Abstract
Transcription of the arginine biosynthetic gene ARG1 is activated by Gcn4p, a transcription factor induced by starvation for any amino acid. Previously we showed that Gcn4p binding stimulates the recruitment of Mcm1p and co-activator SWI/SNF to ARG1 in cells via Gcn4p induction through amino acid starvation. Here we report that Gcn4p binding is reduced by point mutations of the Mcm1p binding site and increased by overexpression of Mcm1p. This result suggests that Mcm1p plays a positive role in recruiting activator Gcn4p to ARG1, similar to the previously described cooperative interaction of Mcm1p with sequence-specific transcription factors at their promoters. In addition, the mutational analysis of Mcm1p binding sites showed that recruitment of the co-activator SWI/SNF correlated more closely with binding of Mcm1p than of Gcn4p at ARG1. Consistent with this, SWI/SNF co-immunoprecipitated with Mcm1p, but not with Gcn4p. These results support that Mcm1p increases the SWI/SNF recruitment at ARG1, a Gcn4p target promoter. The interaction between Mcm1p and SWI/SNF was abolished in a snf2 deletion strain containing an intact SWI/SNF sub-complex, suggesting that Mcm1p targets the catalytic subunit, which has ATPase activity, during SWI/SNF recruitment. We propose that Mcm1p contributes to active transcription at the ARG1 promoter by increasing the binding of the activator Gcn4p and by recruiting the co-activator complex SWI/SNF at ARG1 under Gcn4p-induced conditions.
Keywords: Gcn4p, Mcm1p, ARG1, SWI/SNF, snf2p
Introduction
Transcription of most amino acid biosynthesis genes, including four ARG (ARG1, ARG3, ARG5,6, and ARG8) genes, is induced by Gcn4p in cells starved for any amino acid due to increased expression of GCN4 at the translational level [1,2]. Mcm1p is also an important regulator of the ARG gene promoters [1,3]. Mcm1p mediates the repression of the four ARG genes in response to exogenous arginine by forming a repressor complex, ArgR/Mcm1p, through interactions with Arg80p, Arg81p, and Arg82p [1,3]. Indeed, the ArgR/Mcm1p repressor and the transcriptional activator Gcn4p dually regulate the ARG gene promoters.
Mcm1p is an MCM1, AGAMOUS, DEFICIENS, and serum response factor (MADS) box transcription factor protein that regulates the response of yeast to environmental conditions [3]. Mcm1p also plays an important role in DNA synthesis by binding at the replication origin [4]. Mcm1p cooperates with diverse sequence-specific transcription factors such as α1p, α2p, Ste12p, Yox1p, Yhp1, and Fkh2p at their promoters, stimulating or impeding the function of transcription factors that regulate cell type specification or the cell cycle [3,5,6]. Previously, we showed that binding of Gcn4p transcription factor stimulates the recruitment of Mcm1p to ARG1 [7]. Therefore, it is conversely possible that binding of Mcm1p transcription factor might increase Gcn4p recruitment.
The yeast SWI/SNF complex is an ATP-dependent chromatin remodeling complex that contains 11 different subunits. Deletion of Snf2p, the ATPase subunit of SWI/SNF, is not lethal but leads to altered transcription of a subset (1-2%) of genes in nutrient-rich medium [8,9]. Snf2p can also mediate repression of SER3 without the cooperation of other SWI/SNF subunits [9,10]. Genetic studies suggest that many of these subunits are required for the chromatin remodeling function of the complex [9]. The SWI/SNF complex is recruited to ARG1 in a Gcn4p-dependent manner [11]. Mcm1p recruits the SWI/SNF complex to the STE6 promoter and contributes to the activation of the complex [12]. Therefore, Mcm1p binding might contribute to recruitment of the SWI/SNF complex to the ARG1 promoter.
Here we report that Gcn4p binding is increased by Mcm1p binding. In addition, we provide strong evidence that Mcm1p binding at ARG1 contributes to the recruitment of SWI/SNF. Our results indicate that Mcm1p and Gcn4p cooperatively play a role in recruiting SWI/SNF to ARG1.
Materials and Methods
Yeast strains and plasmids
The strains used in this study are listed in Table 1 in the Supporting Information. The wild-type (WT) parent strain BY4741 (MATa his3Δ1 leu2Δ0 met15Δ0 ura3Δ0) and deletion derivatives, generated by the Saccharomyces Genome Deletion Project, were purchased from Research Genetics. The strain SS5 (MATαura3ΔargΔ-PH) was a gift from Marjolaine Crabeel [13]. The construction of novel strains to generate myc epitope tags, mutations of the ARG1 promoter, or gcn4Δ strains is described in the Supporting Information.
Plasmids pHQ1293, pHQ1239, pED40 and pSK1 were described previously [7,11,14]. The empty vectors were the URA3 CEN4 plasmid YCp50, the LEU2 plasmid pRS315, and pRS425. The details of the construction of novel plasmids are provided in the Supporting Information.
Chromatin immunoprecipitations
Cells were grown to OD600 of ∼0.8 in an appropriate synthetic complete (SC) media (SC, SC-Ura, SC-Leu, SC-Ura-Leu). Cells were starved for isoleucine/valine at 30°C, treated for 2 h with SM (0.6 μg/ml), and cross-linked with formaldehyde. Chromatin immunoprecipitation (ChIP) assays were conducted using anti-myc antibodies as previously described [7,11,14]. PCR analysis was conducted as described in the Supporting Information.
Co-immunoprecipitation assays
Cells were grown as described for ChIP analysis. Co-immunoprecipitation analysis was performed as described previously [7,11]. Briefly, whole cell extracts (WCEs) were prepared as described for GST-pull down assays [15]. The WCEs were incubated for 1 hr at 4°C with 1 μg anti-myc antibody, 100 μg BSA dissolved in PBS, and buffer MTB. The immune complexes were washed three times with 1 ml MTB, dissolved in SDS-PAGE loading buffer, and subjected to Western blot analysis.
Results and Discussion
Gcn4p binding is reduced by point mutations in the MADS box
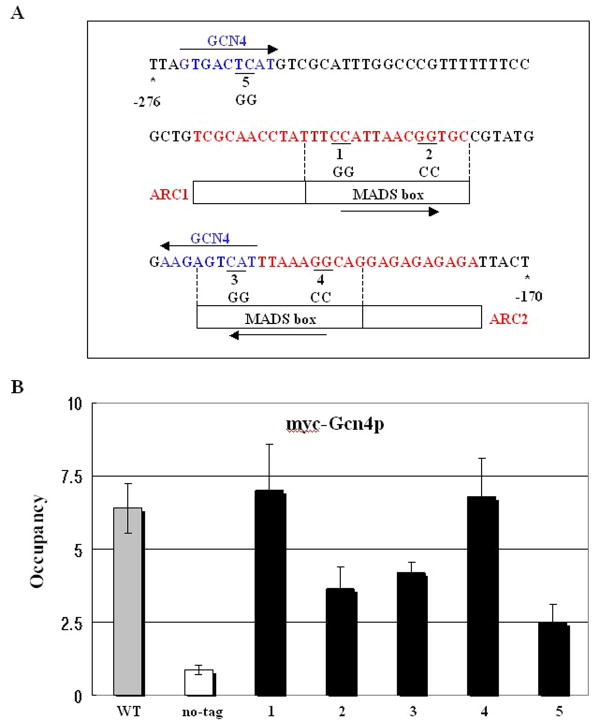
The ARG1 promoter contains two arginine control elements (ARC1 and ARC2) and two Gcn4p binding sites, which are designated by blue and red letters in Fig. 1A, respectively. Genetic and biochemical analyses of the ARG1 promoter have shown that two ARC elements are required for efficient repression of ARG1 expression by the ArgR/Mcm1p complex in medium containing arginine. Two Gcn4p binding sites contribute to efficient activation of ARG1 by Gcn4p [1,3,13,16]. The consensus sequence for Gcn4p contains nine nucleotides, ATGACTCAT. Both the 5′-most Gcn4p binding site and the second Gcn4p binding site (present in reverse orientation) contain eight consensus nucleotides. The half sequences of the ARC elements contain the MADS box, which is the binding site for Mcm1p [13,16]. The consensus sequence for the MADS box, called the P box, contains 16 nucleotides, TTTCCCTATTAGGTAA [17]. The first MADS box in ARC1 includes a greater number of consensus nucleotides (nine) than the second MADS box in ARC2 (six nucleotides). The two MADS boxes are also in opposite orientations, as designated by the arrows in Fig. 1A. Six base pairs in the second Gcn4p site overlap the second MADS box.
Figure 1. Gcn4p binding is reduced by point mutations in the MADS box.
(A) Nucleotide sequence of the ARG1 promoter region. The sequence is numbered (-276 to -170, indicated by asterisks) relative to the main transcription start site. The Gcn4p binding sites (GTGACTCAT and AAGAGTCAT) are overlined and designated with blue. The site-directed mutagenized sequences are underlined with black and indicated below by numbers (1 to 5). There are two arginine control elements (ARC1 and ARC2), which are binding sites for the ArgR/Mcm1p repressor complex, indicated by red letters and boxes. The putative Mcm1p binding sites are boxed within the ARC elements and designated as MCM1, AGAMOUS, DEFICIENS, and serum response factor (MADS) boxes. The two Gcn4p binding sites and MADS boxes are in opposite orientations and designated with arrows. (B) The gcn4Δ SS5 strains containing mutations in the ARG1 alleles are indicated below the histogram; WT (SY722), mutant 1 (SY772), mutant 2 (SY723), mutant 3 (SY583), mutant 4 (SY724), and mutant 5 (SY773). The strains were transformed with a single-copy GCN4-myc plasmid (pSK1). For the non-tagging (no-tag) condition, gcn4Δ (SY722) strains were transformed with an empty vector. ChIP analysis of the transformants was conducted to measure binding of myc-Gcn4p to ARG1, as described in Materials and Methods and Supporting Information. The percentage of ARG1 UAS that immunoprecipitated with myc antibodies was measured for each strain and normalized to the nonspecific immunoprecipitation of POL1 ORF sequences (Occupancy).
Mcm1p binding has been shown to enhance recruitment of other transcription factors [17]. In order to determine whether Mcm1p also plays a role in Gcn4p binding at the ARG1 promoter, we introduced point mutations into the predicted MADS box binding sites in the ARC elements. To eliminate Mcm1p binding at each ARC element, we replaced the conserved CC residues in each predicted half-site with GG residues in each of the two ARC elements (Fig. 1A). In the crystal structure, these residues in the MADS box are important for Mcm1p binding with Matoα2 transcription factor to STE6 promoter [17]. They make base-specific contacts with Lys-38 of Mcm1p, and substitution of these CC residues with GG residues strongly impairs the transcriptional repression mediated by Mcm1p/Matoα2 binding to the STE6 promoter [17].
We also made GG mutations in one of the half-sites in the Gcn4p binding sites to determine the effect of mutating Gcn4p binding sites on myc-Gcn4p binding (Fig. 1A). Note that the GG mutation in the second Gcn4p binding site also mutated two residues in the MADS box in ARC2. We designated the mutants as numbers 1 to 5 (Fig. 1A). The mutations were introduced into the chromosomal copy of ARG1. The gcn4 deletion was generated in each mutant and transformed with a GCN4-myc plasmid encoding a functional myc-tagged form of Gcn4p [11,14]. After inducing myc-Gcn4p protein by incubating cells with sulfometuron methyl (SM), which produces starvation for isoleucine and valine, we performed ChIP analysis to measure myc-Gcn4p occupancy of the ARG1 promoter.
As shown in Fig. 1B, mutants 2, 3, and 5 exhibited reduced myc-Gcn4p binding. Mutant 5 produced the most severe binding defect, whereas mutants 2 and 3 reduced the binding of myc-Gcn4p to similar levels (Fig.1B). Even though mutation 3 altered the Gcn4p binding site and the ARC2 element, we assume that the reduction in myc-Gcn4p binding in mutant 3 resulted from the defective Gcn4p binding site, not from defective Mcm1p binding in the ARC element. Interestingly, mutation 2 also reduced myc-Gcn4p binding, suggesting that Mcm1p binding was involved in the increased myc-Gcn4p binding at ARG1. However, myc-Gcn4p binding was not affected by mutants 1 and 4, suggesting that the conserved 5′-distal CC nucleotides sequences in the MADS box were more critical for Gcn4p binding than the 5′-proximal CC nucleotides. This mutational analysis revealed that the MADS box in the ARC1 element is important for Gcn4p binding. This implies that binding of Mcm1p to ARC1 stimulates Gcn4p binding to the ARG1 promoter and that Mcm1p binding to ARC1 plays a positive role in ARG1 transcription through increased Gcn4p binding.
Levels of Gcn4p binding positively correlate with Mcm1p binding levels at Gcn4p target promoters
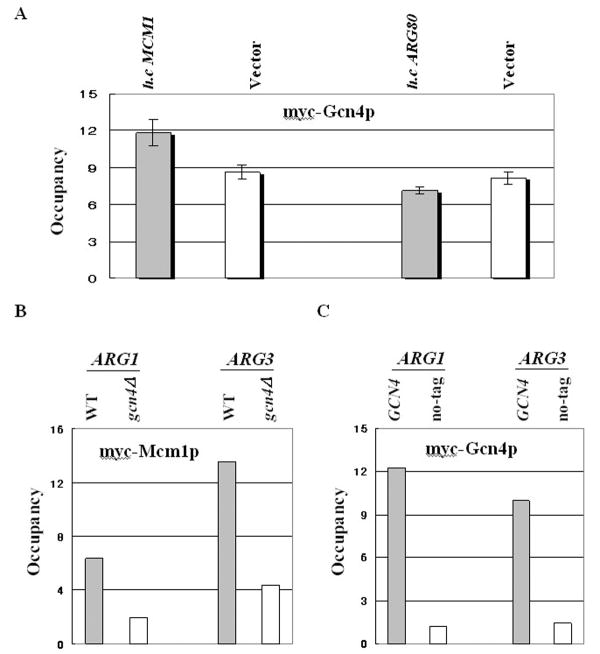
We further assessed the positive relationship between Mcm1p binding and recruitment of Gcn4p by testing whether increased Mcm1p levels led to enhanced Gcn4p binding. After transformation with a high-copy MCM1 plasmid, we measured the binding of myc-Gcn4p (Fig. 2A). The binding of myc-Gcn4p increased with Mcm1p overexpression compared to that with empty vector transfection (ANOVA, P=0.015). Arg80p is a component of the ArgR/Mcm1p complex and MADS box protein, which is recruited by Gcn4p [7]. But, overexpression of Arg80p protein did not result in increased binding of myc-Gcn4p (Fig. 2A), suggesting that Mcm1p could increase recruitment of Gcn4p at the ARC element regardless of the ArgR/Mcm1p complex.
Figure 2. Mcm1p binding levels positively correlate with Gcn4p binding levels at Gcn4p target promoters.
(A) The transformants of gcn4Δ (249) strains containing myc-tagged GCN4 plasmid (pSK1) were re-transformed with an empty vector, a high-copy plasmid pED40 harboring MCM1, or high-copy plasmid pSY365 harboring ARG80. ChIP analysis of the transformants was conducted to measure binding of myc-Gcn4p to ARG1 UAS, as described in Fig. 1B. (B) The target genes (ARG1 and ARG3) of Gcn4p are indicated above the histogram. High-copy plasmid pHQ1239 harboring the GCN4-HA allele was introduced into WT MCM1-myc (SY337) strains. An empty vector was introduced into the gcn4Δ MCM1-myc (SY339) strains. ChIP analysis of the transformants was conducted to measure binding of myc-Mcm1p to ARG1 UAS or to ARG3. Signals for ARG1 UAS or ARG3 in the immunoprecipitate (IP) were normalized to the corresponding POL1 signal and plotted in the histogram (Occupancy). (C) The gcn4Δ (249) strains were transformed with an empty vector (no-tag) or myc-tagged GCN4 plasmid (pSK1). ChIP analysis of the transformants was conducted to measure binding of myc-Gcn4p to ARG1 UAS or ARG3, as described in Fig. 2B.
We also tested whether Gcn4p binding positively correlated with Mcm1p binding at two different Gcn4p target promoters, ARG1 and ARG3. These two ARG genes are dependent on Gcn4p for their transcription and are known to bind ArgR/Mcm1p [1,7,11,14]. As shown in Fig. 2A-B, ARG1 and ARG3 both exhibit high Mcm1p and myc-Gcn4p binding. Consistent with our previous results [7], myc-Mcm1p occupancy is greatly reduced in gcn4Δ cells (Fig. 2A). These results fit with the idea that Gcn4p binding is enhanced by Mcm1p binding at ARG gene promoters, which would imply that Gcn4p and Mcm1p mutually enhance their binding to these promoters. Considering that this is similar to the previously described interaction of Mcm1p with other transcriptional partners such as α1p, Ste12p, and Fkh2p in target promoters [3,5,6], we assumed that Gcn4p is newly identified as a transcription partner for promoters including ARC elements.
SWI/SNF recruitment depends on Mcm1p binding more than Gcn4p binding
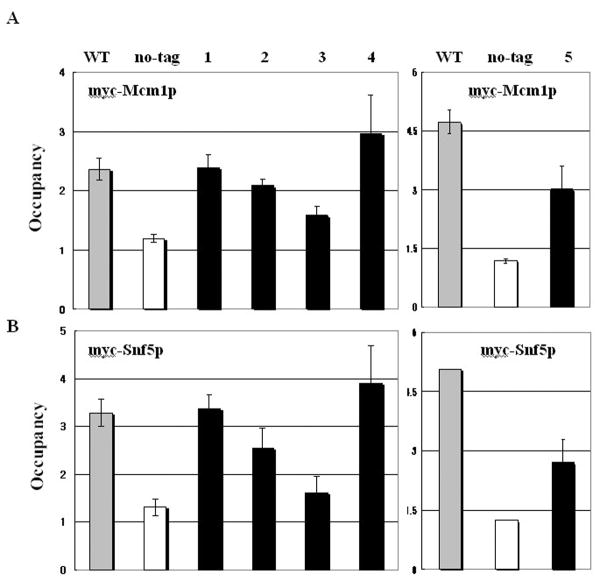
Mcm1p contributes to recruitment of the co-activator SWI/SNF complex at STE6 [17]. Since SWI/SNF complex is recruited to ARG1 in a Gcn4p-dependent manner [11], we tested the relationship between Mcm1p and recruitment of the SWI/SNF complex at ARG1 under Gcn4p-induced conditions by comparing Mcm1p binding and recruitment of Snf5p in mutants 1-5 (Fig. 1A). We were able to measure detectable and distinguishable levels of their recruitment with a high-copy GCN4 plasmid, which increased Gcn4p protein levels and recruitment of SWI/SNF 4- to 5-fold more than single-copy GCN4 plasmid at ARG1 [11]. Mutants 3 and 5 exhibited greatly reduced binding of both Mcm1p and Snf5p (Fig. 3A-B), suggesting that their recruitment was primarily affected by the mutations in the Gcn4p binding sites. Mutant 2 showed a lesser decrease in recruitment of both Mcm1p and Snf5p, while mutation 4 modestly increased the recruitment of both Mcm1p and Snf5p. We observed a similar pattern between Mcm1p and Snf5p by mutational analysis, suggesting that the SWI/SNF recruitment largely depends on Mcm1p binding at ARG1.
Figure 3. Recruitment of SWI/SNF more closely depends on Mcm1p binding than Gcn4p binding.
(A-B) The strains contained a non-tagged allele (no-tag: SY539) or 13 myc-tagged alleles (MCM1-myc and SNF5-myc), designated above each histogram (Table 1 in the Supporting Information). The MCM1-myc strains containing mutations in the ARG1 alleles are indicated at the top of each graph; WT (SY476), mutant 1 (SY650), mutant 2 (SY494), mutant 3 (SY499), mutant 4 (SY504), and mutant 5 (SY760). The SNF5-myc strains containing mutations in the ARG1 alleles are indicated at the top of each graph; WT (SY480), mutant 1 (SY663), mutant 2 (SY498), mutant 3 (SY503), mutant 4 (SY508), and mutant 5 (SY759). High-copy GCN4 plasmid pHQ1239 was introduced into the strains. ChIP analysis of the transformants was conducted to measure binding of myc-Mcm1p or myc-Snf5p to ARG1 UAS, as described in Fig. 1B.
Since SWI/SNF recruitment also depends on Gcn4p at ARG1 [11], we compared Snf5p recruitment (Fig. 3B) with Gcn4p binding (Fig. 1B). Based on the recruitment of Gcn4p, Mcm1p, and Snf5p in mutants 2, 3, and 4, we concluded that SWI/SNF recruitment more closely correlated with Mcm1p binding than with Gcn4p binding, which indicates that Mcm1p binding contributes more to SWI/SNF recruitment than Gcn4p binding. It was puzzling that we did not observe greatly reduced recruitment of both Mcm1p and Snf5p in mutant 2, since Gcn4p binding was significantly reduced in this mutant. Presumably, the residual Gcn4p binding in mutant 2 was sufficient to support nearly WT occupancies of Mcm1p and SWI/SNF. Taken together with the previous finding that SWI/SNF recruitment depends on Gcn4p [11], SWI/SNF recruitment was apparently enhanced by both Mcm1p and Gcn4p at ARG1. As myc-Gcn4p binding was unaffected by deletions of SWI/SNF subunits SWI3 or SNF5 at ARG1 [11], we found that myc-Mcm1p occupancy was also unaffected by their deletions (Supplementary Fig. 1A). This result indicates that binding of both Gcn4p and Mcm1p occurs before SWI/SNF recruitment.
The interaction between Mcm1p and SWI/SNF requires the Snf2p subunit
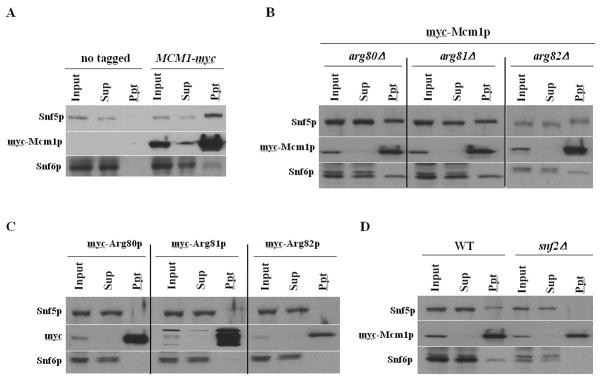
It is possible that protein-protein interactions contribute to the positive relationship between Mcm1p binding and the recruitment of SWI/SNF. However, the interaction between SWI/SNF and Mcm1p has not been identified. To determine whether Mcm1p interacts with the SWI/SNF complex, we performed co-immunoprecipitation experiments. Precipitation of Snf5p and Snf6p subunits was not detected in a non-tagged control strain, while the SWI/SNF subunits co-imunoprecipitated with myc-Mcm1p (Fig. 4A). To the best of our knowledge, this is the first report demonstrating an interaction between Mcm1p and SWI/SNF complex. This suggests that Mcm1p directly interacts with the SWI/SNF complex in solution. However, using a similar approach, we did not detect an interaction between Gcn4p and the SWI/SNF complex in cells containing either high- or single-copy GCN4-myc plasmid (Supplementary Fig. 1B and 1C). Considering that recombinant GST-Gcn4p interacted with SWI/SNF complex in yeast extracts in previous GST-pull down assays [15], our results suggest that the interaction between Gcn4p and SWI/SNF is weaker than the interaction between SWI/SNF and Mcm1p and support our ChIP data that Mcm1p contributes more to the recruitment of SWI/SNF complex at ARG1 than does Gcn4p.
Figure 4. Mcm1p interacts with the SWI/SNF complex and requires Snf2p subunit for its interaction.
(A) Non-tagged allele (BY4741) or myc-tagged MCM1 (SY337) were cultured. WCEs were immunoprecipitated with myc antibodies, as described in Materials and Methods. Ten percent of the input samples (Input), 100% of the immunoprecipitates (Ppt), and 10% of the supernatant (Sup) fractions were subjected to Western blot analysis using myc antibodies or rabbit polyclonal antibodies against Snf5p and Snf6p. (B) GCN4 strains containing arg80Δ MCM1-myc (SY366), arg81ΔMCM1-myc (SY372), and Arg82ΔMCM1-myc (SY367) were grown and subjected to co-immunoprecipitation with myc antibodies as described in Fig 4A. They were then subjected to Western blot analysis using antibodies against myc, Snf5p, and Snf6p. (C) GCN4 strains containing Arg80-myc (SY373), Arg81-myc (SY375), and Arg82-myc (SY377) were grown and subjected to co-immunoprecipitation with myc antibodies as described in Fig 4A. They were then subjected to Western blot analysis using myc antibodies to detect myc-Arg80p, myc-Arg81p, or myc-Arg81p. In addition, rabbit polyclonal antibodies against Snf5p and Snf6p were used for the analysis. (D) MCM1-myc (SY337) and snf2Δ MCM1-myc (SY358) strains were grown and subjected to co-immunoprecipitation with myc antibodies as described in Fig 4A. They were then subjected to Western blot analysis using antibodies against myc, Snf5p, and Snf6p.
Mcm1p forms the ArgR/Mcm1p complex with Arg80p, Arg81p, and Arg82p, which are also recruited to ARG1 [7]. We tested whether the SWI/SNF complex could still interact with Mcm1p when the other subunits of ArgR/Mcm1p complex were absent. Mcm1p precipitated with both Snf5p and Snf6p after deletion of ARG80, ARG81, or ARG82 (Fig. 4B). We also tested whether SWI/SNF interacted with myc-tagged forms of Arg80p, Arg81p, and Arg82p, but found no interaction (Fig. 4C). These results indicate that the interaction between Mcm1p and the SWI/SNF complex did not involve the ArgR/Mcm1p complex. Taken together, our findings suggest that Mcm1p alone may contribute to the increased SWI/SNF binding to ARC elements at ARG1, regardless of the ArgR/Mcm1p complex. Gcn4p binding was not increased by Arg80p overexpression (Fig. 2A), which also supports our conclusion.
Since Snf2p is the catalytic subunit in the SWI/SNF complex, we tested whether Mcm1p interacted with the rest of the SWI/SNF complex in a SNF2 deletion strain. We found that the SWI/SNF complex and Mcm1p no longer interacted when the Snf2p subunit was deleted (Fig. 4D). It was important to determine that Snf2p subunit-dependence in the interaction was not from complex-disruption in the SNF2 deletion strain. We confirmed that the SNF2 deletion did not disrupt the rest of the complex, since the SNF2 deletion did not disrupt the interactions among Snf5p, Snf6p, Swi1p, and Swp73p (Supplementary Fig. 1D and 1E). We conclude that the Snf2p subunit is important for the interaction between Mcm1p and the SWI/SNF complex. Our results reflect that SWI/SNF recruitment by Mcm1p might also depend on Snf2p subunit at STE6 promoter [12]. Previously, we showed that SWI/SNF recruitment at ARG1 was not reduced by the snf2 mutation [11], which was ostensibly at odds with the idea that Mcm1p contributes to SWI/SNF recruitment at ARG1. However, Gcn4p-myc binding was increased almost 2.5-fold in the snf2 strain [14]. This may indicate that enhanced, direct recruitment of SWI/SNF by Gcn4p can compensate for the impaired interaction of Mcm1p with SWI/SNF complex in the snf2 strain.
Our results suggest that Mcm1p binding to the MADS boxes contributes to binding of the activator Gcn4p and recruitment of the co-activator complex SWI/SNF at ARG1 under Gcn4p-induced conditions. The strong interaction between Mcm1p and the SWI/SNF complex could explain the proposed contribution of Mcm1p to SWI/SNF recruitment at ARG1.
Supplementary Material
Acknowledgments
We thank Marjolaine Crabeel for her gifts of the SS5 strain and M13mp7-ARG1, Francine Messenguy and Evelyn Dubois for plasmid pED40, Joseph Reese for Snf2p antibodies and Brehon Laurent for Snf5p, Snf6p, and Swp73p antibodies. We thank members of the Hinnebusch and Dever laboratories for helpful suggestions.
Abbreviation
- ARC
arginine control
- MADS
MCM1, AGAMOUS, DEFICIENS, and serum response factor
- ChIP
Chromatin immunoprecipitations
- UAS
upstream activating sequence
- SM
sulfometuron methyl
- h.c.
high-copy
- s.c.
single-copy
- WT
wild type
- WCE
whole cell extract
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Hinnebusch AG. In: The Molecular and Cellular Biology of the Yeast Saccharomyces: Gene Expression. Broach JR, Jones EW, Pringle JR, editors. Cold Spring Harbor Laboratory Press; Cold Spring Harbor, NY: 1992. pp. 319–414. [Google Scholar]
- 2.Natarajan K, Meyer MR, Jackson BM, Slade D, Roberts C, Hinnebusch AG, Marton MJ. Transcriptional profiling shows that Gcn4p is a master regulator of gene expression during amino acid starvation in yeast. Mol Cell Biol. 2001;21:4347–4368. doi: 10.1128/MCB.21.13.4347-4368.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Messenguy F, Dubois E. Role of MADS box proteins and their cofactors in combinatorial control of gene expression and cell development. Gene. 2003;316:1–21. doi: 10.1016/s0378-1119(03)00747-9. [DOI] [PubMed] [Google Scholar]
- 4.Chang VK, Fitch MJ, Donato JJ, Christensen TW, Merchant AM, Tye BK. Mcm1 binds replication origins. J Biol Chem. 2003;278:6093–6100. doi: 10.1074/jbc.M209827200. [DOI] [PubMed] [Google Scholar]
- 5.Carr EA, Mead J, Vershon AK. Alpha1-induced DNA bending is required for transcriptional activation by the Mcm1-alpha1 complex. Nucleic Acids Res. 2004;32:2298–2305. doi: 10.1093/nar/gkh560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Darieva Z, Pic-Taylor A, Boros J, Spanos A, Geymonat M, Reece RJ, Sedgwick SG, Sharrocks AD, Morgan BA. Cell cycle-regulated transcription through the FHA domain of Fkh2p and the coactivator Ndd1p. Curr Biol. 2003;13:1740–1745. doi: 10.1016/j.cub.2003.08.053. [DOI] [PubMed] [Google Scholar]
- 7.Yoon S, Govind CK, Qiu H, Kim SJ, Dong J, Hinnebusch AG. Recruitment of the ArgR/Mcm1p repressor is stimulated by the activator Gcn4p: a self-checking activation mechanism. Proc Natl Acad Sci USA. 2004;91:11713–11718. doi: 10.1073/pnas.0404652101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Holstege FC, Jennings EG, Wyrick JJ, Lee TI, Hengartner CJ, Green MR, Golub TR, Lander ES, Young RA. Dissecting the regulatory circuitry of a eukaryotic genome. Cell. 1998;95:717–728. doi: 10.1016/s0092-8674(00)81641-4. [DOI] [PubMed] [Google Scholar]
- 9.Sudarsanam P, Iyer VR, Brown PO, Winston F. Whole-genome expression analysis of snf/swi mutants of Saccharomyces cerevisiae. Proc Natl Acad Sci USA. 2000;97:3364–3369. doi: 10.1073/pnas.050407197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Martens JA, Winston F. Evidence that Swi/Snf directly represses transcription in S. cerevisiae. Genes & Dev. 2002;16:2231–2236. doi: 10.1101/gad.1009902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yoon S, Qiu H, Swanson MJ, Hinnebusch AG. Recruitment of SWI/SNF by Gcn4p does not require Snf2p or Gcn5p but depends strongly on SWI/SNF integrity, SRB mediator, and SAGA. Mol Cell Biol. 2003;23:8829–8845. doi: 10.1128/MCB.23.23.8829-9945.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gavin IM, Kladde MP, Simpson RT. Tup1p represses Mcm1p transcriptional activation and chromatin remodeling of an a-cell-specific gene. EMBO J. 2000;19:5875–5883. doi: 10.1093/emboj/19.21.5875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Crabeel M, de Rijcke M, Seneca S, Heimberg H, Pfeiffer I, Matisova A. Further definition of the sequence and position requirements of the arginine control element that mediates repression and induction by arginine in Saccharomyces cerevisiae. Yeast. 1995;11:1367–1380. doi: 10.1002/yea.320111405. [DOI] [PubMed] [Google Scholar]
- 14.Qiu H, Hu C, Yoon S, Natarajan K, Swanson MJ, Hinnebusch AG. An array of coactivators is required for optimal recruitment of TATA binding protein and RNA polymerase II by promoter-bound Gcn4p. Mol Cell Biol. 2004;24:4104–4117. doi: 10.1128/MCB.24.10.4104-4117.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Drysdale CM, Jackson BM, McVeigh R, Klebanow ER, Bai Y, Kokubo T, Swanson M, Nakatani Y, Weil PA, Hinnebusch AG. The Gcn4p activation domain interact specifically in vitro with RNA polymerase II holoenzyme, TFIID, and the Adap-Gcn5p coactivator complex. Mol Cell Biol. 1998;18:1711–1724. doi: 10.1128/mcb.18.3.1711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.De Rijcke M, Seneca S, Punyammalee B, Glansdorff N, Crabeel M. Characterization of the DNA target site for the yeast ARGR regulatory complex, a sequence able to mediate repression or induction by arginine. Mol Cell Biol. 1992;12:68–81. doi: 10.1128/mcb.12.1.68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tan S, Richmond TJ. Crystal structure of the yeast MATalpha2/MCM1/DNA ternary complex. Nature. 1998;391:660–666. doi: 10.1038/35563. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.