Abstract
The ability of transcription factors to gain entrance to the nucleus is critical to their role in gene expression. Signal transducers and activators of transcription (STATs) are latent DNA binding factors activated by specific tyrosine phosphorylation. There are seven mammalian STAT genes encoding proteins that display constitutive nuclear localization and/or conditional nuclear localization. This review will focus on STAT1 and STAT2 that are activated in response to interferon and exhibit conditional nuclear localization. The dynamic redistribution of STAT1 and STAT2 between the cytoplasm and the nucleus is coordinate with their gain of ability to bind DNA.
Keywords: nuclear import, tyrosine phosphorylation, transcription factor, DNA binding
Introduction
Signal transducers and activators of transcription (STAT) possess the ability to both sense environmental cues and to transmit those cues to regulate specific gene expression. The founding members of this family, STAT1 and STAT2, were identified as latent DNA-binding factors activated in response to type I interferons (IFNs)(1, 2). Following IFN binding to cell surface receptors, the receptor-associated Janus kinases phosphorylate STATs on a specific tyrosine residue (3–8). Tyrosine phosphorylation induces a conformational change that generates STAT dimers via reciprocal phosphotyrosine and SH2 domain interaction (9–11). The dimer conformation confers their ability to recognize specific DNA targets in the promoters of responsive genes, and the products of these genes contribute to the biological effects of IFNs on viral resistance, proliferation, and immune cell activation (12–15).
STAT-mediated gene expression can have dramatic effects on cellular function, and for this reason it is not surprising that STAT activity is regulated by various means including receptor activated Janus kinases, cytoplasmic and nuclear tyrosine phosphatases, protein inhibitors of activated STATs (PIAS), and suppressors of cytokine signaling (SOCS) (16–18). To affect gene transcription the STATs must gain access to the nucleus, and consequently nuclear localization is yet another mechanism of STAT regulation (19, 20). Proteins as large as the STATs are restricted from passive diffusion into the nucleus, and so transport must be facilitated. Transport is an active energy-requiring process usually mediated by association with soluble transporters. Since nuclear trafficking has a significant impact on STAT function, understanding the mechanisms that regulate STAT localization should provide information valuable to enhance or prevent their action.
1. Properties of STAT Molecules
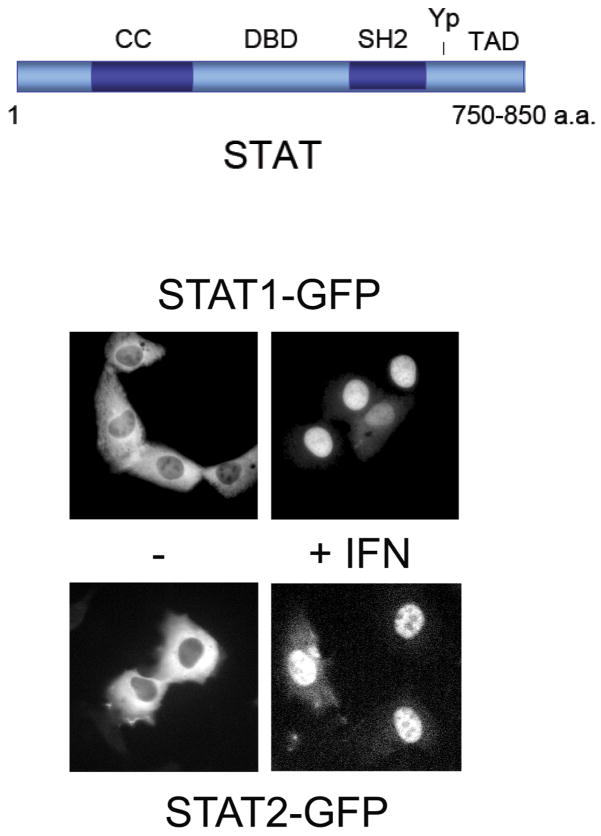
Seven mammalian STAT genes have been identified, and although the encoded proteins share many properties, they respond uniquely to specific stimuli and confer distinct biological responses. The STATs have a similar structural arrangement of functional motifs (Figure 1)(12). These include an amino terminus that plays a role in dimerization, a coiled coil domain that can be involved in interactions with other proteins, a central DNA-binding domain (DBD), a Src homology 2 (SH2) domain, a conserved tyrosine residue that is phosphorylated in response to stimuli, and a carboxyl transcriptional activation domain (TAD). Recent evidence indicates that the STAT molecules can exist as dimers in a latent state, however following tyrosine phosphorylation homodimers or heterodimers form via reciprocal SH2-phosphotyrosine interactions (9–11). The conformational change that accompanies tyrosine phosphorylation provides the dimers with the ability to bind specific target DNA.
Figure 1.
STAT cellular redistribution. Top) Schematic consensus of STAT domain arrangement with coiled coil domain (CC), DNA binding domain (DBD), Src homology domain 2 (SH2), tyrosine target of phosphorylation (Yp), and transcriptional activation domain (TAD). Bottom) Fluorescence microscopy of cells expressing STAT1-GFP or STAT2-GFP. Cells were untreated (−) or treated with 1000U/ml IFNα for 1 hour (+). Figure is modified from previous publications (24, 45).
1.1 Interferon Signaling with STAT1 and STAT2
STAT proteins can be tyrosine phosphorylated by receptor associated Janus kinases, by growth factor receptor tyrosine kinases, or by non-receptor tyrosine kinases (19). STAT1 and STAT2 are the founding members of the STAT family, identified as latent DNA-binding factors activated in response to IFN stimulation. As signal transducers, STAT1 and STAT2 respond to extracellular cues in the cytoplasm, and then move to the nucleus to regulate gene expression. The dynamic redistribution of STAT1 and STAT2 following IFN treatment can be visualized by fluorescence microscopy with GFP tagged STAT proteins (Figure 1). Unphosphorylated STAT1-GFP is primarily in the cytoplasm, although if overexpressed a portion of STAT1 can be found in the nucleus (21–23). Following tyrosine phosphorylation in response to IFN, STAT1-GFP clearly accumulates in the nucleus. Similarly unphosphorylated STAT2-GFP resides primarily in the cytoplasm and accumulates in the nucleus following IFN stimulation (24).
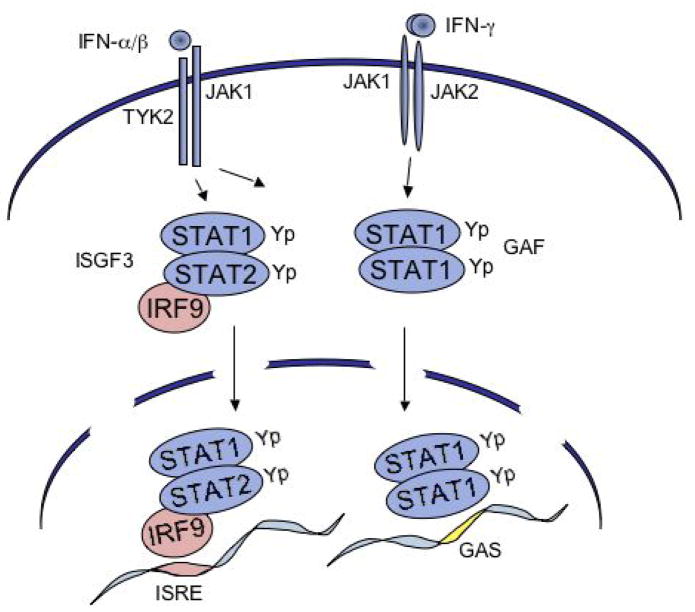
The Janus kinases TYK2 and JAK1 are associated with type I IFN receptor subunits IFNARI and IFNARII, and are activated in response to binding type I IFN (primarily IFNα/β)(Figure 2). These kinases phosphorylate tyrosine residues on the receptors and on STAT1 and STAT2. STAT2 is unique among the STAT proteins because it is constitutively associated with a non-STAT protein, IFN regulatory factor 9 (IRF-9)(25–27). IRF-9 is a member of a family of DNA-binding factors that play diverse roles in the innate immune response (28). The IRFs share a similar amino terminal DNA binding domain, but they have distinct carboxyl domains. The carboxyl domain of IRF-9 associates with the coiled coil domain of STAT2 (25). Following IFNα stimulation, STAT1 and STAT2 heterodimerize via phosphotyrosine and SH2 domains. Because IRF-9 is associated with STAT2, a tyrosine phosphorylated STAT1-STAT2-IRF-9 multimeric complex forms and has been designated the IFN stimulated gene factor 3 (ISGF3)(1, 29). ISGF3 binds to a specific DNA sequence in type I IFN induced genes called the IFN stimulated response element (ISRE) that contains a direct GAAA repeat spaced by 2 nucleotides (29–32).
Figure 2.
Schematic of JAK/STAT signal pathways stimulated by IFNs. Left) Type I IFNs (primarily α/β) bind to the IFNAR activating JAK1 and TYK2. The JAKs phosphorylate the receptors and recruited STAT1 and STAT2. The STATs heterodimerize and with IRF-9 form ISGF3 to bind to the ISRE in target genes. STAT1 homodimers are also formed and can bind to the GAS. Right) Type II IFN (γ) binds to a distinct IFNGR activating JAK1 and JAK2. These JAKs phosphorylate sites on the receptor and the recruited STAT1. STAT1 factors dimerize and gain the ability to bind to the GAS in target genes.
The type II IFN receptor subunits IFNGR1 and IFNGR2 are associated with JAK1 and JAK2, and are activated in response to type II IFN (IFNγ)(Figure 2). In response to IFNγ, STAT1 is tyrosine phosphorylated and forms dimers via phosphotyrosine and SH2 domains (7, 12, 32, 33). IFNγ binding leads to tyrosine phosphorylation of STAT1, but not STAT2. The STAT1 dimer was originally called the gamma-IFN activated factor (GAF). It recognizes a DNA sequence termed the gamma-IFN activated site (GAS) that contains an inverted repeat of GAAA residues spaced by 2–4 nucleotides. Depending on the cell type, IFNs can also stimulate the tyrosine phosphorylation of other STATs such as STAT3 and STAT5 that can bind the GAS target.
2. Nuclear Trafficking
The genomic information of eukaryotic cells is partitioned from the cytoplasm by a membrane bound nucleus. The movement of molecules in and out of the nucleus occurs through discrete passageways known as nuclear pore complexes (NPCs) that span the nuclear membrane (34, 35). Small molecules can freely diffuse through the NPCs, however the movement of large molecules is restricted. To pass through the NPC, macromolecules must be able to directly interact with the proteins that comprise the NPC, the nucleoporins, or interact with transport carrier proteins. The transport carriers are usually members of the karyopherin-β family that play a primary role either in nuclear import or nuclear export and are therefore referred to as importins and exportins (36–42). The nuclear import or export of a protein can be constitutive or can be conditional depending on post-translational modifications such as phosphorylation or dependent on association with other proteins or DNA.
The NPC is composed of approximately 30 different nucleoporins present in multiple copies. Many of the nucleoporins that line the channel of the NPC contain repeats of phenyalanine-glycine (FG) that interact with karyopherin-β transporters to facilitate translocation of the protein cargo. The mechanism by which the complex navigates through the pore is an area of active investigation, but the direction of protein transport is known to be influenced by the ability of the transporter to bind the Ran GTPase.
2.1 Nuclear Import
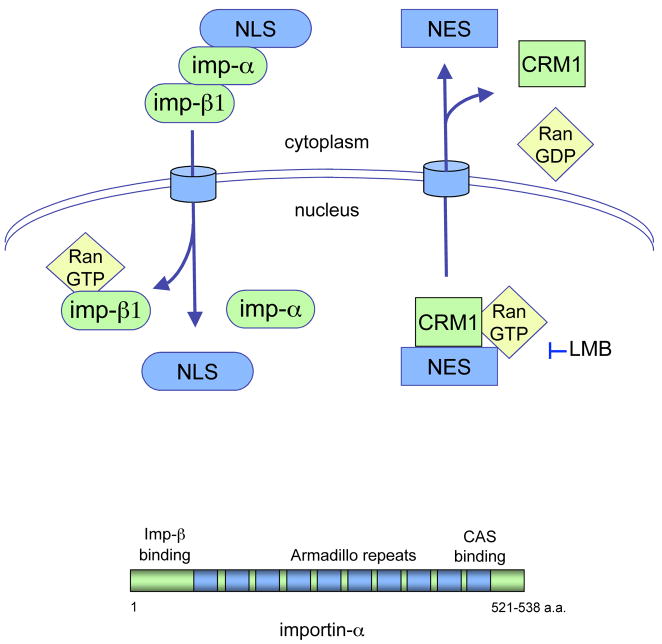
Active transport of proteins into the nucleus requires that they possess an amino acid sequence or structure that serves as a nuclear localization signal (NLS). The best characterized NLSs are rich in the basic amino acids lysine and arginine and function either as a single stretch of residues or as a bipartite sequence (43). The classical basic NLS in proteins can directly bind to adapter proteins in the importin-α family (Figure 3). There are six characterized importin-α proteins and they share similar structural features; a central domain with 8–10 Armadillo repeats that can bind the NLS; a carboxyl terminus that binds the exportin CAS; and an amino terminus that binds to a karyopherin-β, importin-β1. Importin-β1 mediates passage of the complex through the NPC, and following entrance to the nucleus it binds Ran-GTP causing the release of importin-α and the NLS-containing protein.
Figure 3.
Illustration of nuclear trafficking mediated by soluble transporters. Nuclear import can be mediated by the importin-α/importinβ1 heterodimer via recognition of the NLS in proteins. Nuclear export can be mediated by the CRM1 exportin and recognition of the NES in proteins. Leptomycin B (LMB) can specifically inhibit CRM1-mediated export.
2.2 Nuclear Export
Export from the nucleus shares many properties of import. Active export requires the presence of a nuclear export signal (NES) in the protein destined for the cytoplasm. A frequently occurring NES is a hydrophobic sequence rich in leucine amino acids that is recognized by the exportin transporter CRM1 (Figure 3). CRM1 binds the NES-containing protein and binds Ran-GTP in the nucleus. Following transport through the NPC and entrance into the cytoplasm, Ran-GTP is hydrolyzed and CRM1 subsequently dissociates from the NES-containing protein. An antibiotic inhibitor of CRM1, leptomycin B (LMB), has provided much to our understanding of CRM1-mediated export (44).
3. STAT1 Dynamics
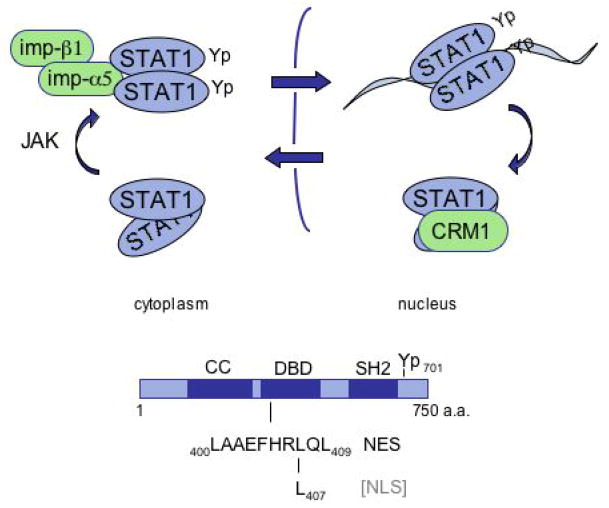
Regulated cellular localization can function as a molecular switch to turn on or turn off a signal. Similarly, the regulated ability to bind DNA can serve as a molecular switch to turn on or turn off gene expression. The tyrosine phosphorylation of STAT1 activates both of these molecular switches. The dimer that is generated by reciprocal phosphotyrosine-SH2 domain interaction between STAT1 monomers gains the ability to recognize a specific importin-α adapter, and to recognize a specific DNA sequence (Figure 4).
Figure 4.
Conceptual diagram of STAT1 dynamics. STAT1 resides primarily in the cytoplasm prior to tyrosine phosphorylation, but following phosphorylation the STAT1 dimer is recognized by importin-α5. The active STAT1 dimer is imported into the nucleus and binds specific DNA targets. Bound to DNA the NES is masked, but following dissociation from DNA the NES is accessible to CRM1 and the protein is exported from the nucleus. The location of the NES and sequences required for NLS function are indicated in the DNA binding domain.
3.1 STAT1 nuclear import
Imaging analyses have revealed the dramatic change in STAT1 localization after tyrosine phosphorylation (7, 21, 23, 45–47) (Figure 1). Most of the studies to evaluate nuclear trafficking of STAT1 have been performed with IFN-γ as the activating ligand. Although stimulation with IFN-γ can activate other STATs that can dimerize with STAT1, it generates STAT1 homodimers primarily. Therefore, altered behavior of STAT1 mutants best reflects properties intrinsic to STAT1.
Unphosphorylated STAT1 in the cytoplasm exists primarily as a freely diffusible protein. FLIP (fluorescence loss in photobleaching) and FRAP (fluorescence recovery after photobleaching) analyses on cytoplasmic STAT1 reveal that it moves throughout the cytoplasm with high mobility (46). In response to an activating cytokine such as IFN-γ, STAT1 is recruited via its SH2 domain to phosphorylated residues on the receptor where JAKs phosphorylate STAT1 on tyrosine 701 (6). Following tyrosine phosphorylation, STAT1 dimers quickly accumulate in the nucleus. Mutations that prevent tyrosine phosphorylation by substituting alanine for tyrosine 701, or mutations that prevent dimerization by substituting leucine for arginine 605 in the SH2 domain, prevent translocation to the nucleus (48). The conformational change triggered by tyrosine phosphorylation is therefore critical for conditional nuclear import of STAT1.
Identifying the region within STAT1 that functions as a NLS was challenging since the NLS is conditional and dependent on the conformation of the tyrosine phosphorylated dimer. Single point mutations in STAT1 can disrupt structure in fundamental ways to produce a protein that cannot be tyrosine phosphorylated or does not form dimers, and these mutations would only indirectly be related to nuclear import. Yet the effects of several mutations within the DNA binding domain suggested that nuclear import of STAT1 co-evolved with its ability to bind DNA (21, 47, 49). A single mutation in leucine 407 (L407A) or a double mutation in lysines 410 and 413 (KK410/413AA) produced a STAT1 protein that is tyrosine phosphorylated following IFN-γ stimulation, but remains in the cytoplasm. In addition, the L407A mutant is tyrosine phosphorylated and forms dimers that retain the ability to bind DNA. Therefore this leucine is part of a region required for conditional nuclear import.
Although the requirement of a leucine residue is unconventional relative to a basic NLS, the nuclear import of tyrosine phosphorylated STAT1 was found to be facilitated by the importin-α5/importin-β1 heterodimer (50). There are six characterized importin-α adapters, but the active STAT1 dimer binds specifically and directly to importin-α5 (21, 51). The binding of tyrosine phosphorylated STAT1 to importin-α5 is unconventional in that it binds to the carboxyl region Arm repeats 8–10 that overlap with the CAS exportin binding site, whereas classical basic NLSs bind within importin-α Arm repeats 2–8. To test whether the import defect of the STAT1 L407A mutant is a consequence of its lack of ability to bind importin-α5, in vitro assays were performed. The results clearly indicated that the tyrosine phosphorylated L407A mutant was not able to bind importin-α5, coordinate with its lack of ability to enter the nucleus (21). This region of the DNA binding domain therefore plays a critical role in the function of a conditional NLS.
Several other aspects of STAT1 nuclear import are noteworthy. Although it is clear that unphosphorylated STAT1 resides primarily in the cytoplasm, there is a detectable constitutive presence of overexpressed STAT1 in the nucleus. This may be due to dimerization with other tyrosine phosphorylated STATs such as STAT3, or association with a non-STAT factor such as IRF-1, or due to direct interaction of STAT1 with the NPC (52, 53). In addition, there has also been another mechanism proposed for the nuclear import of tyrosine phosphorylated STAT1 that depends on a NLS in IFN-γ cytokine (54). In this model the IFN-γ cytokine is translocated into the nucleus in a complex with the IFNGR1 receptor subunit and phosphorylated STAT1. However, this may not be a key mechanism since the STAT1 L407A mutant is tyrosine phosphorylated in response to IFN-γ, but does not localize to the nucleus.
3.2 STAT1 nuclear export
Although tyrosine phosphorylated STAT1 accumulates in the nucleus, this accumulation is transient and within hours STAT1 reappears in the cytoplasm (45, 47, 55). Since the CRM1 exportin is a common transporter, its contribution to STAT1 export was tested with the use of the CRM1 inhibitor, LMB. LMB clearly inhibited the export of STAT1, indicating that STAT1 is actively exported from the nucleus and that CRM1 is a primary exportin for STAT1. Because tyrosine phosphorylation is necessary for the inducible nuclear import of STAT1, the phosphorylation state of STAT1 was evaluated with export. These studies determined that STAT1 was dephosphorylated in the nucleus (45, 47, 56). The nuclear form of the T cell protein tyrosine phosphatase TCPTP has been shown to play a major role in this dephosphorylation (16).
The NES sequences that are recognized by CRM1 are rich in leucine residues, but do not conform to a strict consensus. Scanning the STAT1 sequence there are more than two dozen candidate sequences, however testing the function of these peptides outside the context of STAT1 can be misleading, and evaluating mutations in STAT1 can inhibit other properties of STAT1. For these reasons we used an in vitro CRM1/Ran binding assay to identify the NES in STAT1 that is recognized by CRM1 (45). Bacterially expressed unphosphorylated STAT1 was found to bind directly to CRM1 via a region within the STAT1 DNA binding domain. Binding studies coupled with mutational analyses identified the NES within amino acids 399–410. This region was demonstrated to function as an NES outside the context of STAT1, and to share sequence similarity with other characterized NESs.
The location of the STAT1 NES within the DNA binding domain suggests that the NES is masked when the STAT1 phosphorylated dimer is bound to DNA. To test this premise we evaluated the behavior of tyrosine phosphorylated STAT1 that lacks the ability to bind DNA. Results with fluorescence microscopy and LMB indicated that the phosphorylated STAT1 dimer was imported to the nucleus but efficiently exported from the nucleus (45). These studies support a model in which the NES is masked when tyrosine phosphorylated STAT1 is bound to DNA, but when STAT1 dissociates from DNA, either with or without dephosphorylation, CRM1 can bind the NES and return the STAT1 protein to the cytoplasm.
Regulated nuclear trafficking of STAT1 appears to have co-evolved with its ability to bind DNA (Figure 4). The nuclear import of STAT1 is dependent on the conditional recognition of the tyrosine phosphorylated dimer by importin-α5, and the recognition region of importin-α5 is within the STAT1 DNA binding domain. Following import to the nucleus, specific DNA targets successfully compete with importin-α5 for association with STAT1 (21). Bound to DNA STAT1 recruits transcriptional co-activators to induce expression of target genes. When STAT1 is bound to DNA the NES appears to be masked, but following dissociation from DNA either by tyrosine dephosphorylation or by protein inhibitors of activated STATs (PIAS) (18) or inherent dissociation kinetics, the STAT1 NES can be recognized by the CRM1 exportin. STAT1 is exported out of the nucleus and may be a target of cytoplasmic phosphatases to silence the signal, or it can respond to receptor-kinase signals and reactivate the signal pathway.
4. STAT2 Dynamics
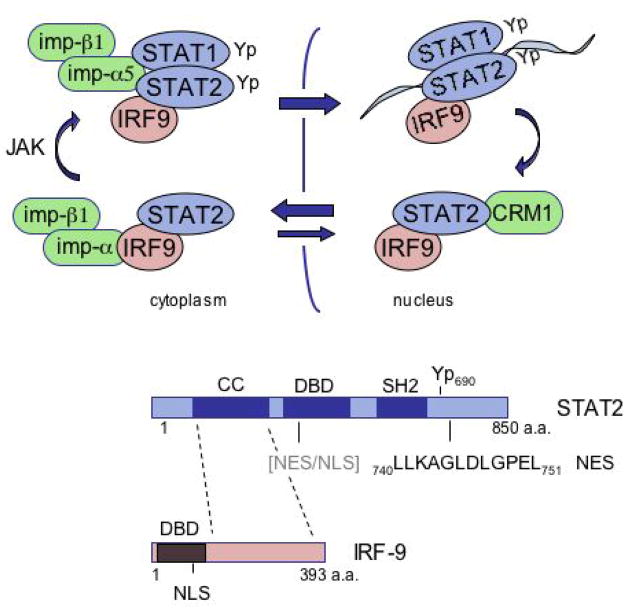
STAT2 is tyrosine phosphorylated in response to type I IFNs, and it dimerizes with phosphorylated STAT1 to form heterodimers that are actively imported to the nucleus. STAT2 is distinct among the STATs in its ability to bind to IRF-9 constitutively, and this property suggested that STAT2 localization may be regulated differently from STAT1 (25). Evidence now indicates that the unphosphorylated STAT2 molecule constitutively shuttles in and out of the nucleus, the import mediated by IRF-9 and the export mediated by a NES intrinsic to STAT2 (Figure 5).
Figure 5.
Conceptual diagram of STAT2 dynamics. Unphosphorylated STAT2 in association with IRF-9 is imported into the nucleus via the NLS in IRF-9, however the strong constitutive NES in the carboxyl terminus of STAT2 redistributes the unphosphorylated STAT2 back to the cytoplasm. The unphosphorylated STAT2 is thereby constitutively shuttling in and out of the nucleus. Following tyrosine phosphorylation, STAT2 heterodimerizes with STAT1 and this leads to the gain of a NLS that is recognized by importin-α5 and the gain of an ability to bind DNA. Following dissociation from DNA STAT2 is actively exported from the nucleus.
4.1. STAT2 import
Static fluorescent images of latent STAT2 clearly show its residence in the cytoplasm. However, following treatment with LMB in the absence of any cytokine addition, unphosphorylated STAT2 accumulates in the nucleus (24). This was the first indication that unphosphorylated STAT2 dynamically shuttles into and out of the nucleus.
STAT2 differs from the other STATs in that it is associated with the non-STAT transcription factor IRF-9. IRF-9 binds to the coiled coil region of STAT2 independent of tyrosine phosphorylation (25, 27). IRF-9 also has a constitutive NLS within its amino terminus that is recognized by several members of the importin-α adapter family, importins-α3, -α5, and -α7 (24). For this reason we tested the possibility that the STAT2 is imported into the nucleus via complex formation with IRF-9. Evaluation of U2A cells that lack IRF-9 indicated that endogenous STAT2 remained in the cytoplasm of these cells following treatment with LMB (24). It should be noted that others have reported nuclear import of overexpressed STAT2 in cells that lack IRF-9, however it is possible that this import is mediated by interaction with phosphorylated STAT1 (57). We have found that mutations in the coiled coil domain of STAT2 that disrupt interaction with IRF-9 eliminate the nuclear import of unphosphorylated STAT2. Therefore nuclear entry of unphosphorylated STAT2 appears to be mediated by its association with IRF-9.
Following tyrosine phosphorylation, STAT2 gains the ability to heterodimerize with phosphorylated STAT1 and accumulate in the nucleus. Because STAT2 does not form tyrosine phosphorylated homodimers, heterodimerization with STAT1 is required for its ability to bind the ISRE or the GAS. Cells lacking STAT1 are defective in nuclear accumulation of phosphorylated STAT2 (24). The change in conformation that occurs with tyrosine phosphorylation of STAT1 and STAT2 and their heterodimerization leads to the gain of a functional NLS. This NLS is recognized by importin-α5, the same adapter that is specific for the STAT1 tyrosine phosphorylated homodimer (24, 51, 58). STAT2 therefore has an intrinsic conditional NLS that functions in the context of a tyrosine phosphorylated heterodimer with STAT1, and this property of STAT2 allows it to rescue a STAT1 molecule defective for nuclear import (24).
4.2 STAT2 export
Unphosphorylated STAT2 is imported into the nucleus by association with IRF-9, but subsequently it is transported back to the cytoplasm. STAT2 must therefore have a mechanism for export, and since this export is inhibited by LMB it is mediated by CRM1.
To identify a region of STAT2 responsible for nuclear export, amino and carboxyl terminal deletion mutations were linked to GFP and their cellular distribution was evaluated. The results defined a hydrophobic sequence in the carboxyl terminus critical for STAT2 nuclear export. Site-directed mutagenesis was performed and LL740/741/AA, L745A, and L751A ablated the nuclear export function. The results indicated a functional and constitutive NES in the carboxyl terminus of STAT2 amino acids 740–751 (24).
The carboxyl terminal NES of STAT2 clearly plays an active role in nuclear export of the unphosphorylated STAT2-IRF-9 complex prior to IFN-α signaling. To determine whether this NES contributes to the export of STAT2 following tyrosine phosphorylation and dimerization with STAT1, we analyzed the cellular localization of full length STAT2 with specific NES point mutations. The localization of wild type STAT2 or the STAT2 NES mutant was evaluated by fluorescence microscopy in response to IFN-α (24). Both proteins accumulated in the nucleus within minutes of IFN-α treatment. There were no significant differences in phosphorylation between the wild type and the NES mutation as judged by a Western blot to evaluate tyrosine phosphorylated STAT2 following IFN-α. But there was a difference in nuclear export. Following removal of IFN-α, wild type STAT2 redistributed to the cytoplasm by 60 minutes whereas the STAT2 NES mutant remained in the nucleus. However, after several hours the STAT2 NES mutant did redistribute back to the cytoplasm. These results indicated that the carboxyl NES plays a significant role in STAT2 export, but that there was another weaker NES that was also functional. Subsequent experiments identified a weaker NES within the DNA binding domain of STAT2 located in a similar region as that of the NES in STAT1.
In summary, STAT2 appears to have two distinct modes of nuclear entry, via IRF-9 in its unphosphorylated form, and as a heterodimer with STAT1 in its tyrosine phosphorylated and DNA binding form. However, the primary mode of nuclear export both prior to and following tyrosine phosphorylation appears to be mediated by a strong NES within the carboxyl terminus of STAT2 (Figure 5).
Summary
A successful innate immune response to viral infection requires the action of IFNs and the induced expression of genes by ISGF3 (STAT1:STAT2:IRF-9) and GAF (STAT1:STAT1). The physiological significance of these transcription factors is clearly evident in animals lacking STAT1 or STAT2, as these deficient animals succumb to infection (59–61). It is therefore not unexpected that the STATs are regulated by various mechanisms including tyrosine phosphorylation by JAKs, inhibition indirectly by SOCS, dephosphorylation of STATs by PTPases, nuclear inactivation of STATs by PIAS proteins, and in addition, regulation of STAT cellular localization by mechanisms both dependent and independent of tyrosine phosphorylation. Understanding the cellular dynamics of STAT proteins will provide information essential for the clinical intervention of diseases that are impacted by misregulation of STAT factors.
Acknowledgments
This review could not have been written without the thoughtful and skillful contributions of current and past laboratory members. Thank you all. Grant support from N.I.H. to N.C.R. is gratefully acknowledged (PO1CA2814 and RO1 CA122910).
Biography
 Nancy C. Reich, Ph.D. is a Professor of Molecular Genetics and Microbiology at Stony Brook University, New York. She received her Ph.D. from Stony Brook University studying viral-host interactions and the p53 tumor suppressor with Dr. Arnold J. Levine. Subsequently she joined Dr. James. E. Darnell, Jr. at The Rockefeller University as a Postdoctoral Associate and entered the field of interferon research. Her research is funded by N.I.H. and she has served on numerous N.I.H. advisory panels. She has been an active member in ISICR and was the 2005 co-recipient of the Milstein Award for exceptional contributions to this field. Her research interests have long centered on innate immunity and the response of cells to viral infections. Some of her professional highlights include discovery of a novel DNA binding factor activated in response to viral infection identified as IRF-3; discovery of tyrosine phosphorylation in the activation of transcription factors STAT1 and STAT2; discovery of the ability of Ras to activate STAT3 indirectly; development of a transgenic JAK/STAT reporter system in Drosophila that can be used to screen for pathway modulators; and elucidation of the mechanisms that distinguish nuclear trafficking of STATs.
Nancy C. Reich, Ph.D. is a Professor of Molecular Genetics and Microbiology at Stony Brook University, New York. She received her Ph.D. from Stony Brook University studying viral-host interactions and the p53 tumor suppressor with Dr. Arnold J. Levine. Subsequently she joined Dr. James. E. Darnell, Jr. at The Rockefeller University as a Postdoctoral Associate and entered the field of interferon research. Her research is funded by N.I.H. and she has served on numerous N.I.H. advisory panels. She has been an active member in ISICR and was the 2005 co-recipient of the Milstein Award for exceptional contributions to this field. Her research interests have long centered on innate immunity and the response of cells to viral infections. Some of her professional highlights include discovery of a novel DNA binding factor activated in response to viral infection identified as IRF-3; discovery of tyrosine phosphorylation in the activation of transcription factors STAT1 and STAT2; discovery of the ability of Ras to activate STAT3 indirectly; development of a transgenic JAK/STAT reporter system in Drosophila that can be used to screen for pathway modulators; and elucidation of the mechanisms that distinguish nuclear trafficking of STATs.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Fu XY, Schindler C, Improta T, Aebersold R, Darnell JE., Jr The proteins of ISGF-3, the interferon alpha-induced transcriptional activator, define a gene family involved in signal transduction. Proc Natl Acad Sci U S A. 1992;89(16):7840–3. doi: 10.1073/pnas.89.16.7840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Schindler C, Fu XY, Improta T, Aebersold R, Darnell JE., Jr Proteins of transcription factor ISGF-3: one gene encodes the 91-and 84- kDa ISGF-3 proteins that are activated by interferon alpha. Proc Natl Acad Sci U S A. 1992;89(16):7836–9. doi: 10.1073/pnas.89.16.7836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Fu XY. A transcription factor with SH2 and SH3 domains is directly activated by an interferon alpha-induced cytoplasmic protein tyrosine kinase(s) Cell. 1992 Jul 24;70(2):323–35. doi: 10.1016/0092-8674(92)90106-m. [DOI] [PubMed] [Google Scholar]
- 4.Schindler C, Shuai K, Prezioso VR, Darnell JE., Jr Interferon-dependent tyrosine phosphorylation of a latent cytoplasmic transcription factor [see comments] Science. 1992;257(5071):809–13. doi: 10.1126/science.1496401. [DOI] [PubMed] [Google Scholar]
- 5.Barbieri G, Velazquez L, Scrobogna M, Fellous M, Pellegrini S. Activation of the protein tyrosine kinase tyk2 by interferon alpha/beta. Eur J Biochem. 1994 Jul 15;223(2):427–35. doi: 10.1111/j.1432-1033.1994.tb19010.x. [DOI] [PubMed] [Google Scholar]
- 6.Greenlund AC, Farrar MA, Viviano BL, Schreiber RD. Ligand-induced IFN gamma receptor tyrosine phosphorylation couples the receptor to its signal transduction system (p91) Embo J. 1994 Apr 1;13(7):1591–600. doi: 10.1002/j.1460-2075.1994.tb06422.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Shuai K, Stark GR, Kerr IM, Darnell JE., Jr A single phosphotyrosine residue of Stat91 required for gene activation by interferon-gamma [see comments] Science. 1993;261(5129):1744–6. doi: 10.1126/science.7690989. [DOI] [PubMed] [Google Scholar]
- 8.Gutch MJ, Daly C, Reich NC. Tyrosine phosphorylation is required for activation of an alpha interferon-stimulated transcription factor. Proc Natl Acad Sci U S A. 1992;89(23):11411–5. doi: 10.1073/pnas.89.23.11411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mao X, Ren Z, Parker GN, Sondermann H, Pastorello MA, Wang W, et al. Structural bases of unphosphorylated STAT1 association and receptor binding. Mol Cell. 2005 Mar 18;17(6):761–71. doi: 10.1016/j.molcel.2005.02.021. [DOI] [PubMed] [Google Scholar]
- 10.Chen X, Vinkemeier U, Zhao Y, Jeruzalmi D, Darnell JE, Jr, Kuriyan J. Crystal structure of a tyrosine phosphorylated STAT-1 dimer bound to DNA. Cell. 1998 May 29;93(5):827–39. doi: 10.1016/s0092-8674(00)81443-9. [DOI] [PubMed] [Google Scholar]
- 11.Becker S, Groner B, Muller CW. Three-dimensional structure of the Stat3beta homodimer bound to DNA. Nature. 1998;394(6689):145–51. doi: 10.1038/28101. [DOI] [PubMed] [Google Scholar]
- 12.Levy DE, Darnell JE., Jr Stats: transcriptional control and biological impact. Nat Rev Mol Cell Biol. 2002 Sep;3(9):651–62. doi: 10.1038/nrm909. [DOI] [PubMed] [Google Scholar]
- 13.Leonard WJ, O’Shea JJ. Jaks and STATs: biological implications. Annu Rev Immunol. 1998;16:293–322. doi: 10.1146/annurev.immunol.16.1.293. [DOI] [PubMed] [Google Scholar]
- 14.van Boxel-Dezaire AH, Rani MR, Stark GR. Complex modulation of cell type-specific signaling in response to type I interferons. Immunity. 2006 Sep;25(3):361–72. doi: 10.1016/j.immuni.2006.08.014. [DOI] [PubMed] [Google Scholar]
- 15.Ihle JN. The Stat family in cytokine signaling. Curr Opin Cell Biol. 2001 Apr;13(2):211–7. doi: 10.1016/s0955-0674(00)00199-x. [DOI] [PubMed] [Google Scholar]
- 16.ten Hoeve J, de Jesus Ibarra-Sanchez M, Fu Y, Zhu W, Tremblay M, David M, et al. Identification of a nuclear Stat1 protein tyrosine phosphatase. Mol Cell Biol. 2002 Aug;22(16):5662–8. doi: 10.1128/MCB.22.16.5662-5668.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wormald S, Hilton DJ. Inhibitors of cytokine signal transduction. J Biol Chem. 2004 Jan 9;279(2):821–4. doi: 10.1074/jbc.R300030200. [DOI] [PubMed] [Google Scholar]
- 18.Shuai K. Regulation of cytokine signaling pathways by PIAS proteins. Cell Res. 2006 Feb;16(2):196–202. doi: 10.1038/sj.cr.7310027. [DOI] [PubMed] [Google Scholar]
- 19.Reich NC, Liu L. Tracking STAT nuclear traffic. Nat Rev Immunol. 2006 Aug;6(8):602–12. doi: 10.1038/nri1885. [DOI] [PubMed] [Google Scholar]
- 20.McBride KM, Reich NC. The ins and outs of STAT1 nuclear transport. Sci STKE. 2003 Aug 12;2003(195):RE13. doi: 10.1126/stke.2003.195.re13. [DOI] [PubMed] [Google Scholar]
- 21.McBride KM, Banninger G, McDonald C, Reich NC. Regulated nuclear import of the STAT1 transcription factor by direct binding of importin-alpha. Embo J. 2002 Apr 2;21(7):1754–63. doi: 10.1093/emboj/21.7.1754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Meyer T, Begitt A, Lodige I, van Rossum M, Vinkemeier U. Constitutive and IFN-gamma-induced nuclear import of STAT1 proceed through independent pathways. Embo J. 2002 Feb 1;21(3):344–54. doi: 10.1093/emboj/21.3.344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Koster M, Hauser H. Dynamic redistribution of STAT1 protein in IFN signaling visualized by GFP fusion proteins. Eur J Biochem. 1999;260(1):137–44. doi: 10.1046/j.1432-1327.1999.00149.x. [DOI] [PubMed] [Google Scholar]
- 24.Banninger G, Reich NC. STAT2 nuclear trafficking. J Biol Chem. 2004 Sep 17;279(38):39199–206. doi: 10.1074/jbc.M400815200. [DOI] [PubMed] [Google Scholar]
- 25.Martinez-Moczygemba M, Gutch MJ, French DL, Reich NC. Distinct STAT structure promotes interaction of STAT2 with the p48 subunit of the interferon-alpha-stimulated transcription factor ISGF3. J Biol Chem. 1997;272(32):20070–6. doi: 10.1074/jbc.272.32.20070. [DOI] [PubMed] [Google Scholar]
- 26.Veals SA, Schindler C, Leonard D, Fu XY, Aebersold R, Darnell JE, Jr, et al. Subunit of an alpha-interferon-responsive transcription factor is related to interferon regulatory factor and Myb families of DNA-binding proteins. Mol Cell Biol. 1992;12(8):3315–24. doi: 10.1128/mcb.12.8.3315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lau JF, Parisien JP, Horvath CM. Interferon regulatory factor subcellular localization is determined by a bipartite nuclear localization signal in the DNA-binding domain and interaction with cytoplasmic retention factors. Proc Natl Acad Sci U S A. 2000;97(13):7278–83. doi: 10.1073/pnas.97.13.7278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Taniguchi T, Ogasawara K, Takaoka A, Tanaka N. IRF family of transcription factors as regulators of host defense. Annu Rev Immunol. 2001;19:623–55. doi: 10.1146/annurev.immunol.19.1.623. [DOI] [PubMed] [Google Scholar]
- 29.Levy D, Reich N, Kessler D, Pine R, Darnell JE., Jr Transcriptional regulation of interferon-stimulated genes: a DNA response element and induced proteins that recognize it. Cold Spring Harb Symp Quant Biol. 1988;53(Pt 2):799–802. doi: 10.1101/sqb.1988.053.01.090. [DOI] [PubMed] [Google Scholar]
- 30.Reich N, Evans B, Levy D, Fahey D, Knight E, Jr, Darnell JE., Jr Interferon-induced transcription of a gene encoding a 15-kDa protein depends on an upstream enhancer element. Proc Natl Acad Sci U S A. 1987;84(18):6394–8. doi: 10.1073/pnas.84.18.6394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Reich NC, Darnell JE., Jr Differential binding of interferon-induced factors to an oligonucleotide that mediates transcriptional activation. Nucleic Acids Res. 1989;17(9):3415–24. doi: 10.1093/nar/17.9.3415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gilmour KC, Reich NC. Signal transduction and activation of gene transcription by interferons. Gene Expr. 1995;5(1):1–18. [PMC free article] [PubMed] [Google Scholar]
- 33.Decker T, Lew DJ, Mirkovitch J, Darnell JE., Jr Cytoplasmic activation of GAF, an IFN-gamma-regulated DNA-binding factor. Embo J. 1991 Apr;10(4):927–32. doi: 10.1002/j.1460-2075.1991.tb08026.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rout MP, Aitchison JD. The nuclear pore complex as a transport machine. J Biol Chem. 2001 May 18;276(20):16593–6. doi: 10.1074/jbc.R100015200. [DOI] [PubMed] [Google Scholar]
- 35.Davis LI. The nuclear pore complex. Annu Rev Biochem. 1995;64:865–96. doi: 10.1146/annurev.bi.64.070195.004245. [DOI] [PubMed] [Google Scholar]
- 36.Pemberton LF, Paschal BM. Mechanisms of receptor-mediated nuclear import and nuclear export. Traffic. 2005 Mar;6(3):187–98. doi: 10.1111/j.1600-0854.2005.00270.x. [DOI] [PubMed] [Google Scholar]
- 37.Macara IG. Transport into and out of the Nucleus. Microbiol Mol Biol Rev. 2001;65(4):570–94. doi: 10.1128/MMBR.65.4.570-594.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gorlich D, Kutay U. Transport between the cell nucleus and the cytoplasm. Annu Rev Cell Dev Biol. 1999;15:607–60. doi: 10.1146/annurev.cellbio.15.1.607. [DOI] [PubMed] [Google Scholar]
- 39.Bednenko J, Cingolani G, Gerace L. Nucleocytoplasmic transport: navigating the channel. Traffic. 2003 Mar;4(3):127–35. doi: 10.1034/j.1600-0854.2003.00109.x. [DOI] [PubMed] [Google Scholar]
- 40.Chook YM, Blobel G. Karyopherins and nuclear import. Curr Opin Struct Biol. 2001 Dec;11(6):703–15. doi: 10.1016/s0959-440x(01)00264-0. [DOI] [PubMed] [Google Scholar]
- 41.Yoneda Y. Nucleocytoplasmic protein traffic and its significance to cell function. Genes Cells. 2000 Oct;5(10):777–87. doi: 10.1046/j.1365-2443.2000.00366.x. [DOI] [PubMed] [Google Scholar]
- 42.Mattaj IW, Englmeier L. Nucleocytoplasmic transport: the soluble phase. Annu Rev Biochem. 1998;67:265–306. doi: 10.1146/annurev.biochem.67.1.265. [DOI] [PubMed] [Google Scholar]
- 43.Lange A, Mills RE, Lange CJ, Stewart M, Devine SE, Corbett AH. Classical nuclear localization signals: definition, function, and interaction with importin alpha. J Biol Chem. 2007 Feb 23;282(8):5101–5. doi: 10.1074/jbc.R600026200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kudo N, Wolff B, Sekimoto T, Schreiner EP, Yoneda Y, Yanagida M, et al. Leptomycin B inhibition of signal-mediated nuclear export by direct binding to CRM1. Exp Cell Res. 1998;242(2):540–7. doi: 10.1006/excr.1998.4136. [DOI] [PubMed] [Google Scholar]
- 45.McBride KM, McDonald C, Reich NC. Nuclear export signal located within theDNA-binding domain of the STAT1transcription factor. Embo J. 2000;19(22):6196–206. doi: 10.1093/emboj/19.22.6196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lillemeier BF, Koster M, Kerr IM. STAT1 from the cell membrane to the DNA. Embo J. 2001 May 15;20(10):2508–17. doi: 10.1093/emboj/20.10.2508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Begitt A, Meyer T, van Rossum M, Vinkemeier U. Nucleocytoplasmic translocation of Stat1 is regulated by a leucine-rich export signal in the coiled-coil domain. Proc Natl Acad Sci U S A. 2000;97(19):10418–23. doi: 10.1073/pnas.190318397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Shuai K, Horvath CM, Huang LH, Qureshi SA, Cowburn D, Darnell JE., Jr Interferon activation of the transcription factor Stat91 involves dimerization through SH2-phosphotyrosyl peptide interactions. Cell. 1994;76(5):821–8. doi: 10.1016/0092-8674(94)90357-3. [DOI] [PubMed] [Google Scholar]
- 49.Melen K, Kinnunen L, Julkunen I. Arginine/lysine-rich structural element is involved in IFN-induced nuclear import of STATs. J Biol Chem. 2001;9:9. doi: 10.1074/jbc.M008821200. [DOI] [PubMed] [Google Scholar]
- 50.Sekimoto T, Imamoto N, Nakajima K, Hirano T, Yoneda Y. Extracellular signal-dependent nuclear import of Stat1 is mediated by nuclear pore-targeting complex formation with NPI-1, but not Rch1. Embo J. 1997;16(23):7067–77. doi: 10.1093/emboj/16.23.7067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Melen K, Fagerlund R, Franke J, Kohler M, Kinnunen L, Julkunen I. Importin alpha nuclear localization signal binding sites for STAT1, STAT2, and influenza A virus nucleoprotein. J Biol Chem. 2003 Jul 25;278(30):28193–200. doi: 10.1074/jbc.M303571200. [DOI] [PubMed] [Google Scholar]
- 52.Marg A, Shan Y, Meyer T, Meissner T, Brandenburg M, Vinkemeier U. Nucleocytoplasmic shuttling by nucleoporins Nup153 and Nup214 and CRM1-dependent nuclear export control the subcellular distribution of latent Stat1. J Cell Biol. 2004 Jun 21;165(6):823–33. doi: 10.1083/jcb.200403057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Chatterjee-Kishore M, Wright KL, Ting JP, Stark GR. How Stat1 mediates constitutive gene expression: a complex of unphosphorylated Stat1 and IRF1 supports transcription of the LMP2 gene. Embo J. 2000 Aug 1;19(15):4111–22. doi: 10.1093/emboj/19.15.4111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ahmed CM, Johnson HM. IFN-gamma and its receptor subunit IFNGR1 are recruited to the IFN-gamma-activated sequence element at the promoter site of IFN-gamma-activated genes: evidence of transactivational activity in IFNGR1. J Immunol. 2006 Jul 1;177(1):315–21. doi: 10.4049/jimmunol.177.1.315. [DOI] [PubMed] [Google Scholar]
- 55.Mowen K, David M. Regulation of STAT1 nuclear export by Jak1. Mol Cell Biol. 2000;20(19):7273–81. doi: 10.1128/mcb.20.19.7273-7281.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Haspel RL, Darnell JE., Jr A nuclear protein tyrosine phosphatase is required for the inactivation of Stat1. Proc Natl Acad Sci U S A. 1999;96(18):10188–93. doi: 10.1073/pnas.96.18.10188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Frahm T, Hauser H, Koster M. IFN-type-I-mediated signaling is regulated by modulation of STAT2 nuclear export. J Cell Sci. 2006 Mar 15;119(Pt 6):1092–104. doi: 10.1242/jcs.02822. [DOI] [PubMed] [Google Scholar]
- 58.Fagerlund R, Melen K, Kinnunen L, Julkunen I. Arginine/lysine-rich nuclear localization signals mediate interactions between dimeric STATs and importin alpha 5. J Biol Chem. 2002 Aug 16;277(33):30072–8. doi: 10.1074/jbc.M202943200. [DOI] [PubMed] [Google Scholar]
- 59.Park C, Li S, Cha E, Schindler C. Immune response in Stat2 knockout mice. Immunity. 2000 Dec;13(6):795–804. doi: 10.1016/s1074-7613(00)00077-7. [DOI] [PubMed] [Google Scholar]
- 60.Durbin JE, Hackenmiller R, Simon MC, Levy DE. Targeted disruption of the mouse Stat1 gene results in compromised innate immunity to viral disease. Cell. 1996 Feb 9;84(3):443–50. doi: 10.1016/s0092-8674(00)81289-1. [DOI] [PubMed] [Google Scholar]
- 61.Meraz MA, White JM, Sheehan KC, Bach EA, Rodig SJ, Dighe AS, et al. Targeted disruption of the Stat1 gene in mice reveals unexpected physiologic specificity in the JAK-STAT signaling pathway. Cell. 1996 Feb 9;84(3):431–42. doi: 10.1016/s0092-8674(00)81288-x. [DOI] [PubMed] [Google Scholar]