many of the T1-dependent bands would represent false positives from cleavage at guanosines. However, true ADAR substrates (I cleavages) were easily distinguished from false positives (G cleavages) by examining genomic sequence. Because ADARs convert adenosines to inosines, T1 cleavage sites [5′ to the added poly(A) tail] within true ADAR substrates appear as adenosines within genomic sequence; however false positives from cleavage at guanosines will be guanosines in the genome.
The genes for five of 46 candidates showed an adenosine at the RNase T1 cleavage site, indicating that these RNAs, 1TC, 9A, 14TG, 16G, and 52G (see Fig. 2 legend for nomenclature), were likely to be true ADAR substrates. The remaining 41 genomic sequences contained a guanosine at the cleavage site as expected for false positives from T1 cleavage at guanosines.
Confirmation and Characterization of Candidate Substrates.
To eliminate the possibility that any of the remaining five sequences were false positives resulting from errors in the sequence database, or RNase T1 cleavage at adenosine, we conducted further analyses. We verified each genomic sequence and analyzed additional cDNAs to look for known characteristics of ADAR substrates. All but one of the sequences showed characteristics typical of ADAR substrates. For the one exception, 14TG, we did not see additional evidence of A to G changes, at the observed cleavage site or elsewhere. Possibly, 14TG was mistakenly identified as an ADAR substrate because of an artifactual RNase T1 cleavage after adenosine. Alternatively, 14TG may be edited, but inefficiently.
The results of our characterization of 1TC, 9A, 16G, and 52G are summarized in Fig. 3. All of the substrates had the potential to form remarkably stable stem-loop structures. The secondary structures shown in Fig. 3 are based on lowest free-energy predictions using the program mfold (www.ibc.wustl.edu/∼zuker/rna/), and further analyses have not been performed. However, given the stability of the predicted structures, many aspects of the structures are likely correct. Genomic sequences downstream of the RNase T1 cleavage site of 16G recently have appeared in the database and show the potential for a highly base-paired structure similar to those of the other substrates. However, we have not yet verified that this region is expressed as part of the 16G transcript.
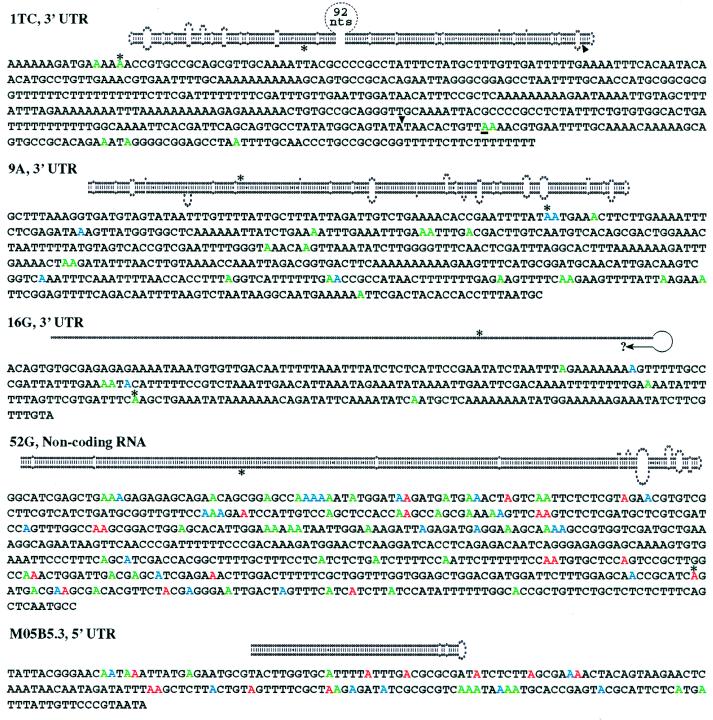
Figure 3.
The predicted structure for each ADAR substrate is shown above its sequence, with observed editing sites shown in color. Ten cDNAs were sequenced for 1TC and 52G and six for all others. Adenosines edited in >70% of the cDNAs are red, those in 40–70% blue, and less than 40% green. For each cDNA, the percentage of total adenosines that appeared as guanosines was determined. 1TC (0–3%), 9A (2–7%), and 16G (0–4%) were selectively edited, whereas 52G (10–22%) and M05B5.5 (23–32%) were deaminated more promiscuously. ∗, RNase T1 cleavage sites detected by differential display; arrowhead (1TC), Tc1 insertion site; question mark (16G), unfinished sequence not yet confirmed by our independent sequencing. The most efficiently edited A in 1TC is underlined. Conclusions about editing site locations in M05B5.3, 9A, and 16G are based on ORFs predicted by genefinder and on our cDNA sequencing. cDNA sequences revealed that two introns predicted by genefinder are not removed by splicing. These are the predicted first intron of M05B5.3 and the predicted last intron of ZC239.6 (9A). Although the edited region in the pop-1 (1TC) message was previously reported to lie within an intron (23), our PCR and Northern blot analyses indicate the region is in the 3′ UTR. cDNA sequences corresponding to 7339–8109 of F55A4 showed 7339 is an SL2 trans-splicing site and 7681–7225 is an intron. A likely polyadenylation signal begins at 8178 that would generate a transcript of a size consistent with Northern analyses. The number of nucleotides sequenced beyond the ends of the indicated structures were as follows (5′, 3′): 1TC (167, 203), 9A (370, 91), and MO5B5.3 (26, 74).
Multiple cDNAs were sequenced for each ADAR substrate, and Fig. 3 shows the observed A to G transitions in the context of the unedited sequence. As expected for ADAR substrates, many cDNAs contained multiple A to G changes, indicative of multiple deamination events within a single molecule. As observed in previously characterized substrates (2), RNAs that were predicted to be almost completely double-stranded (52G; M05B5.3, see below) were deaminated at more sites than those whose structures frequently were interrupted by mismatches, bulges, and loops (1TC, 9A). Further, editing sites showed a 5′ neighbor preference (A:U:C:G = 3:5:2:1) similar to that observed in previously characterized ADAR substrates (18). ADARs require double-stranded substrates, and all of the A to G changes detected by cDNA sequencing occurred within the boundaries of the predicted structures (see legend Fig. 3).
We wanted to verify that the A to G changes observed in the cDNAs were not spurious but correlated with deamination of a significant fraction of the steady-state RNA population. (ADARs do not usually deaminate a given site in all molecules of the population, a characteristic that is important for synthesizing multiple isoforms from a single transcript.) We chose a single editing site from each substrate and performed a limited primer extension assay on cDNA derived from total poly(A)+ RNA. Fig. 4 shows a primer extension assay of the original RNase T1 cleavage site that led to the identification of substrate 9A; ≈48% of the steady-state RNA population is edited at this site. Each site assayed was edited in a significant fraction of the population, and importantly, primer extension of genomic DNA verified each site was an adenosine in the genome.
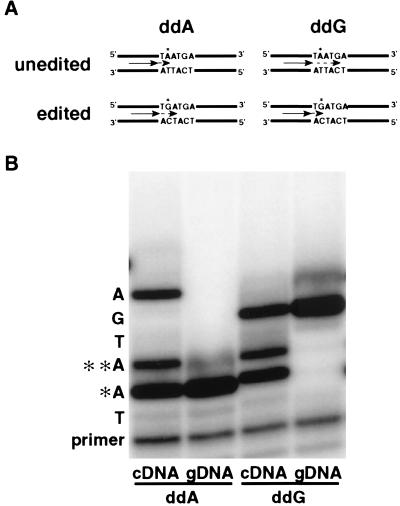
Figure 4.
Limited primer extension assay. (A) Principle: 5′ end-labeled primers (arrows) are extended in the presence of three deoxynucleotides and one dideoxynucleotide (see Methods). With ddA, primers annealed to unedited molecules are extended to the editing site (∗), whereas primers annealed to edited molecules continue to the next A (sense strand). With ddG, edited molecules show a new stop compared with unedited sequences. Dashed arrows indicate primer extension. (B) An autoradiogram showing primer extension products from 9A cDNA or genomic DNA (gDNA). Additional bands in the cDNA lanes verified that the RNase T1 cleavage site (∗) and the next adenosine (∗∗) were edited. Quantification indicated 48% of the 9A RNA molecules were edited at the T1 cleavage site (∗). In similar experiments we quantified the T1 cleavage site of 16G (5%) and the most efficiently edited site in 1TC (25%; underlined, Fig. 3). Assays of the T1 cleavage site in 52G gDNA and cDNA verified the editing site and showed it was edited ≥25%. However, the 52G structure made cDNA synthesis so inefficient that trace amounts of contaminating gDNA were coamplified to become a significant fraction of the sample; thus an exact number could not be obtained.
Inverted Repeat Elements as ADAR Substrates.
Database searches identified substrate 9A as an IR-3 element, and this information led to the identification of the fifth substrate shown in Fig. 3. IR elements are inverted repeats that are found in multiple copies throughout the C. elegans genome (19). Reasoning that other IR elements might be edited, we cloned cDNA corresponding to an IR-5 sequence found within the C. elegans M05B5.3 gene. The cDNA clones showed multiple A to G changes throughout the IR-5 element (Fig. 3). Although we are currently examining other IR elements for editing, assaying inverted repeats likely will reveal only a particular type of substrate. For example, the secondary structures required for editing the Q/R sites of mammalian GluR-5 and GluR-6 pre-mRNAs are formed from highly imperfect inverted repeats that are separated by ≈1,900 nt (20), which would be very hard to identify.
DISCUSSION
As hoped, the ADAR substrates identified with our method suggest new biological roles for ADARs. None of the editing sites occur within codons. The edited regions of 1TC, 9A, and 16G occur within 3′ untranslated regions (UTRs), and that of M05B5.3 in a 5′ UTR (Fig. 3). In these cases, various regulatory mechanisms come to mind. For example, in the absence of deamination, the highly structured 5′ UTR of M05B5.3 would be expected to inhibit translation by impeding scanning of the 40S ribosomal subunit. Because ADARs convert AU base pairs to less stable IU pairs, editing could destabilize the structure to allow translation. For the substrates edited in their 3′ UTRs (1TC, 9A, and 16G), ADARs may act to modulate binding by proteins that regulate translation, localization, or stability (21).
Northern analyses showed that 52G is an abundant polyadenylated RNA (L. Tonkin, D.P.M., and B.L.B., unpublished data), and several observations suggest that it is noncoding. It encodes only short ORFs, each of which is preceded by multiple start codons, and its double-stranded structure encompasses almost the entire ≈1-kb transcript. In addition, sucrose gradient centrifugation experiments showed that 52G RNA was not associated with polysomes (unpublished data). Interestingly, 52G is located on the X chromosome, ≈10 kb away from an ORF that encodes a putative dsRNA binding protein closely related to the Drosophila maternal effect protein, staufen (see F55A4.3 and F55A4.5 in U67949).
M05B5.3 and the genes for 9A and 16G contain predicted ORFs encoding proteins of unknown function. However, 1TC occurs within the 3′ UTR of the pop-1 mRNA, which encodes a transcription factor required for cell-fate decisions during C. elegans development (22, 23). Interestingly, a maternal-effect lethal allele, pop-1 (zu189), contains a Tc1 transposon insertion within the 3′ UTR (23), very close to its most efficiently deaminated adenosine (Fig. 3). The transposon blocks expression of the maternally encoded protein but has no effect on the zygotic protein. Because the Tc1 insertion undoubtedly would inhibit the formation of the double-stranded structure, and thus deamination, we are exploring the possibility that RNA editing is required for normal expression of maternal POP-1.
In closing, we note that even without considering their editing sites, the predicted structures of the ADAR substrates are quite remarkable. Previously, such long, intramolecular, rod-like structures have been observed in plant viroids and viruses such as hepatitis delta virus, but the molecules we observed are within cellular RNA. In mammals, viral dsRNA activates dsRNA binding proteins involved in the interferon response, and it has been speculated that cells have dsRNA that can activate these proteins in the absence of viral infection (24–28). Possibly, C. elegans also has mechanisms for regulation by dsRNA, as suggested by the recent discovery of RNA interference (29). In addition, the dsRNA molecules we identified in C. elegans could have a correlate in mammals. Interestingly, our studies indicate these molecules may be hard to find. Because of their extensive structures, they are difficult to reverse transcribe and thus are underrepresented in cDNA libraries.
During the 12 years since ADARs were discovered, about a dozen endogenous substrates have been stumbled on. Although the differential display method described here took a long time to develop, its application resulted in the identification of five additional substrates in only 2 months. The search for C. elegans substrates is far from saturated, and our technique is applicable to any tissue, of any organism. In particular, it will become feasible to apply our method to mammals as their genome sequences become available. We are hopeful that the substrates we identified, and the method we developed, will lead to rapid progress in understanding the various in vivo roles of ADARs.
Acknowledgments
We thank L. Urness and L. Landweber for helpful discussions, L. Tonkin for assistance with PCR and Northern analyses, V. Maricq, S. Mango, and E. Jorgensen for advice on worm protocols, M. Robertson and her staff for sequencing a large number of cDNA clones, and E. Meenen for synthesizing numerous primers. DNA was sequenced by the University of Utah Health Sciences Sequencing Facility supported by the National Cancer Institute (Grant 5P30CA42014); primers were synthesized by the Howard Hughes Medical Institute oligonucleotide synthesis facility at the University of Utah supported by the Department of Energy (Grant DE-FG03-94ER61817). This work was supported by funds to B.L.B. from the National Institute of General Medical Sciences (GM 44073, National Institutes of Health), and the David and Lucile Packard Foundation. B.L.B. is a Howard Hughes Medical Institute Associate Investigator. D.P.M. was supported by National Institutes of Health Training Grant CA 09602 and a postdoctoral fellowship from the American Cancer Society (PF 3891).
ABBREVIATIONS
- ADAR
adenosine deaminase that acts on RNA
- dsRNA
double-stranded RNA
- UTR
untranslated region
References
- 1.Rueter S, Emeson R. In: Modification and Editing of RNA. Grosjean H, Benne R, editors. Washington, DC: Am. Soc. Microbiol.; 1998. pp. 343–361. [Google Scholar]
- 2.Bass B L. Trends Biochem Sci. 1997;22:157–162. doi: 10.1016/s0968-0004(97)01035-9. [DOI] [PubMed] [Google Scholar]
- 3.Bass B L, Nishikura K, Keller W, Seeburg P H, Emeson R B, O’Connell M A, Samuel C E, Herbert A. RNA. 1997;3:947–949. [PMC free article] [PubMed] [Google Scholar]
- 4.Paul M S, Bass B L. EMBO J. 1998;17:1120–1127. doi: 10.1093/emboj/17.4.1120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bass B L, Weintraub H. Cell. 1987;48:607–613. doi: 10.1016/0092-8674(87)90239-x. [DOI] [PubMed] [Google Scholar]
- 6.Rebagliati M R, Melton D A. Cell. 1987;48:599–605. doi: 10.1016/0092-8674(87)90238-8. [DOI] [PubMed] [Google Scholar]
- 7.Bass B L, Weintraub H. Cell. 1988;55:1089–1098. doi: 10.1016/0092-8674(88)90253-x. [DOI] [PubMed] [Google Scholar]
- 8.Wagner R W, Smith J E, Cooperman B S, Nishikura K. Proc Natl Acad Sci USA. 1989;86:2647–2651. doi: 10.1073/pnas.86.8.2647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Burns C M, Chu H, Rueter S M, Hutchinson L K, Canton H, Sanders-Bush E, Emeson R B. Nature (London) 1997;387:303–308. doi: 10.1038/387303a0. [DOI] [PubMed] [Google Scholar]
- 10.Higuchi M, Single F N, Kohler M, Sommer B, Sprengel R, Seeburg P H. Cell. 1993;75:1361–1370. doi: 10.1016/0092-8674(93)90622-w. [DOI] [PubMed] [Google Scholar]
- 11.Egebjerg J, Heinemann S F. Proc Natl Acad Sci USA. 1993;90:755–759. doi: 10.1073/pnas.90.2.755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lomeli H, Mosbacher J, Melcher T, Hoger T, Geiger J R, Kuner T, Monyer H, Higuchi M, Bach A, Seeburg P H. Science. 1994;266:1709–1713. doi: 10.1126/science.7992055. [DOI] [PubMed] [Google Scholar]
- 13.Polson A G, Bass B L, Casey J L. Nature (London) 1996;380:454–456. doi: 10.1038/380454a0. [DOI] [PubMed] [Google Scholar]
- 14.Bantle J A, Maxwell I H, Hahn W E. Anal Biochem. 1976;72:413–427. doi: 10.1016/0003-2697(76)90549-2. [DOI] [PubMed] [Google Scholar]
- 15.Morse D P, Bass B L. Biochemistry. 1997;36:8429–8434. doi: 10.1021/bi9709607. [DOI] [PubMed] [Google Scholar]
- 16.Liang P, Pardee A B. Science. 1992;257:967–971. doi: 10.1126/science.1354393. [DOI] [PubMed] [Google Scholar]
- 17.Liang P, Bauer D, Averboukh L, Warthoe P, Rohrwild M, Muller H, Strauss M, Pardee A B. Methods Enzymol. 1995;254:304–321. doi: 10.1016/0076-6879(95)54022-9. [DOI] [PubMed] [Google Scholar]
- 18.Polson A G, Bass B L. EMBO J. 1994;13:5701–5711. doi: 10.1002/j.1460-2075.1994.tb06908.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Devine S E, Chissoe S L, Eby Y, Wilson R K, Boeke J D. Genome Res. 1997;7:551–563. doi: 10.1101/gr.7.5.551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Herb A, Higuchi M, Sprengel R, Seeburg P H. Proc Natl Acad Sci USA. 1996;93:1875–1880. doi: 10.1073/pnas.93.5.1875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Decker C J, Parker R. Curr Opin Cell Biol. 1995;7:386–392. doi: 10.1016/0955-0674(95)80094-8. [DOI] [PubMed] [Google Scholar]
- 22.Lin R, Hill R J, Priess J R. Cell. 1998;92:229–239. doi: 10.1016/s0092-8674(00)80917-4. [DOI] [PubMed] [Google Scholar]
- 23.Lin R, Thompson S, Priess J R. Cell. 1995;83:599–609. doi: 10.1016/0092-8674(95)90100-0. [DOI] [PubMed] [Google Scholar]
- 24.Pratt G, Galpine A, Sharp N, Palmer S, Clemens M J. Nucleic Acids Res. 1988;16:3497–3510. doi: 10.1093/nar/16.8.3497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Belhumeur P, Lanoix J, Blais Y, Forget D, Steyaert A, Skup D. Mol Cell Biol. 1993;13:2846–2857. doi: 10.1128/mcb.13.5.2846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jacobs B L, Langland J O. Virology. 1996;219:339–349. doi: 10.1006/viro.1996.0259. [DOI] [PubMed] [Google Scholar]
- 27.Nicholson A W. Prog Nucleic Acid Res Mol Biol. 1996;52:1–65. doi: 10.1016/s0079-6603(08)60963-0. [DOI] [PubMed] [Google Scholar]
- 28.Proud C G. Trends Biochem Sci. 1995;20:241–246. doi: 10.1016/s0968-0004(00)89025-8. [DOI] [PubMed] [Google Scholar]
- 29.Fire A, Xu S, Montgomery M K, Kostas S A, Driver S E, Mello C C. Nature (London) 1998;391:806–811. doi: 10.1038/35888. [DOI] [PubMed] [Google Scholar]