Abstract
Psoriasis is a common, immunologically-mediated, inflammatory and hyperproliferative disease of the skin and joints, with a multifactorial genetic basis. We previously mapped PSORS1, the major psoriasis susceptibility gene in the major histocompatibility complex, to within or very near HLA-Cw6. In an effort to identify non-MHC psoriasis genes, we carried out a collaborative genome-wide association study. After initial follow-up genotyping of 21 SNPs from 18 loci showing strong evidence of association in the initial scan, we confirmed evidence of association at seven loci. Three of these loci confirm previous reports of association (HLA-C, IL12B, IL23R) and four identify novel signals located near plausible candidate genes (IL23A, IL4/IL13, TNFAIP3, and TNIP1). In other work, we have also shown that interferon-γ (IFN-γ) treatment induces IL-23 mRNA and protein in antigen-presenting cells (APC), leading to the proliferation of CD4+ and CD8+ memory T-cells expressing IL-17. While functional variants remain to be identified, we speculate that genetic variants at the IL4/IL13 locus contribute to the Th1 bias that is characteristic of psoriasis, that Th1-derived IFN-γ supports expansion of IL-17+ T-cells via APC-derived IL-23, and that negative regulation of inflammatory signaling through the NF-κB axis is impaired due to genetic variants of TNFAIP3 and TNIP1.
Keywords: Psoriasis, dermatology, human genetics, interleukins, NF-κB, immunology
Epidemiology of Psoriasis: An Overview
Psoriasis vulgaris (PsV) is a common inflammatory and hyperproliferative skin disease, affecting over 4 million Americans (about 2%) at an estimated cost of $1.6 to $3.2 billion annually 1. The cutaneous manifestations of psoriasis are unpleasant and obvious, with a very negative impact on quality of life 2. The majority of the 150,000 new U.S. cases diagnosed annually arise in individuals <30 years of age, and 10,000 of these are in individuals <10 years old 3. Moreover, up to 40% of psoriatics develop psoriatic arthritis (PsA), and in 5% of them the arthritis is severe and deforming 4.
The clinical and genetic epidemiology of psoriasis has been reviewed in detail 5-8 and will be only briefly considered here. Two forms of psoriasis differing in age of onset have been proposed, with early onset disease (onset ≤ 40 years) more likely to be familial, severe, and associated with HLA-Cw6 9. The peak age of disease onset is the early twenties. Twin studies, pedigree studies, and recurrence risk analysis support a multifactorial model of inheritance, with a major susceptibility locus (psoriasis susceptibility 1 or PSORS1) residing within the MHC, with other loci throughout the genome. There is clearly a role for environmental factors such as trauma, stress, and infections such as streptococcal pharyngitis (for review, see 5). PsA is even more strongly influenced by genes, than is PsV 10; 11.
The clinical variants of psoriasis include chronic plaque psoriasis, guttate psoriasis, localized pustular psoriasis, inverse psoriasis, sebopsoriasis and generalized pustular psoriasis, as well as palmoplantar pustulosis. Chronic plaque disease is by far the most common form. The clinical manifestations of psoriasis can change over time in any given person. Nail changes (pitting, onychodystrophy, and/or “oil drop” spotting) are found in around 50% of psoriasis patients. Guttate psoriasis is characterized by the sudden appearance of hundreds of small papules, with spontaneous resolution in approximately half of cases, with the other half progressing to chronic plaque psoriasis. The association of guttate psoriasis with HLA-Cw6 is even stronger than it is for chronic plaque psoriasis 12. Generalized pustular psoriasis manifests the same HLA associations found in plaque-type psoriasis 13. In contrast, palmoplantar pustulosis is not associated with HLA-Cw6 14 and is only rarely associated with typical psoriatic plaques. Therefore, it appears to be a distinct entity. PsA typically presents between the ages of 35 and 45, usually but not always after onset of skin disease. Disease is oligoarticular and asymmetrical in over 80% of patients.
The Th1 —Th17 axis: New Insights into Psoriasis Pathogenesis
Recently, there has been a major expansion knowledge that may provide specific insights into the link between immunocytic infiltration and epidermal hyperplasia. A new subset of T-cells expressing IL-17 appears to play a major role in psoriasis 15 as well as other inflammatory autoimmune disorders including multiple sclerosis 16 and Crohn’s disease (CD) 17. IL-23 drives this novel immune circuit. It is produced by myeloid cells and acts on T-cells via its cognate receptor. IL-12 and IL-23 share the p40 subunit, encoded by the IL12B gene. The p40 subunit heterodimerizes with p19 to for IL-12 and with p35 to form IL-23. The IL-12 and IL-23 receptors share the common IL12rRβ1 subunit, which binds to IL12Rβ2 to form the IL-12 receptor and to the product of the IL23R gene to form the IL-23 receptor. In mice, injection of IL-23 leads to epidermal hyperplasia mediated by IL-22 produced by IL-17-expressing T-cells 18; 19, and consistent if not identical phenomena are observed in humans 20. Keratinocytes express high levels of IL-22 receptors, and are highly responsive to IL-22 as well as other cytokines of the IL-20 subfamily including IL-19, IL-20, and IL-24 21; 22. IL-22 is distinctive among these cytokines in that it is primarily expressed by activated T-cells and not keratinocytes 23 and is therefore well-positioned as a bridge between the two. We have recently implicated IFN-γ as a key stimulus causing CD14+ macrophages to stimulate the proliferation of IL-17+ T-cells, via their production of IL-1 and IL-23 24 (Figure 1). We also identified a population of CD8+ IL-17-expressing cells in the epidermis of psoriatic lesions. Essentially all of the IL-17 producing T-cells in psoriatic epidermis were CD8+, whereas there were no such cells in normal epidermis 24. these cells may play a causal role in provoking epidermal hyperplasia in psoriasis, as recent studies in xenografted mice have shown that entry of T-cells into the epidermis is necessary for development of the epidermal hyperplastic response 25.
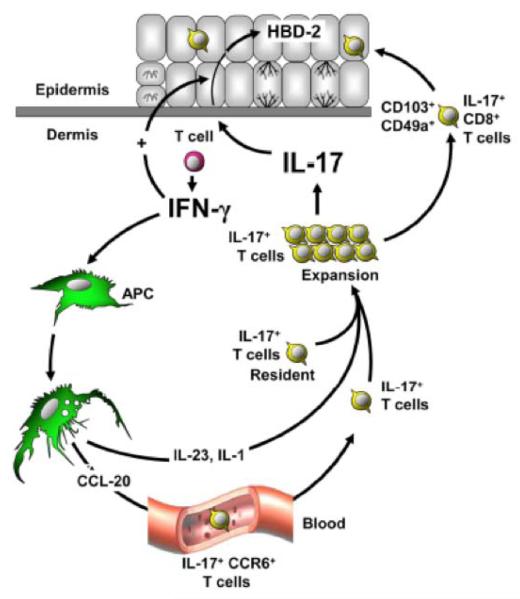
Figure 1.
Interplay between IFN-γ producing Th1 cells and IL-17-producing T-cells in psoriatic lesions. Interferon-γ produced by Th1 cells stimulates myeloid APCs and/or macrophages to produce IL-23, which together with IL-1, stimulates the survival and expansion of T-cells expressing IL-17 and/or IL-22. The entry of CD8+ T-cells expressing these cytokines into the epidermis is associated with increased epidermal hyperplasia and the production of innate immune peptides such as human β—defensin 2 (HBD-2). From 24, with permission.
Genome-wide Association Scan of Psoriasis
With the advent of the HapMap 26, we now have a dense map of millions of SNPs to choose from, and massive throughput genotyping technologies allow 100,000-1,000,000 SNPs to be characterized economically in thousands of individuals. Anticipating these developments, in 2003 we refocused our experimental approach from linkage to association, opting to collect large numbers of cases and controls rather than families. In 2006 we formed a multicenter collaboration with Dr. Anne Bowcock at Washington University of St. Louis and Dr. Gerald Krueger of the University of Utah, to carry out a GWAS of psoriasis(the Collaborative Association Study of Psoriasis, or CASP). We were funded by the Genetic Association Information Network (GAIN), a public-private partnership formed to facilitate the execution of GWAS and the rapid dissemination of their results 27. We also linked up with five additional groups of collaborators interested in PsV and PsA, in order to carry out a powerful replication study based on the GAIN results. The results of this study have recently been published 28 but will be briefly reviewed here.
We carried out a GWAS of 1,409 Caucasian psoriasis cases and 1,436 Caucasian controls, making use of a Perlegen Sciences microarray platform that types a large number of SNPs with known effects on protein structure and gene expression, and allows for efficient incorporation of HapMap tag SNPs. After removing SNPs and samples with low genotype call rates, screening for related individuals, and removing markers with strong deviations from Hardy-Weinberg equilibrium (HWE), we carried out our initial association analysis on 438,670 SNPs in 1,539 cases and 1,400 controls. By far, the strongest association signals mapped to the MHC (Figure 2). SNP rs12191877, the marker demonstrating the strongest association with psoriasis (fcontrol = 0.15, fcase = 0.30, ORfollow-up = 2.64, pcombined << 10-100), was in linkage disequilibrium (LD) with HLA-Cw6 (r2 = 0.63). Nearly equally strong signals were observed in the vicinity of CDSN, as expected from our earlier studies of the MHC in psoriasis families 29. In a subset of cases and controls with HLA-Cw6 typing, this allele was more strongly associated with psoriasis than any genotyped or imputed SNP, but could not fully account for all observed association signals. We used a forward selection procedure to assess the possibility that multiple psoriasis susceptibility alleles might exist within the MHC. This analysis resulted in a model with three imputed SNPs. The first two of these (rs12204500 and rs13191343, forward selection p-values of 8×10-57 and 2×10-10, respectively) are close to and in strong LD with HLA-Cw6 (r2 = 0.78 and 0.52, respectively). However, the third SNP (rs2022544, p-value = 10-7) maps closer to the HLA-DR gene cluster in the C6orf10 gene and exhibits only weak LD with HLA-Cw6 (r2 = 0.01). These results confirm the predominance of the PSORS1 in terms of the magnitude of its genetic effect, and suggest that at least one additional determinant of psoriasis susceptibility resides within the MHC. This would be consistent with studies demonstrating a higher risk associated with an extended ancestral haplotype carrying HLA-Cw6, HLA-B57, and HLA-DR4, relative to other HLA-Cw6-bearing haplotypes 29; 30.
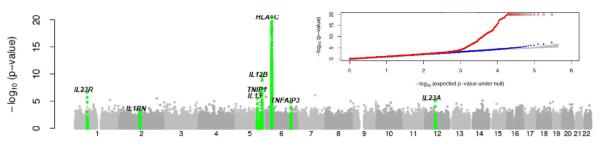
Figure 2.
CASP GWAS results plotted against chromosomal position; the inset presents quantile-quantile plots. Loci that were followed up and showed convincing evidence of association in the replication study are labeled in green. In the inset, red represents all the SNPs; blue symbols represent results after excluding SNPs at replicated loci and the gray area corresponds to the 90% confidence region for a null distribution of p-values. All panels are truncated at -log10(p-value) = 20, markers near HLA-C exceed this threshold. Adapted from 28, with permission.
Based on our initial genome-scan results, 21 SNPs representing 18 independent loci were genotyped in independent samples totaling 5,048 cases and 5,051 controls (1,642 cases and 1,101 Caucasian controls from the Michigan, 718 cases and 1,464 controls from Kiel, Germany) 981 cases and 925 controls from Celera Genomics, 302 cases and 500 controls from St. Louis, 691 cases and 217 controls from Toronto, and 368 cases and 358 controls from Newfoundland) as well as a pedigree-based collection from France (1130 individuals from 45 families ranging from 5 to 60 members). Follow-up genotyping results confirmed association at 7 loci (with p < 10-3 in the replication study and p < 10-8 overall), including three loci previously associated with psoriasis, HLA-C, IL12B, IL23R 28; 31-33, and four new loci located near plausible psoriasis candidate genes: IL23A, IL4/IL13, TNFAIP3, and TNIP1. We next assessed the risk of PsA conferred by these replicated loci. As shown in Table 1, three loci reached genome-wide significance for PsA compared to normal controls (HLA-C, IL12B, and TNIP1), and one came close (IL4 / IL13). Three loci manifested a statistically significant difference (p < 0.05) between PsA and purely cutaneous psoriasis: HLA-C, IL12B, and IL23R.
Table 1.
Association of replicated CASP GWAS signals with PsA.
| PsC vs. control (3523 cases, 5942 controls) |
PsA vs. control (1755 cases, 5942 controls) |
PsA vs. PsC (1755 cases, 3523 controls) |
|||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Marker | Chr | Position (bld 36.1) |
Nearby Notable Gene |
Alleles (risk/ nonrisk) |
· | freq cases |
freq controls |
meta OR |
meta allelic p-value |
freq cases |
freq controls |
meta OR |
meta allelic p-value |
freq cases |
freq controls |
meta OR |
meta allelic p-value |
| rs12191877 | 6 | 3.1E+O7 | HLA-C | T/C | 0.316 | 0.139 | 2.87 | 2.98E-178 | 0.260 | 0.139 | 2.34 | 6.26E-62 | 0.260 | 0.316 | 0.87 | 6.14E-03 | |
| rs2082412 | S | I.6E+O8 | 1L12B | G/A | 0.846 | 0.797 | 1.42 | 4.57E-18 | 0.861 | 0.797 | 1.63 | 3.90E-16 | 0.861 | 0.846 | 1.19 | 9.51E-03 | |
| rs17728338 | 5 | 1.5E+O8 | TNIP1 | A/G | 0.083 | 0.054 | 1.56 | 1.40E-13 | 0.100 | 0.054 | 1.78 | 2.32E-14 | 0.100 | 0.083 | 1.15 | 7.43E-02 | |
| rs20541 | 5 | 1.3E+O8 | IL13 | G/A | 0.821 | 0.787 | 1.24 | 3.77E-08 | 0.833 | 0.787 | 1.34 | 1.35E-07 | 0.833 | 0.821 | 1.10 | 1.13E-01 | |
| rs610604 | 6 | 1.4E+08 | TNFAIP3 | G/T | 0.364 | 0.318 | 1.23 | 3.O7E-1O | 0.362 | 0.318 | 1.18 | 2.94E-04 | 0.362 | 0.364 | 0.98 | 6.70E-01 | |
| rs2O668O7 | 12 | 5.5E+O7 | IL23A | C/G | 0.949 | 0.932 | 1.33 | 1.9OE-O5 | O.955 | 0.932 | 1.55 | 9.49E-06 | 0.955 | 0.949 | 1.15 | 2.25E-01 | |
| rs2201841 | 1 | 6.7E+07 | IL23R | G/A | 0.340 | 0.295 | 1.23 | 1.75E-1O | 0.318 | 0.295 | 1.07 | 1.42E-01 | 0.318 | 0.340 | 0.89 | 1.50E-02 | |
With the probable exception of HLA-Cw6 29, the precise genetic variants responsible for the remaining six observed associations remain to be determined. Nevertheless, our results suggest roles for several key immunologic pathways in disease susceptibility.
HLA-Cw6
HLA-C plays a key role in the presentation of antigens to CD8+ T-cells, which predominate in the epidermis of psoriatic lesions. Guttate eruptions frequently follow attacks of Streptococcal pharyngitis 5. Other streptococcal skin infections such as erysipelas, impetigo, or cellulitis do not seem to trigger psoriasis, suggesting a critical role for the tonsils. Tonsillar T-cells recognize activated skin endothelium 34, and home to the skin. Indeed, the same skin-homing T-cell clones present in the tonsils are also found in the lesional skin of psoriatic patients 35. Many of these clonally exapanded T-cells are CD8+ 36. Several other studies have also identified oligoclonal TCR rearrangements in psoriasis 35-39. These findings support the notion that T-cells originally stimulated in the tonsils may traffic into the skin, where some of them become clonally expanded due to antigenic stimulation. However, the identity of the antigen(s) has remains elusive. One study identified several potential psoriasis antigens by expression cloning from psoriatic skin RNA 40. However, peripheral blood T-cells from normal controls reacted as strongly as T-cells derived from psoriatic patients 40, making the results hard to interpret. In another study, HLA-Cw6 preferentially presented cross-reactive peptides derived from Streptococcal M protein and the hyperproliferative keratin K17 to skin-homing CD8+ T cells 41, suggesting that the evolution of guttate into chronic plaque psoriasis might reflect a transition from a self-limited response to Streptococcus initiated in the tonsils, to a sustained response to homologous peptides derived from hyperproliferative skin keratins 41. It has also been suggested that peptidoglycan (PG), the major constituent of the streptococcal cell wall, acts as a T cell activator in psoriasis 42. PG-containing cells were detected in CD68+ macrophages in the dermal papillae and cellular infiltrates of guttate and chronic plaque skin lesions 42. Based on these results, it has been proposed that macrophages may serve as a vehicle for transportation of PG from the tonsils to the skin, where it may serve both as an antigen and as a stimulus for Toll-like receptor (TLR) mediated stimulation of innate immunity 43. Whatever the antigen(s) may be, it is important to realize that most T-cells present in psoriatic skin are not clonally expanded, indicative of important roles for additional mechanisms to maintain the psoriatic infiltrate.
HLA-C also plays a role in binding to killer immunoglobulin-like receptors (KIRs) on natural killer (NK) cells, and KIR genes have previously been associated with PsA44; 45. NK cells are major producers of interferons and serve as a bridge between innate and acquired immunity. KIR molecules can either inhibit or stimulate NNK cells, and inhibitory KIR genes interact with a dimorphic allotype (Asn80/Lys80) present on HLA-C molecules 46. HLA-Cw6 is one of several “group 2” alleles carrying Lys at position 80, thus one might expect that a combination of all “group 2” alleles would provide a stronger association signal than does HLA-Cw6, but this is not the case (unpublished data). Thus, at present, the role of HLA-Cw6 as a mediator of in NK cell activity in psoriasis remains to be genetically clarified through typing of both KIRs and HLA-C in a large dataset.
IL-23 signaling
Three SNPs exhibiting strong evidence of association map near IL12B (encoding the p40 subunit of IL-23 and IL-12), IL23A (encoding the p19 subunit of IL-23), and IL23R (encoding a subunit of the IL-23 receptor) (see 28 for details). Our study implicated genetic variants in the IL23A locus for the first time in psoriasis and for that matter in any human autoimmune disorder. Interestingly, our GWAS identified no associations with either of the IL-12-specific genes IL12B or IL12RB2, and psoriasis lesions markedly overexpress IL-12/23 p40 and IL-23 p19, but not IL-12 p35 47. IL-23 signaling promotes cellular immune responses by promoting the survival and expansion of a recently identified subset of T-cells expressing IL-17 that protects epithelia against microbial pathogens 48. Dysregulated IL-23 signaling could lead to inappropriate, chronic immune responses that target epithelial cells, perhaps helping to explain the relatively skin-specific inflammation seen in psoriasis. One of the same genetic variations in the IL23R gene that increases risk for psoriasis also confers risk for Crohn’s Disease 49, a disorder that has long been known to be clinically associated with psoriasis 50. It is possible that this clinical association reflects the similarities between the epithelial linings of the skin and the gut.
NF-κB signaling
The products of the TNFAIP3 and TNIP1 genes (A20 and ABIN1/Naf1α, respectively), physically interact with each other to influence the ubiquitin-mediated destruction of IKKγ / NEMO, a central nexus of NF-κB signaling 51. A20 also regulates the degradation of several other components of the TNF signaling pathway 51. TNF-α blockade improves symptoms in a mouse model of psoriasis induced by administration of IL-23 52 and a region of mouse chromosome 10 encompassing Tnfaip3 promotes psoriasis in a TNF-α dependent manner in another mouse model 53. This region of the mouse genome has been also associated with atherosclerosis 54, a major co-morbidity of psoriasis 55.
Of particular interest in the context of this meeting, common polymorphisms near TNFAIP3 have recently been associated with systemic lupus erythematosus (SLE) 56; 57 and with rheumatoid arthritis RA 58; 59. Notably, the polymorphisms implicated in RA and SLE show no association with psoriasis in our sample (all p > 0.30) and are not in linkage disequilibrium (LD, all r2 < 0.03) with the psoriasis associated alleles, suggesting that each of these common autoimmune diseases is driven by a different variant of the TNFAIP3 gene. Some of the polymorphisms implicated in RA and SLE reside far upstream from the gene (∼200 kb), suggesting that they might reflect the existence of regulatory variants. Of note, we have also found an association between PsA and SNPs in this upstream region (unpublished data).
Th2-predisposing genes
IL-13 and IL-4 are products of Th2 cells that play a multifaceted role in allergic reactions and in the immune response to extracellular pathogens. Both IL-13 and IL-4 are expressed at high levels in atopic dermatitis, but only at very low levels in psoriasis 60. IL-4 treatment has led to significant clinical improvement of psoriasis 61. The IL13 and IL4 genes are located within 12.5 kb of each other on human chromosome 5q31.1, just telomeric to the RAD50 gene, within a block of linkage disequilibrium (LD). While our most positive signals reside near IL4 and IL13, positive signals are also found in RAD50. Thus, the functional variant could influence not just IL13 but alternatively or in addition might influence IL4 and RAD50. Interestingly, a locus control region that regulates the transcription of both IL13 and IL4 within the RAD50 gene 62.
Immuno-Genetic Model for the Development of Psoriasis
During an initial flare of guttate psoriasis following a strep throat, we envision that T-cells encounter streptococcal antigens presented in the context of HLA-Cw6 in the tonsils, where they proliferate, differentiate into an effector / memory phenotype, and acquire skin homing capability (i.e., become CLA+). Upon entering the skin, these cells encounter a locally activated dermal environment characterized by capillary dilatation and edema 63-66 and the presence of plasmacytoid dendritic cells 67; 68. We suspect that this “pre-psoriatic environment” may be initiated by focal mast cell degranulation and activation of plasmacytoid dendritic cells and macrophages, with release of TNF-α and interferon leading to induction of adhesion molecules on the endothelial cell surface, facilitating entry of T-cells into the dermis. These events might be triggered by circulating pathogen-derived factors such as PG 43 and/or innate immune mediators induced by them. Many of the signaling pathways are mediated by TNF-α, toll-like receptors, or other ligands and receptors of the TNF receptor family that pass through NF-κB, often by way of IKK-γ. Thus, genetically-mediated defects in TNFAIP3 and TNIP1 could enhance this early inflammatory stage of lesion development by interfering with normal negative feedback regulation of NF-κB signaling.
At this early stage of lesional evolution epidermal changes are subtle, but include a modest increase in keratinocyte DNA synthesis, widened extracellular spaces between keratinocytes, and biochemical alterations in the stratum corneum indicative of altered differentiation, despite a lack of visible parakeratosis 69. These epidermal changes may be provoked by macrophage-derived proteases creating holes in the epidermal basement membrane 70; 71, allowing permeation of fibronectin 72 and cytokine-laden mast cell granules into the epidermis 73. These events could be hyperactive in psoriasis due to defective negative feedback regulation of signaling by TNFAIP3 and TNIP1. Keratinocyte hyperplasia results the activation of a set of genes involved in regenerative hyperplasia, including keratins K6, K16 and K17 74; 75, along with many other genes involved in innate immunity, including human β-defensin-2 (hBD-2), psoriasin (S100A7), S100A8, and S100A9, small proline-rich region (SPRR) proteins, and late cornified envelope (LCE) proteins 76; 77. Many of the most strongly up-regulated genes in psoriasis are located in the epidermal differentiation complex on chromosome 1q21.3, also known as PSORS4 because of several reports of genetic linkage and association of this region to psoriasis 78-82. It is also notable that the defensin gene cluster on human chromosome 8p exists in different numbers of copies in different individuals, and psoriasis has been associated with increasing defensin gene copy number 83. Once their expression has been turned on, peptides derived from these regenerative hyperplasia-associated proteins might serve as neoantigens on the surface of epidermal keratinocytes in the context of HLA Class I molecules, such as HLA-Cw6. It is also possible that Strep-derived PG could be delivered from the tonsils to the skin by macrophages, where they would be recognized as foreign antigens and/or promote innate immune responses via binding to TLR2 43. In the latter setting, downstream inflammatory signals could again be amplified as a consequence of TNFAIP3 and/or TNIP1 hypofunction.
As a consequence of Streptococcal infection, the pool of T-cells responding to Streptococcal antigens will be expanded and activated. CD4+ and CD8+ T-cells will enter the dermis via the inflamed endothelium. A subset of both CD4+ and CD8+ cells will express IL-17 and/or IL-22 due to stimulation by IL-23 and IL-1. Genetically-mediated hyperfunction of IL-23 and/or of its receptor could enhance the production of IL-17-expressing T-cells. Moreover, development of Th1 bias might be facilitated genetically-mediated hypofunction of the IL4 and IL13 genes. This would be predicted to lead to overproduction of IFN-γ, a major product of Th1 cells. We have shown that IFN-γ markedly stimulates the production of IL-23 by myeloid APC 24, and IL-23 in turn supports the development of T-cells expressing IL-17 and/or IL-22 (Figure 1).
Many of the CD8+ cells entering the skin will selectively traffic to the epidermis, because they express VLA-1 as well as integrin aEβ7, which binds to E-cadherin expressed by keratinocytes 25; 84. Once in the epidermis, a subset of Strep-reactive CD8+ T-cells is predicted to recognize self-derived or Streptococcal peptides in the context of HLA-Cw6, thus maintaining immunologic activation in an antigen-driven manner. We envision that a transition of reactivity from Strep-derived to self proteins might be necessary for the transition from guttate to chronic plaque psoriasis. Whatever the nature of the antigen, entry of CD8+ T-cells into the epidermis would trigger more extensive epidermal hyperplasia, possibly by means of cytokines such as IL-17 and/or IL-22 produced by epidermal CD8+ T-cells 18-20; 24. Macrophages may also participate in this process, as suggested by two different mouse models of psoriasis 85; 86, but it is important to remember that the development of extensive epidermal hyperplasia in the human skin xenograft model requires entry of T-cells into the epidermis 25. CD8+ T-cells could also trigger the local release a variety of soluble factors other than or in addition to IL-17 and/or IL-22, including cytokines such as TNF-α, chemokines such as IL-8 and CCL20, eicosanoids, and/or other innate immune mediators, which could further increase local inflammation and stimulate keratinocyte proliferation 87. Whether mediated by soluble factors or by actual physical damage, keratinocytes could respond to T-cell insult by elaborating growth factors such as amphiregulin, thereby encouraging their own proliferation and survival 88. Concomitant with the induction of increased epidermal hyperplasia, there will be further up-regulation of keratinocyte-derived innate immune peptides with antimicrobial and chemotactic activity such as human β-defensin 2, CCL20, S100A7, S100A8, and S100A9, thus further increasing the number of immune and inflammatory cells entering the lesion, including neutrophils.
As summarized in Figure 3, we envision the psoriatic tissue reaction as a multi-stage process, with the recognition of antigen in the context of HLA-Cw6 by CD8+ T-cells playing a major role. This recognition probably takes place not only on antigen-presenting cells, but also on the surface of keratinocytes. Genetic alterations in the TNFAIP3 and TNIP1 genes may play very important roles in APC and macrophages due to the central role of these genes in regulating NF-κB signaling. Genetic variation in IL12B, IL23A, and IL23R may contribute to hyperexpansion of T-cells expressing IL-17 and/or IL-22, and this process is further supported by IFN-γ-producing Th1 cells whose polarization is influenced by genetically-mediated hypofunction of IL-4 and/or IL-13. Entry of CD8+ T-cells expressing IL-17 and/or IL-22 into the epidermis may provoke epidermal hyperplasia, leading to the overexpression by keratinocytes of many proteins involved in innate immunity. At least some of these may serve a source of antigen as well. Thus, the epidermal response “feeds back” into all earlier stages of the psoriatic developmental process, amplifying the psoriatic tissue response. Based on the model shown in Figure 3, the psoriasis susceptibility genes that we and others have identified so far seem to fit very well with current concepts of its immunopathogenesis.
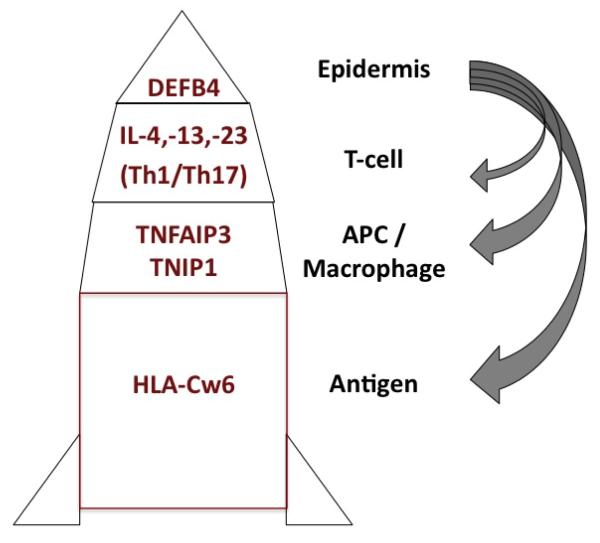
Figure 3.
Multi-stage model integrating genetics and immunology of psoriasis. See text for details.
Future Prospects
Much additional work is required to identify the actual disease-predisposing variants of these genes, and to understand how they contribute to pathology. Moreover, as can be seen in Figure 2, additional genes remain to be discovered. Currently, we are carrying out a deeper follow-up scan of approximately 10,000 SNPs in approximately 3,000 cases and 3,000 controls to identify additional psoriasis loci. Moreover, we expect that the number of psoriasis genes that can be found will increase substantially as sample size grows through collaboration and continued subject enrollment. For example, work on lipids progressed from one new locus in a ∼2,800 sample GWAS 89, to 7 new loci in an ∼8,800 sample GWAS 90; 91 and now 7-9 additional loci from a ∼20,000 sample GWAS (unpublished data). Another example is human height, where two initial GWAS identified one gene each 92; 93 and subsequent larger GWAS and meta-analysis 92; 94 expanded the number of loci to nearly 40. Similar stories are playing out for other traits, notably Crohn’s disease where a recent study of ∼14,000 samples identified a total of 32 confirmed loci, 21 of which were new 95. Even if the risk conferred by the variants uncovered by larger GWAS is relatively small, these genes may constitute very good therapeutic targets. We already know that monoclonal antibodies directed against TNF-α and IL/12/23 p40 provide highly efficacious therapeutic regimens for many psoriasis patients 96; 97, meaning that five of the genes implicated in our GWAS (IL12B, IL23A, IL23R, TNFAIP3, and TNIP1) play key roles in pathways targeted by therapeutic interventions. We expect that as new psoriasis susceptibility genes are identified, that many of them will further illuminate these pathways, and discover new ones amenable to therapeutic intervention. Moreover, once the full catalog of psoriasis genes have been identified, it may be possible to generate a “psoriasis gene profile” that can accurately predict one’s risk of developing psoriasis. We have already been able to demonstrate a 25-fold difference in risk depending upon how many disease alleles one inherits at the HLA-C and IL12B loci 98. Finally, as illustrated by IL23R in Crohn’s disease and TNFAIP3 in RA and SLE, genetic insights into psoriasis may contribute to the better understanding of a variety of autoimmune and inflammatory disorders. Thus, continuing the search for psoriasis genes seems well worthwhile.
References
- 1.Sander HM, Morris LF, Phillips CM, Harrison PE, Menter A. The annual cost of psoriasis. J Am Acad Dermatol. 1993;28(3):422–425. doi: 10.1016/0190-9622(93)70062-x. [DOI] [PubMed] [Google Scholar]
- 2.Gupta MA, Schork NJ, Gupta AK, Kirkby S, Ellis CN. Suicidal ideation in psoriasis. Int J Dermatol. 1993;32(3):188–190. doi: 10.1111/j.1365-4362.1993.tb02790.x. [DOI] [PubMed] [Google Scholar]
- 3.Krueger GG, Bergstresser PR, Lowe NJ, Voorhees JJ, Weinstein GD. Psoriasis. J Am Acad Dermatol. 1984;11(5 Pt 2):937–947. doi: 10.1016/s0190-9622(84)80018-3. [DOI] [PubMed] [Google Scholar]
- 4.Gladman DD. Natural history of psoriatic arthritis. Baillieres Clin Rheumatol. 1994;8(2):379–394. doi: 10.1016/s0950-3579(94)80024-3. [DOI] [PubMed] [Google Scholar]
- 5.Gudjonsson JE, Elder JT. Psoriasis. In: Wolff K, Goldsmith LA, Katz SI, Gilchrest BA, Paller AM, Leffell DJ, editors. Fitzpatrick’s Dermatology in General Medicine. vol. 1. McGraw-Hill; New York: 2007. pp. 169–194. [Google Scholar]
- 6.Gudjonsson JE, Elder JT. Psoriasis: epidemiology. Clin Dermatol. 2007;25(6):535–546. doi: 10.1016/j.clindermatol.2007.08.007. [DOI] [PubMed] [Google Scholar]
- 7.Elder JT, Nair RP, Guo SW, Henseler T, Christophers E, Voorhees JJ. The genetics of psoriasis. Arch Dermatol. 1994;130(2):216–224. [PubMed] [Google Scholar]
- 8.Elder JT, Nair RP, Henseler T, Jenisch S, Stuart P, Chia N, et al. The genetics of psoriasis 2001: the odyssey continues. Arch Dermatol. 2001;137(11):1447–1454. doi: 10.1001/archderm.137.11.1447. [DOI] [PubMed] [Google Scholar]
- 9.Henseler T, Christophers E. Psoriasis of early and late onset: characterization of two types of psoriasis vulgaris. J Am Acad Dermatol. 1985;13(3):450–456. doi: 10.1016/s0190-9622(85)70188-0. [DOI] [PubMed] [Google Scholar]
- 10.Moll JM, Wright V, O’Neill T, Silman AJ. Familial occurrence of psoriatic arthritis. Ann Rheum Dis. 1973;32(3):181–201. doi: 10.1136/ard.32.3.181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chandran V, Pellett FJ, Shanmugarajah S, Schentag CT, Brockbank J, Toloza S, et al. Recurrence risk of psoriatic arthritis (PsA) and psoriasis (Ps) in relatives of patients with PsA (abstract) Arthritis Rheum. 2007;56(Suppl 9):S798. [Google Scholar]
- 12.Mallon E, Bunce M, Savoie H, Rowe A, Newson R, Gotch F, et al. HLA-C and guttate psoriasis. Br J Dermatol. 2000;143(6):1177–1182. doi: 10.1046/j.1365-2133.2000.03885.x. [DOI] [PubMed] [Google Scholar]
- 13.Ozawa A, Miyahara M, Sugai J, Iizuka M, Kawakubo Y, Matsuo I, et al. HLA class I and II alleles and susceptibility to generalized pustular psoriasis: significant associations with HLA-Cw1 and HLA-DQB1 *0303. J Dermatol. 1998;25(9):573–581. doi: 10.1111/j.1346-8138.1998.tb02461.x. [DOI] [PubMed] [Google Scholar]
- 14.Torii H, Nakagawa H, Ishibashi Y, Tokunaga K, Juji T. Genetic polymorphisms in HLA-A, -B, -C and -DR antigens in Japanese patients with palmoplantar pustulosis. Dermatology. 1994;188(4):290–292. doi: 10.1159/000247168. [DOI] [PubMed] [Google Scholar]
- 15.Lowes MA, Kikuchi T, Fuentes-Duculan J, Cardinale I, Zaba LC, Haider AS, et al. Psoriasis vulgaris lesions contain discrete populations of Th1 and Th17 T cells. J Invest Dermatol. 2008 doi: 10.1038/sj.jid.5701213. doi:10.1038/sj.jid.5701213. [DOI] [PubMed] [Google Scholar]
- 16.Kebir H, Kreymborg K, Ifergan I, Dodelet-Devillers A, Cayrol R, Bernard M, et al. Human TH17 lymphocytes promote blood-brain barrier disruption and central nervous system inflammation. Nat Med. 2007;13(10):1173–1175. doi: 10.1038/nm1651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Neurath MF. IL-23: a master regulator in Crohn disease. Nat Med. 2007;13(1):26–28. doi: 10.1038/nm0107-26. [DOI] [PubMed] [Google Scholar]
- 18.Zheng Y, Danilenko DM, Valdez P, Kasman I, Eastham-Anderson J, Wu J, et al. Interleukin-22, a T(H)17 cytokine, mediates IL-23-induced dermal inflammation and acanthosis. Nature. 2007;445(7128):648–651. doi: 10.1038/nature05505. [DOI] [PubMed] [Google Scholar]
- 19.Ma HL, Liang S, Li J, Napierata L, Brown T, Benoit S, et al. IL-22 is required for Th17 cell-mediated pathology in a mouse model of psoriasis-like skin inflammation. J Clin Invest. 2008;118(2):597–607. doi: 10.1172/JCI33263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wilson NJ, Boniface K, Chan JR, McKenzie BS, Blumenschein WM, Mattson JD, et al. Development, cytokine profile and function of human interleukin 17-producing helper T cells. Nat Immunol. 2007;8(9):950–957. doi: 10.1038/ni1497. [DOI] [PubMed] [Google Scholar]
- 21.Sa SM, Valdez PA, Wu J, Jung K, Zhong F, Hall L, et al. The effects of IL-20 subfamily cytokines on reconstituted human epidermis suggest potential roles in cutaneous innate defense and pathogenic adaptive immunity in psoriasis. J Immunol. 2007;178(4):2229–2240. doi: 10.4049/jimmunol.178.4.2229. [DOI] [PubMed] [Google Scholar]
- 22.Wolk K, Witte E, Wallace E, Docke WD, Kunz S, Asadullah K, et al. IL-22 regulates the expression of genes responsible for antimicrobial defense, cellular differentiation, and mobility in keratinocytes: a potential role in psoriasis. Eur J Immunol. 2006;36(5):1309–1323. doi: 10.1002/eji.200535503. [DOI] [PubMed] [Google Scholar]
- 23.Wolk K, Kunz S, Asadullah K, Sabat R. Cutting edge: immune cells as sources and targets of the IL-10 family members? J Immunol. 2002;168(11):5397–5402. doi: 10.4049/jimmunol.168.11.5397. [DOI] [PubMed] [Google Scholar]
- 24.Kryczek I, Bruce AT, Gudjonsson JE, Johnston A, Vatan L, Szeliga W, et al. Induction of memory IL-17+ T cell trafficking and expansion by IFN-gamma: Mechanism and pathological relevance. Journal of Immunology. 2008;181:4733–4741. doi: 10.4049/jimmunol.181.7.4733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Conrad C, Boyman O, Tonel G, Tun-Kyi A, Laggner U, de Fougerolles A, et al. Alpha1beta1 integrin is crucial for accumulation of epidermal T cells and the development of psoriasis. Nat Med. 2007;13(7):836–842. doi: 10.1038/nm1605. [DOI] [PubMed] [Google Scholar]
- 26.Altshuler D, Brooks LD, Chakravarti A, Collins FS, Daly MJ, Donnelly P. A haplotype map of the human genome. Nature. 2005;437(7063):1299–1320. doi: 10.1038/nature04226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Manolio TA, Rodriguez LL, Brooks L, Abecasis G, Ballinger D, Daly M, et al. New models of collaboration in genome-wide association studies: the Genetic Association Information Network. Nat Genet. 2007;39(9):1045–1051. doi: 10.1038/ng2127. [DOI] [PubMed] [Google Scholar]
- 28.Nair RP, Duffin K Callis, Helms C, Ding J, Stuart PE, Goldgar D, et al. Genome-wide scan reveals association of psoriasis with IL-23 and NF-κB pathways. Nat Genet. 2008 doi: 10.1038/ng.311. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nair RP, Stuart PE, Nistor I, Hiremagalore R, Chia NV, Jenisch S, et al. Sequence and haplotype analysis supports HLA-C as the psoriasis susceptibility 1 gene. Am J Hum Genet. 2006;78(5):827–851. doi: 10.1086/503821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gudjonsson JE, Karason A, Runarsdottir EH, Antonsdottir AA, Hauksson VB, Jonsson HH, et al. Distinct clinical differences between HLA-Cw*0602 positive and negative psoriasis patients--an analysis of 1019 HLA-C-and HLA-B-typed patients. J Invest Dermatol. 2006;126(4):740–745. doi: 10.1038/sj.jid.5700118. [DOI] [PubMed] [Google Scholar]
- 31.Tsunemi Y, Saeki H, Nakamura K, Sekiya T, Hirai K, Fujita H, et al. Interleukin-12 p40 gene (IL12B) 3′-untranslated region polymorphism is associated with susceptibility to atopic dermatitis and psoriasis vulgaris. J Dermatol Sci. 2002;30(2):161–166. doi: 10.1016/s0923-1811(02)00072-5. [DOI] [PubMed] [Google Scholar]
- 32.Capon F, Di Meglio P, Szaub J, Prescott NJ, Dunster C, Baumber L, et al. Sequence variants in the genes for the interleukin-23 receptor (IL23R) and its ligand (IL12B) confer protection against psoriasis. Hum Genet. 2007;122(2):201–206. doi: 10.1007/s00439-007-0397-0. [DOI] [PubMed] [Google Scholar]
- 33.Cargill M, Schrodi SJ, Chang M, Garcia VE, Brandon R, Callis KP, et al. A large-scale genetic association study confirms IL12B and leads to the identification of IL23R as psoriasis-risk genes. Am J Hum Genet. 2007;80(2):273–290. doi: 10.1086/511051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Akagi Y, Kimura T, Kunimoto M, Kuki K, Tabata T. A role of tonsillar lymphocyte for focal infection. With special reference to lymphocyte adhesion to vessels in dermis. Adv Otorhinolaryngol. 1992;47:129–133. [PubMed] [Google Scholar]
- 35.Diluvio L, Vollmer S, Besgen P, Ellwart JW, Chimenti S, Prinz JC. Identical TCR beta-Chain Rearrangements in Streptococcal Angina and Skin Lesions of Patients with Psoriasis Vulgaris. J Immunol. 2006;176(11):7104–7111. doi: 10.4049/jimmunol.176.11.7104. [DOI] [PubMed] [Google Scholar]
- 36.Chang JC, Smith LR, Froning KJ, Schwabe BJ, Laxer JA, Caralli LL, et al. CD8+ T cells in psoriatic lesions preferentially use T-cell receptor V beta 3 and/or V beta 13.1 genes. Proc Natl Acad Sci USA. 1994;91(20):9282–9286. doi: 10.1073/pnas.91.20.9282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Prinz JC, Vollmer S, Boehncke WH, Menssen A, Laisney I, Trommler P. Selection of conserved TCR VDJ rearrangements in chronic psoriatic plaques indicates a common antigen in psoriasis vulgaris. Eur J Immunol. 1999;29(10):3360–3368. doi: 10.1002/(SICI)1521-4141(199910)29:10<3360::AID-IMMU3360>3.0.CO;2-G. [DOI] [PubMed] [Google Scholar]
- 38.Vollmer S, Menssen A, Prinz JC. Dominant lesional T cell receptor rearrangements persist in relapsing psoriasis but are absent from nonlesional skin: evidence for a stable antigen-specific pathogenic T cell response in psoriasis vulgaris. J Invest Dermatol. 2001;117(5):1296–1301. doi: 10.1046/j.0022-202x.2001.01494.x. [DOI] [PubMed] [Google Scholar]
- 39.Lin WJ, Norris DA, Achziger M, Kotzin BL, Tomkinson B. Oligoclonal expansion of intraepidermal T cells in psoriasis skin lesions. J Invest Dermatol. 2001;117(6):1546–1553. doi: 10.1046/j.0022-202x.2001.01548.x. [DOI] [PubMed] [Google Scholar]
- 40.Jones DA, Yawalkar N, Suh KY, Sadat S, Rich B, Kupper TS. Identification of autoantigens in psoriatic plaques using expression cloning. J Invest Dermatol. 2004;123(1):93–100. doi: 10.1111/j.0022-202X.2004.22709.x. [DOI] [PubMed] [Google Scholar]
- 41.Johnston A, Gudjonsson JE, Sigmundsdottir H, Love TJ, Valdimarsson H. Peripheral blood T cell responses to keratin peptides that share sequences with streptococcal M proteins are largely restricted to skin-homing CD8(+) T cells. Clin Exp Immunol. 2004;138(1):83–93. doi: 10.1111/j.1365-2249.2004.00600.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Baker BS, Laman JD, Powles A, van der Fits L, Voerman JS, Melief MJ, et al. Peptidoglycan and peptidoglycan-specific Th1 cells in psoriatic skin lesions. J Pathol. 2006;209(2):174–181. doi: 10.1002/path.1954. [DOI] [PubMed] [Google Scholar]
- 43.Baker BS, Powles A, Fry L. Peptidoglycan: a major aetiological factor for psoriasis? Trends Immunol. 2006;27(12):545–551. doi: 10.1016/j.it.2006.10.001. [DOI] [PubMed] [Google Scholar]
- 44.Williams F, Meenagh A, Sleator C, Cook D, Fernandez-Vina M, Bowcock AM, et al. Activating Killer Cell Immunoglobulin-Like Receptor Gene KIR2DS1 Is Associated With Psoriatic Arthritis. Hum Immunol. 2005;66(7):836–841. doi: 10.1016/j.humimm.2005.04.005. [DOI] [PubMed] [Google Scholar]
- 45.Nelson GW, Martin MP, Gladman D, Wade J, Trowsdale J, Carrington M. Cutting edge: heterozygote advantage in autoimmune disease: hierarchy of protection/susceptibility conferred by HLA and killer Ig-like receptor combinations in psoriatic arthritis. J Immunol. 2004;173(7):4273–4276. doi: 10.4049/jimmunol.173.7.4273. [DOI] [PubMed] [Google Scholar]
- 46.Long EO, Rajagopalan S. HLA class I recognition by killer cell Ig-like receptors. Semin Immunol. 2000;12(2):101–108. doi: 10.1006/smim.2000.0212. [DOI] [PubMed] [Google Scholar]
- 47.Lee E, Trepicchio WL, Oestreicher JL, Pittman D, Wang F, Chamian F, et al. Increased expression of interleukin 23 p19 and p40 in lesional skin of patients with psoriasis vulgaris. J Exp Med. 2004;199(1):125–130. doi: 10.1084/jem.20030451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bettelli E, Oukka M, Kuchroo VK. T(H)-17 cells in the circle of immunity and autoimmunity. Nat Immunol. 2007;8(4):345–350. doi: 10.1038/ni0407-345. [DOI] [PubMed] [Google Scholar]
- 49.Duerr RH, Taylor KD, Brant SR, Rioux JD, Silverberg MS, Daly MJ, et al. A genome-wide association study identifies IL23R as an inflammatory bowel disease gene. Science. 2006;314(5804):1461–1463. doi: 10.1126/science.1135245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Najarian DJ, Gottlieb AB. Connections between psoriasis and Crohn’s disease. J Am Acad Dermatol. 2003;48(6):805–821. doi: 10.1067/mjd.2003.540. quiz 822-804. [DOI] [PubMed] [Google Scholar]
- 51.Mauro C, Pacifico F, Lavorgna A, Mellone S, Iannetti A, Acquaviva R, et al. ABIN-1 binds to NEMO/IKKgamma and co-operates with A20 in inhibiting NF-kappaB. J Biol Chem. 2006;281(27):18482–18488. doi: 10.1074/jbc.M601502200. [DOI] [PubMed] [Google Scholar]
- 52.Chan JR, Blumenschein W, Murphy E, Diveu C, Wiekowski M, Abbondanzo S, et al. IL-23 stimulates epidermal hyperplasia via TNF and IL-20R2-dependent mechanisms with implications for psoriasis pathogenesis. J Exp Med. 2006;203(12):2577–2587. doi: 10.1084/jem.20060244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wang H, Kess D, Lindqvist AK, Peters T, Sindrilaru A, Wlaschek M, et al. A 9-centimorgan interval of chromosome 10 controls the T cell-dependent psoriasiform skin disease and arthritis in a murine psoriasis model. J Immunol. 2008;180(8):5520–5529. doi: 10.4049/jimmunol.180.8.5520. [DOI] [PubMed] [Google Scholar]
- 54.Idel S, Dansky HM, Breslow JL. A20, a regulator of NFkappaB, maps to an atherosclerosis locus and differs between parental sensitive C57BL/6J and resistant FVB/N strains. Proc Natl Acad Sci USA. 2003;100(24):14235–14240. doi: 10.1073/pnas.1835672100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Gelfand JM, Neimann AL, Shin DB, Wang X, Margolis DJ, Troxel AB. Risk of myocardial infarction in patients with psoriasis. Jama. 2006;296(14):1735–1741. doi: 10.1001/jama.296.14.1735. [DOI] [PubMed] [Google Scholar]
- 56.Graham RR, Cotsapas C, Davies L, Hackett R, Lessard CJ, Leon JM, et al. Genetic variants near TNFAIP3 on 6q23 are associated with systemic lupus erythematosus. Nat Genet. 2008;40:1059–1061. doi: 10.1038/ng.200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Musone SL, Taylor KE, Lu TT, Nititham J, Ferreira RC, Ortmann W, et al. Multiple polymorphisms in the TNFAIP3 region are independently associated with systemic lupus erythematosus. Nat Genet. 2008;40:1062–1064. doi: 10.1038/ng.202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Plenge RM, Cotsapas C, Davies L, Price AL, de Bakker PI, Maller J, et al. Two independent alleles at 6q23 associated with risk of rheumatoid arthritis. Nat Genet. 2007;39(12):1477–1482. doi: 10.1038/ng.2007.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Thomson W, Barton A, Ke X, Eyre S, Hinks A, Bowes J, et al. Rheumatoid arthritis association at 6q23. Nat Genet. 2007;39(12):1431–1433. doi: 10.1038/ng.2007.32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Griffiths CE, Barker JN. Pathogenesis and clinical features of psoriasis. Lancet. 2007;370(9583):263–271. doi: 10.1016/S0140-6736(07)61128-3. [DOI] [PubMed] [Google Scholar]
- 61.Ghoreschi K, Thomas P, Breit S, Dugas M, Mailhammer R, van Eden W, et al. Interleukin-4 therapy of psoriasis induces Th2 responses and improves human autoimmune disease. Nat Med. 2003;9(1):40–46. doi: 10.1038/nm804. [DOI] [PubMed] [Google Scholar]
- 62.Lee GR, Fields PE, Griffin TJ, Flavell RA. Regulation of the Th2 cytokine locus by a locus control region. Immunity. 2003;19(1):145–153. doi: 10.1016/s1074-7613(03)00179-1. [DOI] [PubMed] [Google Scholar]
- 63.Brody I. Alterations of clinically normal skin in early eruptive guttate psoriasis. J Cutan Pathol. 1978;5:219–233. doi: 10.1111/j.1600-0560.1978.tb00216.x. [DOI] [PubMed] [Google Scholar]
- 64.Ragaz A, Ackerman AB. Evolution, maturation, and regression of lesions of psoriasis. New observations and correlation of clinical and histologic findings. Am J Dermatopathol. 1979;1(3):199–214. doi: 10.1097/00000372-197900130-00002. [DOI] [PubMed] [Google Scholar]
- 65.Schubert C, Christophers E. Mast cells and macrophages in early relapsing psoriasis. Arch Dermatol Res. 1985;277(5):352–358. doi: 10.1007/BF00509232. [DOI] [PubMed] [Google Scholar]
- 66.Braun-Falco O, Schmoeckel C. The dermal inflammatory reaction in initial psoriatic lesions. Arch Dermatol Res. 1977;258(1):9–16. doi: 10.1007/BF00582862. [DOI] [PubMed] [Google Scholar]
- 67.Wollenberg A, Wagner M, Gunther S, Towarowski A, Tuma E, Moderer M, et al. Plasmacytoid dendritic cells: a new cutaneous dendritic cell subset with distinct role in inflammatory skin diseases. J Invest Dermatol. 2002;119(5):1096–1102. doi: 10.1046/j.1523-1747.2002.19515.x. [DOI] [PubMed] [Google Scholar]
- 68.Nestle FO, Conrad C, Tun-Kyi A, Homey B, Gombert M, Boyman O, et al. Plasmacytoid predendritic cells initiate psoriasis through interferon-{alpha} production. J Exp Med. 2005;202(1):135–143. doi: 10.1084/jem.20050500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Braun-Falco O. Dynamics of growth and regression in psoriatic lesions: Alterations in the skin from normal into a psoriatic lesion, and during regression of psoriatic lesions. In: Farber EM, Cox AJ, editors. Psoriasis: Proceedings of the International Symposium, Stanford University, 1971. Stanford University Press; Stanford, CA: 1971. pp. 215–237. [Google Scholar]
- 70.Boehncke WH, Wortmann S, Kaufmann R, Mielke V, Sterry W. A subset of macrophages located along the basement membrane (“lining cells”) is a characteristic histopathological feature of psoriasis. Am J Dermatopathol. 1995;17(2):139–144. doi: 10.1097/00000372-199504000-00005. [DOI] [PubMed] [Google Scholar]
- 71.van den Oord JJ, de Wolf-Peeters C. Epithelium-lining macrophages in psoriasis. Br J Dermatol. 1994;130(5):589–594. doi: 10.1111/j.1365-2133.1994.tb13104.x. [DOI] [PubMed] [Google Scholar]
- 72.Bata-Csorgo Z, Cooper KD, Ting KM, Voorhees JJ, Hammerberg C. Fibronectin and alpha5 integrin regulate keratinocyte cell cycling. A mechanism for increased fibronectin potentiation of T cell lymphokine-driven keratinocyte hyperproliferation in psoriasis. J Clin Invest. 1998;101(7):1509–1518. doi: 10.1172/JCI171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Brody I. Mast cell degranulation in the evolution of acute eruptive guttate psoriasis vulgaris. J Invest Dermatol. 1984;82(5):460–464. doi: 10.1111/1523-1747.ep12260955. [DOI] [PubMed] [Google Scholar]
- 74.Stoler A, Kopan R, Duvic M, Fuchs E. Use of monospecific antisera and cRNA probes to localize the major changes in keratin expression during normal and abnormal epidermal differentiation. J Cell Biol. 1988;107(2):427–446. doi: 10.1083/jcb.107.2.427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Leigh IM, Navsaria H, Purkis PE, McKay IA, Bowden PE, Riddle PN. Keratins (K16 and K17) as markers of keratinocyte hyperproliferation in psoriasis in vivo and in vitro. Br J Dermatol. 1995;133(4):501–511. doi: 10.1111/j.1365-2133.1995.tb02696.x. [DOI] [PubMed] [Google Scholar]
- 76.Zhou X, Krueger JG, Kao MC, Lee E, Du F, Menter A, et al. Novel mechanisms of T-cell and dendritic cell activation revealed by profiling of psoriasis on the 63,100-element oligonucleotide array. Physiol Genomics. 2003;13(1):69–78. doi: 10.1152/physiolgenomics.00157.2002. [DOI] [PubMed] [Google Scholar]
- 77.Gudjonsson JE, Ding J, Li X, Nair RP, Stuart PE, Tejasvi T, et al. Global gene expression analysis reveals evidence for decreased lipid biosynthesis and increased innate immunity in uninvolved psoriatic skin. Proc Natl Acad Sci USA. 2009 doi: 10.1038/jid.2009.173. submitted. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Capon F, Novelli G, Semprini S, Clementi M, Nudo M, Vultaggio P, et al. Searching for psoriasis susceptibility genes in Italy: genome scan and evidence for a new locus on chromosome 1. J Invest Dermatol. 1999;112(1):32–35. doi: 10.1046/j.1523-1747.1999.00471.x. [DOI] [PubMed] [Google Scholar]
- 79.Capon F, Semprini S, Chimenti S, Fabrizi G, Zambruno G, Murgia S, et al. Fine mapping of the PSORS4 psoriasis susceptibility region on chromosome 1q21. J Invest Dermatol. 2001;116(5):728–730. doi: 10.1046/j.1523-1747.2001.01311.x. [DOI] [PubMed] [Google Scholar]
- 80.Bowcock AM. Genetic association in psoriasis. Eighth International Psoriasis Genetics Committee Meeting; Paris, France. 2005. Unpublished. [Google Scholar]
- 81.Bhalerao J, Bowcock AM. The genetics of psoriasis: a complex disorder of the skin and immune system. Hum Mol Genet. 1998;7(10):1537–1545. doi: 10.1093/hmg/7.10.1537. [DOI] [PubMed] [Google Scholar]
- 82.de Cid R, Riveira-Munoz E, Zeeuwen PLJM, Robarge J, Liao W, Dannhauser E, et al. Deletion of the late cornified envelope (LCE) 3C and 3B genes as a susceptibility factor for psoriasis. Nat Genet. 2008 doi: 10.1038/ng.313. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Hollox EJ, Huffmeier U, Zeeuwen PL, Palla R, Lascorz J, Rodijk-Olthuis D, et al. Psoriasis is associated with increased beta-defensin genomic copy number. Nat Genet. 2008;40(1):23–25. doi: 10.1038/ng.2007.48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Rottman JB, Smith TL, Ganley KG, Kikuchi T, Krueger JG. Potential role of the chemokine receptors CXCR3, CCR4, and the integrin alphaEbeta7 in the pathogenesis of psoriasis vulgaris. Lab Invest. 2001;81(3):335–347. doi: 10.1038/labinvest.3780242. [DOI] [PubMed] [Google Scholar]
- 85.Stratis A, Pasparakis M, Rupec RA, Markur D, Hartmann K, Scharffetter-Kochanek K, et al. Pathogenic role for skin macrophages in a mouse model of keratinocyte-induced psoriasis-like skin inflammation. J Clin Invest. 2006;116(8):2094–2104. doi: 10.1172/JCI27179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Wang H, Peters T, Kess D, Sindrilaru A, Oreshkova T, Van Rooijen N, et al. Activated macrophages are essential in a murine model for T cell-mediated chronic psoriasiform skin inflammation. J Clin Invest. 2006;116(8):2105–2114. doi: 10.1172/JCI27180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Bata-Csorgo Z, Hammerberg C, Voorhees JJ, Cooper KD. Kinetics and regulation of human keratinocyte stem cell growth in short-term primary ex vivo culture. Cooperative growth factors from psoriatic lesional T lymphocytes stimulate proliferation among psoriatic uninvolved, but not normal, stem keratinocytes. J Clin Invest. 1995;95(1):317–327. doi: 10.1172/JCI117659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Iordanov MS, Sundholm AJ, Simpson EL, Hanifin JM, Ryabinina OP, Choi RJ, et al. Cell death-induced activation of epidermal growth factor receptor in keratinocytes: implications for restricting epidermal damage in dermatitis. J Invest Dermatol. 2005;125(1):134–142. doi: 10.1111/j.0022-202X.2005.23804.x. [DOI] [PubMed] [Google Scholar]
- 89.Saxena R, Voight BF, Lyssenko V, Burtt NP, de Bakker PI, Chen H, et al. Genome-wide association analysis identifies loci for type 2 diabetes and triglyceride levels. Science. 2007;316(5829):1331–1336. doi: 10.1126/science.1142358. [DOI] [PubMed] [Google Scholar]
- 90.Willer CJ, Sanna S, Jackson AU, Scuteri A, Bonnycastle LL, Clarke R, et al. Newly identified loci that influence lipid concentrations and risk of coronary artery disease. Nat Genet. 2008;40(2):161–169. doi: 10.1038/ng.76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Kathiresan S, Melander O, Guiducci C, Surti A, Burtt NP, Rieder MJ, et al. Six new loci associated with blood low-density lipoprotein cholesterol, high-density lipoprotein cholesterol or triglycerides in humans. Nat Genet. 2008;40(2):189–197. doi: 10.1038/ng.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Weedon MN, Lettre G, Freathy RM, Lindgren CM, Voight BF, Perry JR, et al. A common variant of HMGA2 is associated with adult and childhood height in the general population. Nat Genet. 2007;39(10):1245–1250. doi: 10.1038/ng2121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Sanna S, Jackson AU, Nagaraja R, Willer CJ, Chen WM, Bonnycastle LL, et al. Common variants in the GDF5-UQCC region are associated with variation in human height. Nat Genet. 2008;40(2):198–203. doi: 10.1038/ng.74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Weedon MN, Lango H, Lindgren CM, Wallace C, Evans DM, Mangino M, et al. Genome-wide association analysis identifies 20 loci that influence adult height. Nat Genet. 2008;40(5):575–583. doi: 10.1038/ng.121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Barrett JC, Hansoul S, Nicolae DL, Cho JH, Duerr RH, Rioux JD, et al. Genome-wide association defines more than 30 distinct susceptibility loci for Crohn’s disease. Nat Genet. 2008 doi: 10.1038/NG.175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Chaudhari U, Romano P, Mulcahy LD, Dooley LT, Baker DG, Gottlieb AB. Efficacy and safety of infliximab monotherapy for plaque-type psoriasis: a randomised trial. Lancet. 2001;357(9271):1842–1847. doi: 10.1016/s0140-6736(00)04954-0. [DOI] [PubMed] [Google Scholar]
- 97.Krueger GG, Langley RG, Leonardi C, Yeilding N, Guzzo C, Wang Y, et al. A human interleukin-12/23 monoclonal antibody for the treatment of psoriasis. N Engl J Med. 2007;356(6):580–592. doi: 10.1056/NEJMoa062382. [DOI] [PubMed] [Google Scholar]
- 98.Nair RP, Ruether A, Stuart PE, Jenisch S, Tejasvi T, Hiremagalore R, et al. Polymorphisms of the IL12B and IL23R genes are associated with psoriasis. J Invest Dermatol. 2008;128(7):1653–1661. doi: 10.1038/sj.jid.5701255. [DOI] [PMC free article] [PubMed] [Google Scholar]