Abstract
Effective vaccines should confer long-term protection against future outbreaks of severe acute respiratory syndrome (SARS) caused by a novel zoonotic coronavirus (SARS-CoV) with unknown animal reservoirs. We conducted a cohort study examining multiple parameters of immune responses to SARS-CoV infection, aiming to identify the immune correlates of protection. We used a matrix of overlapping peptides spanning whole SARS-CoV proteome to determine T cell responses from 128 SARS convalescent samples by ex vivo IFN-γ ELISPOT assays. Approximately 50% of convalescent SARS patients were positive for T cell responses, and 90% possessed strongly neutralizing Abs. Fifty-five novel T cell epitopes were identified, with spike protein dominating total T cell responses. CD8+ T cell responses were more frequent and of a greater magnitude than CD4+ T cell responses (p < 0.001). Polychromatic cytometry analysis indicated that the virus-specific T cells from the severe group tended to be a central memory phenotype (CD27+/CD45RO+) with a significantly higher frequency of polyfunctional CD4+ T cells producing IFN-γ, TNF-α, and IL-2, and CD8+ T cells producing IFN-γ, TNF-α, and CD107a (degranulation), as compared with the mild-moderate group. Strong T cell responses correlated significantly (p < 0.05) with higher neutralizing Ab. The serum cytokine profile during acute infection indicated a significant elevation of innate immune responses. Increased Th2 cytokines were observed in patients with fatal infection. Our study provides a roadmap for the immunogenicity of SARS-CoV and types of immune responses that may be responsible for the virus clearance, and should serve as a benchmark for SARS-CoV vaccine design and evaluation.
Although a huge public health initiative successfully contained the original severe acute respiratory syndrome (SARS)4 outbreak of 2002–2003 caused by a novel coronavirus (SARS-CoV), many concerns remain over the possibility of its re-emergence, either naturally or accidentally, as is evidenced by sporadic SARS cases in late 2003/early 2004 and several laboratory-acquired infections after the outbreak. Phylogenetic analysis indicates that SARS-CoV is a zoonotic virus that crossed the species barrier and evolved in palm civets and humans (1). However, the failure to isolate SARS-CoV from wild civets or farmed civets from nonepidemic areas argues against the civets being the natural reservoir of the virus (2). Recently, several SARS-CoV-like viruses have been isolated from wild bats, and if the bats are the natural reservoir, it is unlikely that we can prevent further spread of this virus to the human population (3). Given that SARS has a significant impact on health and economics, there is an urgent need to develop effective treatments as well as prophylactic vaccines against any future outbreak of SARS.
The clinical outcomes of SARS infection were highly variable. So far, there has been no consensus regarding whether any treatment benefited SARS patients during the outbreak (4). In addition, it is not clear what role host immunity against SARS-CoV played in viral clearance or tissue damage. High initial viral load was shown to be independently associated with severity of the disease, and may be influenced by host immune responses (5). However, recent studies have suggested that type I IFN played a key role in the switch from innate immunity to adaptive immunity during the acute phase of SARS, and patients with poor outcomes showed type I IFN-mediated immunopathological events and deficient adaptive immune responses (6, 7).
Several studies have shown that most recovered SARS patients have higher and sustainable levels of neutralizing Ab responses, whereas patients with a longer illness showed a lower neutralizing Ab activity than patients with a shorter illness duration (8, 9), suggesting that Ab responses are likely to play an important role in determining the ultimate disease outcome of SARS-CoV infection. Several forms of possible vaccines, such as attenuated or inactivated SARS-CoV, DNA, and viral vector-based vaccines have been evaluated in a number of animal models, including nonhuman primates (10). Neutralizing Abs to SARS-CoV spike protein are the major components of protective immunity (11, 12). However, these animal models, including nonhuman primates, lack the severe clinical disease features observed in humans (13). Hence, it is difficult to evaluate whether these vaccines will prevent the disease in humans. It is possible that a vaccine could be harmful, because immune-mediated enhancement of pathology has been reported in other animal coronavirus infections (14) as well as in animals vaccinated with a modified vaccinia virus expressing SARS-CoV spike protein (15). Some variants of SARS-CoV were resistant to Ab neutralization, and the infection was enhanced by the Abs (16). Thus, without full understanding of the mechanism underlying protective immunity, many fear that some vaccines might worsen the disease rather than prevent it, echoing the respiratory syncytial virus vaccine disaster between 1960 and 1970 (17).
A major obstacle to accurate and rapid development of vaccines for SARS is the scarcity of basic information about epitopes recognized by adaptive immune responses to SARS-CoV infection in humans. This lack of knowledge also hampers studies to determine what types of immune responses are involved in protection or pathology during the course of the infection, with a view to designing an effective vaccine to stimulate only protective immunity. In this study, we aimed to characterize the entire SARS-CoV-specific T cell response in a cohort of patients (n = 128) who had recovered from SARS-CoV infection during the 2003 outbreak in China, using a matrix of 1843 overlapping peptides spanning the whole SARS-CoV proteome and fresh PBMC in IFN-γ ELISPOT assays. We also used polychromatic flow cytometry to evaluate T cell effector functions at the single-cell level (18). Additionally, we investigated a similar number of high-risk health care workers who were in close contact with SARS infected patients during the outbreak. Epidemiological data suggested SARS had its origins in Guangdong province, China. To investigate the change of immune response following the evolution of SARS-CoV, we collected samples from South China (Guangdong province) for the early outbreak and North China (Beijing) for the later, more severe outbreak. In addition, we analyzed the serum cytokine profile in another cohort of the acute phase infection from the early phase of the outbreak. Our study provides the first roadmap of the immunogenicity of SARS-CoV in humans and identifies types of immune responses that may be, at least partially, responsible for virus clearance or disease progression. The results may serve as a benchmark for SARS-CoV vaccine design and evaluation.
Materials and Methods
Demographics of study populations
Two cohorts of SARS infection containing a total of 226 SARS-infected patients were used in the present study: a convalescent cohort to study protective immune responses and an acute cohort to study acute infection through serum cytokine responses. This study was approved by the ethical research committees of the Medical School of Oxford University (U.K.), Beijing Youan Hospital, and Guangzhou Nan-Fang Hospital. The convalescent cohort contained a total of 128 recovered SARS patients (12 mo postinfection, 79% female and 21% male). It included 49 individuals from South China (Guangdong) and 79 from North China (Beijing). In control groups, 113 high-risk health care workers from South China (n = 62) and North China (n = 51) during the 2003 outbreak were also included. The illness onset time of the patients recruited was between March and April 2003 during the peak of the SARS outbreak, and the blood sampling time of convalescent samples was between April and September 2004, a year after the infection. In brief, 61 and 89% of patients were treated with steroids in South and North China, respectively.
In the acute cohort, a total of 98 patients from South China (Guangdong) were admitted between January 30 and April 27, 2003. In the control groups, 21 high-risk healthy individuals and 32 non-SARS pneumonia patients were included. The non-SARS pneumonia group was patients who had x-ray evidence of pneumonia, and they all responded to treatment with antibiotics, suggesting that most or all were suffering from bacterial pneumonia rather than viral pneumonia. The average time of sera collection following disease onset (fever and respiratory symptoms) was 7.5 days ± 4.4 SD. For the acute cohort, most of the sera (>90%) were collected within 2 days of hospital admission, before any specific drug treatment, and stored at −80°C. None of the patients received steroid therapy before sample collection.
The clinical diagnosis of SARS was made according to the World Health Organization case definitions (19), and disease severity was further classified according to Centre for Disease Control and Prevention criteria (20). In mild-to-moderate illness, the cases were defined by fever (>38°C), one or more clinical findings of lower respiratory illness (e.g., cough, shortness of breath, difficulty in breathing), and radiographic evidence of pneumonia. In addition to the above clinical features of mild-to-moderate illness, the severe cases were defined by evidence of respiratory failure (pO2 < 60 mmHg and pCO2 > 40 mmHg). All patients had a history of exposure to SARS patients. During hospital admission, all patients had evidence of SARS-CoV infection detected by ELISA for anti-SARS-CoV-specific IgG Ab in the serum and/or RT-PCR for the SARS virus, as described previously (21). In the convalescent cohort study, PBMC were isolated from whole blood by Ficoll-Hypaque density gradient centrifugation for an immediate IFN-γ ELISPOT assay. Paired serum samples were collected for determining SARS-specific Ab responses. DNA from the blood was extracted from each patient sample for in-house HLA typing by PCR with sequence-specific primers. In the acute cohort study, serum for cytokine analysis was collected from 5 ml blood samples taken for routine diagnostic purposes. A flow diagram illustrating how the studies were performed is presented in the Fig. 2.5
FIGURE 2.
Flow chart for determining T cell responses to SARS-CoV. Freshly isolated PBMC were stimulated with SARS-specific peptides spanning across the entire SARS-CoV proteome (1,843 overlapping peptides), and the immediate IFN-γ release by T cell responses were measured by IFN-γ ELISPOT assay as described in the Materials and Methods. The 2-dimensional matrices were set up with a total of 88 pools (1-dimensional = 43 pools; 2-dimensional = 44 pools; up to 45 peptides/pool) so that each peptide was present in two different pools. Peptides were used at a final concentration of 2 μM each.
SARS-IgG ELISA and virus neutralization assays
SARS-specific IgG in the serum samples was determined by ELISA based on purified whole virus lysates (S200300004, Hua Da Company). A positive result was defined as OD ≥ 0.14 (0.13 + mean of negatives (0.01)) as described previously (22). Neutralizing Ab against spike protein was measured by a pseudotype retrovirus-based neutralization assay as described previously (8). In brief, serum samples were heat inactivated at 56°C for 30 min, 2-fold serially diluted from 1/10 in culture medium, and mixed with murine leukemia virus pseudotyped with SARS spike protein, murine leukemia virus (SARS), and virions (≈100 IU) at a 1:1 v/v ratio. After incubation at 37°C for 1 h, 100 μl of each dilution was added to QT6/ACE2 cells seeded at 1 × 104 cells per well in 96-well flat-bottom tissue culture plates seeded 24 h previously. GFP-positive cells were counted 48 h later by fluorescence microscopy. Neutralizing Ab titers are presented as geometric mean titers of assays performed in triplicate. IC90 was used as the end-point titer, and titers ≥1:10 were considered to be positive, as described previously.
Ex vivo IFN-γ ELISPOT
To identify T cell epitopes from SARS-CoV in the cohort, we used overlapping synthetic peptides (total 1843 peptides, 15–18mers overlapping by 10 amino acid residues) spanning the whole proteome of SARS-CoV (Tor-2, Accession number AY- 274119). The peptides were manufactured by New England Peptides and the quality of each peptide was checked by MALDI-TOF mass spectrometry, identifying the correct mass at >75% of purity. As previously described (23), we simultaneously tested all 1,843 overlapping peptides in each individual using 2-dimensional matrices with a total of 88 pools (1st D = 43 pools; 2nd D = 44 pools; up to 45 peptides/pool) so that each peptide was present in two different pools. Peptides were used at a final concentration of 2 μM each. The internal negative control was no peptide in triplicates, and positive controls were FEC (a mixture of Flu, EBV, and CMV T cell epitope peptides), PPD, or PHA. Fresh PBMCs were isolated from 50 ml of blood collected in heparin containing tubes (BD Vacutainer) by venipunture and added into 96-well plates at 300,000 cells/well for overnight incubation. All ELISPOT assays were performed using the human IFN-γ ELISPOT kit (Mabtech) according to the manufacturer’s instructions. The spots on each well were counted using an AID-ELISPOT reader (Autoimmune Diagnostika). Any true positive response was determined by two separated ELISPOT assays: the matrix screening (first ELISPOT) and a confirmatory test examining the individual peptide (second ELISPOT). Wells containing spot numbers greater than the mean + 3 SD of three negative control wells (no peptide) were regarded as positives in each individual, provided that the total was greater than 18 spot forming cells (SFC)/million PBMC, to rule out false positives where background was very low. In all assays, values of no peptide control wells were 6.7 ± 0.9 SFC/million PBMC for healthy subjects and 8.3 ± 0.7 SFC/million PBMC for SARS patients. To determine whether T cells were CD4 or CD8, in the second ELISPOT assay, cell depletion was also conducted by Dynal CD8 beads, as described in the manufacturer’s instructions (Invitrogen), before the ELISPOT assay. Undepleted PBMC were used as positive controls.
Intracellular cytokine staining (ICS)
T cell lines were generated as effector cells to confirm SARS peptides and the CD4 or CD8+ property of each peptide by ICS and flow cytometry, as described previously (24). In brief, frozen samples of PBMC were thawed and stimulated with 100 μM of each peptide for 1 h. Cells were cultured in RPMI 1640 supplemented with 10% human serum and 25 ng/ml IL-7 (PeproTech) for 3 days, and then 100 U of IL-2/ml was added every 3 to 4 days thereafter. In assay, T cell lines were stimulated with 100 μM peptide-pulsed EBV-transformed autologous B cell lines in the presence of anti-CD28 and anti-CD49d mAbs (each at 1 μg/ml; BD Biosciences) for 1 h at 37°C and before the addition of brefeldin A (10 μg/ml, Sigma-Aldrich). After a further 5-h incubation, cells were washed in PBS with 1% FCS and sodium azide, then fixed and permeabilized in permeabilizing buffer (BD Biosciences). Cells were stained with mAbs against human IFN-γ (BD Biosciences), CD4 (DakoCytomation), and CD8 (BD Biosciences). Lymphocytes were gated on a FACSCalibur flow cytometer (BD Biosciences), and cells stimulated with medium alone were used as negative controls.
Polychromatic flow cytometric analysis
Eleven-parameter flow cytometric analysis was performed using a LSRII flow cytometer (BD Biosciences). FITC, PE, Cy7PE, Cy5.5PE, allophycocyanin, Cy7 allophycocyanin, Texas Red PE, violet amine reactive dye, and Quantum-dot 705 (QD705) were used as the fluorophores. At least 300,000 live lymphocytes were collected. The list-mode data files were analyzed using FlowJo (Tree Star). Functional capacity was determined after Boolean gating and subsequent analysis was performed using Simplified Presentation of Incredibly Complex Evaluations (SPICE, version 2.9, Mario Roederer, VRC, NIAID, National Institutes of Health). All values used for analyzing proportionate representation of responses are background-subtracted.
To study ex vivo peptide-specific polyfunctional responses of memory T lymphocytes from recovered SARS patients, a 6-h short-term stimulation was performed on fresh or frozen lymphocytes as described elsewhere (25). Freshly isolated or thawed lymphocytes were resuspended at 106/ml in R10 supplemented with 1 μg/ml anti-CD28 and anti-CD49d Abs. Peptides 15 amino acids in length, overlapping by 11 amino acids, and encompassing SARS spike protein, were used to stimulate SARS-specific T cells in the presence of brefeldin-A (1 μg/ml, Sigma-Aldrich) for 6 h at 37°C. All cells were surface-stained for phenotypic markers of interest and intracellularly stained for cytokines (ICS).
mAbs used for phenotypic and functional characterization of T cell subsets were anti-CD3 Cy7 allophycocyanin, anti-CD45RO Texas Red PE, anti-CD27 Cy5PE, anti-CD4 Cy5.5PE, anti-CD8 QD705, anti-IFN-γ FITC, anti-IL-4 PE and anti-TNF-α Cy7PE, and anti-IL-2 allophycocyanin (BD Pharmingen). As naive T cells do not express CCR5, and as SARS-specific T cells are not detectable in the naive T cell pool, we report these data as percentages of memory T cells. We first gated for memory CD4 and CD8 T cells based upon characteristic expression patterns of CD45RO and CD27.
Cytokine cytometric bead array
Serum concentrations of the cytokines IL-1, IL-2, IL-4, IL-5, IL-6, IL-8, IL-10, IL-12, IFN-γ, and TNF-α in acute samples (50 μl) were measured using cytometric bead-array assays (BD Biosciences) according to the manufacturer’s instructions. The sensitivities of cytometric bead array for each cytokine were 7.2 pg/ml (IL-1), 2.6 pg/ml (IL-2), 2.6 pg/ml (IL-4), 2.4 pg/ml (IL-5), 2.5 pg/ml (IL-6), 3.6 pg/ml (IL-8), 2.8 pg/ml (IL-10), 1.9 pg/ml (IL-12), 7.1 pg/ml (IFN-γ), and 2.8 pg/ml (TNF-α).
Statistical analysis
Graphs were presented by Prism 4 (GraphPad) software. All statistical analysis was conducted independently by a statistician using SPSS 12.0 software. The T cell response (breadth and magnitude) was analyzed as described previously (23). Categorical variables were compared using Fisher’s exact test, and group comparison of numerical data was analyzed by Mann-Whitney U test. To compare the immune response in different disease groups, multiple regression analysis was conducted for each response measure separately, all adjusted for age, sex, and location. In the regression model, natural logarithm transformation was performed for each response variable, and a t test was used to test the difference of the log-transformed mean immune response between the mild/moderate group and the severe group. A two-tailed t test was used to compare two different groups taking into account unequal variances, and a p value <0.05 was considered to be significant. For acute cytokine analysis, continuous variables were compared using the independent samples t test. Recovered and fatal SARS groups were described using the geometric mean ratio and 95% confidence intervals. ANOVA with age as a covariate was used for comparison of variables between groups of patients.
Results
SARS patients
PBMC samples from a cohort of 128 convalescent SARS patients were collected 12 mo postinfection from South (n = 49) and North (n = 79) China. As shown in Table I, 29% of patients from the cohort had severe disease progression compared with 71% of patients who had mild to moderate progression. In agreement with previous reports (7, 26), age was also associated with severe disease outcome among survivors (mean age of mild to moderate vs severe illness groups: 31 ± 10 vs 42 ± 13, p = 0.01) as well as mortality (mean age of survival vs death: 43 ± 2 vs 59 ± 5, p = 0.007). Medical complications were observed in both groups including liver, kidney, and cardiac damage. Seventy-eight percent of the patients in the cohort were treated with steroids. In the acute SARS cohort (n = 98), 11 patients died of respiratory failure, representing a mortality of 11.2%. The mean time of blood serum collections was 2 ± 3 days from admission to hospital.
Table I.
Characteristics of patients with SARS
| Cohort for Convalescent Samples |
|||
|---|---|---|---|
| Mild-to-Moderate SARS | Severe SARS | Total | |
| No. of patients (%) | 91 (71) | 37 (29) | 128 |
| Male/female (n) | 18/73 | 9/28 | 27/101 |
| Age (year), mean ± SDa | 31 ± 10 | 42 ± 13 | 34 ± 12 |
| Clinical complications: n (%) | |||
| Liver | 28 (31) | 20 (54) | 48 |
| Kidney | 1 (1) | 1 (5) | 2 |
| Cardiac | 0 (0) | 2 (5) | 2 |
| Treatment with steroid: n (%) | 68 (75) | 32 (86) | 100 (78) |
| Cohort for Acute Samples |
|||
| Recovered | Fatal | Significance | |
|
| |||
| No. of patients | 87 | 11 | |
| Male/female (n) | 46/41 | 7/4 | NS |
| Age (year), mean ± SD | 43 ± 2 | 59 ± 5 | p = 0.007 |
| Aspartate aminotransferase | 162 ± 84 | 44 ± 4 | p < 0.001 |
| Alanine aminotransferase | 129 ± 58 | 183 ± 13 | NS |
p-value is equal to 0.01.
Anti-SARS-CoV spike Ab responses
Many studies have demonstrated that the spike protein is highly immunogenic and induces neutralizing Abs enabling protection against reinfection in animal models (6). We profiled anti-SARS-CoV Ab responses in sera from the cohort using ELISA as well as a pseudotype-based SARS-CoV neutralizing Ab (Nab) assay. Over 90% of the convalescent SARS patients retained detectable levels of Spike-specific IgG as well as Nab activities after 1 year postinfection (Fig. 1, A and B). However, the level of SARS spike-specific IgG measured by ELISA did not correlate with the titer of Nab from each individual (data not shown).
FIGURE 1.
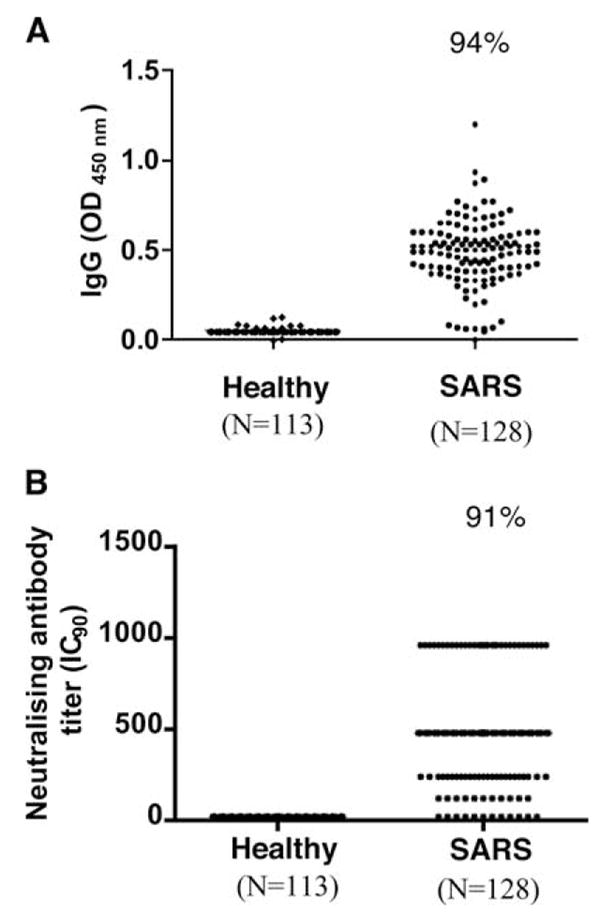
SARS-CoV-specific Ab responses in patients recovered from SARS-CoV infection. A, Response of SARS specific-IgG between high-risk healthy group and SARS pneumonia by ELISA. The assay was based on purified whole virus lysates, and positive was defined by an OD value >0.14. B, Response of neutralizing Ab against spike protein of SARS-CoV using pseudotype-based virus neutralization test. In this assay, heat-inactivated serum samples were mixed with murine leukemia virus (MLV) pseudotyped with SARS spike protein MLV (SARS) virions before infecting QT6 cells expressing ACE2, a receptor for SARS-CoV virus for 48 h. The pseudotype virus had a GFP-reporter, and infected cells turned green and were counted by fluorescence microscopy. IC90 was used as end-point titer, and titers ≥1:10 were considered to be positive.
T cell immune responses
The evaluation of immune responses to individual peptides spanning the entire SARS-CoV genome requires a large number of PBMC. We used peptide pools and a 2-dimensional matrix-based approach for initial screening, to reduce the level of specimen usage; the candidate peptides were then reconfirmed individually in a second assay as described in previous studies (23). In addition, to increase the sensitivity of detection, freshly isolated PBMC were used to perform IFN-γ ELISPOT assays in each individual. As illustrated in Fig. 2, in the first ELISPOT assay (Matrix screening), we found that >50% (67/128) of patients were positive as determined by greater than the mean + 3 SD of negative control wells (8.3 ± 0.7 SFC/million PBMC, background with no peptide), whereas none of the individuals from the control groups had a spot number greater than the mean + 3 SD of their background controls (6.7 ± 0.9 SFC/million PBMC). In the second ELISPOT assay (confirmatory test), we confirmed that 46 patients, representing nearly 40% of the SARS cohort (119 SARS-CoV Ab positive), had detectable levels of SARS-CoV-peptide specific memory T cell responses after 1 year postinfection. In addition, the lack of detectable T cell immune responses or virus-specific Ab (data not shown) in the high-risk health care workers (n = 113) suggested that subclinical infection with SARS-CoV during the outbreak was rare. Interestingly, patients from North China, the late phase of outbreak, had a significantly higher level of both Ab and T cell responses compared with patients from South China (the early phase of outbreak) (Fig. 3, A and B).
FIGURE 3.

SARS-specific Ab and T cell responses from convalescent SARS patients between South and North cohorts. A, Serum samples were tested for SARS-specific IgG and neutralizing Ab, and the corrected neutralizing Ab response over the total SARS-specific IgG is shown on the y-axis. B, Freshly isolated PBMC were stimulated with peptides from SARS proteome, and the IFN-γ response was measured by ELISPOT assay.
Of 1,843 overlapping peptides, 55 (3%) were recognized by T cells in the cohort (Table II). There was a large variation in the percentage of peptides recognized by T cells within the individual proteins. Eight of fourteen SARS proteins were able to stimulate T cell responses such as replicase, spike, Orf3, Orf4, envelope, membrane, Orf13, and nucleocapsid. However, most responses (70%, 39/55) were focused on the structural proteins (spike, envelope, membrane, and nucleocapsid), whereas 30% were distributed in nonstructural proteins (replicase and Orfs). Spike protein induced the most dominant T cell response (41%, 23/55), whereas replicase was much less immunogenic (13%, 7/55) despite covering 2/3 of the SARS-CoV proteome. The frequency and the magnitude of individual T cell peptide responses were highly variable between peptides from different proteins (Fig. 4, A and B). Although the majority of the peptides were only recognized by one individual, some were targeted by several individuals (Table III). The three most frequently recognized T cell epitopes were located in Orf3 (PLQASLPFGWLVIGV) and the spike protein (NYNYKYRYL RGKLRPF and AGCLIGAEHVDTSYECDI).
Table II.
Number of peptides recognized per SARS-CoV protein subunit
| Protein Subunit | No. of Overlapping Peptides Recognized per SARS-CoV Protein Subunit (%) |
|---|---|
| Replicase | 7/1386 (0.5) |
| Spike | 23/169 (14) |
| Orf3 | 3/27 (11) |
| Orf4 | 1/29 (3) |
| Envelope | 2/14 (14) |
| Membrane | 8/43 (19) |
| Orf7 | 2/11 (18) |
| Orf8 | 0 (0) |
| Orf9 | 0 (0) |
| Orf10 | 0 (0) |
| Orf11 | 0 (0) |
| Orf13 | 3/18 (17) |
| Orf14 | 0/12 (0) |
| Nucleocapsid | 6/83 (7) |
| Total | 55/1843 (3) |
FIGURE 4.
Distribution of peptide recognition and magnitude of SARS-specific T cell responses across the entire expressed SARS-CoV genome in convalescent SARS patients, as determined by immediate IFN-γ ELISPOT assay. Freshly isolated PBMC from convalescent SARS samples were stimulated overnight with overlapping peptides of different proteins across SARS genome, and the IFN-γ response was measured by ELISPOT assay. The individual 1,843 overlapping peptides of different proteins are represented on the x-axis. A, The y-axis represents the percentage recognition of the study subjects to the individual peptide. B, The y-axis represents the average magnitude of response by the study subjects to individual peptides.
Table III.
Summary of the most frequently recognized peptides in the study cohort
| Protein | Amino Acid Position |
Peptide Sequence | Study Subjects with Response (%) |
|---|---|---|---|
| Orf3 | 36–50 | PLQASLPFGWLVIGV | 24 |
| Spike | 435–451 | NYNYKYRYLRGKLRPF | 22 |
| Spike | 633–650 | AGCLIGAEHVDTSYECDI | 17 |
| Nucleocapsid | 211–225 | GETALALLLLDRLNQ | 11 |
| Membrane | 146–160 | GHLRMAGHSLGRCDI | 9 |
| Spike | 528–545 | NFNGLTGTGVLTPSSKRF | 7 |
| Spike | 649–666 | DIPIGAGICASYHTVSLL | 7 |
| Spike | 1082–1097 | SWFITQRNFFSPQII | 7 |
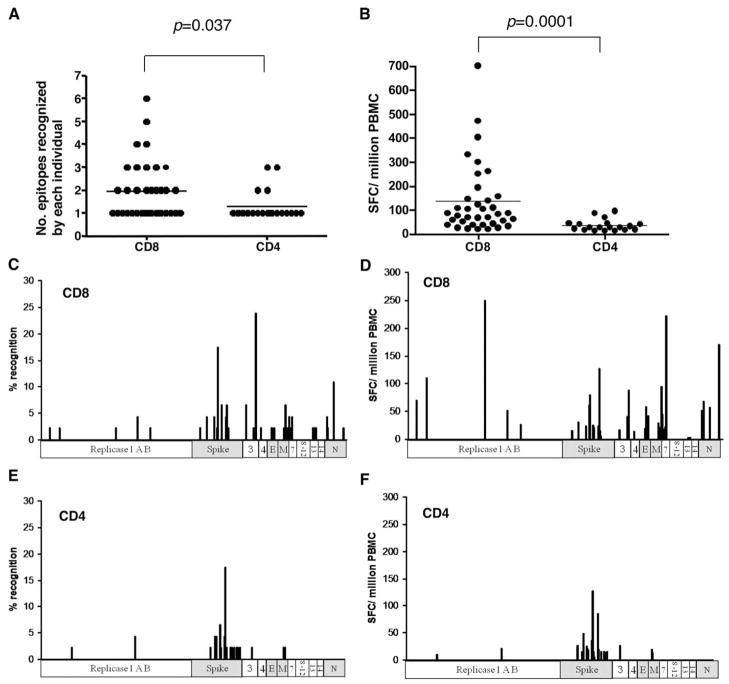
Strong T cell responses correlated significantly (p < 0.05) with higher neutralizing Ab activity (Table IV), consistent with T cells playing an important role in the generation of Ab responses during the course of SARS-CoV infection. A full list of all SARS T cell peptides, including their sequence and subsets of T cell responses, is summarized in supplementary Table S1. We did not observe any significant effect of steroid treatment on the levels of T cell or Ab responses in this study (data not shown). Furthermore, both CD4+ and CD8+ T cell responses were identified in this study, and the CD8+ response predominated over CD4 in terms of frequency and magnitude of the responses (Fig. 5, A and B). Interestingly, the CD8+ T cell responses were widely distributed across the SARS proteome, whereas the CD4+ T cell responses were clustered mainly in the spike protein (Fig. 5, C–F).
Table IV.
Ab response between different ELISPOT groups
| Antibody Response | ELISPOT−, Mean ± SD (SE) | ELISPOT+, Mean ± SD (SE) | p |
|---|---|---|---|
| No. individuals | 53 | 40 | |
| Total SARS-specific IgG | 0.48 ± 0.18 (0.02) | 0.50 ± 0.20 (0.03) | 0.592 |
| Neutralizing Aba | 425.66 ± 305.55 (41.97) | 580.50 ± 329.02 (52.02) | 0.040 |
| Neutralizing Ab/IgG | 8.76 ± 6.60 (0.91) | 12.41 ± 8.19 (1.29) | 0.024 |
Natural logarithm transformation of the raw data was performed for the purpose of statistical testing. Unequal variances were taken into account by the t test for comparison of two samples.
FIGURE 5.
Breadth and magnitude of SARS-specific T cell responses between CD8+ and CD4+ responses in convalescent SARS samples by IFN-γ ELISPOT assays. A, Number of epitopes recognized by each study subject, as represented on the y-axis, is compared between CD8+ and CD4+ responses. B, Magnitude of response as measured by SFC per million PBMC, as represented by the y-axis, is compared between CD8+ and CD4+ responses. C, Breadth of CD8+ response as measured by the percent recognition of the peptide response across the SARS-CoV proteome. D, Magnitude of CD8+ response as measured by the SFC per million PBMC. E, Breadth of CD4+ response as measured by the percent recognition of the peptide response across the SARS-CoV proteome. F, Magnitude of CD4+ response as measured by the SFC per million PBMC. p values represent the results of a Mann-Whitney U test.
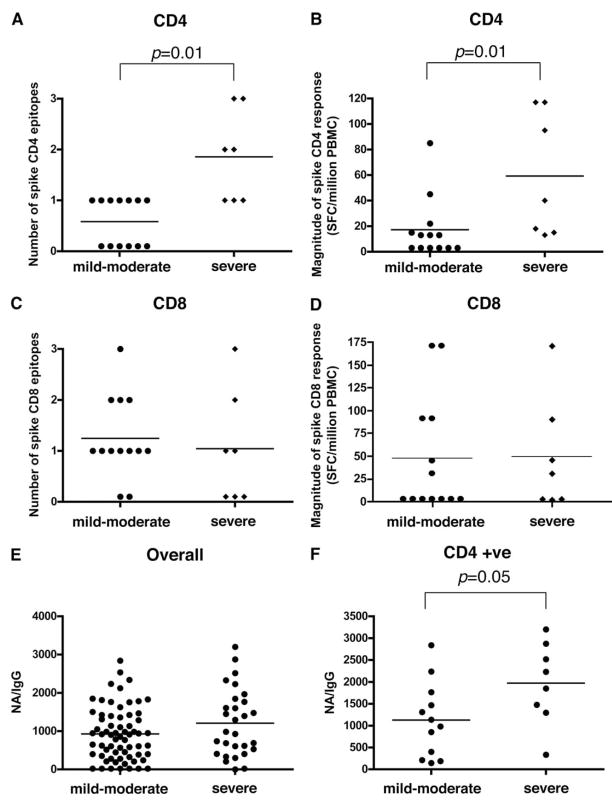
Next, we used multivariate regression analysis to investigate retrospectively whether there was any association between disease severity and T cell immune responses. The severe SARS patient group had significantly stronger CD4+ T cell responses to the spike protein in terms of the number of epitopes recognized (p = 0.01) and the magnitude of the response (p = 0.01) (Fig. 6, A and B) by each study subject. In contrast, there was no difference in the CD8+ T cell responses (Fig. 6, C and D), making it unlikely that the differences in CD4+ T cell responses simply reflected levels of virus Ags. We also found that the overall level of neutralizing Ab was not associated with disease severity (Fig. 6E), but if we examined patients who were CD4 ELISPOT positive, there was a significant correlation between severe illness and a higher level of neutralizing Ab (p = 0.05) (Fig. 6F). This may simply reflect the association between the CD4+ T cell responses and severity as described above.
FIGURE 6.
T cell and Ab responses between mild-to-moderate and severe groups of convalescent SARS patients. A, Number of CD4+ spike responses, as represented on the y-axis, is compared between mild-to-moderate and severe groups, as measured by IFN-γ ELISPOT assay. B, Magnitude of CD4+ spike response as represented by SFC per million PBMC is compared between mild-to-moderate and severe groups. C, Number of CD8+ spike responses as represented on the y-axis is compared between mild-moderate and severe groups. D, Magnitude of CD8+ spike response as represented by SFC per million PBMC between mild-to-moderate and severe groups. E, The corrected neutralizing Ab response in all SARS samples studied, as represented on the y-axis, from all samples between mild-to-moderate and severe groups, as measured by SARS IgG and neutralizing Ab. F, The corrected neutralizing Ab response as represented on the y-axis from SARS patients (mild-to-moderate or severe groups) who have spike-specific CD4+ T cell responses. p values represent the results of a Mann-Whitney U test.
To further characterize and compare functional response profiles in individuals suffering from mild-to-moderate or severe SARS, we stimulated PBMC with antigenic peptides corresponding to SARS proteins identified by IFN-γ ELISPOT assays. We then assessed the frequency, function, and memory phenotype of SARS-specific T cells by multicolor flow cytometry, with the gating strategy as shown in Supplementary Figure S1. In doing so, we found comparable phenotypes and functionality of SARS-specific CD4+ T cells in individuals with mild-to-moderate and severe SARS (Fig. 7, A and B). The largest component of the CD4+ T cell response was comprised of CD4+ T cells that produced only one cytokine. In addition, SARS-specific CD4 T cells from individuals with mild-to-moderate and severe SARS tended to be of a central memory phenotype (CD27+ and CD45RO+). However, the frequency of memory CD4+ T cells producing IFN-γ, IL-2, and TNF-α was significantly higher in individuals with severe SARS compared with individuals with mild-to-moderate SARS (p = 0.04, Fig. 6C). Similar analysis was performed on SARS-specific CD8+ T cells (Fig. 8). This analysis revealed similar functionality, memory phenotype, and frequencies of SARS-specific CD8+ T cells in individuals with mild-to-moderate and severe SARS. Although the majority of the CD8+ T cell response contained CD8+ T cells capable of producing only IFN-γ, a significant proportion of the responding cells could also produce TNF-α and degranulate (based upon mobilization of CD107a to the cell membrane).
FIGURE 7.
Phenotype, functionality, and frequency of SARS-specific CD4+ T cells in blood. Lymphocytes were stimulated with overlapping SARS peptides and stained as described in the Materials and Methods. A, The memory phenotype of SARS-specific CD4+ T cells in individuals with mild-to-moderate and in individuals with severe SARS, as defined by the expression of CD27 and CD45RO. B, Functionality of SARS-specific CD4+ T cells in individuals with moderate and in individuals with severe SARS. C, Frequency of SARS-specific CD4+ T cells. The response is shown as the percentage of memory CD4+ T cells that are SARS-specific. The frequency of the total SARS-specific responses producing one, two, three, four, or five functions was determined using SPICE as described in methods. p values represent the results of a Mann-Whitney U test.
FIGURE 8.
Phenotype, functionality, and frequency of SARS-specific CD8+ T cells in blood. Lymphocytes were stimulated with overlapping SARS peptides and stained as described in the Materials and Methods. A, The memory phenotype of SARS-specific CD8+ T cells in individuals with mild-to-moderate and in individuals with severe SARS, as defined by the expression of CD27 and CD45RO. B, Functionality of SARS-specific CD8+ T cells in individuals with moderate and in individuals with severe SARS. C, Frequency of SARS-specific CD8+ T cells. The response is shown as the percentage of memory CD8+ T cells that are SARS-specific. The frequency of the total SARS-specific responses producing one, two, three, four, or five functions was determined using SPICE as described in methods. p values represent the results of a Mann-Whitney U test.
Cytokine profiles in acute phase SARS and their association with disease progression
For safety reasons, it was not possible to gain access to PBMC from patients with acute SARS-CoV infection. However, we were able to study cytokine profiles in an independent cohort of serum from acute SARS patients (n = 98). Proinflammatory cytokines (IL-1, IL-6, IL-8), Th1 cytokines (IFN-γ, TNF-α, IL-2, IL-12), and Th2 cytokines (IL-4, IL-5, IL-10) were evaluated in sera collected within 2 days of hospital admission without any specific treatment. A substantial elevation of IFN-γ, IL-6, IL-8, IL-10, and IL-12 was observed in SARS patients as compared with normal sera (Fig. 9). These cytokines represented mainly the activation of innate immune responses such as monocyte and NK cells. A reduction of these cytokines was associated with recovery from SARS pneumonia (data not shown). Interestingly, non-SARS pneumonia had a lower level of IFN-γ but a much higher level of IL-8 compared with SARS patients at the early time of infection. Thus, monitoring IFN-γ and IL-8 in sera may have high predictive value for SARS vs non-SARS pneumonia. Finally, there was a significant increase in Th2 cytokines (IL-4, IL-5, IL-10) in fatal SARS cases (Table V). Of note, similar phenomena were observed in influenza infection in elderly humans (27), suggesting that an imbalance of Th1/Th2 cytokines might be a key influence in the outcome of severe respiratory viral infection in the elderly.
FIGURE 9.
Serum cytokine profile between acute SARS and non-SARS pneumonia by cytometric beads arrays. Serum samples (>90%) were collected within 2 days of hospital admission before any specific drug treatment, and none of the patients received steroid therapy before sample collection. The average time of sera collection following disease onset was 7.5 days ± 4.4 SD. p values represent the results of a Mann-Whitney U test.
Table V.
Serum cytokine profile between fatal and recovered SARSa
| Fatal SARS | Recovered SARS | p | |
|---|---|---|---|
| Patients (n) | 11 | 87 | |
| Th1 cytokines (pg/ml) | |||
| IFN-γ | 22.4 ± 8.0 | 28.2 ± 6.50 | NS |
| IL-2 | 0.92 ± 0.30 | 0.87 ± 0.20 | NS |
| TNF-α | 1.85 ± 0.33 | 1.24 ± 0.53 | NS |
| IL-12 | 6.33 ± 2.29 | 19.4 ± 9.83 | NS |
| Th2 cytokines (pg/ml) | |||
| IL-4 | 6.63 ± 0.84 | 3.42 ± 0.66 | <0.001 |
| IL-5 | 2.81 ± 0.97 | 0.64 ± 0.12 | 0.04 |
| IL-10 | 13.2 ± 5.16 | 9.05 ± 1.97 | 0.05 |
| Inflammatory cytokines (pg/ml) | |||
| IL-1 | 9.01 ± 2.09 | 29.4 ± 8.41 | 0.04 |
| IL-6 | 27.7 ± 11.9 | 47.7 ± 8.99 | NS |
| IL-8 | 159.7 ± 116 | 48.8 ± 7.15 | 0.03 |
Recovered and fatal SARS groups were described using the geometric mean ratio and 95% confidence intervals. ANOVA with age as a covariate (ANCOVA) was used for comparisons of variables between groups of patients. Unequal variances were taken into account by the t test for comparison of two samples.
Discussion
SARS-CoV causes the most severe coronavirus-associated disease that has been described in humans so far. Little is known about its pathogenesis, in particular how host immune responses influence the disease outcome. Therefore, an understanding of the molecular details of immune responses to SARS-CoV and their contribution to protection and disease progression will in turn lead to the discovery and development of new therapeutics and vaccines for SARS. Most previous studies on SARS-CoV-specific T cell responses have focused on single epitopes or single SARS proteins (28–30). Our study is the first to measure SARS-CoV-specific T cell responses directed against the entire expressed SARS-CoV genome in a substantial study population, using a highly sensitive and specific peptide matrix system and flow cytometry-based intracellular cytokine staining. We used PBMC from recovered SARS patients to establish detailed maps of T cell immune responses to SARS-CoV, with the exact epitopic regions targeted. We then used the information to evaluate breadth and magnitude of the virus-specific T cell responses in association with the level of anti-SARS-CoV Nabs or disease severity in the early and late phase of the outbreak.
Our data showed that structural proteins such as spike, membrane, and envelope were the most immunogenic to T cells, as compared with the nonstructural proteins. In the detailed analysis of epitopic regions, the most frequently targeted peptides were detected in the spike and Orf 3 proteins, with 22 or 24% of individuals recognizing the sequence NYNYKYRYLRGKLRPF (spike) and PLQASLPFGWLVIGV (Orf 3), respectively. In contrast, the SARS replicase genes which represent two thirds of the SARS-CoV genome were poorly immunogenic (0.5%, 7 of 1386 peptides). Among the 55 T cell epitopes identified, more than 70% elicited CD8+ T cell responses, with much broader responses compared with the CD4+ T cells. Interestingly, the CD4+ T cell responses were mainly clustered in the spike protein, presumably helping B cells to produce Nabs, because helper CD4+ T cells and neutralizing Abs are interdependent, and the production of Abs by plasma B cells requires the help of CD4+ T cells specific to the same protein (31).
We also found that overall immune responses (both Ab and T cells) in patients from the late phase of the outbreak (Beijing) were much stronger when compared with those from the early phase (Guangdong). It was not clear whether this was due to virus mutation from the early to the late phase of the outbreak. Recent epidemiological studies suggested that SARS-CoV became more virulent following human-to-human transmission (1), which may be associated with increasing immunogenicity of the virus. Despite no significant correlation between the total T cell responses and disease progression, multivariate regression analysis showed that the disease severity correlates strongly with high level CD4+ T cell responses and the titer of Nabs against the spike protein. By contrast, the level of the memory CD8+ T cell response is independent of disease severity. Further characterization of virus-specific T cells by polychromatic flow cytometry indicated that although the largest component of the virus-specific CD4+ T cells produced only one cytokine, the frequency of polyfunctional memory CD4+ T cells (producing IFN-γ, IL-2, and TNF-α) was significantly higher in individuals with severe SARS compared with individuals with mild-to-moderate SARS. However, SARS-specific CD8+ T cells revealed similar functionality, memory phenotype, and frequencies between moderate and severe SARS. Although the majority of these CD8+ T cells produced only IFN-γ, a significant proportion of the virus-specific CD8+ T cells could also produce TNF-α and degranulate (CD107a+).
Phylogenetic analysis indicated that SARS-CoV was distinct from other human coronaviruses (32). However, any potential cross-reactivity of T cells between these viruses may have significant impact on overall T cell responses and clinical outcomes. We therefore searched the amino acid sequences of known human coronaviruses (NL63, OC43, 229E, and HKU1) against the 55 T cell epitopes identified in this study. We did not find any meaningful sequence homology to the SARS-CoV epitope sequences (data not shown). It is therefore unlikely that pre-existing cross-reactive T cells played a significant role in SARS-CoV pathogenesis. Although the involvement of immunopathology in SARS-CoV infection or disease enhancement following the virus infection in previously vaccinated animals has been reported (14), in our retrospective study it was difficult to determine how strong immune responses related to disease severity as all samples in the present study were taken from convalescent phase of infection. It would require analysis of the kinetics of the immune responses and correlation with the virus load and clinical settings during the acute phase of the infection to address this issue. As it is not possible to gain access to acute PBMC samples, we investigated cytokine profiles in serum collected during the acute phase SARS. We found that, within the first week of illness, several cytokines were elevated in the serum of SARS patients. Many of these cytokines were proinflammatory, reflecting innate immune responses associated with NK cells or monocyte/macrophage activation, which is in agreement with previous studies (7). However, this pattern of cytokines was also observed in other severe viral pneumonias such as acute avian influenza (Ref. 33 and our manuscript in preparation). We observed that patients who died from SARS showed a significant increase in the Th2 cytokines IL-4, IL-5, and IL-10 compared with patients who recovered; this could be influenced by aging, which was associated with severe disease (27).
It has been unclear whether the adaptive immune response contributes to recovery or disease. The development of severe disease as the viremia declines suggests the latter, as do some studies with other coronaviruses (14, 34). However, we found that both neutralizing Ab (B cells) responses and T cell responses to the Spike were dominant in patients recovered from SARS, indicating that a spike-based vaccine maybe sufficient for a prophylactic vaccine, as previously demonstrated in a mouse model (11). The association between the Th2 cytokine pattern in acute infection and death suggests that it may be the quality of immune response, rather than magnitude, which is critical. A definitive answer could not be derived from the current study, but this work does give us the tools to resolve this rapidly should SARS-CoV infection return or a similar respiratory virus infection appear. Serial analysis of innate, Ab and T cell responses during the critical first 2–3 wk when patients die or recover should give a clear answer of whether they require immunosuppressing therapy. However, the immune responses studied in this study were in patients who recovered fully, so they are therefore likely to have been beneficial, and should serve as a benchmark for SARS-CoV vaccine design and evaluation.
Acknowledgments
First, we thank Drs Xiaoning Wang, Bingzhong Shi, and Chunhui Zhao for help in organizing SARS cohorts in Guangzhou and in Beijing. We are also grateful for advice and technical support from Drs. Tao Dong, Hongchao Pan, and Nilu Goonetilleke.
Footnotes
This work was support by Beijing Municipal Government, National Science Fund for Distinguished Young Scholars, Medical Research Council U.K., and the Euro-Asian SARS-DTV Network (SP22-CT-2004–511064) from the European Commission specific research and technological development Program “Integrating and strengthening the European Research area.”
Abbreviations used in this paper: SARS, severe acute respiratory syndrome; CoV, coronavirus; PBMC, peripheral blood mononuclear cell; ICS, intracellular cytokine staining; SFC, spot forming cell; Nab, neutralizing Ab.
The online version of this article contains supplemental data.
Disclosures
The authors have no financial conflict of interest.
References
- 1.Zhao GP. SARS molecular epidemiology: a Chinese fairy tale of controlling an emerging zoonotic disease in the genomics era. Philos Trans R Soc Lond B. 2007;362:1063–1081. doi: 10.1098/rstb.2007.2034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Shi Z, Hu Z. A review of studies on animal reservoirs of the SARS coronavirus. Virus Res. 2007;133:74–87. doi: 10.1016/j.virusres.2007.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Li W, Shi Z, Yu M, Ren W, Smith C, Epstein JH, Wang H, Crameri G, Hu Z, Zhang H, et al. Bats are natural reservoirs of SARS-like corona-viruses. Science. 2005;310:676–679. doi: 10.1126/science.1118391. [DOI] [PubMed] [Google Scholar]
- 4.Stockman LJ, Bellamy R, Garner P. SARS: systematic review of treatment effects. PLoS Med. 2006;3:e343. doi: 10.1371/journal.pmed.0030343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chu CM, Poon LL, Cheng VC, Chan KS, Hung IF, Wong MM, Chan KH, Leung WS, Tang BS, Chan VL, et al. Initial viral load and the outcomes of SARS. Canadian Medical Association Journal. 2004;171:1349–1352. doi: 10.1503/cmaj.1040398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chen J, Subbarao K. The immunobiology of SARS*. Annu Rev Immunol. 2007;25:443–472. doi: 10.1146/annurev.immunol.25.022106.141706. [DOI] [PubMed] [Google Scholar]
- 7.Cameron MJ, Ran L, Xu L, Danesh A, Bermejo-Martin JF, Cameron CM, Muller MP, Gold WL, Richardson SE, Poutanen SM, et al. Interferon-mediated immunopathological events are associated with atypical innate and adaptive immune responses in patients with severe acute respiratory syndrome. J Virol. 2007;81:8692–8706. doi: 10.1128/JVI.00527-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Temperton NJ, Chan PK, Simmons G, Zambon MC, Tedder RS, Takeuchi Y, Weiss RA. Longitudinally profiling neutralizing antibody response to SARS coronavirus with pseudotypes. Emerg Infect Dis. 2005;11:411–416. doi: 10.3201/eid1103.040906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ho MS, Chen WJ, Chen HY, Lin SF, Wang MC, Di J, Lu YT, Liu CL, Chang SC, Chao CL, et al. Neutralizing antibody response and SARS severity. Emerg Infect Dis. 2005;11:1730–1737. doi: 10.3201/eid1111.040659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Roberts A, Lamirande EW, Vogel L, Jackson JP, Paddock CD, Guarner J, Zaki SR, Sheahan T, Baric R, Subbarao K. Animal models and vaccines for SARS-CoV infection. Virus Res. 2007;133:20–32. doi: 10.1016/j.virusres.2007.03.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yang ZY, Kong WP, Huang Y, Roberts A, Murphy BR, Subbarao K, Nabel GJ. A DNA vaccine induces SARS coronavirus neutralization and protective immunity in mice. Nature. 2004;428:561–564. doi: 10.1038/nature02463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhu Z, Chakraborti S, He Y, Roberts A, Sheahan T, Xiao X, Hensley LE, Prabakaran P, Rockx B, Sidorov IA, et al. Potent cross-reactive neutralization of SARS coronavirus isolates by human monoclonal antibodies. Proc Natl Acad Sci USA. 2007;104:12123–12128. doi: 10.1073/pnas.0701000104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hogan RJ. Are nonhuman primates good models for SARS? PLoS Med. 2006;3:e411. doi: 10.1371/journal.pmed.0030411. author reply e415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Perlman S, Dandekar AA. Immunopathogenesis of coronavirus infections: implications for SARS. Nat Rev Immunol. 2005;5:917–927. doi: 10.1038/nri1732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Weingartl H, Czub M, Czub S, Neufeld J, Marszal P, Gren J, Smith G, Jones S, Proulx R, Deschambault Y, et al. Immunization with modified vaccinia virus Ankara-based recombinant vaccine against severe acute respiratory syndrome is associated with enhanced hepatitis in ferrets. J Virol. 2004;78:12672–12676. doi: 10.1128/JVI.78.22.12672-12676.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yang ZY, Werner HC, Kong WP, Leung K, Traggiai E, Lanzavecchia A, Nabel GJ. Evasion of antibody neutralization in emerging severe acute respiratory syndrome coronaviruses. Proc Natl Acad Sci USA. 2005;102:797–801. doi: 10.1073/pnas.0409065102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kim HW, Canchola JG, Brandt CD, Pyles G, Chanock RM, Jensen K, Parrott RH. Respiratory syncytial virus disease in infants despite prior administration of antigenic inactivated vaccine. Am J Epidemiol. 1969;89:422–434. doi: 10.1093/oxfordjournals.aje.a120955. [DOI] [PubMed] [Google Scholar]
- 18.Chattopadhyay PK, Price DA, Harper TF, Betts MR, Yu J, Gostick E, Perfetto SP, Goepfert P, Koup RA, De Rosa SC, et al. Quantum dot semiconductor nanocrystals for immunophenotyping by polychromatic flow cytometry. Nat Med. 2006;12:972–977. doi: 10.1038/nm1371. [DOI] [PubMed] [Google Scholar]
- 19.WHO. Case Definitions for Surveillance of Severe Acute Respiratory Syndrome (SARS) 2003 casedefinition/en/.WHO, http://www.who.int/csr/sars/
- 20.CDC. Public Health Guidance for Community-Level Preparedness and Response to Severe Acute Respiratory Syndrome (SARS) 2003 CDC, Version 2 2003 at: http://www.cdc.gov/mmwr/preview/mmwrhtml/mm5217a5.ht.
- 21.Chen X, Zhou B, Li M, Liang X, Wang H, Yang G, Wang H, Le X. Serology of severe acute respiratory syndrome: implications for surveillance and outcome. J Infect Dis. 2004;189:1158–1163. doi: 10.1086/380397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Li T, Xie J, He Y, Fan H, Baril L, Qiu Z, Han Y, Xu W, Zhang W, You H, et al. Long-term persistence of robust antibody and cytotoxic T cell responses in recovered patients infected with SARS coronavirus. PLoS ONE. 2006;1:e24. doi: 10.1371/journal.pone.0000024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Addo MM, Yu XG, Rathod A, Cohen D, Eldridge RL, Strick D, Johnston MN, Corcoran C, Wurcel AG, Fitzpatrick CA, et al. Comprehensive epitope analysis of human immunodeficiency virus type 1 (HIV-1)-specific T-cell responses directed against the entire expressed HIV-1 genome demonstrate broadly directed responses, but no correlation to viral load. J Virol. 2003;77:2081–2092. doi: 10.1128/JVI.77.3.2081-2092.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Goulder PJ, Addo MM, Altfeld MA, Rosenberg ES, Tang Y, Govender U, Mngqundaniso N, Annamalai K, Vogel TU, Hammond M, et al. Rapid definition of five novel HLA-A*3002-restricted human immunodeficiency virus-specific cytotoxic T-lymphocyte epitopes by elispot and intracellular cytokine staining assays. J Virol. 2001;75:1339–1347. doi: 10.1128/JVI.75.3.1339-1347.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pitcher CJ, Quittner C, Peterson DM, Connors M, Koup RA, Maino VC, Picker LJ. HIV-1-specific CD4+ T cells are detectable in most individuals with active HIV-1 infection, but decline with prolonged viral suppression. Nat Med. 1999;5:518–525. doi: 10.1038/8400. [DOI] [PubMed] [Google Scholar]
- 26.Donnelly CA, Ghani AC, Leung GM, Hedley AJ, Fraser C, Riley S, Abu-Raddad LJ, Ho LM, Thach TQ, Chau P, et al. Epidemiological determinants of spread of causal agent of severe acute respiratory syndrome in Hong Kong. Lancet. 2003;361:1761–1766. doi: 10.1016/S0140-6736(03)13410-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Han SN, Meydani SN. Antioxidants, cytokines, and influenza infection in aged mice and elderly humans. J Infect Dis. 2000;182(Suppl 1):S74–S80. doi: 10.1086/315915. [DOI] [PubMed] [Google Scholar]
- 28.Wang B, Chen H, Jiang X, Zhang M, Wan T, Li N, Zhou X, Wu Y, Yang F, Yu Y, et al. Identification of an HLA-A*0201-restricted CD8+ T-cell epitope SSp-1 of SARS-CoV spike protein. Blood. 2004;104:200–206. doi: 10.1182/blood-2003-11-4072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhou M, Xu D, Li X, Li H, Shan M, Tang J, Wang M, Wang FS, Zhu X, Tao H, et al. Screening and identification of severe acute respiratory syndrome-associated coronavirus-specific CTL epitopes. J Immunol. 2006;177:2138–2145. doi: 10.4049/jimmunol.177.4.2138. [DOI] [PubMed] [Google Scholar]
- 30.Zhao J, Huang Q, Wang W, Zhang Y, Lv P, Gao XM. Identification and characterization of dominant helper T-cell epitopes in the nucleocapsid protein of severe acute respiratory syndrome coronavirus. J Virol. 2007;81:6079–6088. doi: 10.1128/JVI.02568-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mitchison NA. T-cell-B-cell cooperation. Nat Rev Immunol. 2004;4:308–312. doi: 10.1038/nri1334. [DOI] [PubMed] [Google Scholar]
- 32.Rota PA, Oberste MS, Monroe SS, Nix WA, Campagnoli R, Icenogle JP, Penaranda S, Bankamp B, Maher K, Chen MH, et al. Characterization of a novel coronavirus associated with severe acute respiratory syndrome. Science. 2003;300:1394–1399. doi: 10.1126/science.1085952. [DOI] [PubMed] [Google Scholar]
- 33.de Jong MD, Simmons CP, Thanh TT, Hien VM, Smith GJ, Chau TN, Hoang DM, Chau NV, Khanh TH, Dong VC, et al. Fatal outcome of human influenza A (H5N1) is associated with high viral load and hypercytokinemia. Nat Med. 2006;12:1203–1207. doi: 10.1038/nm1477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Peiris JS, Chu CM, Cheng VC, Chan KS, Hung IF, Poon LL, Law KI, Tang BS, Hon TY, Chan CS, et al. Clinical progression and viral load in a community outbreak of coronavirus-associated SARS pneumonia: a prospective study. Lancet. 2003;361:1767–1772. doi: 10.1016/S0140-6736(03)13412-5. [DOI] [PMC free article] [PubMed] [Google Scholar]