Abstract
Many biologically active proteins are intrinsically disordered. A reasonable understanding of the disorder status of these proteins may be beneficial for better understanding of their structures and functions. The disorder contents of disordered proteins vary dramatically, with two extremes being fully ordered and fully disordered proteins. Often, it is necessary to perform a binary classification and classify a whole protein as ordered or disordered. Here, an improved error estimation technique was applied to develop the cumulative distribution function (CDF) algorithms for several established disorder predictors. A consensus binary predictor, based on the artificial neural networks, NN-CDF, was developed by using output of the individual CDFs. The consensus method outperforms the individual predictors by 4~5% in the averaged accuracy.
Keywords: Intrinsically Disordered Protein, Prediction, Accuracy, CDF
1. Introduction
A number of proteins lacking rigid 3D structures under physiological conditions in vitro yet fulfilling key biological functions is rapidly increasing [1–10]. These proteins are known as intrinsically disordered proteins (IDPs) among other names. They are highly abundant in nature [11–13], typically involved in signaling, recognition and regulation [7,8,14–18], and are strongly associated with human diseases [19]. IDPs typically possess highly dynamic structures in solution with high mobility at different timescales, and therefore such proteins almost never form crystals. Hence, the existence of these proteins represents a substantial challenge to the structural genomics initiative [20].
IDPs and IDRs differ from structured globular proteins and domains with regard to many attributes, including amino acid composition, sequence complexity, hydrophobicity, charge, flexibility, and type and rate of amino acid substitutions over evolutionary time [4,21–23]. Based on these differences between IDPs and ordered proteins, numerous disorder predictors have been developed (reviewed in [24–26]). Nearly all of the predictive tools developed so far provide disorder prediction on the per-residue basis; i.e., they give the likely disorder status of each amino acid residue. Often, in the analysis of a given dataset, it is useful to carry out a binary classification of whole proteins, indicating whether a protein is likely to fold or likely to remain unstructured. Such a classification is not a simple task, as the extent to which a sequence is ordered or disordered and the nature of disorder vary widely among proteins. In fact, the structural variability of IDPs is extremely high and native coils, native pre-molten globules, and native molten globules were described in literature [4,9,10,14,16,18,27]. The protein can be completely unstructured or contain some elements of tertiary and/or secondary structure. In multi-domain proteins, domains might be connected by highly flexible linkers, and one or several domains might be completely disordered. Some proteins might have long disordered loops or tails. Because of this great variability, there is no strict boundary between ordered and intrinsically disordered proteins.
Two distinct binary classification methods have been reported previously [3,11,13]. One of these approaches uses charge-hydropathy plots (CH-plots), where ordered and disordered proteins are plotted in CH-space, and a linear boundary separates them [3]. The other method is based on predictor of natural disordered regions (PONDR®) VLXT [21,28], which predicts the order-disorder score for every residue in a protein. Cumulative distribution function (CDF) distinguishes ordered and disordered proteins based on the distribution of prediction scores [11,13]. CDF curve gives the fraction of the outputs that are less than or equal to a given value. According to the CDF analysis, fully disordered proteins have very low percentage of residues with low predicted disorder scores, as the majority of their residues possess high predicted disorder scores. On the contrary, the majority of residues in ordered proteins are predicted to have low disorder scores. Hence, theoretically, all the fully disordered proteins should stay at the lower right corner of the CDF plot, whereas all the fully ordered proteins should be located at the upper left corner of this plot [11,13].
Due to the significant improvement in the prediction accuracy observed for several per-residue predictors, it was of interest to determine whether the CDF analysis based on these predictors would give improved binary classifications. An additional question was whether new methods can be used to optimize the CDF boundary line to achieve higher prediction accuracy. In this paper, the CDF method was developed for two other members of the PONDR® family of disorder predictors, VSL2 [29,30] and VL3 [31], for a simplified predictor based on the TOP-IDP scale [32], as well as for IUPred [33,34] and FoldIndex [35]. We also proposed a new method for optimizing the order-disorder boundary line in the CDF plots. Finally, a consensus method was elaborated by using a neural network based on CDF values from the outputs of the PONDR® VXLT, PONDR® VSL2, PONDR® VL3, TOP-IDP, IUPred, and FoldIndex, and this method appears to be more accurate than any of the methods based on individual predictors.
2. Materials and methods
2.1. Dataset construction
Four groups of datasets were used in this study. The first group included the ‘original datasets’ from Ref. [13]: (i) an ordered dataset of 105 wholly ordered proteins and (ii) a disordered dataset of 54 fully disordered proteins. These two datasets were used to take advantage of their high quality, and to provide an unambiguous comparison of the new methods developed in this paper with the previously developed method [13]. The second group was new fully ordered and fully disordered datasets. The new set of fully ordered proteins had 554 chains that were derived from the PDB database as of July 20, 2008 to include sequences of non-homologous single chain non-membrane proteins, which had no ligands, no disulfide bonds, and no missing residues, and which were characterized by unit cells with primitive space groups. The new dataset of fully disordered protein had 84 chains that were extracted from DisProt (release 4.5 of July 17, 2008) [36] to include non-homologous proteins without structured regions. Each of these new datasets was randomly and equally split into training and testing sets. The third group was the datasets of sequences for Escherichia coli K12, Archaeoglobus fulgidus, and Methanobacterium thermoautotrophicum generated from the UniProt database after removing all the fragments. The last group was a dataset that included 64 partially disordered proteins with less than 25% of sequence identity which were also extracted from PDB and had missing electron density for at least 30 residues, as in Ref. [13].
2.2 Individual disorder predictors and CDF
PONDR® VLXT [21,28] is composed of three neural networks, two for the termini of the sequence and one for internal region. The final output is an average over above three outs. The inputs of the neural networks are residue composition-related quantities. PONDR® VL3 [31] employs majority-voting over a bunch of neural networks which also take composition, complexity, and entropy as the inputs. PONDR® VSL2 [29,30] is built up on support vector machine with sequence composition, evolution information, and predicted secondary structure as the inputs. TOP-IDP [32] is a new amino acid scale developed to discriminate ordered and disordered residues with the highest accuracy. IUPred [33,34] applies the sequence-based pair-wise potential energy evaluated from the globular proteins to distinguish disordered residues/proteins from the ordered ones. FoldIndex [35] takes the relative relation of net charges and normalized hydrophobicity scale which is originated from CH plot to partition ordered and disordered residues.
CDF analysis summarizes the per-residue predictions by plotting predicted disorder scores against their cumulative frequency, which allows ordered and disordered proteins to be distinguished based on the distribution of prediction scores [11,13]. At any given point on the CDF curve, the ordinate gives the proportion of residues with a disorder score less than or equal to the abscissa. To develop corresponding CDF algorithms, the outputs of all the above-mentioned predictors were unified to produce the per-residue disorder scores ranging from 0 (ordered) to 1 (disordered). In this way, CDF curves for various disorder predictors always began at the point (0, 0) and ended at the point (1, 1) because disorder predictions were defined only in the range [0, 1] with values less than 0.5 indicating a propensity for order and values greater than or equal to 0.5 indicating a propensity for disorder. As a result, fully ordered proteins yield convex curves because a high proportion of the prediction outputs are below 0.5, while fully disordered proteins typically yield concave curves because a high proportion of the prediction outputs are above 0.5. In practice, the range of prediction score (from 0 to 1) was divided into 20 bins [11,13]. It is expected therefore that there should be an approximately diagonal boundary line that could be used to separate the ordered and disordered proteins with an acceptable accuracy.
The original datasets were divided into training sets and testing sets. The boundary line for each CDF was optimized in the training set, and tested in the testing set. Bootstrap sampling of 1000 times was also applied to validate the confidence region of the accuracy.
A quantity termed CDF distance was also applied to assess whether the protein is ordered or disordered. The CDF distance is defined as:
| (1) |
where dCDF is the averaged CDF distance of the protein from the CDF boundary line. Ks and Ke are the starting and ending bins of the CDF boundary line. CDFi is the CDF value of i-th bin, while CDF0i is the value of CDF boundary at that bin.
2.3. Consensus prediction based on neural networks
By combining the CDFs based on PONDR® VLXT, PONDR® VSL2, PONDR® VL3, TopIDP, IUPred, and FoldIndex, a neural network-based consensus method of predicting the order/disorder status was developed. The neural network was fully connected with twenty inputs (three from the PONDR® VLXT-based CDF, four from the PONDR® VSL2-based CDF, three from the PONDR® VL3-based CDF, three from TopIDP-based CDF, four from IUPred-based CDF, and three from FoldIndex-based CDF), one hidden layer with ten hidden units, and one output. A sigmoidal curve was used as the activation function at each node. Inputs from the CDF of each predictor were selected from the bins having the highest separating accuracies. The above mentioned fully disordered and fully ordered datasets were randomly separated into eight groups with each group having one eighth of both the original training and testing sets. At each time, seven groups were used for training, while one group was taken for testing. The training sets were further randomly split into two parts. One, with 90% of the original dataset, was used for the training. Another 10% was used for protection against over-fitting. Weight parameters in the neural networks were chosen by maximizing the accuracy in these 10% of samples. The accuracy was evaluated by using testing datasets. This process was repeated for eight times to implement the eight-fold cross-validation. The final accuracy was the average over eight times on the testing sets.
3. Results and discussion
3.1. Finding the CDF-based boundary line between the ordered and disordered proteins
Originally, a statistical method, where the accuracy of separation is calculated by the summation over both ordered and disordered proteins, was applied to locate the CDF boundary line [11]. Here we describe an alternative approach. First, the average CDF values of ordered and disordered proteins were calculated separately for 20 bins along the X-axis. Next, for each bin, the vertical distance between the averaged ordered and disordered CDF values was divided into 30 parts irrespectively of the distances between the two values. Then, the position of the boundary point was varied and the prediction accuracies of both the ordered and disordered proteins were determined for each choice of boundary point. The accuracies of ordered and disordered proteins for all the boundary choices for all the bins gave an accuracy distribution matrix. Based on this matrix, the location and length of the boundary line was found.
To identify a boundary line made up of one continuous segment for which the low accuracy ends are removed and the high accuracy central region is kept, the following criteria were used:
At each selected boundary point, the accuracy of both ordered and disordered proteins should be above 80% or the highest and should be as close to being equal to each other as possible;
The selected boundary points should be consecutive over the CDF bins, the number of points should be odd, and all the boundary points should have the highest accuracy according to the first criterion.
3.2. Evaluation of the CDF boundary accuracy
Table 1 shows that the new PONDR® VLXT-based boundary achieved averaged accuracies of 88% and 89% for ordered and disordered datasets, respectively. The new boundary outperforms the previous boundary [13] by 2% for disordered proteins but was 2% less accurate for ordered proteins. However, the difference in accuracy between ordered and disordered datasets was only 1% for the new method, compared to 3% for the previous method. This decreased discrepancy means an improved balance between ordered and disordered protein predictions, which is useful for reducing the overall false positive rate. Although this statement is less prominent after the errors are taken into account, the new results are still comparable to the previous ones. The PONDR® VLS2-based boundary reached the similar accuracy as the PONDR® VLXT-based boundary, whereas VL3-based boundary surpasses PONDR® VLXT-based boundary by 2% on the ordered dataset. IUPred-based boundary had the highest accuracy of 91% in disordered dataset which is about 6% higher than that in ordered dataset. The TOP-IDP-based CDF boundary was the least accurate one. FoldIndex-based boundary showed slightly better results than that for TOP-IDP (see Table 1). However, in partially disordered dataset, all the accuracies decreased significantly. For this dataset, PONDR® VSL2-based CDF had the best accuracy of 84% followed by PONDR® VL3 CDF of 81%. FoldIndex was ranked the third at 80%. All other CDFs accuracies were around 70% or below (Table 1).
Table 1.
Accuracies of various CDF boundaries at three different datasets for six disorder predictors, PONDR® VLXT, VSL2, VL3, TOP-IDP, IUPred, and FoldIndex.
| VLXTa | VLXTb | VLS2b | VL3b | TOP-IDPb | IUPredb | FoldIndexb | |
|---|---|---|---|---|---|---|---|
| Ordered Dataset | 90 | 87.7±1.5 | 87.8±1.5 | 89.5±2.3 | 83.7±1.3 | 85.0±1.6 | 86.7±1.6 |
| Disordered Dataset | 87 | 89.0±1.4 | 88.9±1.5 | 88.9±1.5 | 83.3±1.4 | 90.7±1.3 | 83.3±1.7 |
| Partially Disordered dataset | 70 | 73.5±2.0 | 84.4±1.7 | 81.3±4.1 | 62.5±1.7 | 72.0±2.1 | 79.7±1.9 |
Indicates the boundary is calculated by the old method of Ref. [25].
Refers to the data obtained by the new error-estimation method.
The accuracy and error are from the average of 1000 times of bootstrap sampling.
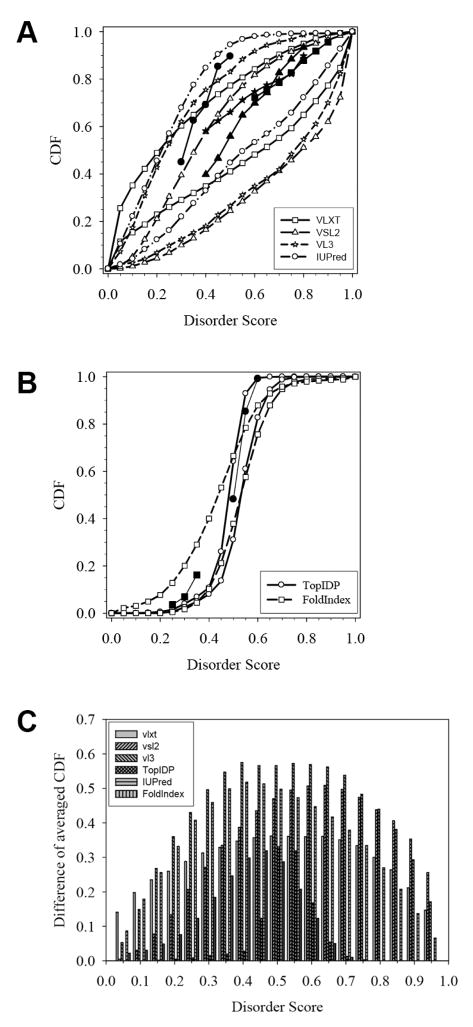
The reasons of why some boundaries achieved the higher accuracy are explored in Figure 1, which represents all the averaged CDF curves from each dataset and corresponding boundaries. Figure 1A shows that for the disordered proteins, the shapes of PONDR® VSL2-CDF and PONDR® VL3-CDF curves are almost identical. The averaged PONDR® VLXT-CDF curve for the disordered proteins starts with noticeably higher values. This implies that the percentage of residues predicted to be ordered by PONDR® VLXT is relatively high, suggesting that this predictor has a tendency to over-predict order. IUPred-CDF is lower than PONDR® VLXT-CDF at small prediction scores but higher than PONDR® VLXT-CDF at scores larger than 0.4. That is to say IUPred predicted many fully disordered residues to have scores of 0.4 or so. For the ordered dataset, PONDR® VSL2 CDF is always at the lowest location. When the prediction score is higher than 0.25, IUPred CDF ranks the highest followed by the PONDR® VL3 CDF. This is expected results because IUPred was created using data obtained from globular proteins. However, when the prediction score is less than 0.25, PONDR® VLXT CDF is ranked the highest, whereas IUPred CDF and PONDR® VL3 CDF are similar to each other. Figure 1B represents the averaged CDF curves and the boundaries for TOP-IDP and FoldIndex for fully ordered and fully disordered datasets. It is clear that CDF curves for these two predictors possess very unusual sigmoidal shapes. Therefore, these two predictors intended to assign intermediate score to all the residues and had the poor separation over ordered and disordered proteins. This indicates that both TOP-IDP and FoldIndex are not very suitable for the binary classification individually.
Figure 1.
(a) Average CDF values of fully ordered (upside curves) and fully disordered (downside curves) datasets, for PONDR® VLXT (open squares), PONDR® VSL2 (open triangles), PONDR® VL3 (open stars), and IUPred (open circles). The filled symbols and bold lines are the optimized boundary lines for PONDR® VLXT CDF (filled squares), PONDR® VSL2 CDF (filled triangles), VL3 CDF (filled stars), and IUPred (black circles). (b) TOP-IDP-based (open circles) and FoldIndex-based CDF (open squares) analysis of fully ordered (upside curves) and disordered proteins (downside curves). The filled circles and filled squares correspond to the optimized boundary line for TOPIDP and FoldIndex, accordingly. (c) Distribution of differences of averaged CDF scores for various predictors over the disorder score. Differences are calculated between the average CDF values of fully ordered and fully disordered datasets.
Figure 1C represents the distribution of the distances between the ordered and disordered CDF curves for six predictors. It is seen that the PONDR® VLXT data are skewed toward the low disorder scores, the PONDR® VSL2 data are somehow skewed toward the high disorder scores, the TOP-IDP and FoldIndex data are distributed in a very narrow interval, IUPred also shifts to the low score region, whereas the PONDR® VL3 data are the most evenly distributed through the entire interval of disorder scores. This clearly shows that the PONDR® VL3 could produce one of the best separations. In agreement with this conclusion, the average CDF differences between the ordered and disordered datasets were 0.33, 0.47, 0.54, 0.06, 0.49, and 0.24 in the boundary bins for PONDR® VLXT, PONDR® VSL2, PONDR® VL3, TOP-IDP, IUPred, and FoldIndex, respectively. By taking into consideration all these observations, it is obvious that PONDR® VL3 has the most accurate boundary for the separation of the ordered and disordered dataset.
The data shown in Figure 1 were used to generate CDF boundary points, which were then fit by the following linear equations:
| (2) |
| (3) |
| (4) |
| (5) |
| (6) |
| (7) |
were CDFVLXT, CDFVSL2, and CDFVL3, CDFTOP-IDP, CDFIUPred, and CDFFoldIndex correspond to the CDF boundary values based on the PONDR® VLXT, PONDR® VSL2, PONDR® VL3, TOP-IDP, IUPred, and FoldIndex predictors, respectively, whereas DO corresponds to the disorder score. Compared to the PONDR® VLXT-based CDF boundary, PONDR® VL3-based boundary is parallel to PONDR® VLXT boundary but is also shifted to the lower disorder scores, all other boundaries are steeper and are shifted to the lower disorder scores. The values of disordered score at the low-end of each boundary line are 0.6, 0.4, 0.4, 0.5, 0.3, and 0.25 for the PONDR® VLXT-, PONDR® VSL2-, PONDR® VL3-, TOP-IDP-, IUPred-, and FoldIndex-CDFs, respectively.
Figure 2A represents the PONDR® VLXT-, PONDR® VSL2-, PONDR® VL3-, TOP-IDP-, IUPred-, and FoldIndex-based CDF curves for partially disordered proteins. It is important to emphasize that all the partially disordered proteins in this study were collected from PDB. As a result, all of them have significant amount of ordered residues, suggesting that the current set of partially disordered proteins is highly biased toward order. Based on these observations, one can expect that the majority of partially disordered proteins in the current dataset will be predicted by CDF analyses as ordered. In agreement with this hypothesis, all CDF curves in Figure 2A are rather similar to CDF curves calculated for the fully ordered proteins (cf. Figure 1).
Figure 2.

(a) Distribution of averaged CDF values for a set of partially disordered proteins (partially disordered dataset, PDD) calculated by six CDF predictors, PONDR® VLXT (solid line), PONDR® VSL2 (long dash line), PONDR® VL3 (short dash line), TOP-IDP (dash-dot-dot line), IUPred (dot line), and FoldIndex (dash-dot line). (b) Averaged CDF curves calculated for PDD-L (thin lines) and PDD-S (bold lines) by CDF predictors based on PONDR® VLXT (solid line), PONDR® VSL2 (long dash line), PONDR® VL3 (short dash line), TOP-IDP (dash-dot-dot line), IUPred (dot line), and FoldIndex (dash-dot line).
Next, to understand whether there is a difference in the prediction tendencies for partially disordered proteins with long disordered regions and for proteins with several short disordered regions, an original partially disordered dataset (PDD) was divided in two groups, one with proteins having disordered regions longer than 50aa (PDD-L), and another one with proteins having shorter disordered regions (PDD-S). Results of the analysis of these subsets by various CDFs are represented in Figure 2B and Table 3, which clearly show that proteins in the PDD-S set are predicted to be more ordered than proteins in the PDD-L set. This conclusion follows from the fact that partially disordered proteins with long disordered regions are generally located closer to the boundary than proteins with several short disordered regions (see Table 3).
Table 3.
Statistics and CDF distances for partially disordered proteins with disordered regions longer than 50aa (partially disordered dataset – long, PDD-L) and for partially disordered proteins with short disordered regions (PDD-S)
| No. of Seq. | Avg. length | No. of IDR | Avg. length of IDR | VLXT | VSL2 | VL3 | TOP-IDP | IUPred | Fold-Index | |
|---|---|---|---|---|---|---|---|---|---|---|
| PDD-L | 18 | 411 | 80 | 42 | 0.034 | 0.089 | 0.120 | 0.018 | 0.010 | −0.016 |
| PDD-S | 46 | 362 | 123 | 17 | 0.048 | 0.121 | 0.143 | 0.035 | 0.037 | −0.027 |
At the final stage, the outputs from the PONDR® VLXT, PONDR® VSL2, PONDR® VL3, TOPIDP, IUPred, and FoldIndex CDFs were used to build a neural network-based consensus method, NN-CDF, for the binary disorder classifications. The data were divided into 8 subsets to implement 8-fold cross validation. Table 2 illustrates that compared to the individual PONDR® VLS2, PONDR® VL3, and IUPred CDF predictions, this new consensus predictor showed ~4% increment in the averaged prediction accuracy over both fully ordered and fully disordered datasets. The accuracy on ordered dataset is 2% higher than PONDR® VL3 CDF predictor which is the second best in all the methods. For disordered dataset, this method has the same similar accuracy with IUPred CDF which is around 90%. The larger error observed in the consensus NN may be a result of insufficient samples in the testing subsets. And for partially disordered proteins, the accuracy of consensus NN is around 10% higher the second best PONDR® VSL2 CDF.
Table 2.
Accuracy of six individual CDF predictors and the consensus method on large datasets.
| VLXT | VSL2 | VL3 | TOP-IDP | IUPred | FoldIndex | Consensus NN | |
|---|---|---|---|---|---|---|---|
| Ordered Dataset | 82.6±1.4 | 87.0±1.5 | 89.1 ± 1.5 | 86.6 ± 1.3 | 83.4±1.7 | 83.1±1.8 | 92.1±2.9 |
| Disordered Dataset | 83.0±1.7 | 88.1±1.5 | 85.8 ± 1.5 | 75.5 ± 1.6 | 90.1±1.4 | 85.4±1.7 | 90.1±5.1 |
| Partially Disordered Dataset | 64.1±2.2 | 87.5±1.5 | 82.8±3.8 | 64.1±1.7 | 65.8±.1 | 74.8±2.1 | 96.1±1.8 |
The accuracy of individual CDF predictor is from the average over 1000 times bootstrapping, while the accuracy of consensus method is evaluated by 8-fold cross validation.
3.3. Application of CDF predictors for the disorder evaluation in entire genomes
Table 4 represents the percentages of fully disordered proteins in three genomes, Escherichia coli K12, Archaeoglobus fulgidus, and Methanobacterium thermoautotrophicum, as evaluated by CDF predictors based on PONDR® VLXT, PONDR® VSL2, PONDR® VL3, TOP-IDP, IUPred, FoldIndex, and NN. PONDR® VLXT-based CDF predicts 2 to3 times more disordered sequences in all three species than PONDR® VSL2-, PONDR® VL3-, TOPIDP-, IUPred-, and FoldIndex-based CDF methods. Even in the case when whole CDF curve is completely below the boundary line (data in brackets of Table 4), the PONDR® VLXT CDF still has much more disordered sequences, especially for Archaeoglobus fulgidus and Methanobacterium thermoautotrophicum. The results for PONDR® VSL2, PONDR® VL3, TOP-IDP, and FoldIndex are more or less similar to each other, although TOP-IDP has slightly lower percentage of disordered proteins for Escherichia coli and higher values for Archaeoglobus fulgidus, IUPred has higher percentage of disordered proteins on Escherichia coli and extremely low disordered ration on Archaeoglobus fulgidus. By applying the consensus method, the percentage of disordered protein is further decreased to 4~9%.
Table 4.
Analysis of whole genomes using various CDF binary classifiers.
| Kingdom | Species | No. of proteins | Avg. Length | VLXTa | VSL2 | VL3 | TOP-IDP | IUpred | FoldIndex | NN |
|---|---|---|---|---|---|---|---|---|---|---|
| Bacteria | Escherichia coli K12 | 4393 | 315 | 18.5(10.9) | 9.7 | 7.9 | 7.7 | 13.2 | 11.9 | 4.2 |
| Archaea | Archaeoglobus fulgidus | 2416 | 275 | 27.6(16.5) | 9.0 | 11.1 | 14.7 | 3.3 | 12.1 | 7.5 |
| Archaea | Methanobacterium thermoautotrophicum | 1971 | 279 | 43.6(24.2) | 13.2 | 14.0 | 15.5 | 10.5 | 15.3 | 8.4 |
Data in brackets for the VLXT-based CDF are based on the cases when whole CDF curve was completely below the boundary line.
4. Concluding remarks
We developed a new error-estimation method for the identification of boundary line in CDF graphs containing CDF curves for both ordered and disordered proteins. This method does not need the pre-assumption on the normal distribution of CDF values around the average in the corresponding datasets. By using this new method, we generated CDF-based prediction tools for PONDR® VLXT, PONDR® VSL2, PONDR® VL3, TOP-IDP, IUPred, and FoldIndex predictors. All of them achieved reasonable prediction accuracy. We also developed the neural network-based consensus method that used the output of all mentioned above CDF outputs. This consensus method was 4~5% more accurate than any of the individual predictors. We further implemented a series of experiments by removing one or two less-accurate CDF predictors from the input of the consensus method. To our surprise, even the less-accurate predictors were useful for the improvement of the final prediction accuracy. The influence of various components for the performance of the final tool will be further analyzed in future. It is also worthwhile to notice that although the consensus method achieved high accuracy on partially disordered dataset, the identification and classification of partially disordered proteins are not a trivial task. By definition, the partially disordered proteins should have an “evenly increased” curve or “flat central region” on the CDF plots. The peculiarities of the CDF predictions for partially disordered proteins need to be more carefully studied.
The numbers of predicted wholly disordered proteins in Escherichia coli K12, Archaeoglobus fulgidus, and Methanobacterium thermoautotrophicum by PONDR® VLXT-based CDF were higher than previously reported [13]. Furthermore, the PONDR® VLXT-CDF predictor identified significantly larger number of disordered sequences in all the three species, compared to other CDF predictors. This is because the new PONDR® VLXT boundary line was located higher than the PONDR® VLXT-based CDF boundary line calculated in the previous study [13]. This shift was determined by the need of balancing the false positives in both wholly ordered and fully disordered sets. Since the same method was used in other CDF predictions, it could be expected that other boundary lines are also shifted to higher positions. The final consensus prediction reveale that the percentages of disordered proteins in Escherichia coli K12, Archaeoglobus fulgidus, and Methanobacterium thermoautotrophicum are 4.2%, 7.5%, and 8.4%, respectively. These results are very similar to previous reported ratios of 4.6%, 6.3%, and 8.0% [13]. The discrepancy among individual predictors indicates that there is still an urgent need for the new prediction protocols and the precise estimation of the disordered content on whole genome.
Acknowledgments
This work was supported in part by the grants R01 LM007688-01A1 (to A.K.D and V.N.U.) and GM071714-01A2 (to A.K.D and V.N.U.) from the National Institutes of Health and the Program of the Russian Academy of Sciences for the “Molecular and cellular biology” (to V. N. U.). We gratefully acknowledge the support of the IUPUI Signature Centers Initiative.
Abbreviations
- IDP
intrinsically disordered protein
- IDR
intrinsically disordered region
- CDF
cumulative distribution function
- PONDR
predictor of natural disordered regions
- PDD
partially disordered dataset
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Dunker AK, et al. Protein disorder and the evolution of molecular recognition: theory, predictions and observations. Pac Symp Biocomput. 1998:473–84. [PubMed] [Google Scholar]
- 2.Wright PE, Dyson HJ. Intrinsically unstructured proteins: re-assessing the protein structure-function paradigm. J Mol Biol. 1999;293:321–31. doi: 10.1006/jmbi.1999.3110. [DOI] [PubMed] [Google Scholar]
- 3.Uversky VN, Gillespie JR, Fink AL. Why are “natively unfolded” proteins unstructured under physiologic conditions? Proteins. 2000;41:415–27. doi: 10.1002/1097-0134(20001115)41:3<415::aid-prot130>3.0.co;2-7. [DOI] [PubMed] [Google Scholar]
- 4.Dunker AK, et al. Intrinsically disordered protein. J Mol Graphics Modelling. 2001;19:26–59. doi: 10.1016/s1093-3263(00)00138-8. [DOI] [PubMed] [Google Scholar]
- 5.Dunker AK, Brown CJ, Lawson JD, Iakoucheva LM, Obradovic Z. Intrinsic disorder and protein function. Biochemistry. 2002;41:6573–82. doi: 10.1021/bi012159+. [DOI] [PubMed] [Google Scholar]
- 6.Dyson HJ, Wright PE. Coupling of folding and binding for unstructured proteins. Curr Opin Struct Biol. 2002;12:54–60. doi: 10.1016/s0959-440x(02)00289-0. [DOI] [PubMed] [Google Scholar]
- 7.Iakoucheva LM, Brown CJ, Lawson JD, Obradovic Z, Dunker AK. Intrinsic disorder in cell-signaling and cancer-associated proteins. J Mol Biol. 2002;323:573–84. doi: 10.1016/s0022-2836(02)00969-5. [DOI] [PubMed] [Google Scholar]
- 8.Tompa P. Intrinsically unstructured proteins. Trends Biochem Sci. 2002;27:527–533. doi: 10.1016/s0968-0004(02)02169-2. [DOI] [PubMed] [Google Scholar]
- 9.Uversky VN. Natively unfolded proteins: A point where biology waits for physics. Protein Sci. 2002;11:739–56. doi: 10.1110/ps.4210102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Uversky VN. What does it mean to be natively unfolded? Eur J Biochem. 2002;269:2–12. doi: 10.1046/j.0014-2956.2001.02649.x. [DOI] [PubMed] [Google Scholar]
- 11.Dunker AK, Obradovic Z, Romero P, Garner EC, Brown CJ. Intrinsic protein disorder in complete genomes. Genome Inform Ser Workshop Genome Inform. 2000;11:161–71. [PubMed] [Google Scholar]
- 12.Ward JJ, Sodhi JS, McGuffin LJ, Buxton BF, Jones DT. Prediction and functional analysis of native disorder in proteins from the three kingdoms of life. J Mol Biol. 2004;337:635–45. doi: 10.1016/j.jmb.2004.02.002. [DOI] [PubMed] [Google Scholar]
- 13.Oldfield CJ, Cheng Y, Cortese MS, Brown CJ, Uversky VN, Dunker AK. Comparing and combining predictors of mostly disordered proteins. Biochemistry. 2005;44:1989–2000. doi: 10.1021/bi047993o. [DOI] [PubMed] [Google Scholar]
- 14.Dyson HJ, Wright PE. Intrinsically unstructured proteins and their functions. Nat Rev Mol Cell Biol. 2005;6:197–208. doi: 10.1038/nrm1589. [DOI] [PubMed] [Google Scholar]
- 15.Dunker AK, Cortese MS, Romero P, Iakoucheva LM, Uversky VN. Flexible nets. The roles of intrinsic disorder in protein interaction networks. Febs J. 2005;272:5129–48. doi: 10.1111/j.1742-4658.2005.04948.x. [DOI] [PubMed] [Google Scholar]
- 16.Uversky VN, Oldfield CJ, Dunker AK. Showing your ID: intrinsic disorder as an ID for recognition, regulation and cell signaling. J Mol Recognit. 2005;18:343–84. doi: 10.1002/jmr.747. [DOI] [PubMed] [Google Scholar]
- 17.Dunker AK, et al. The unfoldomics decade: an update on intrinsically disordered proteins. BMC Genomics 9 Suppl. 2008;2:S1. doi: 10.1186/1471-2164-9-S2-S1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dunker AK, Obradovic Z. The protein trinity--linking function and disorder. Nat Biotechnol. 2001;19:805–6. doi: 10.1038/nbt0901-805. [DOI] [PubMed] [Google Scholar]
- 19.Uversky VN, Oldfield CJ, Dunker AK. Intrinsically disordered proteins in human diseases: introducing the D2 concept. Annu Rev Biophys. 2008;37:215–46. doi: 10.1146/annurev.biophys.37.032807.125924. [DOI] [PubMed] [Google Scholar]
- 20.Oldfield CJ, Ulrich EL, Cheng Y, Dunker AK, Markley JL. Addressing the intrinsic disorder bottleneck in structural proteomics. Proteins. 2005;59:444–53. doi: 10.1002/prot.20446. [DOI] [PubMed] [Google Scholar]
- 21.Romero P, Obradovic Z, Li X, Garner EC, Brown CJ, Dunker AK. Sequence complexity of disordered protein. Proteins. 2001;42:38–48. doi: 10.1002/1097-0134(20010101)42:1<38::aid-prot50>3.0.co;2-3. [DOI] [PubMed] [Google Scholar]
- 22.Williams RM, et al. The protein non-folding problem: amino acid determinants of intrinsic order and disorder. Pac Symp Biocomput. 2001;6:89–100. doi: 10.1142/9789814447362_0010. [DOI] [PubMed] [Google Scholar]
- 23.Radivojac P, Iakoucheva LM, Oldfield CJ, Obradovic Z, Uversky VN, Dunker AK. Intrinsic disorder and functional proteomics. Biophys J. 2007;92:1439–56. doi: 10.1529/biophysj.106.094045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ferron F, Longhi S, Canard B, Karlin D. A practical overview of protein disorder prediction methods. Proteins. 2006;65:1–14. doi: 10.1002/prot.21075. [DOI] [PubMed] [Google Scholar]
- 25.Dosztanyi Z, Sandor M, Tompa P, Simon I. Prediction of protein disorder at the domain level. Curr Protein Pept Sci. 2007;8:161–71. doi: 10.2174/138920307780363406. [DOI] [PubMed] [Google Scholar]
- 26.Dosztanyi Z, Tompa P. Prediction of protein disorder. Methods Mol Biol. 2008;426:103–15. doi: 10.1007/978-1-60327-058-8_6. [DOI] [PubMed] [Google Scholar]
- 27.Uversky VN. Protein folding revisited. A polypeptide chain at the folding-misfolding-nonfolding cross-roads: which way to go? Cell Mol Life Sci. 2003;60:1852–71. doi: 10.1007/s00018-003-3096-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Li X, Romero P, Rani M, Dunker AK, Obradovic Z. Predicting Protein Disorder for N-, C-, and Internal Regions. Genome Inform Ser Workshop Genome Inform. 1999;10:30–40. [PubMed] [Google Scholar]
- 29.Peng K, Vucetic S, Radivojac P, Brown CJ, Dunker AK, Obradovic Z. Optimizing long intrinsic disorder predictors with protein evolutionary information. J Bioinform Comput Biol. 2005;3:35–60. doi: 10.1142/s0219720005000886. [DOI] [PubMed] [Google Scholar]
- 30.Obradovic Z, Peng K, Vucetic S, Radivojac P, Dunker AK. Exploiting heterogeneous sequence properties improves prediction of protein disorder. Proteins. 2005;61(Suppl 7):176–82. doi: 10.1002/prot.20735. [DOI] [PubMed] [Google Scholar]
- 31.Obradovic Z, Peng K, Vucetic S, Radivojac P, Brown CJ, Dunker AK. Predicting intrinsic disorder from amino acid sequence. Proteins. 2003;53(Suppl 6):566–72. doi: 10.1002/prot.10532. [DOI] [PubMed] [Google Scholar]
- 32.Campen AM, Williams RM, Brown CJ, Meng J, Uversky VN, Dunker AK. TopIDP-Scale: A new amino acid scale measuring propensity for intrinsic disorder. Protein & Peptide Lett. 2008;15:956–963. doi: 10.2174/092986608785849164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dosztanyi Z, Csizmok V, Tompa P, Simon I. The pairwise energy content estimated from amino acid composition discriminates between folded and intrinsically unstructured proteins. J Mol Biol. 2005;347:827–39. doi: 10.1016/j.jmb.2005.01.071. [DOI] [PubMed] [Google Scholar]
- 34.Dosztanyi Z, Csizmok V, Tompa P, Simon I. IUPred: web server for the prediction of intrinsically unstructured regions of proteins based on estimated energy content. Bioinformatics. 2005;21:3433–4. doi: 10.1093/bioinformatics/bti541. [DOI] [PubMed] [Google Scholar]
- 35.Prilusky J, Felder CE, Zeev-Ben-Mordehai T, Rydberg EH, Man O, Beckmann JS, Silman I, Sussman JL. FoldIndex: a simple tool to predict whether a given protein sequence is intrinsically unfolded. Bioinformatics. 2005;21:3435–8. doi: 10.1093/bioinformatics/bti537. [DOI] [PubMed] [Google Scholar]
- 36.Sickmeier M, et al. DisProt: the Database of Disordered Proteins. Nucleic Acids Res. 2007;35:D786–93. doi: 10.1093/nar/gkl893. [DOI] [PMC free article] [PubMed] [Google Scholar]