Abstract
Tamoxifen (TAM) is a selective estrogen receptor modulator widely used in the prevention and treatment of breast cancer. A major mode of metabolism of the major active metabolites of TAM, 4-OH-TAM and endoxifen, is by glucuronidation via the UDP-glucuronosyltransferase (UGT) family of enzymes. To examine whether polymorphisms in the UGT enzymes responsible for the glucuronidation of active TAM metabolites play an important role in inter-individual differences in TAM metabolism, cell lines over-expressing wild-type or variant UGTs were examined for their activities against TAM metabolites in vitro. For variants of active extra-hepatic UGTs, the UGT1A8173Ala/277Tyr variant exhibited no detectable glucuronidation activity against the trans isomers of either 4-OH-TAM or endoxifen. No difference in TAM glucuronidating activity was observed for the UGT1A8173Gly/277Cys or UGT1A10139Lys variants as compared to their wild-type counterparts. For active hepatic UGTs, the UGT2B7268Tyr variant exhibited significant (p<0.01) 2- and 5-fold decreases in activity against the trans isomers of 4-OH-TAM and endoxifen, respectively, as compared to wild-type UGT2B7268His. In studies of 111 human liver microsomal specimens, the rate of O-glucuronidation against trans-4-OH-TAM and trans-endoxifen was 28% (p<0.001) and 27% (p=0.002) lower, respectively, in individuals homozygous for the UGT2B7 Tyr268Tyr genotype as compared to subjects with the UGT2B7 His268His genotype, with a significant (p<0.01) trend of decreasing activity against both substrates with increasing numbers of the UGT2B7268His allele. These results suggest that functional polymorphisms in TAM-metabolizing UGTs, including UGT2B7 and potentially UGT1A8, may be important in inter-individual variability in TAM metabolism and response to TAM therapy.
INTRODUCTION
TAM (1-[4-(2-dimethylaminoethoxy)-phenyl]-1,2-diphenylbut-1(Z)-ene)a is a non-steroidal anti-estrogen that has been commonly used for the treatment and prevention of estrogen-dependent breast cancer (1–4). Adjuvant TAM treatment increases recurrence-free survival and overall survival in breast cancer patients with hormone receptor-positive tumors irrespective of their nodal status, menopausal status or age (5). In addition to its anti-estrogenic properties, which have been related to symptoms such as hot flashes, vaginal bleeding and pruritus vulvae (2, 6), TAM also has partial estrogen-agonistic effects that may be linked to reduced risk for ischemic heart disease and osteoporosis (7, 8), but may also increase the risk for endometrial cancer (9, 10) and venous thromboembolism (11). Although TAM is generally well-tolerated, there is significant inter-individual variability in the clinical efficacy as well as toxicities of TAM. For instance, about 30% of patients acquire TAM resistance and relapse (12). In addition, the relative risk of endometrial cancer in patients treated with TAM is estimated to be two- to three-fold that of controls (10, 13–15). The mechanisms underlying variability in response to TAM and to TAM-related toxicities remains obscure. Since there is compelling evidence that TAM is converted to anti-estrogenic metabolites that are more potent than TAM itself, altered patterns of metabolism of TAM and/or its active metabolites might contribute to this inter-individual variability.
TAM is metabolized via cytochrome P450s, primarily CYP2D6 and CYPs 3A4/3A5, into several metabolites after oral administration including the hydroxylated TAM metabolites, 4-OH-TAM and endoxifen. Both 4-OH-TAM and endoxifen are abundant in the serum of TAM-treated patients, with the levels of serum endoxifen approaching 6- to 12-fold that observed for 4-OH-TAM. Since the trans isomers of both 4-OH-TAM and endoxifen exhibit up to 100-fold the levels of anti-estrogenic activity as compared to TAM (16–21), it is thought that they may be the major contributors to TAM’s anti-estrogenic properties. While cis-4-OH-TAM exhibits anti-estrogenic activity in vitro in the presence of estradiol, it has also been suggested to possess some estrogen agonist activity (22–24). The trans isomers of 4-OH-TAM and endoxifen are more abundant than the cis isomers, possibly at a ratio of 70:30, at physiological pH (25, 26).
An important route of elimination and detoxification of TAM and its metabolites is via glucuronidation. TAM is excreted predominantly through the bile primarily by conjugation to glucuronic acid (27), with most of the 4-OH-TAM found in the bile of TAM-treated patients as a glucuronide conjugate (27, 28). TAM glucuronides have also been identified in the urine and serum of TAM-treated patients (27, 28), and it has been suggested that glucuronidation within target tissues like the adipose tissue of the breast may also be important in terms of TAM metabolism and overall TAM activity (29).
N-glucuronidation occurs for both TAM and 4-OH-TAM while O-glucuronidation occurs for 4-OH-TAM and endoxifen (30, 31). In vitro studies have demonstrated that the hepatic UGT1A4 is the only active enzyme responsible for the N-glucuronidation of TAM and 4-OH-TAM while UGT2B7 and, to a lesser extent UGT1A1, are the major hepatic enzymes involved in the O-glucuronidation of the trans isomers of 4-OH-TAM and endoxifen (31). UGT2B7 exhibited higher levels of activity against the trans isomers of 4-OH-TAM and endoxifen; other hepatic UGTs (including UGTs 1A3, 1A9, 2B15, and 2B17) were significantly more active against cis TAM metabolites (31). The extra-hepatic UGTs 1A10 and 1A8 are expressed in target tissues including breast and were also demonstrated to be highly active against isomers of 4-OH-TAM and endoxifen in vitro (31). While previous studies have demonstrated that the UGT1A448Val variant exhibits increased N-glucuronidation activity in vitro against 4-OH-TAM as compared with the wild-type UGT1A448Leu isoform (30), no studies have been performed examining UGT variants and O-glucuronidation activity against 4-OH-TAM or endoxifen. The goal of the present study was to examine whether prevalent missense SNPs in the most active TAM metabolite O-glucuronidating enzymes alters their activity against the trans isomers of 4-OH-TAM and endoxifen and could therefore potentially play an important role in patient response to TAM.
MATERIALS AND METHODS
Chemicals and materials
trans-4-OH-TAM (98% pure), UDPGA, alamethicin, β-glucuronidase and bovine serum albumin were purchased from Sigma-Aldrich (St. Louis, MO). Endoxifen was synthesized in the Organic Synthesis Facility at the Penn State College of Medicine, with the trans-endoxifen isomer purified as described previously (31). HPLC-grade ammonium acetate, acetonitrile and peptide synthesis grade triethylamine were purchased from Fisher Scientific (Pittsburgh, PA) and were used after filtration. Dulbecco’s modified Eagles medium, Dulbecco’s phosphate-buffered saline (minus calcium-chloride and magnesium-chloride), fetal bovine serum, penicillin-streptomycin and geneticin (G418) were purchased from Gibco (Grand Island, NY). The Platinum® Pfx DNA polymerase and the pcDNA3.1/V5-His-TOPO mammalian expression vector were obtained from Invitrogen (Carlsbad, CA) while the restriction enzymes DpnI and StuI were purchased from New England Biolabs (Beverly, MA). The BCA protein assay kit was purchased from Pierce (Rockford, IL) while the QIAEX® II gel extraction kit was purchased from Qiagen (Valencia, CA). The human UGT1A western blotting kit and anti-UGT1A antibody were purchased from Gentest (Woburn, MA). All other chemicals used were purchased from Fisher Scientific (Pittsburgh, PA) unless otherwise specified.
UGT-over-expressing cell lines
The HEK293 cell lines over-expressing the wild-type UGT1A10139Glu, UGT2B7268His and UGT1A8173Ala/277Cys isoforms and the UGT1A10139Gly and UGT2B7268Tyr variants used in this study have been described previously (32–34). The UGT1A8173Gly/277Cys and UGT1A8173Ala/277Tyr variants were generated by site-directed mutagenesis of the pcDNA3.1/V5-His-TOPO plasmid expressing wild-type the UGT1A8 gene as previously described (31, 33) using the QuikChange Site-Directed Mutagenesis Kit (Stratagene). The primers used to change UGT1A8 codon 173 from Ala to Gly were: sense, 5′-TTTAACTTATTTTTTTCGCATTGCAGGAG-3′, and antisense, 5′-CTCCTGCAATGCGAAAAAAATAAGTTAAA-3′, corresponding to nucleotides +349 to +377 relative to the translation start site. The primers used to change UGT1A8 codon 277 from Cys to Tyr were sense, 5′-GTGGTATCAACTACCATCAGGGAAAGCC-3′, and antisense, 5′-GGCTTTCCCTGATGGTAGTTGATACCAC-3′, corresponding to nucleotides +815 to +843 relative to the translation start site. The underlined base for each primer indicates the base-pair change. Similar to that described previously for site-directed mutagenesis-generated UGT variants (31, 33), the UGT1A8173Gly/277Cys and UGT1A8173Ala/277Tyr cDNA sequences were confirmed by dideoxy sequencing prior to transfection by electroporation into the HEK293 (human embryonic kidney fibroblast) cell line as previously described (31, 33). Cells were grown in Dulbecco’s Modified Eagle’s medium to 80% confluence prior to the preparation of cell homogenates as previously described (34). Total homogenate protein concentrations were measured using the BCA protein assay.
UGT protein levels were determined by Western blot analysis for all UGT-over-expressing cell lines examined in this study as previously described (33). For UGT1A-over-expressing cells, the UGT1A antibody from Gentest was utilized; for UGT2B-over-expressing cells, a previously described UGT2B-specific antibody was used (31). Relative UGT protein levels were expressed as the mean of three independent experiments, and all activity assays were normalized relative to UGT expression in the respective UGT-over-expressing cell line.
HLM
Normal human liver tissue specimens (n=111) were obtained from the Tissue Procurement Facility at the H. Lee Moffitt Cancer Center (Tampa, FL) and include 78 liver specimens that were examined in previous studies (34, 35). Microsomes (HLM) were prepared as previously described (34) and stored at 10–20 mg protein/mL at −80°C. Matching genomic DNA was prepared from nuclei isolated during the microsomal differential centrifugation preparation procedure for each specimen using standard phenol chloroform techniques. Microsomal protein concentrations were measured using the BCA protein assay. Matching total RNA was obtained for each specimen directly from Moffitt’s Tissue Procurement Facility and was stored at −80°C.
Glucuronidation assays
The glucuronidation activities of homogenates from HEK293 cell lines over-expressing UGTs 1A8, 1A10 and 2B7 against trans-4-OH-TAM and trans-endoxifen were performed as previously described (30, 31). Briefly, after an initial incubation of cell homogenate protein (100–1000 μg) or HLM (2.5 μg total protein) with alamethicin (50 μg/mg protein) for 15 min in an ice bath, glucuronidation reactions were performed in a final reaction volume of 100 μL (cell homogenates) or 25 μL (HLM) at 37°C in 50 mM Tris-HCl (pH 7.4), 10 mM MgCl2, 4 mM UDPGA, and 1 to 250 μM trans-4-OH-TAM or 8 to 725 μM trans-endoxifen for cell homogenate glucuronidation assays, and 4 μM trans-4-OH-TAM or 30 μM trans-endoxifen for HLM glucuronidation assays. Kinetic analysis of HLM from subjects with varying UGT2B7 genotypes was performed using 0.5–15 μM trans-4-OH-TAM and 2–60 μM trans-endoxifen in five HLM specimens from individual subjects from each UGT2B7 genotype group (15 HLM specimens total). Reactions were terminated by the addition of 100 μl cold methanol on ice. Mixtures were centrifuged for 10 min at 4°C at 16,100 g prior to the collection of the supernatant. All glucuronidation assays were performed in triplicate for cell homogenate assays and duplicate for HLM assays in independent experiments.
TAM metabolites were assessed for cell homogenate glucuronidation assays by HPLC identical to that described previously (30, 31). Assays of TAM metabolite formation in HLM were analyzed by UPLC/MS/MS using a Waters (Milford, MA) Aquity UPLC consisting of a binary gradient pump, an autosampler (maintained at 4°C), and a UV detector operated at 254 nm, attached to a Waters TQD triple quadrupole mass spectrometer that was purchased after the analysis of UGT-over-expressing cell lines (described above). Similar to that described previously (26), each sample was injected onto an Aquity UPLC BEH C18 1.7 μM, 2.1X100 mm column (Waters) with the following gradient elution conditions for trans-4-OH-TAM: starting with 69% buffer A (0.01mol/L NH4Ac, pH 5.0)/31% acetonitrile for 2 min with a subsequent linear gradient to 75% acetonitrile over 2 min. The gradient elution conditions for trans-endoxifen, using the same buffers, were as follows: starting with 30% acetonitrile for 4 min and a subsequent linear gradient to 75% acetonitrile for 2 min. The elution flow rate was 0.5 mL/min and 5 μL of the reaction was injected for all assays. Electrospray ionization mass spectrometry (ESI-MS) daughter scans of 564 and 550 (m+1/z) verified the glucuronides of trans-4-OH-TAM and trans-endoxifen. The formation of both O- and N+-glucuronides of 4-OH-TAM and endoxifen were quantified by UPLC based on the ratio of the glucuronide versus free trans-4-OH-TAM or trans-endoxifen. HLM assays without TAM metabolite were regularly analyzed as negative controls for glucuronidation activity as previously described (34).
For 4-MU, glucuronidation assays were performed as described above with an incubation time of 120 min. HPLC analysis utilized the following gradient program: starting with 98% buffer A (100 mM NH4Ac, pH 5.0)/2% acetonitrile for 5 min, a linear gradient to 70% acetonitrile over 17.5 min was performed and then maintained at 70% for 10 min.
UGT Genotyping
Genomic DNA from the 111 liver specimens examined for glucuronidation activity in this study were used to genotype the UGT2B7 codon 268 (His>Tyr) polymorphism, the UGT1A1*28 TATAA box polymorphism (see GenBank wild-type accession number NM_000463 and SNP rs8175347), and the UGT1A4 codon 24 (Pro>Thr) and codon 48 (Leu>Val) polymorphisms. UGT2B7 and UGT1A4 genotypes were determined by direct sequencing of PCR-amplified PCR products spanning the codon 268 polymorphism for UGT2B7, and codons 24 and 48 for UGT1A4. The same primers were used for both PCR amplification and sequencing; UGT2B7: sense, 5′-CTATAGTGCTTTACTTTGACTTTTGGTTCG-3′ and antisense, 5′-GCTAGAAAAGCAAAGAAGGGAAAAAATGATTAGTTATATCTGA-3′, corresponding to nucleotides +642–+670 and +1555–+1597, respectively, relative to the UGT2B7 translation start site (Genbank accession #NM_001074); UGT1A4: sense, 5′-GGCTTCTGCTGAGATGGCCAG-3′, and antisense, 5′-CCTTGAGTGTAGCCCAGCGT-3′, corresponding to nucleotides located −13 to +8 and +277 to +306, respectively, relative to the UGT1A4 translation start site (Genbank accession #NM_007120). Sequencing was performed using an ABI 3130 Capillary Sequencer at the Functional Genomics Core Facility at the Penn State College of Medicine.
The UGT1A1*28 polymorphism was genotyped utilizing DNA fragment analysis by capillary electrophoresis on the ABI 3130 Capillary Sequencer at the Penn State Molecular Genetics Core Facility using primers and PCR conditions similar to those described previously (36) using 0.5 μL of a size standard (GeneScan 500 LIZ Size Standard, Applied Biosystems, Foster City, CA) as a DNA size marker. Informative results were obtained for 105 of the 111 liver specimens examined in this study.
UGT2B7 codon 268 genotypes were determined primarily by real-time PCR assays using the TaqMan Drug Metabolism Genotyping Assay C_32449742_20 (Applied Biosystems, Foster City, CA) in the ABI 7900HT sequence detection system equipped with an autoloader in the Functional Genomics Core Facility at the Penn State College of Medicine. Forty percent of all samples within each of the three potential UGT2B7 genotype groups (His268His, His268Tyr and Tyr268Tyr; as identified by real-time PCR) were further confirmed by standard PCR and direct sequencing. The same primers were used for both PCR and sequencing: sense, 5′-CTATAGTGCTTTACTTTGACTTTTGGTTCG-3′, located +642–+670 from the UGT2B7 translation start site; and antisense, 5′-GCTAGAAAAGCAAAGAAGGGAAAAAATGATTAGTTATATCTGA-3′, located +1555–+1597 from the UGT2B7 translation start site. For those samples for which real-time assays of UGT2B7 genotypes were inconclusive (n=13), PCR/direct sequencing analysis was performed as described above. For eight of these samples, both real-time and direct sequencing failed to provide informative results, and direct sequencing of UGT2B7 cDNA was then performed for these samples using matching total RNA as template. Briefly, after reverse transcription-PCR was performed using standard conditions with 5 μg total liver RNA as template and oligo(dT) as primer, PCR was performed using 2 μL of cDNA as template and sense (5′-TGCAGATGCTATTTTTCCCTGTA-3′) and antisense (5′-GAACCTTTTGTGGGATCTGGGCC-3′) primers located +456 to +478 and +984 to +1006, respectively, from the UGT2B7 translation start site. Direct sequencing of these RT-PCR-amplified fragments was then performed using the same primers used for PCR.
UGT2B7 expression analysis
Matching total RNA was available for expression analysis for 99 of the 111 liver specimens analyzed in this study. Five ug of RNA was used for cDNA synthesis using standard reverse transcription methods as described above, with 20 ng of cDNA used for expression analysis using the ABI gene expression kit assay for UGT2B7 (Hs02556232_s1; Applied Biosystems, Foster City, CA) with the ABI 7900HT detection system equipped with an autoloader in the Functional Genomics Core Facility at the Penn State College of Medicine. Expression assays were performed in triplicate with expression normalized relative to the expression levels of the housekeeping gene PPIA within the same samples.
Statistical analysis
The Student’s t-test (2-sided) was used for comparing kinetic values of glucuronidation formation for UGT wild-type versus variant over-expressing cell lines, and for HLM glucuronidation rates stratified by UGT genotypes. The one-way ANOVA trend test was used to compare HLM glucuronidation rates across multiple UGT genotypes. Kinetic constants were determined using Graphpad Prism4 software.
RESULTS
Kinetic studies of TAM metabolite glucuronidation by UGT variants
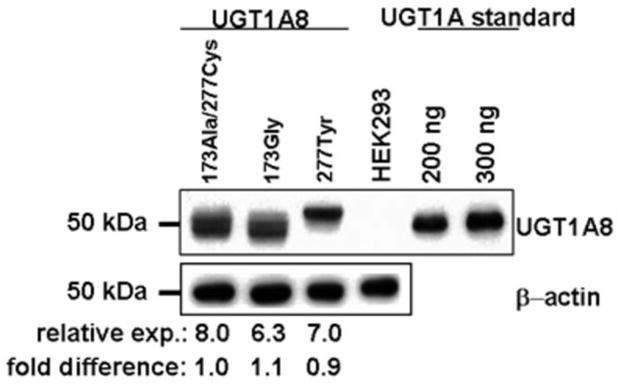
Results from previous studies demonstrated that the hepatic UGT2B7 and the extra-hepatic UGTs 1A8 and 1A10 exhibited the highest overall glucuronidating activities against trans-4-OH-TAM and trans-endoxifen (31). Known missense polymorphisms have been identified for each of these UGTs, including a highly-prevalent SNP at codon 268 of the UGT2B7 gene (32), a codon 139 SNP in the UGT1A10 gene that is most prevalent in African Americans (37), and two coding region SNPs that result in amino acid changes of Ala to Gly at codon 173 and Cys to Tyr at codon 277 of the UGT1A8 gene (38). To determine whether any of these SNPs result in differential activities against the trans isomers of 4-OH-TAM or endoxifen, in vitro kinetic analysis of HEK293 cells over-expressing the wild-type or variant isoforms of each of these three UGT enzymes was performed. While the levels of expression of wild-type versus variant UGT1A10 protein was determined for UGT1A10 over-expressing cell lines in previous studies (33), semi-quantitative Western blot analysis was performed for the UGT1A8- and UGT2B7-over-expressing cell lines. As shown in Figure 1, each of the UGT1A8-over-expressing cell lines developed for the present analysis exhibited similar levels of expression (8.0, 6.3 and 7.0 ng UGT1A8/μg total protein for the UGT1A8173Ala/277Cys-, UGT1A8173Gly/277Tyr- and UGT1A8173Ala/277Tyr-over-expressing lines, respectively). As described previously in other laboratories for the UGT2B7-over-expressing cell lines (32), UGT2B7268His was consistently expressed at a level that was 3.3-fold that of the UGT2B7268Tyr variant in the present analysis (results not shown). These values were used for normalization of UGT1A8 and UGT2B7 levels in their respective UGT-over-expressing cell lines in in vitro kinetic analysis.
Figure 1. Western blot analysis of UGT 1A8 variants from UGT-overexpressing cell lines.
Equal amounts of protein (30 μg) from untransfected HEK293 cells, and from UGT1A8173Ala/277Cys-, UGT1A8173Gly/277Cys-, and UGT1A8173Ala/277Tyr-over-expressing HEK293 cells, were loaded into each lane for Western blot analysis. β-Actin was examined as an internal control for protein loading for each lane, and 200 or 300 ng of UGT1A protein standards (Gentest) were loaded as a gel-loading reference for UGT protein quantification. The relative ratios of UGT1A8 : β-actin protein expression are from three independent experiments and their expression relative to the wild-type UGT1A8173Ala/277Cys are indicated below each lane.
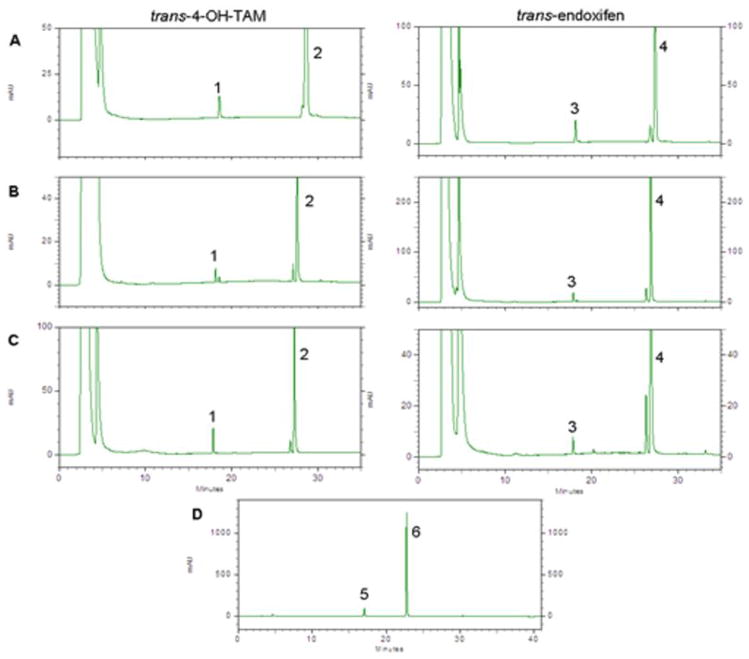
Representative HPLC traces of glucuronidation formation by UGT-over-expressing cell homogenates are shown in Figure 2. As described previously for cell lines over-expressing wild-type UGTs 1A8, 1A10 or 2B7 (31), significant levels of O-glucuronidation of the trans isomers of 4-OH-TAM or endoxifen were observed in the present studies; no N-glucuronidation of these TAM metabolites was observed for these UGTs. The UGT1A8173Gly/277Cys variant exhibited no difference in overall glucuronidation activity (Vmax/KM) against trans-4-OH-TAM and exhibited a small (1.25-fold) but significant (p< 0.05) decrease in overall activity (manifested primarily by a higher KM) against trans-endoxifen as compared to wild-type UGT1A8173Ala/277Cys (Table 1). In contrast, the UGT1A8173Ala/277Tyr variant exhibited no detectable glucuronidation against the trans isomers of either 4-OH-TAM or endoxifen (Table 1). However, the UGT1A8173Ala/277Tyr variant did exhibit detectable levels of activity against 4-MU (Figure 2, panel D). No difference in overall glucuronidation activity was observed for the UGT1A10139Lys variant versus wild-type UGT1A10 against the trans isomers of 4-OH-TAM and endoxifen.
Figure 2. HPLC analyses of over-expressing cell line glucuronidating activities.
Shown are HPLC traces of trans-TAM-4-O-glucuronide and trans-endoxifen-O-glucuronide formation by, (A) UGT1A8173Ala/277Cys-, (B) UGT1A10139Glu-, and (C) UGT2B7268His-over-expressing HEK293 cells using 500 μg homogenate protein in glucuronidation activity assays as described in the Materials and Methods. Panel D shows 4-MU-O-glucuronide formation using 1 mg UGT1A8173Ala/277Tyr-over-expressing cell homogenate. Over-expressing cell homogenates were incubated at 37°C for 60 or 120 min for assays with TAM metabolites or 4-MU, respectively, prior to analysis by HPLC as described in the Materials and Methods. Peak 1, trans-4-TAM-O-glucuronide; peak 2, trans-4-OH-TAM; peak 3, trans-endoxifen-O-glucuronide; peak 4, trans-endoxifen; peak 5, 4-MU-O-glucuronide; peak 6, 4-MU.
Table 1.
Kinetic analyses of O-glucuronidation of the trans isomers of 4-OH-TAM and endoxifen by UGT variants.a
|
trans-4-OH-TAM |
trans-endoxifen |
|||||
|---|---|---|---|---|---|---|
| UGT Variant | Vmax (pmol•min−1•μg−1)b |
KM (μM) |
Vmax/KM (μl•min−1•μg−1)b |
Vmax (pmol•min−1•μg−1)b |
Km (μM) |
Vmax/KM (μl•min−1•μg−1)b |
| UGT2B7268His | 0.55 ± 0.18 | 3.7 ± 0.6 | 0.15 ± 0.03 | 3.0 ± 0.44 | 101 ± 17 | 0.030 ± 0.004 |
| UGT2B7268Tyr | 0.54 ± 0.09* | 8.7 ± 0.8** | 0.062 ± 0.01** | 0.55 ± 0.01** | 101 ± 15 | 0.006 ± 0.001** |
| UGT1A10139Glu | 4.7 ± 0.3 | 96 ± 8 | 0.049 ± 0.006 | 5.7 ± 0.7 | 40 ± 3 | 0.14 ± 0.005 |
| UGT1A10139Lys | 2.1 ± 0.2** | 52 ± 6** | 0.040 ± 0.006 | 1.9 ± 0.2** | 13 ± 2** | 0.14 ± 0.004 |
| UGT1A8173Ala/277Cys | 2.3 ± 0.1 | 23 ± 2 | 0.10 ± 0.02 | 5.4 ± 0.2 | 98 ± 9 | 0.060 ± 0.004 |
| UGT1A8173Gly/277Cys | 5.4 ± 0.2** | 43 ± 7** | 0.13 ± 0.03 | 5.9 ± 0.4 | 135 ± 26 | 0.040 ± 0.005* |
| UGT1A8173Ala/277Tyr | no detectable activity | no detectable activity | ||||
All data are the mean ± S.D. based on three independent experiments. Homogenates from cells over-expressing UGT1A8173Ala/277Tyr exhibited no detectable activity against trans-4-OH-TAM and trans-endoxifen.
Data are expressed per μg UGT protein as determined by Western blot analysis.
p ≤ 0.05;
p<0.01.
Kinetic analysis demonstrated that significantly higher glucuronidation activities were observed for the wild-type UGT2B7268His as compared to the UGT2B7268Tyr variant against the trans isomers of both 4-OH-TAM (p<0.05) and endoxifen (p<0.01; Table 1). This was manifested by a higher KM (2.4-fold) and a lower Vmax/KM (2.4-fold) for 4-OH-TAM, as well as a lower Vmax (5.5-fold) and lower Vmax/KM (5.0-fold) for endoxifen.
Glucuronidation activities of HLM against the trans isomers of 4-OH-TAM and endoxifen stratified by UGT genotypes
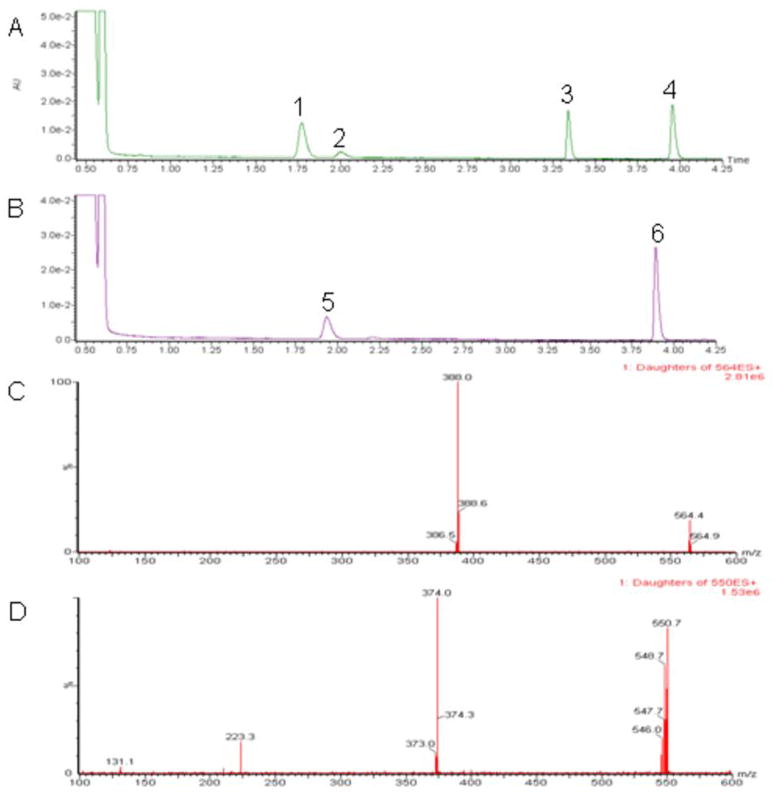
To determine the rate of glucuronidation of trans-4-OH-TAM and trans-endoxifen, glucuronidation assays were performed for 111 HLMs and analyzed by UPLC/MS/MS. The concentrations of 4-OH-TAM or endoxifen used in the HLM glucuronidation activity assays was determined after kinetic analysis of three randomly-chosen HLM specimens - the resulting KM’s were 4 μM and 30 μM for trans-4-OH-TAM and trans-endoxifen, respectively (data not shown). Using 4 μM trans-4-OH-TAM as substrate in HLM glucuronidation activity assays, two major putative glucuronide peaks were observed, the TAM-4-O-glucuronide and the 4-OH-TAM-N+-glucuronide, which exhibited retention times of 1.76 and 3.35 min, respectively, distinct from free trans-4-OH-TAM which eluted at 3.95 min (Figure 3, panel A). Using 30 μM trans-endoxifen as substrate in HLM glucuronidation activity assays, a single major putative glucuronide peak was observed at a retention time of 1.95 min which was distinct from the endoxifen peak eluting at 3.90 min (Figure 3, panel B). All putative glucuronide peaks were sensitive to treatment with β-glucuronidase (results not shown), eluted at retention times identical to previously characterized TAM-glucuronide standards (26); results not shown), and were confirmed by tandem MS (Figure 3, panels C–D; the MS/MS pattern observed for trans-TAM-4-O-glucuronide and trans-4-OH-TAM-N+-glucuronide are identical). For 4-OH-TAM glucuronidation assays, a third, smaller peak eluting at a retention time of 2.02 min was confirmed to be cis-TAM-4-O-glucuronide, likely formed due to spontaneous interconversion between the trans and cis 4-OH-TAM isomers (34). Similar to that observed in previous studies for three HLM specimens (31), no endoxifen-N-glucuronide was observed with any of the 111 HLM examined in the present analysis. The mean rate of formation of TAM-4-O-glucuronide, 4-OH-TAM-N+-glucuronide and endoxifen-O-glucuronide in HLM was 141 ± 45, 175 ± 52 and 168 ± 66 pmol•min−1•mg−1, respectively. A 4.5-, 10-, and 17-fold range in glucuronide formation was observed for TAM-4-O-glucuronide, 4-OH-TAM-N+-glucuronide, and endoxifen-O- glucuronide, respectively. The range of the ratio of TAM-4-O-glucuronide : 4-OH-TAM-N+-glucuronide in the HLM samples was 8.0-fold.
Figure 3. UPLC analyses of HLM glucuronidating activities against TAM metabolites.
Representative UPLC traces of glucuronidation assays incubated with HLM (40 μg) and either (A) trans-4-OH-TAM (4 μM) or (B) trans-endoxifen (30 μM). Tandem MS/MS traces of putative glucuronide peaks from HLM glucuronidation activity assays are shown in panels (C) for trans-TAM-4-O-glucuronide, and (D) for trans-endoxifen-O-glucuronide. Peak 1, trans-TAM-4-O-glucuronide; peak 2, cis-TAM-4-O-glucuronide; peak 3, trans-4-OH-TAM-N+-glucuronide; peak 5, trans-endoxifen-O-glucuronide; peak 6, trans-endoxifen.
As described above, previous studies have indicated that UGT2B7 is the major hepatic enzyme that performs O-glucuronidation of the trans isomers of both 4-OH-TAM and endoxifen (31). When stratifying HLM O-glucuronidation activities by UGT2B7 codon 268 genotype, there was a near-significant (p=0.059) 13% decrease in TAM-4-O-glucuronide formation in HLM with the UGT2B7 (His268Tyr) genotype and a significant (p<0.001) 28% decrease in TAM-4-O-glucuronide formation in HLM with the UGT2B7 (Tyr268Tyr) genotype as compared to HLM with the UGT2B7 (His268His) genotype (Figure 4, panel A). A significant (p=0.01) 17% decrease in TAM-4-O-glucuronide formation was observed in HLM with the UGT2B7 His268Tyr genotype versus HLM with the UGT2B7 (Tyr268Tyr) genotype. A significant trend of decreasing O-glucuronidation of trans-4-OH-TAM was observed in HLM with increasing numbers of the UGT2B7268Tyr allele (p<0.001).
Figure 4. Analysis of glucuronidation activities against trans-4-OH-TAM and trans-endoxifen in HLM stratified by UGT2B7 genotypes.
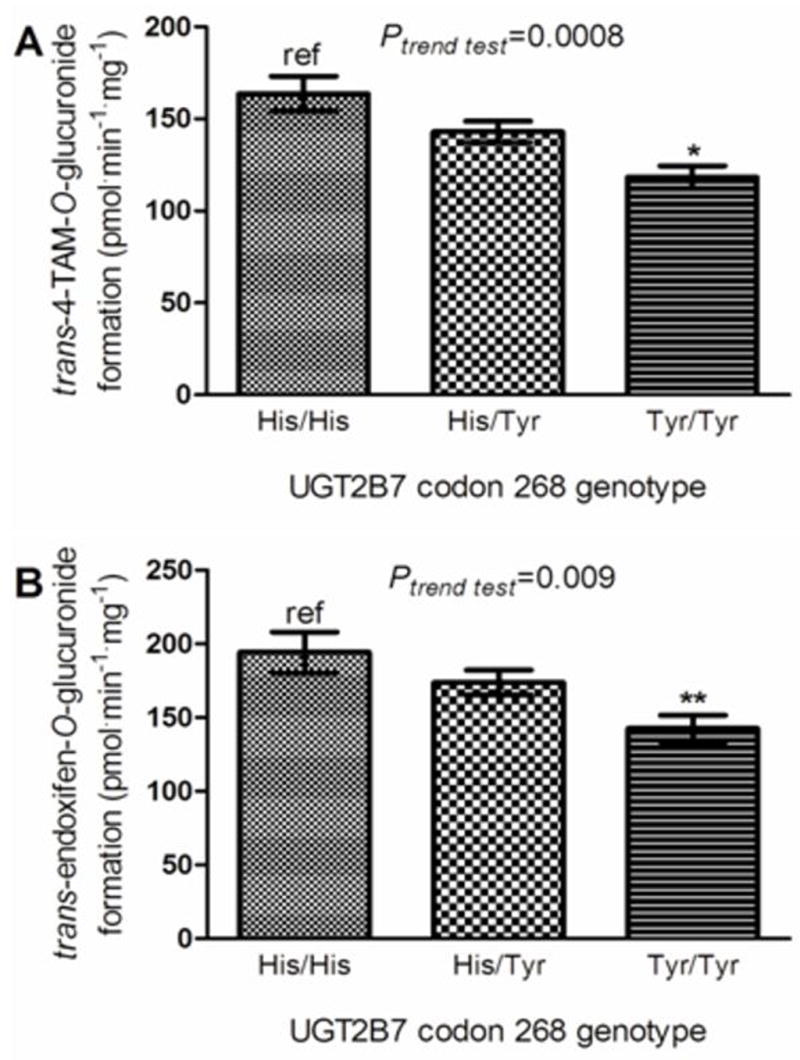
Glucuronidation activity assays were performed and 4-OH-TAM- and endoxifen-glucuronides were separated by UPLC as described in the Materials and Methods. Shown are the rate of O-glucuronide formation from, (A) trans-4-OH-TAM, stratified by UGT2B7 codon 268 genotypes; and (B) trans-endoxifen, stratified by UGT2B7 codon 268 genotypes. Comparative analysis was performed using the wild-type UGT2B7268His as the referent; * denotes p< 0.001; ** denotes p < 0.002, and error bars represent standard error.
Similar to that observed for trans-4-OH-TAM, a significant (p=0.002) 27% decrease in O-glucuronidation of trans-endoxifen was observed in HLM with the UGT2B7 (Tyr268Tyr) genotype as compared to HLM with the UGT2B7 (His268His) genotype (Figure 4, panel B). In addition, a significant trend of decreasing O-glucuronidation of trans-endoxifen was observed in HLM with increasing numbers of the UGT2B7268Tyr allele (p=0.009). The mean level of UGT2B7 expression was similar in liver specimens within each of the UGT2B7 genotype groups, with specimens with the UGT2B7 (His268His) genotype exhibiting a 4% higher level of expression than liver specimens with the UGT2B7 (Tyr>Tyr) genotype (data not shown). In an analysis of five randomly-chosen HLM specimens from subjects from each UGT2B7 genotype group (ie, 5 HLM per genotype = 15 total HLM specimens), the resulting KM’s for trans-4-OH-TAM followed a non-significant decreasing trend of 9, 7, and 5 μM, respectively, for the UGT2B7 His268His, His268Tyr, and Tyr268Tyr genotypes (data not shown). The resulting KM’s (16 μM) for trans-endoxifen were similar across UGT2B7 genotypes (data not shown).
To determine whether genotypes in other hepatic UGTs shown previously to exhibit activity against the trans isomers of 4-OH-TAM or endoxifen were similarly linked to altered HLM glucuronidation phenotype, HLM glucuronidation activities were stratified by UGT1A1 and UGT1A4 genotypes (30, 31). Non-significant decreases in O-glucuronidation activity of 14 and 11% were observed against the trans isomers of 4-OH-TAM and endoxifen, respectively, in HLM with the UGT1A1 (*28/*28) genotype as compared to HLM with the UGT1A1 (*1/*1) genotype; this decrease remained non-significant when combining HLM with either the UGT1A1 (*28/*28) or (*1/*28) genotypes (results not shown). No significant associations were observed for either the UGT1A4 codons 24 (Pro>Thr) or 48 (Leu>Val) polymorphisms and HLM N-glucuronidation activity against 4-OH-TAM (results not shown).
Discussion
The present study examines the potential role of UGT polymorphisms on the metabolism of the trans isomers of 4-OH-TAM and endoxifen, the major active metabolites of tamoxifen. Several UGTs were previously shown to be active against these metabolites, with UGT2B7 the most active hepatic UGT. As UGT2B7 expression has been detected in a variety of tissues including liver, the gastrointestinal tract and breast (39–43), variations in UGT2B7 function or expression could potentially significantly impact individual response to drugs or chemotherapeutic agents. The data presented in this study demonstrate that O-glucuronidation of both trans-4-OH-TAM and trans-endoxifen in HLM was significantly associated with UGT2B7 genotype, with lower activities correlated with increasing numbers of the UGT2B7268Tyr allele. These data were consistent with the observation that HEK293 cells that over-expressed the UGT2B7268Tyr variant exhibited lower activity in vitro against both TAM metabolites as compared to cells over-expressing wild-type UGT2B7268His. These results are also consistent with a functional role for this polymorphism against other substrates including tobacco carcinogen metabolites like 4-(methylnitrosamino)-1-(3-pyridyl)-1-butanol [NNAL; (34)].
Previous studies have observed a SNP located at −161(T>C) relative to the ATG transcriptional start site in the promoter of UGT2B7 that is in complete linkage disequilibrium with the SNP at UGT2B7 codon 268 (42). In the present study, UGT2B7 genotype was also not associated with a change in UGT2B7 expression, as there was less than a 4% difference in expression levels between HLM with the UGT2B7 (His268His) versus (Tyr268Tyr) genotypes, a pattern similar to that observed in previous studies (44). While recent studies have indicated that the intronic SNP IVS1 + 985A>G is associated with altered UGT2B7 expression and the formation of morphine glucuronide metabolites (44), in an analysis of a subset of the liver specimens examined in the present study (n=45), specimens containing the IVS1 + 985G variant did not result in a change in UGT2B7 expression or a change in activity against TAM metabolites as compared to HLM containing the wild-type IVS1 + 985A (results not shown). These data suggest that the decrease in O-glucuronidation activity against TAM metabolites in HLM associated with the UGT2B7 codon 268 polymorphism is indeed due to functional changes within the UGT2B7 enzyme.
In addition to UGT2B7, polymorphisms in UGTs 1A1 and 1A4, which were previously shown to be active against TAM, trans--4-OH-TAM and/or trans-endoxifen (30, 31), were examined for their effect on HLM glucuronidation activity. A microsatellite (TA)-repeat polymorphism present in the TATAA-box of the UGT1A1 promoter has been linked to lower UGT1A1 expression (45, 46) and lower activity against a variety of endogenous and exogenous substrates, including bilirubin (45, 47, 48), carcinogens such as metabolites of benzo(a)pyrene (45), and chemotherapeutic agents such as SN-38, the major metabolite of irinotecan (49, 50). The non-significant trend of decreased glucuronidation activity against TAM metabolites that was observed in HLMs from subjects with one or more UGT1A1*28 alleles is consistent with previous studies indicating that UGT1A1 exhibits only weak relative activity against these substrates as compared to UGT2B7 (31) and may therefore play a more minor role in TAM metabolism.
The N-glucuronidation of TAM and 4-OH-TAM was previously shown to be performed exclusively by UGT1A4 (30, 51). While previous studies also demonstrated that the UGT1A4 codon 48 polymorphism was linked to a small but significant alteration in N-glucuronidation activity against TAM and 4-OH-TAM in vitro (30), no significant difference was observed in HLM against trans-4-OH-TAM in the present study. This may be due to the relatively low number of HLM specimens that were heterozygous (n=13) or homozygous (n=1) for the UGT1A448Val variant in this study.
The extra-hepatic UGTs 1A10 and 1A8 exhibited the highest levels of activity in vitro against the trans isomers of 4-OH-TAM and endoxifen in previous studies (31). In the present study, no effect on TAM metabolite glucuronidation activity was observed in vitro for the UGT1A10139Lys variant as compared to wild-type UGT1A10139Glu. Similarly, the UGT1A8173Gly variant exhibited a marginally significant lower overall in vitro glucuronidating activity against trans-endoxifen as determined by Vmax/KM as compared to wild-type UGT1A8173Ala; no significant difference was observed for this variant against trans-4-OH-TAM. This relatively minor effect on TAM metabolite glucuronidating activities is consistent with the fact that the Ala>Gly amino acid substitution at UGT1A8 codon 173 is a conservative non-polar amino acid change and with data from previous in vitro metabolic studies that revealed that UGT1A8173Ala and UGT1A8173Gly exhibit similar catalytic properties (38, 52). Interestingly, the UGT1A8277Tyr variant exhibited no detectable glucuronidating activity against both trans-4-OH-TAM and trans-endoxifen. This is consistent with previous data indicating that this variant exhibited dramatically reduced activity towards other substrates (38, 52). While the prevalence of this polymorphism is low in the population [~2% in Caucasians; (38)], the observation that UGT1A8 is highly active against TAM metabolites and is well-expressed in the breast (53, 54) suggests that, like the UGT2B7 codon 268 polymorphism, the UGT1A8 codon 277 polymorphism could potentially be important in individual response to TAM.
The results described above are consistent with that observed previously for functional polymorphisms in the CYP2D6 gene. Decreased levels of endoxifen were observed in the serum of TAM-treated women after stratifying patients by the CYP2D6 deletion genotype (55), a decrease also observed when CYP2D6 inhibitors were co-administered with TAM (19). These data suggest an important role for endoxifen in TAM therapeutic efficacy. The CYP2D6*4 deletion allele has been associated with time until breast cancer recurrence, relapse-free survival, disease-free survival, and overall survival in patients treated with TAM (19, 55, 56). In addition, patients with the CYP2D6*4 genotype report few, if any, occurrences of hot flashes (55). Despite a strong correlation between CYP2D6 genotype and serum levels of endoxifen in patients treated with TAM, large variability in circulating endoxifen levels are still observed after controlling for CYP2D6 genotype (19, 55). The evidence presented in the present study suggests that inter-individual differences in TAM glucuronidation pathways may help explain this variability.
In summary, results from this study suggest that genetic variants in UGTs that are highly active against TAM metabolites significantly alter TAM metabolism in vitro and, potentially, its elimination in TAM-treated individuals. Similar to that described above for CYP2D6, this could potentially affect overall patient response to TAM. Additional studies examining the effect of UGT1A8 and UGT2B7 genotypes on breast microsomal glucuronidation activity against TAM metabolites, plasma TAM metabolite levels, and overall patient response to TAM will be required to further examine the role of UGT polymorphisms on the therapeutic efficacy of TAM.
Acknowledgments
We thank the Functional Genomics and Molecular Biology Core Facilities at the Penn State University College of Medicine for DNA genotyping, DNA sequencing, and usage of densitometric equipment. These studies were supported by Public Health Service (PHS) grants R01-DE13158 (National Institute for Dental and Craniofacial Research) from the National Institutes of Health, Department of Health and Human Services (to P. Lazarus).
Grant support: USPHS grants R01-DE13158 (National Institute for Dental and Craniofacial Research) from the NIH, Department of Health and Human Services (P. Lazarus), and a formula grant under the Pennsylvania Department of Health’s Health Research Formula Funding Program, State of PA, Act 2001-77–part of the PA Tabacco Settlement Legislation (P. Lazarus).
Footnotes
Abbreviations: TAM, tamoxifen; 4-OH-TAM, 4-hydroxytamoxifen; endoxifen, 4-hydroxy-N-desmethylTAM; UGT, UDP-glucuronosyltransferase; SNP, single nucleotide polymorphism; UDPGA, UDP-glucuronic acid; 4-OH-TAM-N+-Gluc, 4-hydroxytamoxifen quaternary ammonium glucuronide; HLM, human liver microsomes; HPLC, high pressure liquid chromatography; LC-MS, liquid chromatography-mass spectrometry; UPLC/MS/MS, ultra-performance liquid chromatography attached sequentially to tandem mass spectrometry; 4-MU, 4-methylumbelliferone; SNP, single nucleotide polymorphism; KM, Michaelis-Menten equilibrium constant; Vmax, maximal velocity.
References
- 1.Fisher B, Costantino JP, Wickerham DL, et al. Tamoxifen for prevention of breast cancer: report of the National Surgical Adjuvant Breast and Bowel Project P-1 Study. J Natl Cancer Inst. 1998;90:1371–88. doi: 10.1093/jnci/90.18.1371. [DOI] [PubMed] [Google Scholar]
- 2.Osborne CK. Tamoxifen in the treatment of breast cancer. N Engl J Med. 1998;339:1609–18. doi: 10.1056/NEJM199811263392207. [DOI] [PubMed] [Google Scholar]
- 3.Cuzick J, Powles T, Veronesi U, et al. Overview of the main outcomes in breast-cancer prevention trials. Lancet. 2003;361:296–300. doi: 10.1016/S0140-6736(03)12342-2. [DOI] [PubMed] [Google Scholar]
- 4.Howell A, Howell SJ, Evans DG. New approaches to the endocrine prevention and treatment of breast cancer. Cancer Chemother Pharmacol. 2003;52 Suppl 1:S39–44. doi: 10.1007/s00280-003-0645-5. [DOI] [PubMed] [Google Scholar]
- 5.Ghosh SS, Sappal BS, Kalpana GV, Lee SW, Chowdhury JR, Chowdhury NR. Homodimerization of human bilirubin-uridine-diphosphoglucuronate glucuronosyltransferase-1 (UGT1A1) and its functional implications. J Biol Chem. 2001;276:42108–15. doi: 10.1074/jbc.M106742200. [DOI] [PubMed] [Google Scholar]
- 6.Nechushtan H, Peretz T. Tamoxifen and breast cancer. Harefuah. 2002;141:718–20. 61, 60. [PubMed] [Google Scholar]
- 7.McDonald CC, Stewart HJ. Fatal myocardial infarction in the Scottish adjuvant tamoxifen trial. The Scottish Breast Cancer Committee. Bmj. 1991;303:435–7. doi: 10.1136/bmj.303.6800.435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rutqvist LE, Mattsson A. Cardiac and thromboembolic morbidity among postmenopausal women with early-stage breast cancer in a randomized trial of adjuvant tamoxifen. The Stockholm Breast Cancer Study Group. J Natl Cancer Inst. 1993;85:1398–406. doi: 10.1093/jnci/85.17.1398. [DOI] [PubMed] [Google Scholar]
- 9.van Leeuwen FE, Benraadt J, Coebergh JW, et al. Risk of endometrial cancer after tamoxifen treatment of breast cancer. Lancet. 1994;343:448–52. doi: 10.1016/s0140-6736(94)92692-1. [DOI] [PubMed] [Google Scholar]
- 10.Rutqvist LE, Johansson H, Signomklao T, Johansson U, Fornander T, Wilking N. Adjuvant tamoxifen therapy for early stage breast cancer and second primary malignancies. Stockholm Breast Cancer Study Group. J Natl Cancer Inst. 1995;87:645–51. doi: 10.1093/jnci/87.9.645. [DOI] [PubMed] [Google Scholar]
- 11.Meier CR, Jick H. Tamoxifen and risk of idiopathic venous thromboembolism. Br J Clin Pharmacol. 1998;45:608–12. doi: 10.1046/j.1365-2125.1998.00733.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Early Breast Cancer Trialists’ Collaborative Group. Tamoxifen for early breast cancer: an overview of the randomised trials. Lancet. 1998;351:1451–67. [PubMed] [Google Scholar]
- 13.Fisher B, Costantino JP, Redmond CK, Fisher ER, Wickerham DL, Cronin WM. Endometrial cancer in tamoxifen-treated breast cancer patients: findings from the National Surgical Adjuvant Breast and Bowel Project (NSABP) B-14. J Natl Cancer Inst. 1994;86:527–37. doi: 10.1093/jnci/86.7.527. [DOI] [PubMed] [Google Scholar]
- 14.Bernstein L, Deapen D, Cerhan JR, et al. Tamoxifen therapy for breast cancer and endometrial cancer risk. J Natl Cancer Inst. 1999;91:1654–62. doi: 10.1093/jnci/91.19.1654. [DOI] [PubMed] [Google Scholar]
- 15.Bergman L, Beelen ML, Gallee MP, Hollema H, Benraadt J, van Leeuwen FE. Risk and prognosis of endometrial cancer after tamoxifen for breast cancer. Comprehensive Cancer Centres’ ALERT Group. Assessment of Liver and Endometrial cancer Risk following Tamoxifen. Lancet. 2000;356:881–7. doi: 10.1016/s0140-6736(00)02677-5. [DOI] [PubMed] [Google Scholar]
- 16.Jordan VC, Collins MM, Rowsby L, Prestwich G. A monohydroxylated metabolite of tamoxifen with potent antioestrogenic activity. J Endocrinol. 1977;75:305–16. doi: 10.1677/joe.0.0750305. [DOI] [PubMed] [Google Scholar]
- 17.Furr BJ, Jordan VC. The pharmacology and clinical uses of tamoxifen. Pharmacol Ther. 1984;25:127–205. doi: 10.1016/0163-7258(84)90043-3. [DOI] [PubMed] [Google Scholar]
- 18.Katzenellenbogen BS, Norman MJ, Eckert RL, Peltz SW, Mangel WF. Bioactivities, estrogen receptor interactions, and plasminogen activator-inducing activities of tamoxifen and hydroxy-tamoxifen isomers in MCF-7 human breast cancer cells. Cancer Res. 1984;44:112–9. [PubMed] [Google Scholar]
- 19.Borges S, Desta Z, Li L, et al. Quantitative effect of CYP2D6 genotype and inhibitors on tamoxifen metabolism: implication for optimization of breast cancer treatment. Clin Pharmacol Ther. 2006;80:61–74. doi: 10.1016/j.clpt.2006.03.013. [DOI] [PubMed] [Google Scholar]
- 20.Johnson MD, Zuo H, Lee KH, et al. Pharmacological characterization of 4-hydroxy-N-desmethyl tamoxifen, a novel active metabolite of tamoxifen. Breast Cancer Res Treat. 2004;85:151–9. doi: 10.1023/B:BREA.0000025406.31193.e8. [DOI] [PubMed] [Google Scholar]
- 21.Lim YC, Desta Z, Flockhart DA, Skaar TC. Endoxifen (4-hydroxy-N-desmethyltamoxifen) has anti-estrogenic effects in breast cancer cells with potency similar to 4-hydroxy-tamoxifen. Cancer Chemother Pharmacol. 2005;55:471–8. doi: 10.1007/s00280-004-0926-7. [DOI] [PubMed] [Google Scholar]
- 22.Buckley MM, Goa KL. Tamoxifen. A reappraisal of its pharmacodynamic and pharmacokinetic properties, and therapeutic use. Drugs. 1989;37:451–90. doi: 10.2165/00003495-198937040-00004. [DOI] [PubMed] [Google Scholar]
- 23.Chu W, Fyles A, Sellers EM, et al. Association between CYP3A4 genotype and risk of endometrial cancer following tamoxifen use. Carcinogenesis. 2007;28:2139–42. doi: 10.1093/carcin/bgm087. [DOI] [PubMed] [Google Scholar]
- 24.Robertson DW, Katzenellenbogen JA, Long DJ, Rorke EA, Katzenellenbogen BS. Tamoxifen antiestrogens. A comparison of the activity, pharmacokinetics, and metabolic activation of the cis and trans isomers of tamoxifen. J Steroid Biochem. 1982;16:1–13. doi: 10.1016/0022-4731(82)90137-6. [DOI] [PubMed] [Google Scholar]
- 25.Malet C, Spritzer P, Cumins C, Guillaumin D, Mauvais-Jarvis P, Kuttenn F. Effect of 4-hydroxytamoxifen isomers on growth and ultrastructural aspects of normal human breast epithelial (HBE) cells in culture. J Steroid Biochem Mol Biol. 2002;82:289–96. doi: 10.1016/s0960-0760(02)00226-1. [DOI] [PubMed] [Google Scholar]
- 26.Zheng Y, Sun D, Sharma AK, Chen G, Amin S, Lazarus P. Elimination of antiestrogenic effects of active tamoxifen metabolites by glucuronidation. Drug Metab Dispos. 2007;35:1942–8. doi: 10.1124/dmd.107.016279. [DOI] [PubMed] [Google Scholar]
- 27.Lien EA, Solheim E, Lea OA, Lundgren S, Kvinnsland S, Ueland PM. Distribution of 4-hydroxy-N-desmethyltamoxifen and other tamoxifen metabolites in human biological fluids during tamoxifen treatment. Cancer Res. 1989;49:2175–83. [PubMed] [Google Scholar]
- 28.Lien EA, Solheim E, Kvinnsland S, Ueland PM. Identification of 4-hydroxy-N-desmethyltamoxifen as a metabolite of tamoxifen in human bile. Cancer Res. 1988;48:2304–8. [PubMed] [Google Scholar]
- 29.Nowell SA, Ahn J, Rae JM, et al. Association of genetic variation in tamoxifen-metabolizing enzymes with overall survival and recurrence of disease in breast cancer patients. Breast Cancer Res Treat. 2005;91:249–58. doi: 10.1007/s10549-004-7751-x. [DOI] [PubMed] [Google Scholar]
- 30.Sun D, Chen G, Dellinger RW, Duncan K, Fang JL, Lazarus P. Characterization of tamoxifen and 4-hydroxytamoxifen glucuronidation by human UGT1A4 variants. Breast Cancer Res. 2006;8:R50. doi: 10.1186/bcr1539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sun D, Sharma AK, Dellinger RW, et al. Glucuronidation of active tamoxifen metabolites by the human UDP-glucuronosyltransferases (UGTs) Drug Metab Dispos. 2007 doi: 10.1124/dmd.107.017145. [DOI] [PubMed] [Google Scholar]
- 32.Coffman BL, King CD, Rios GR, Tephly TR. The glucuronidation of opioids, other xenobiotics, and androgens by human UGT2B7Y(268) and UGT2B7H(268) Drug Metab Dispos. 1998;26:73–7. [PubMed] [Google Scholar]
- 33.Dellinger RW, Fang JL, Chen G, Weinberg R, Lazarus P. Importance of UDP-glucuronosyltransferase 1A10 (UGT1A10) in the detoxification of polycyclic aromatic hydrocarbons: decreased glucuronidative activity of the UGT1A10139Lys isoform. Drug Metab Dispos. 2006;34:943–9. doi: 10.1124/dmd.105.009100. [DOI] [PubMed] [Google Scholar]
- 34.Wiener D, Fang JL, Dossett N, Lazarus P. Correlation between UDP-glucuronosyltransferase genotypes and 4-(methylnitrosamino)-1-(3-pyridyl)-1-butanone glucuronidation phenotype in human liver microsomes. Cancer Res. 2004;64:1190–6. doi: 10.1158/0008-5472.can-03-3219. [DOI] [PubMed] [Google Scholar]
- 35.Lazarus P, Zheng Y, Aaron Runkle E, Muscat JE, Wiener D. Genotype-phenotype correlation between the polymorphic UGT2B17 gene deletion and NNAL glucuronidation activities in human liver microsomes. Pharmacogenet Genomics. 2005;15:769–78. doi: 10.1097/01.fpc.0000175596.52443.ef. [DOI] [PubMed] [Google Scholar]
- 36.Huang CK, Dulau A, Su-Rick CJ, Pan Q. Validation of rapid polymerase chain reaction-based detection of all length polymorphisms in the UGT 1A1 gene promoter. Diagn Mol Pathol. 2007;16:50–3. doi: 10.1097/01.pdm.0000213467.91139.c9. [DOI] [PubMed] [Google Scholar]
- 37.Elahi A, Bendaly J, Zheng Z, et al. Detection of UGT1A10 polymorphisms and their association with orolaryngeal carcinoma risk. Cancer. 2003;98:872–80. doi: 10.1002/cncr.11587. [DOI] [PubMed] [Google Scholar]
- 38.Huang YH, Galijatovic A, Nguyen N, et al. Identification and functional characterization of UDP-glucuronosyltransferases UGT1A8*1, UGT1A8*2 and UGT1A8*3. Pharmacogenetics. 2002;12:287–97. doi: 10.1097/00008571-200206000-00004. [DOI] [PubMed] [Google Scholar]
- 39.Nakamura A, Nakajima M, Yamanaka H, Fujiwara R, Yokoi T. Expression of UGT1A and UGT2B mRNA in human normal tissues and various cell lines. Drug Metab Dispos. 2008 doi: 10.1124/dmd.108.021428. [DOI] [PubMed] [Google Scholar]
- 40.Ren Q, Murphy SE, Zheng Z, Lazarus P. O-Glucuronidation of the lung carcinogen 4-(methylnitrosamino)-1- (3-pyridyl)-1-butanol (NNAL) by human UDP-glucuronosyltransferases 2B7 and 1A9. Drug Metab Dispos. 2000;28:1352–60. [PubMed] [Google Scholar]
- 41.Strassburg CP, Strassburg A, Nguyen N, Li Q, Manns MP, Tukey RH. Regulation and function of family 1 and family 2 UDP-glucuronosyltransferase genes (UGT1A, UGT2B) in human oesophagus. Biochem J. 1999;338( Pt 2):489–98. [PMC free article] [PubMed] [Google Scholar]
- 42.Turgeon D, Carrier JS, Levesque E, Hum DW, Belanger A. Relative enzymatic activity, protein stability, and tissue distribution of human steroid-metabolizing UGT2B subfamily members. Endocrinology. 2001;142:778–87. doi: 10.1210/endo.142.2.7958. [DOI] [PubMed] [Google Scholar]
- 43.Zheng Z, Fang JL, Lazarus P. Glucuronidation: an important mechanism for detoxification of benzo[a]pyrene metabolites in aerodigestive tract tissues. Drug Metab Dispos. 2002;30:397–403. doi: 10.1124/dmd.30.4.397. [DOI] [PubMed] [Google Scholar]
- 44.Innocenti F, Liu W, Fackenthal D, et al. Single nucleotide polymorphism discovery and functional assessment of variation in the UDP-glucuronosyltransferase 2B7 gene. Pharmacogenet Genomics. 2008;18:683–97. doi: 10.1097/FPC.0b013e3283037fe4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Fang JL, Lazarus P. Correlation between the UDP-glucuronosyltransferase (UGT1A1) TATAA box polymorphism and carcinogen detoxification phenotype: significantly decreased glucuronidating activity against benzo(a)pyrene-7,8-dihydrodiol(-) in liver microsomes from subjects with the UGT1A1*28 variant. Cancer Epidemiol Biomarkers Prev. 2004;13:102–9. doi: 10.1158/1055-9965.epi-03-0070. [DOI] [PubMed] [Google Scholar]
- 46.Hsieh TY, Shiu TY, Huang SM, et al. Molecular pathogenesis of Gilbert’s syndrome: decreased TATA-binding protein binding affinity of UGT1A1 gene promoter. Pharmacogenet Genomics. 2007;17:229–36. doi: 10.1097/FPC.0b013e328012d0da. [DOI] [PubMed] [Google Scholar]
- 47.Bosma PJ, Chowdhury JR, Bakker C, et al. The genetic basis of the reduced expression of bilirubin UDP-glucuronosyltransferase 1 in Gilbert’s syndrome. N Engl J Med. 1995;333:1171–5. doi: 10.1056/NEJM199511023331802. [DOI] [PubMed] [Google Scholar]
- 48.Monaghan G, Ryan M, Seddon R, Hume R, Burchell B. Genetic variation in bilirubin UPD-glucuronosyltransferase gene promoter and Gilbert’s syndrome. Lancet. 1996;347:578–81. doi: 10.1016/s0140-6736(96)91273-8. [DOI] [PubMed] [Google Scholar]
- 49.Iyer L, King CD, Whitington PF, et al. Genetic predisposition to the metabolism of irinotecan (CPT-11). Role of uridine diphosphate glucuronosyltransferase isoform 1A1 in the glucuronidation of its active metabolite (SN-38) in human liver microsomes. J Clin Invest. 1998;101:847–54. doi: 10.1172/JCI915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Iyer L, Hall D, Das S, et al. Phenotype-genotype correlation of in vitro SN-38 (active metabolite of irinotecan) and bilirubin glucuronidation in human liver tissue with UGT1A1 promoter polymorphism. Clin Pharmacol Ther. 1999;65:576–82. doi: 10.1016/S0009-9236(99)70078-0. [DOI] [PubMed] [Google Scholar]
- 51.Ogura K, Ishikawa Y, Kaku T, et al. Quaternary ammonium-linked glucuronidation of trans-4-hydroxytamoxifen, an active metabolite of tamoxifen, by human liver microsomes and UDP-glucuronosyltransferase 1A4. Biochem Pharmacol. 2006;71:1358–69. doi: 10.1016/j.bcp.2006.01.004. [DOI] [PubMed] [Google Scholar]
- 52.Bernard O, Tojcic J, Journault K, Perusse L, Guillemette C. Influence of nonsynonymous polymorphisms of UGT1A8 and UGT2B7 metabolizing enzymes on the formation of phenolic and acyl glucuronides of mycophenolic acid. Drug Metab Dispos. 2006;34:1539–45. doi: 10.1124/dmd.106.010553. [DOI] [PubMed] [Google Scholar]
- 53.Lehmann L, Wagner J. Gene expression of 17beta-estradiol-metabolizing isozymes: comparison of normal human mammary gland to normal human liver and to cultured human breast adenocarcinoma cells. Adv Exp Med Biol. 2008;617:617–24. doi: 10.1007/978-0-387-69080-3_64. [DOI] [PubMed] [Google Scholar]
- 54.Thibaudeau J, Lepine J, Tojcic J, et al. Characterization of common UGT1A8, UGT1A9, and UGT2B7 variants with different capacities to inactivate mutagenic 4-hydroxylated metabolites of estradiol and estrone. Cancer Res. 2006;66:125–33. doi: 10.1158/0008-5472.CAN-05-2857. [DOI] [PubMed] [Google Scholar]
- 55.Goetz MP, Rae JM, Suman VJ, et al. Pharmacogenetics of tamoxifen biotransformation is associated with clinical outcomes of efficacy and hot flashes. J Clin Oncol. 2005;23:9312–8. doi: 10.1200/JCO.2005.03.3266. [DOI] [PubMed] [Google Scholar]
- 56.Goetz MP, Knox SK, Suman VJ, et al. The impact of cytochrome P450 2D6 metabolism in women receiving adjuvant tamoxifen. Breast Cancer Res Treat. 2007;101:113–21. doi: 10.1007/s10549-006-9428-0. [DOI] [PubMed] [Google Scholar]