Abstract
Efficient iron acquisition is critical for an invading microbe’s survival and virulence. Most of the iron in mammals is incorporated into heme, which can be plundered by certain bacterial pathogens as a nutritional iron source. Utilization of exogenous heme by bacteria involves the binding of heme or hemoproteins to the cell surface receptors, followed by the transport of heme into cells. Once taken into the cytosol, heme is presented to heme oxygenases where the tetrapyrrole ring is cleaved in order to release the iron. Some Gram-negative bacteria also secrete extracellular heme-binding proteins called hemophores, which function to sequester heme from the environment. The heme transport genes are often genetically linked as gene clusters under Fur (Ferric uptake repressor) regulation. This review discusses the gene clusters and proteins involved in bacterial heme acquisition, transport and processing processes, with special focus on the heme coordination, protein structures and mechanisms underlying heme transport.
Keywords: Heme, transport, structure, bacteria, iron, pathogen
Heme, iron and bacteria
Iron is an essential element for almost all organisms, including bacteria. As a catalytic center for redox reactions in many enzymes, iron facilitates numerous cellular processes such as electron transport, peroxide reduction and nucleotide biosynthesis [1–4]. Upon entry into the mammalian host, bacterial pathogens have to acquire iron from tissues to survive [5]. Almost all bacterial pathogens require iron to be infectious [6]. However, free iron is scarcely available inside animal hosts (free iron concentration at ~ 10−18 M within the human host) [7, 8], albeit it is the most abundant metal on the Earth [9]. Most of the iron in the human body is sequestered by high affinity iron-binding proteins, such as transferrin (Tf), lactoferrin (Lf) and the iron storage protein ferritin, or by incorporation into a protoporphyrin ring as heme in hemoproteins [5, 10, 11].
Bacteria have developed sophisticated mechanisms for the uptake of iron from the environment. A common mechanism, seen in many bacteria, is to secrete siderophores to chelate iron from their surroundings [3]. A second mechanism, found only in pathogenic bacteria, is to steal iron from the host by capturing iron from Tf, Lf or ferritin via specific outer membrane receptors [3, 4, 10, 12, 13]. A third mechanism, also identified mostly in bacterial pathogens [14–16] is to take up iron either from free heme or from heme-containing proteins, such as hemoglobin or hemopexin.
Heme refers to the ferrous iron (Fe2+) bound to tetrapyrrole, a macrocyclic porphyrin; whereas the oxidized form (Fe3+) is known as hemin. However, both are commonly called “heme”, so we will apply this convention throughout this paper. As a prosthetic group in many proteins and enzymes, heme plays significant roles in many fundamental biological processes[17], including O2 storage and transport (e.g. myoglobin and hemoglobin), respiration and electron transfer (e.g. cytochromes), activation of O-O bond (e.g. P450 enzymes and peroxidase) [18], signal transduction and gas sensing (e.g. CooA, nitric oxide synthase (NOS) and soluble guanylate cyclase (sGC)) [19], and control of gene expression and microRNA processing [20]. Heme can also serve as a source of both iron and porphyrin for the microbes. Recently, it has been discovered that pathogenic bacteria Staphylococcus aureus significantly prefers heme iron over transferrin iron during the early stage of infection [21].
Heme is a lipophilic molecule with low molecular weight (616.48 Da), so it can easily intercalate into membranes and thus impair lipid bilayers and organelles, such as mitochondria and nuclei, and destabilize the cytoskeleton [22]. Furthermore, because heme catalyzes the formation of reactive oxygen species, accumulation of heme in cells will result in cellular damage, oxidative stress and tissue injury [23, 24]. Therefore, free heme is also poorly tolerated by cells. When heme-containing molecules are degraded, released heme will be rapidly bound by molecules such as hemopexin (Kd ~ 10−12 M) and serum albumin (Kd ~ 10−8 M) [25]. In the presence of the scavenging proteins, the concentration of free heme within the animal host is maintained at very low levels [26].
Heme sources
The total body intracellular iron pool (~ 4 g) consists of heme which bound to hemoglobin (2.67 g), myoglobin (0.2 g), some heme enzymes (0.08 g), insoluble ferric irons that were stored in ferritin (1 g), and a poorly defined labile iron compartment (0.07 g) [27]. Hemoglobin and myoglobin are the most abundant heme sources in the body. Free hemoglobin, like free heme, is not tolerated by the body [28], and therefore upon being liberated by intravascular hemolysis from erythrocytes, hemoglobin dimers are bound with high affinity (Kd ~ 10−12 M) by haptoglobin, a serum heterotetrameric glycoprotein [29, 30]. Then the haptoglobin-hemoglobin complexes are subsequently delivered to the reticulo-endothelial system by CD163 receptor-mediated endocytosis [31]. Although not all bacterial pathogens are able to use haptoglobin-bound hemoglobin as an iron source, it has been reported that Neisseria spp., Hemophilus influenzae and Porphyromonas gingivalis are capable of utilizing iron through specific haptoglobin-hemoglobin receptors [32–35].
Upon heme is released from hemoproteins such as hemoglobin, as occurs in the erythrocyte lysis, the free heme will be sequestered immediately by hemopexin, which is a 60-kDa plasma glycoprotein with a high heme affinity (Kd ~ 10−13 M), in an equimolar complex [30, 36, 37]. Hemopexin is a major heme transportation vehicle in the plasma, functioning to prevent heme-mediated oxidative stress and heme-bound iron loss. The heme-hemopexin complex is routed to the liver, where apo-hemopexin is recycled after heme is discharged intracellularly.
Human serum albumin (HSA), the most abundant plasma protein (~ 640 μM), which has a lower affinity (Kd ~ 10−8 M) for heme compared to hemopexin [38], may also contribute to heme transport [39, 40]. HSA consists of three homologous domains (I, II and III). Each of which is made up of two separate subdomains (designated A and B) that are connected by a random coil. The crystal structure shows that heme binds to a hydrophobic, narrow D-shaped cavity in subdomain IB and the propionate side chains of heme are coordinated with a triad of basic residues at the entrance of the pocket. The central iron atom of heme is coordinated by Tyr161 [25, 41].
It is interesting to note that the high and low density lipoproteins (HDL and LDL, respectively), which are the most oxidatively intolerant components in plasma, also bind to heme with high affinity (Kd ~ 10−11 M to 10−10 M), and the binding is kinetically faster than serum albumin and hemopexin [42], although the role of the transient heme binding to HDL and LDL is not clear yet.
The heme-containing proteins, such as hemoglobin, myoglobin, haptoglobin–hemoglobin complex, HSA, HDL, LDL, or peroxidases, catalases, cytochrome P450s and cytochromes, provide multiple potential iron sources to the infectious bacteria seeking this essential nutrient.
An overview of heme uptake systems in Gram-negative bacteria
Three major heme acquisition systems have been identified in Gram-negative bacteria. The well-characterized system involves direct binding of heme or heme proteins (e.g. hemopexin, hemoglobin, hemoglobin-haptoglobin) to specific outer membrane receptors on the bacterial surface (named “direct heme uptake system” here). The heme, is then transported into the cell via ATP-binding cassette (ABC) transporters. The best studied systems in this category consist of the hemR-hemSTUV system in Yersinia enterocolitica [43], the hmuRSTUV system in Yersinia pestis [44], shuASTUV system of Shigella dysenteriae [45, 46], and the phuRSTUVW system in Pseudomonas aeruginosa [47–49]. The second heme uptake system is comprised of an outer membrane receptor, an ABC transporter and extracellular hemophores. Best known in the hasADEB system in Serratia marcescens [16], hemophores function to take up free heme or extract heme from hemoproteins in the external medium and shuttle it to the cell surface, where it interacts with a TonB-dependent hemophore specific outer membrane receptor [50, 51]. A few researchers classified the bipartite hemoglobin receptor HpuAB in Neisseria meningitidis as a third heme acquisition system [52]. HpuAB is composed of a TonB-dependent outer membrane receptor HpuB and an accessory outer membrane lipoprotein HpuA [34, 53], which is analogous to the bipartite transferrin receptor Tbp1 and Tbp2 [54]. It has been demonstrated that the N. meningitidis HpuAB bipartite receptor is capable of transporting intact porphyrin from hemoglobin and haptoglobin-hemoglobin complexes [55]. It is of interest to note that heme can passively diffuse across model lipid bilayers, although the outer membrane permeability in Gram-negative bacteria is less than 600 Da [56].
Hemophore-mediated heme uptake systems
HasA hemophore
Two types of hemophore systems have been reported so far. One is HasA (heme acquisition system), which has been identified in Serratia marcescens (Figure 1) [57–59], Pseudomonas aeruginosa [60, 61], Pseudomonas fluorescens [60, 62], Yersinia pestis [63], and Yersinia enterocolitica [43, 64]. The other type of hemophore is HxuA (heme/hemopexin utilization) which has been described only in Hemophilus influenzae [65, 66], a species that lacks the heme biosynthetic pathway and requires exogenous heme for aerobic growth.
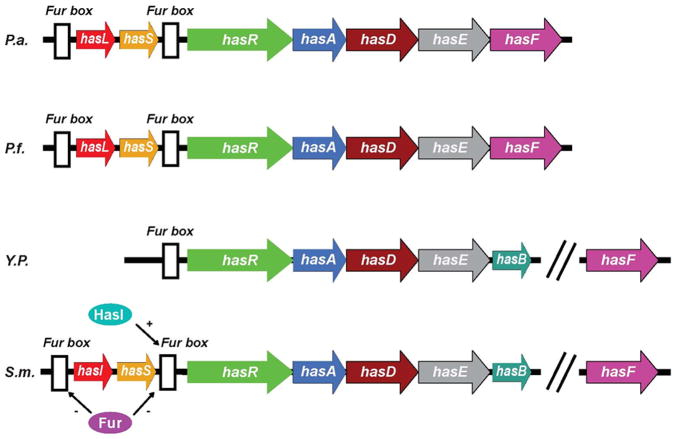
Figure 1.
Genetic organization of the has systems in different bacteria. P.a., Pseudomonas aeruginosa; P.f., Pseudomonas fluorescens; Y.p., Yersinia pestis; S.m., Serratia marcescens. HasI and hasS encode sigma and antisigma factors, respectively. The white boxes indicate consensus Fur boxes. The has operon is directly controlled by Fur (Ferric uptake regulator) [26] and positively regulated by sigma and antisigma factors [158]. HasA is secreted by an ABC transporter consisting of three envelope proteins: HasD, HasE and HasF. HasD, an inner membrane ATPase (the ABC protein) provides the energy for the substrate export; HasE is another inner membrane component and also a membrane fusion protein; HasF, an outer membrane component, is homologous to TolC, which creates a channel in ~ 30Å diameter through the periplasmic space and the outer membrane through which the secreted protein is most likely translocated [159]. Like most proteins secreted by an ABC exporter, hemophores have an α-helical C-terminal secretion signal which can be accessed by SecB, a cytoplasmic chaperone required for hemophore secretion [160]. The secretion signal is recognized by the ABC protein HasD, through modulating its ATPase activity, a HasA-HasDEF multi-protein complex will be formed [161]. Once translocated into the extracellular medium, HasA will bind heme and return it to the HasR receptor. Apo-HasA will be released into the extracellular medium through interaction with TonB-dependent outer membrane receptor.
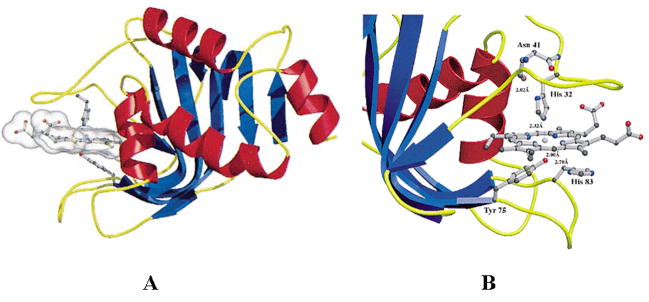
The HasA from S. marcescens (HasASM) was the first identified hemophore. It is a 19 kDa monomer that binds one b-type heme per molecule with high affinity (Kd ~ 10−11 M) [67]. The crystal structure of HasASM has been determined at 1.9 Å resolution [58] (Figure 2a). The HasASM shows an original α/β fold in which seven β-strands form an antiparallel β-sheet on one face of the protein, and the four α-helices are positioned on the other face. The heme is held between two loops and is located at the inferface between the α- and β-fold of the molecule (Figure 2b). In contrast to the bis-histidinyl ligation based on EPR studies [68], heme iron is ligated by His32 and Tyr75 [58]. The possible heme binding and release mechanism has been proposed to be involved in a tight hydrogen bond between the Nδ of His 83 and the phenolate group of Tyr75. If Tyr75 is deprotonated, heme ligation is favored. In contrast, heme can be released by weakening the Fe-Tyr75 bond through H-bonding of the Tyr75 phenolate group with His83 Nδ [58]. His83, which has been confirmed by mutagenesis study to play a critical role in the stability of axial coordination, was suggested to be an alternative iron ligand in the absence of Tyr75 [67, 69]. Recently, it has also been proposed to be involved in heme delivery to the receptor [70].
Figure 2.
Crystal structures of holo-HasASM (PDB ID: 1b2v). A: Ribbon diagram with helices colored in red and strands in blue. Heme and the ligands of heme are shown in ball-and-stick representation. B: View of the residues in the heme binding site. Adapted from [58].
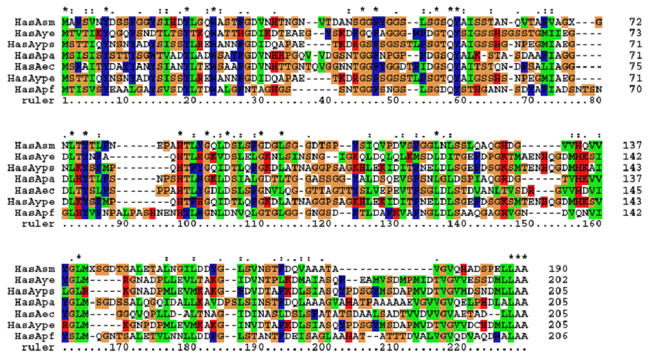
Sequence alignment (Figure 3) reveals that HasAs share about 30–50% identity. The high variable regions are the two heme binding loops H1 (ruler: 30–44) and H2 (ruler: 85–97), and the C-terminal stretch. The relative high flexibility of the heme binding pocket formed by H1 and H2 is suggested to be important for heme recognition and/or for the recognition by the receptor HasR [59]. The last four residues in the C-terminus containing the secretion information is very conserved. However, one of the heme ligation residues, His32, is not conserved and is replaced by either Gln or Asp. His83 and Tyr75 are conserved among all the species. Taken together, the heme ligation environment among HasAs is similar, but may have some slight differences.
Figure 3.
Multiple sequence alignment of HasAs from Serratia marcescens, Yersinia enterocolitica strain 8081, Yersinia pseudotuberculosis, Pseudomonas aeruginosa PAO1, Erwinia carotovora subsp. Atroseptica, Yersinia pestis and Pseudomonas fluorescens.
The receptors of HasA hemophore
Like other outer membrane receptors, the hemophore receptor, HasR, was proposed to be dependent on HasB (a TonB-like protien) for energy [71–73]. Based on sequence and functional similarities with other known TonB-dependent receptors, a HasR structure was predicted to be composed of a 99-residue amino-terminal extension, followed by a plug domain and a β-barrel domain with lumen closed by the plug [74]. HasR is highly homologous to other heme and hemoprotein receptors, in particular the conserved FRAP/NPL sequences in the β-barrel and two conserved histidines (His189 in the plug and His603 in the barrel) [75]. HasR also functions as a heme receptor, taking up free heme (Kd ~ 10−6 M) and hemoglobin-bound heme, albeit less efficiently than taking up hemophore-bound heme (Kd ~ 10−9 M) [26, 69].
HasA has a markedly higher affinity (Kd ~ 10−11 M) for heme than its receptor HasR (Kd ~ 10−6 M) [76], which raises the question of how heme could transfer from a high-affinity protein to a lower-affinity protein. The interactions between HasA, hemoglobin and HasR have been characterized [68, 77]. It has been biochemically demonstrated that heme transfer in vitro from HasA to HasR is energy- and TonB-independent and is only driven by protein-protein interactions [78]. The binding of HasA and HasR is mediated by two independent hemophore-binding regions, β strands 51–60 and 95–105 [79]. Based on the three-dimensional HasR model and mutagenesis studies, three loops L6, L8, and L9 of HasR are proposed to contribute either directly or indirectly to one or several HasA binding sites. By investigating the HasA with direct-detected 13C NMR, Caillet-Saguy proposed that upon binding to HasR, the high-spin/low-spin equilibrium shifts toward the high-spin species. The affinity of HasA for heme in the HasA-HasR complex would become lower than that of HasR, thus allowing heme transfer to HasR [70].
HxuA hemophore
The gene cluster hxuCBA in Hemophilus influenzae type b (Hib), a species that lacks the heme biosynthetic pathway and requires exogenous heme for aerobic growth, has been shown to be necessary for utilization of hemopexin-bound heme [65, 80]. Sequence analysis of the hxuA gene and N-terminal amino acid analysis of the HxuA protein revealed that the mature protein was 99 kDa with a leader peptide removed from a 101 kDa precursor [81]. HxuA was first identified to bind heme-hemopexin [65]. The HxuA-heme-hemopexin complex is presumably bound by the bacterial cell and the heme is internalized; the outer-membrane HxuA receptor has not been identified albeit it may be HxuC [66, 82]. The exact mechanism by which Hib acquires heme from the heme-hemopexin complex is yet unknown.
The 60-kDa HxuB protein was proposed to be involved in the release of the soluble HxuA molecule from the Hib cell. HxuA and HxuB were believed to be involved only in the heme-hemopexin utilization process [82]. While the 78-kDa outer membrane protein HxuC is required for Hib to acquire the very low concentrations of free heme [80], heme-albumin complex [82], or hemoglobin in the absence of the Hgps [83]. All the three genes in hxuABC are essential for Hib to grow on free heme or heme-hemopexin medium. The Hxu system of Hib differs from the Has system of S. marcescens in that HxuA actually extends the range of the substrates rather than simply increasing its efficiency [66].
Direct heme uptake systems in Gram-negative bacteria
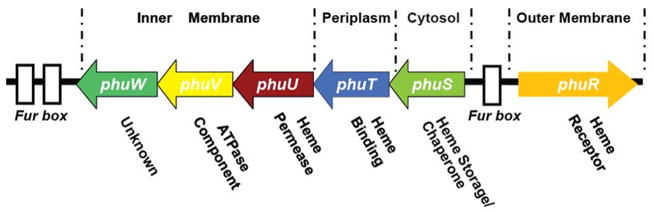
The direct heme uptake systems in Gram-negative bacteria are similar and are encoded by a Fur regulated gene cluster (Figure 4 shows the gene cluster from P. aeruginosa [84]). This system involves a single outer membrane heme receptor (e.g., PhuR) (Figure 5A), a periplasmic heme transport protein (e.g., PhuT) (Figure 5D), inner membrane proteins (e.g., PhuUVW) (see Figure 5C, D and E) typical for an ABC transport system, and a cytoplasmic protein (PhuS, possible for heme storage/chaperone) (See Figure 11, the crystal structure of a PhuS analogue, HemS). Based on the current understanding, a possible heme acquisition and transport pathway is illustrated in Figure 6. Bacterial pathogens secrete hemolysin to lyse red blood cells and release hemoglobin (80–800 nM) into the serum. Hemoglobin is rapidly complexed by the excess quantities of serum haptoglobin (5–20 μM) and delivered to the liver for removal. Heme, after being released from the hemoglobin (mechanism is unclear) is rapidly sequestered by hemopexin (12 μM, with a high affinity to heme, Kd ~ 10−13 M), or by excess quantities of serum albumin (640 μM, Kd ~ 10−8 M). The bacteria heme receptor (PhuR) can recognize heme, hemoglobin, haptoglobin-hemoglobin, hemopexin, and myoglobin, assimilating the heme and passing it via a TonB dependent process to the periplasmic heme transporter (PhuT), which is proposed to shuttle the heme between the outer and inner membrane [49]. Heme is then passed to inner membrane ATP-dependent permeases (PhuUVW) which transfer the heme to a cytoplasmic heme-binding protein (PhuS) [47, 85]. Once internalized into the cytosol, heme is rapidly transferred to heme oxygenase (HO) which opens the porphyrin ring producing biliverdin, carbon monoxide and iron [85, 86].
Figure 4.
Map of the P. aeruginosa heme uptake locus containing the phuR gene and the phuSTUVW operon. Three Fur binding elements are shown as white boxes (adapted from [49]).
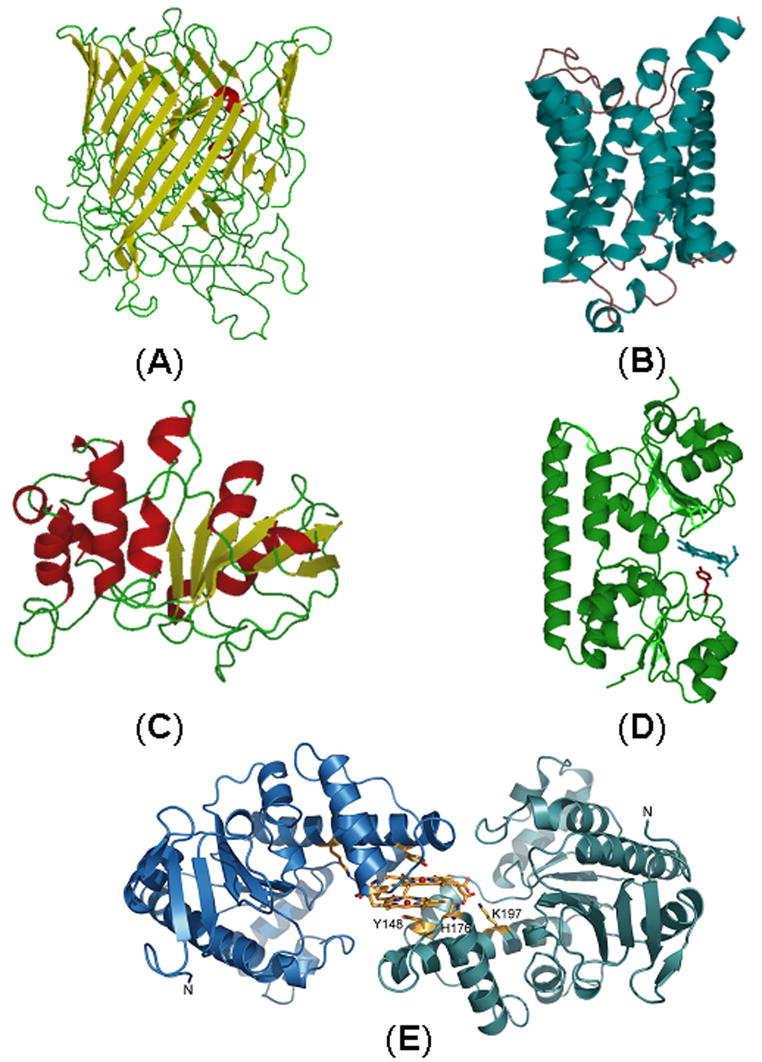
Figure 5.
Homology models and structures of heme transport proteins of P. aeruginosa in the phu locus: PhuR (A); PhuU (B); PhuV (C); Crystal structure of PhuT (D) [48] and Crystal structure of a PhuW analogue, ChaN (adapted from [123]) (E).
Figure 6.
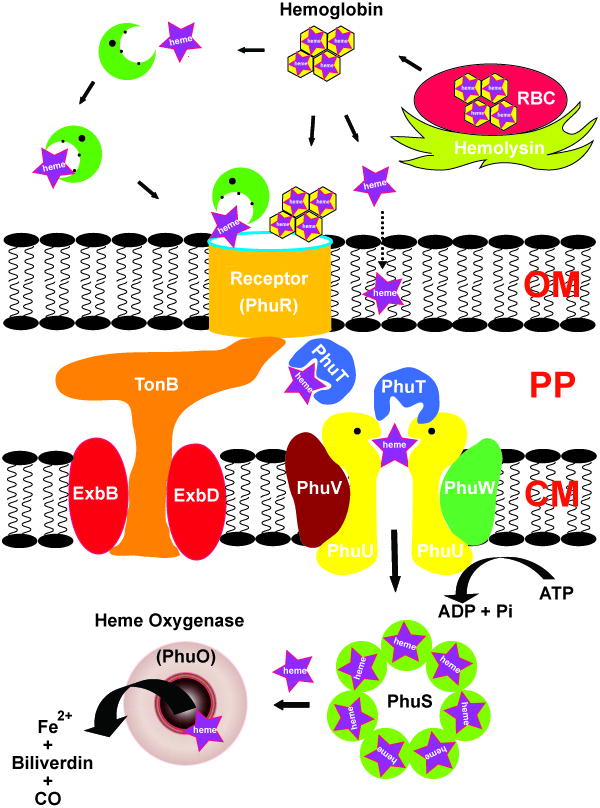
Carton showing the organization and the key elements of heme (purple star) acquisition systems in Gram negative bacteria P. aeruginosa (adpted from [49]).
Heme transport across the outer membrane
In Gram-negative bacteria, specific outer membrane receptors sequester heme from free heme or host hemoproteins and bring it into the periplasmic space via a TonB-dependent process. The receptors identified so far include heme receptors, hemopexin receptors, hemoglobin (Hb) and hemoglobin-haptoglobin (Hb-Hp) receptors. Over thirty outer membrane heme receptors have been reported from a wide variety of Gram-negative bacteria [87]. The receptors range from 70 kDa to 120 kDa in size, and vary in their binding diversity, from interacting with a large number of ligands to only one or a few. Heme receptors have been subdivided into three categories: the first group recognizes heme; the second group binds host hemoproteins; and the third group interacts with hemophores [87].
The heme receptor HemR from Y. enterocolitica has been most investigated [75, 87]. Mutational analysis has postulated that heme-binding involves two conserved histidine residues, His128 and His461 which are located between two conserved motifs, the FRAP and NPNL boxes, identified by comparison of the HemR sequence with other heme receptors [75]. HemR also recognizes other hemoproteins such as, myoglobin, hemoglobin, hemopexin, haptoglobin-hemoglobin and heme-albumin complexes. Similar to HemR, outer membrane heme receptor BhuR in Bordetella pertussis also recognizes heme, hemoglobin, haptoglobin-hemoglobin and the heme-albumin complex. Mutation of the bhuR gene makes B. pertussis incapable of utilizing any of these compounds, suggesting that BhuR is essential for the heme uptake system [52]. However, in contrast to HemR, the BhuR receptor from B. pertussis and B. bronchiseptica as well as other heme receptors PfhR (P. fluorescens) and PhuR (P. aeruginosa) are predicted to lack the conserved histidine residue in the FRAP and NPNL motifs [52, 88], suggesting that the heme binding mechanism between different heme receptors may be somewhat different.
Some hemophore receptors may also function as heme receptors, such as HasR in S. marcescens, not only binding hemophore-bound heme, but also taking up free and hemoglobin-bound heme [26].
Hemopexin receptors are not ubiquitously expressed [89]. It has been reported that hemophore HxuA in Hib, is not only released into the culture supernatant, but also presented on the bacterial cell surface as an outer membrane hemopexin receptor, which is regulated by the availability of iron in the growth medium [66, 80, 90]. In response to iron depleted conditions, H. influenzae 706705 and DL42 do not express the 99-kDa hemopexin receptor [65]. Wong et al. also suggested that some H. influenzae strains could possess at least two hemopexin receptors and the expression of which is determined by the prevailing growth medium [90].
Two Hb receptors have been described in Neisseria meningitidis, HmbR and HpuB. HmbR is an 89.5 kDa single-component TonB-dependent outer membrane receptor. It has been identified that the N. meningitidis hmbR mutant was unable to use Hb but could still use heme as the sole iron source [91]. Further evidence demonstrated that HmbR is not involved in the acquisition of iron from Hb-Hp or heme [55]. Studies also suggested that the two-component receptor HpuAB in N. meningitidis, composed of HpuB (a TonB-dependent transport protein) and the lipoprotein HpuA, was required for the utilization of either Hb or Hb-Hp complex [34, 53, 55].
Three Hb/Hb-Hp receptors, HgpA, HgpB and HgpC, have been identified from H. influenzae HI689. Expression of any one of these three proteins is adequate to support growth with Hb-Hp as the heme source [32, 92, 93]. Simpson and co-workers characterized a TonB-dependent outer membrane receptor HmuR (Heme utilization receptor) in Porphyromonas gingivalis [88]. HmuR, in concert with another outer membrane putative heme-binding lipoprotein HmuY [94], is involved in the utilization of both heme and Hb in P. gingivalis. HmuR shares extensive sequence homology with TonB-dependent outer membrane heme/Hb receptors of other Gram-negative bacteria, such as HemR of Y. enterocolitica and ShuA of S. dysenteriae [88]. The N-terminal regions of the hmuR gene are 100% identical with the previously reported hemR gene, but their C-terminal regions are strikingly different [95]. The role of the hmuR gene in heme uptake has been well studied [88, 96, 97]. It has been proposed that HmuR may bind heme through histidine residues [56]. A three-dimensional homology model of HmuR suggested a β-barrel structure with two distinct domains. The first domain is a β-barrel with long loops on the extracellular side and short turns on the periplasmic side. The second domain is a globular amino-terminal domain that folds into the barrel pore [98]. This barrel-like structure has been suggested to be necessary for the heme transport.
Recently, a 73 kDa outer membrane receptor ShuA of S. dysenteriae has been characterized [46]. It has been proposed that oxidized hemoglobin is the most likely physiological substrate for ShuA. Mutagenesis studies demonstrate that His-86 and His420 are essential for substrate recognition, heme coordination and transfer.
The TonB-ExbB-ExbD complex
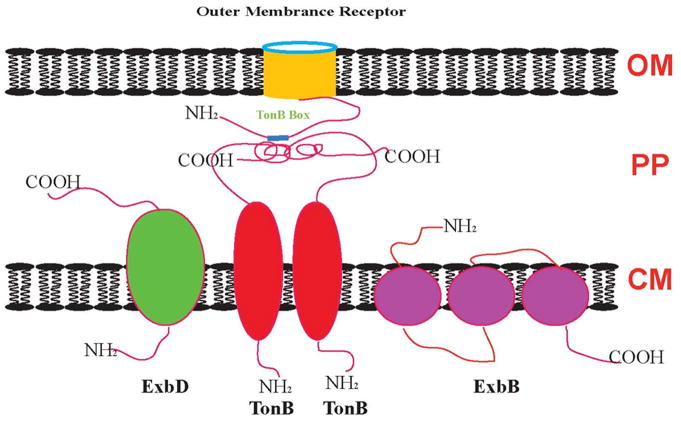
Heme transport across the outer membrane is an energy-requiring process. The energy for transport of heme into periplasmic space in most Gram-negative organisms is provided by cytoplasmic membrane (CM) protein TonB in association with two helper proteins, ExbB and ExbD (Figure 7), which form a complex in a molar ratio of 1:7:2, respectively [99–102]. This is an active process driven by the proton motive force generated from the CM. TonB-homologues are widespread in bacteria where they may have similar functions in heme uptake systems [103]. TonB is composed of three domains [104]: the N-terminal domain which is anchored to the CM, interacts with ExbB and ExbD to form an energy transducing complex; the C-terminal domain directly contacts receptors in the outer membrane; and the intermediate domain which is a proline-rich region, consists of alternating Pro-Glu and Pro-Lys repeats. ExbB possesses three transmembrane segments with the bulk of the protein in the cytoplasm, while ExbD has only one transmembrane segment and most of the protein is located in the periplasm [105].
Figure 7.
Schematic illustration of the protein complex involved in energy transduction from the cytoplasmic membrane to the outer membrane in Gram-negative bacteria. OM: outer membrane; PP: periplasmic space; CM: cytoplasmic membrane (modified from [97]).
Figure 7 schematically illustrates the protein complex involved in energy transduction in Gram-negative bacteria. Since no energy source is available in the outer membrane, TonB-dependent outer membrane receptors must utilize the energy provided by the proton motive force through the TonB-ExbB-ExbD complex. The TonB N-terminal domain contains a histidine residue, which probably binds proton together with ExbB and ExbD, which induces the TonB structural change into an energized form. Then the energized TonB will interact with the outer membrane receptor via TonB box, which is in the N-terminal domain of the receptor and shares amino acid homology in several regions [106–109]. After energy transduction, ExbB and ExbD will retrieve TonB from the outer membrane.
A tonB mutation prevented S. marcescens growth in iron-restricted medium, but not free heme or heme-hemoglobin, indicating a TonB-like protein may be involved in the hemophore-dependent heme acquisition system. Paquelin and co-workers [103] characterized this TonB-like protein (named HasB) and found that those two proteins have partially redundant function with respect to heme uptake. In addition, a second tonB gene (tonB2) in P. aeruginosa was proposed to be linked to the exbB and exbD genes, but tonB2exbBD is not required for heme acquisition [110]. Two sets of tonB, exbB, exbD genes have been reported in V. cholerae [111–113]. They have at least partially overlapped functions in iron or heme utilization. The mechanism of redundant TonB proteins in the bacteria is not yet clear.
Heme transport in the periplasmic space
Once across the outer membrane, heme is internalized into periplasm and is then bound by a heme-specific periplasmic transport protein (HTP), which mediates heme transfer to the cytoplasm through ATP-binding cassette (ABC) transporter.
A few periplasmic HTPs have been identified at genetic level, such as HemT (Y. enterocolitica), HmuT (C. diphtheriae,), ShuT (Shigella dysenteriae), PhuT (P. aeruginosa), HutB (Vibrio cholerae), and ChuT (E. Coli), which share 30 to 90% identical amino acid residues [85]. HTPs have been proposed to bind heme in the periplasm and deliver it to the membrane-bound ABC transporter [49, 114]. It has been considered that heme is bound in a cleft between the two subdomains of HTP, which will induce a conformational change in HTP and this allows the holo-HTP to interact with the transmembrane domain of ABC transporter. As a result, heme will face to the channel of ABC transporter and be delivered across the inner membrane.
HbpA, a 51-kDa lipoprotein from Hib purified in a heme-agarose affinity purification protocol, was initially proposed to possess heme transport or heme binding functions when expressed in E. coli [115, 116], but the role of HbpA in heme utilization was not identified until recently by an insertional mutation of hbpA [117]. However, the protein sequence of HbpA is not homologous to any known HTPs, but instead shares 53% sequence identity with the E. coli periplasmic dipeptide transport protein DppA [116]. Interestingly, two well known periplasmic dipeptide-binding protein DppA and MppA, the L-alanyl-gamma-D-glutamyl-meso-diaminopimelate binding protein, with a low heme-binding affinity (Kd: ~10−5 - 5 × 10−5 M) have been recently demonstrated to function in heme transport in the periplasmic space of E. coli under the condition of the absence of a heme-specific HTP [118].
Two of the better characterized periplasmic heme transport proteins are ShuT from Shigella dysenteriae [45] and PhuT from Pseudomonas aeruginosa [49]. ShuT was expressed as a monomeric recombinant protein of 28.5 KDa, binding one b-type heme per monomer and bearing no significant homology with other known heme proteins [45]. Resonance Raman, MCD, and UV-visible spectra of the hemin-loaded ShuT indicate a five-coordinate high spin heme with a Tyr proximal ligand, which is supported by site-directed mutagenesis studies [45]. Recombinant PhuT from P. aeruginosa was purified as a 33 kDa His-tagged protein with ~ 50% heme-bound in the native form [49]. Similar to that in ShuT, the heme is noncovalently attached as a five-coordinate, high-spin, ferric heme and can be transferred to apo-myoglobin in vitro. The heme in PhuT can be reduced by dithionite but not by either DTT or ascorbate. A Tyr-to-heme charge transfer band was identified in the oxidized form of holo-PhuT, suggesting a Tyr-heme coordination. This Tyr was identified to be Tyr71 in PhuT by site-directed mutagenesis [49]. Apo-PhuT binds heme or protoporphyrin IX at 1:1 ratio with high affinity (Kd ~1.2 nM and 14 nM, respectively).
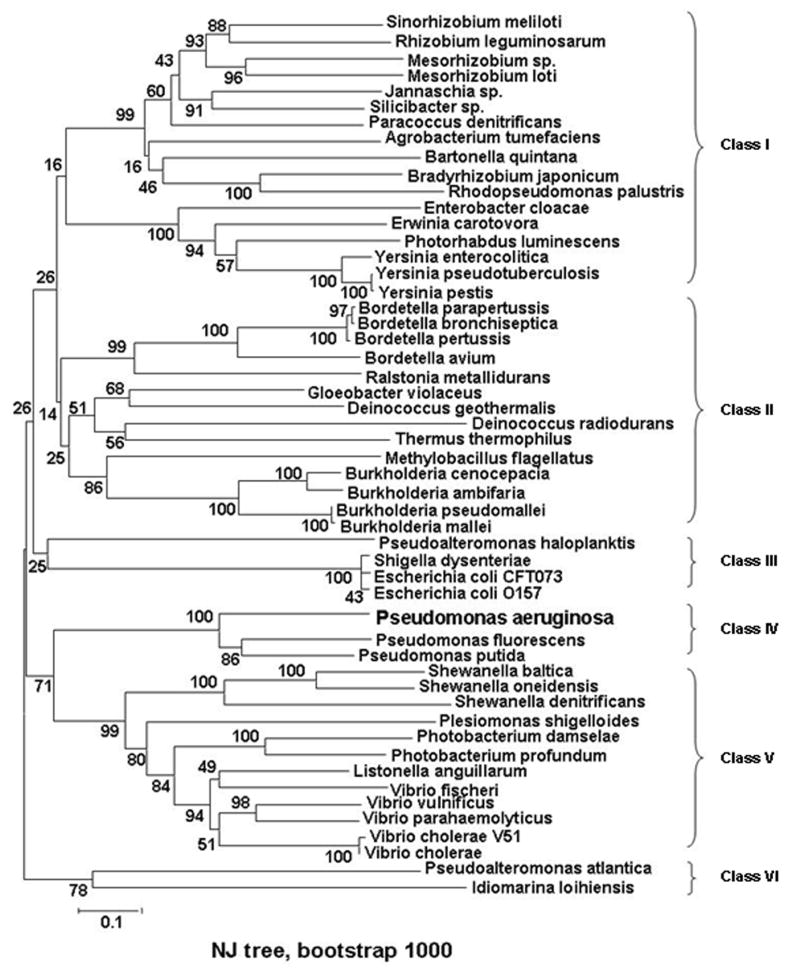
A BLASTP search revealed ca. 52 putative bacterial heme-specific HTPs, which can be grouped into 6 classes (Figure 8), most of which are associated with pathogenic bacteria. Multiple sequence alignment analysis (Supporting info.) reveals that these putative periplasmic heme transporters share low sequence homology and have no conserved common binding motif for heme-ligation. However, a tyrosine residue (Tyr71 in PhuT) is highly conserved in all but one of the 52 HTPs, implying that Tyr is a possible axial heme ligand in these HTPs.
Figure 8.
Unrooted NJ tree of HTP sequences of 52 taxa was conducted using MEGA version 3.1. Eight clusters are indicated and are grouped into 6 classes. Pseudomonas aeruginosa (P.ae), AAC13287; Pseudomonas fluorescens Pf-5, YP_262338; Pseudomonas putida KT2440, NP_746798; Bordetella avium, AAM28270; Burkholderia cenocepacia AU 1054, ZP_00457560; Burkholderia pseudomallei 668 (B.ps), ZP_00488203; Burkholderia ambifaria AMMD, ZP_00689266; Escherichia coli CFT073, NP_756175; Burkholderia mallei SAVP1, ZP_00449888; Shigella dysenteriae (S.dy), AAC27815; Escherichia coli O157:H7 EDL933, NP_290082; Bordetella pertussis Tohama I, NP_879218; Vibrio vulnificus CMCP6, NP_763480; Bordetella parapertussis 12822, NP_886318; Bordetella bronchiseptica RB50, NP_891189; Ralstonia metallidurans CH34 (R.me), ZP_00596240; Gloeobacter violaceus PCC 7421, BAC88521; Vibrio parahaemolyticus RIMD 2210633, NP_799933; Shewanella baltica OS155, ZP_00581516; Vibrio cholerae V51, ZP_00751095; Shewanella oneidensis MR-1, NP_719214; Paracoccus denitrificans PD1222 (P.de), ZP_00628787; Photobacterium damselae subsp. Piscicida, CAE46553; Listonella anguillarum serovar O1, CAF25487; Photobacterium profundum SS9, YP_130306; Erwinia carotovora subsp. Atroseptica SCRI1043, CAG74748; Pseudoalteromonas haloplanktis TAC125, YP_341564; Chloroflexus aurantiacus J-10-fl, ZP_00768862; Sinorhizobium meliloti, CAC47008; Vibrio fischeri ES114 (V. fi), YP_204605; Shewanella denitrificans OS217, EAN70978; Shewanella putrefaciens CN-32, EAO95810; Vibrio cholerae, AAB94547; Jannaschia sp. CCS1, ZP_00557202; Deinococcus geothermalis DSM 11300, ZP_00395505; Rhizobium leguminosarum, CAC34393; Plesiomonas shigelloides, AAK38770; Methylobacillus flagellatus KT, ZP_00566306; Deinococcus radiodurans R1, NP_051557; Yersinia enterocolitica, CAA54866; Yersinia pseudotuberculosis IP 32953, YP_068884; Bradyrhizobium japonicum, CAC38745; Enterobacter cloacae (E. cl), CAD61864; Mesorhizobium sp. BNC1, EAN05942; Silicibacter sp. TM1040, ZP_00621030; Yersinia pestis biovar Medievalis str. 91001, AAS60707; Agrobacterium tumefaciens str. C58, NP_533132; Pseudoalteromonas atlantica T6c, ZP_00775180; Thermus thermophilus HB8, YP_145459; Photorhabdus luminescens subsp. Laumondii TTO1, NP_929869; Rhodopseudomonas palustris CGA009, NP_947465; Mesorhizobium loti MAFF303099, NP_102804; Idiomarina loihiensis L2TR (I.lo), YP_154505; Bartonella quintana str. Toulouse, CAF25913. The values at each node are bootstrap value on 1000 replications.
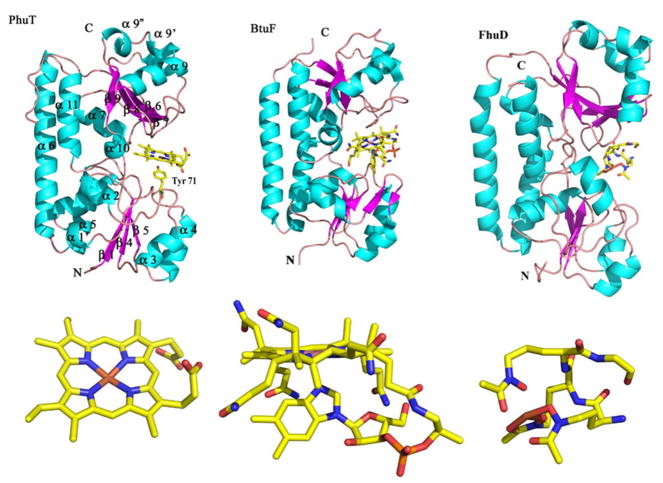
Recently, the crystal structures of PhuT and ShuT have been solved in apo- and heme-bound forms (Figure 9) [48]. They share common structural features with 2 topologically similar globular domains and a long, rigid α-helix as an interdomain linker, typical of a Class III periplasmic binding protein, such as BtuF, a vitamin B12 transporting protein [119, 120] and FhuD, an E. coli periplasmic protein that binds hydroxamate-type siderophores [121]. In terms of ligands, heme, B12 and peptide siderophores, PhuT/ShuT are more closely resemble BtuF, which shows the modest 1Å shrinking of the B12 binding pocket upon ligand binding. It has been proposed that heme binding to PhuT/ShuT may only induce a minor conformational change of the heme binding pocket, including a reorientation of the Tyr heme ligand, which may play a role in the heme binding/release process.
Figure 9.
Structure of PhuT compared with BtuF and FhuD. The major elements of the secondary structure according to the BtuF structure are shown on the PhuT structure. The ligands for each protein are also shown. The PDB accession code for BtuF and FhuD are, 1N2Z and 1K7S, respectively (modified from [48]). Figures were made with PyMOL [162].
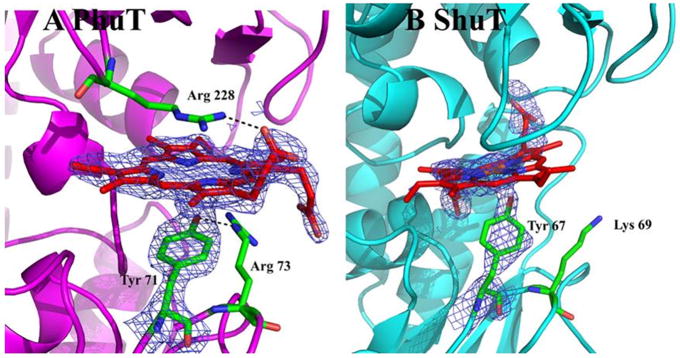
In PhuT, the heme prosthetic group is embedded in a narrow cleft between the 2 globular domains. As we expected, the heme is pentacoordinate with Tyr71 from the C terminal end of the β-3 strand in the N terminal domain serving as the proximal heme ligand [49]. The heme propionate side chains are pointing outward to the surface of the protein, and each is bent to the opposite side of the heme plane, respectively (Figure 10). One propionate side chain is directly hydrogen bonded to Arg228 which is stacked against the distal surface of heme. In the close vicinity of the other propionate side chain, Arg73, on the proximal side of heme, is hydrogen bonded to the ligand Tyr71. However, in ShuT, Lys69, which is the homolog to the PhuT Arg73, is far from the heme ligation residue Tyr67. In order to form a H-bond between Lys69 and Tyr67 to mimic the Tyr71-Arg73 interaction in PhuT, there must be a reorientation of the Lys69 side chain. Therefore, PhuT and ShuT may have similar conformational changes upon binding/releasing heme, but they may have slightly different mechanism.
Figure 10.
Heme binding pockets in PhuT and ShuT. The maps shown are 2Fo−Fc maps contoured at 1.0 σ. (A) In PhuT, Arg228 H-bonds with one heme propionate while Arg73, in the proximal pocket, is H-bonding with the Tyr71 ligand. (B) The partially bound heme in the ShuT structure has Lys69 nearby. In order to form a H-bond with the Tyr ligand, an unfavorable bending of the side chain would be required (adapted from [48]).
Heme transport across the cytoplasmic membrane
In Gram-negative bacteria, once heme is transported into the periplasmic space, it will be translocated across the cytoplasmic membrane by specific ABC transporters. The ABC transporters mediate active translocation of a variety of molecules across cell membranes in both Gram-positive and Gram-negative pathogens [114, 122]. When associated with heme-specific HTP, it forms a complete heme-uptake permease. The first heme permease was identified from Y. enterocolitica, composed of one periplasmic heme-binding protein (HemT), one transmembrane protein (HemU), and one ATPase protein (HemV) [85].
The inner membrane proteins in P. aeruginosa which are proposed to be involved in heme transport include PhuU, PhuV and PhuW. No biochemical studies on these proteins have been reported yet, but the homology models of PhuU, PhuV have been shown in Figure 5. Function of PhuW is not known yet, but recently, an analogue protein ChaN, which is an iron-regulated lipoprotein from Campylobacter jejuni, has been characterized [123]. The crystal structure of ChaN (Figure 5) reveals that it consists of a large parallel β-sheet with flanking α-helices and a smaller domain with α-helices. Interestingly, two cofacial heme groups (~ 3.5 Å apart with an inter-iron distance of 4.4 Å) bind in a pocket formed by a dimer of ChaN monomers. Tyr148 residue was found to be involved in coordination with heme iron (2.47 Å) [123]. The EPR spectrum at acidic pH is consistent with a five-coordinate ferriheme center with a proximal Tyr ligand, while the spectrum at alkaline pH is more complex.
Interestingly, the PhuUV analogues in S. dysenteriae, ShuUV, have recently been cloned and incorporated into artificial liposomes [122]. By encapsulation of ShuT into the lumen of the liposomes, the transport of heme from heme-bound ShuT to apo-ShuS (the cytoplasmic heme-binding protein) was observed. The heme translocation process is found to be coupled with ATP hydrolysis and probably via direct complex formation between ShuT and ShuU [122]. Site-directed mutagenesis studies revealed that 2 histidine residues on ShuU are critical in the translocation of heme across the membrane [122].
Heme transport, degradation and iron releasing in cytosol
Once internalized into the cytoplasm, heme is rapidly catabolized by heme oxygenase (HOs), which opens the heme porphyrin ring. HOs, first identified in mammals, are primarily involved in protecting cells against the toxic effect of heme, whereas bacteria utilize HOs to access heme as a nutrient iron source [124]. HOs are ubiquitous in nature, and function to catalyze the oxidation of heme into biliverdin, carbon monoxide, and free iron [125, 126]. However, before heme is delivered to a HO, it may be sequestered by a PhuS analogue, because the free heme is toxic to cells.
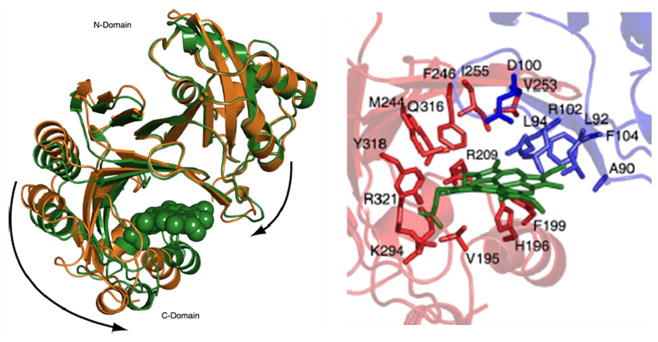
HemS (Figure 11), a 40 kDa protein from Y. enterocolitica, is a PhuS analogue. It is thought to be responsible for oxidative cleavage of the heme, protecting the cellular environment from the heme toxicity effects [64]. However, no biochemistry study on the function of this protein has been reported yet. More recently, Schneider et al. reported the crystal structures of apo- and heme-bound HemS, thus providing penetrating evidence into its mechanism of heme binding and release [127]. HemS associates with heme through a pocket, which is formed between a large β-sheet dome and a three-helix subdomain. The His196 coordinates the iron in the heme-HemS complex, which is also stabilized by a distal Arg102 residue. A “heme-induced fit” binding mechanism based on the striking conformational change was revealed by comparison of apo- and holo-formed HemS crystal structures, indicating that HemS is an intracellular heme transporter rather than a heme oxygenase.
Figure 11.
(A) Crystal structure reveals HemS conformation switches between apo, open state and the heme-bond closed state. The holo-form is shown in green, and the apo-structure is shown in gold. (B) Heme-binding pocket in the heme-HemS complex: His196 coordinates to the heme iron; Arg-102 extends over the porphyrin plane to interact with the heme propionates by electrostatic interactions (adapted from [127]).
In Shigella dysenteriae, the cytoplasmic protein, ShuS, which shares 64% sequence identity with HemS and binds one heme per monomer (Kd ~ 13 μM), has been shown to function as both a heme storage protein and a DNA binding protein for protection against heme-induced oxidative damage, but does not exhibit any ability to break down the heme [128]. A shuS mutant study identified that ShuS improves heme utilization efficiency at low heme concentration and protects cells from growth inhibition by heme at high heme concentration [129]. However, carbon monoxide detection confirmed that the cytoplasmic analogue ChuS from E. coli O157:H7 is a HO, which shares 98% identity with the ShuS sequence [126]. DLS (Dynamic Light Scattering) analysis demonstrated that ChuS and ChuS-heme both form high molecular weight aggregates after incubation for 4 days in Tris buffer at 4°C. Suits et al. suggested that this aggregation could be the reason why ShuS is absent of HO activity [126], while it is also possible that the different phenotype is due to the different genetic backgrounds. The crystal structure of ChuS displays a previously uncharacterized fold and is unique when compared with known mammalian and bacterial HOs. In addition, E. coli ChuS is capable of using ascorbic acid or cytochrome P450 reductase-NADPH as an electron donor for heme degradation.
HmuS in Y. pestis, was originally proposed to be a heme oxygenase [64]. However, inactivation of Y. pestis hmuS showed that these mutants were still able to use hemin and all hemoproteins as an iron source [130, 131]. Therefore, another hypothesis is that HmuS is probably involved in heme storage and/or chaperoning. Since then, heme oxygenases have been found in several pathogenic bacteria including N. meningitides, C. diphtheriae and P. aeruginosa.
The cytoplasmic heme-binding protein PhuS from P. aeruginosa has been characterizated recently [47, 132]. PhuS binds one heme per monomer with alternate His ligands at positions 209 and 212. A second distinct six-coordinated low-spin heme site is present in dimeric PhuS, but the coordination residues have not been identified [47, 132]. In contrast to the previous reports, Block and co-workers [132] provided compelling evidence that PhuS is not a HO but a heme chaperone responsible for passing heme to a pa-HO, which is driven by a specific protein-protein interaction. It has been proposed that the specific and unidirectional heme transfer from PhuS to pa-HO was triggered by a spin-state crossover, according to the experiments conducted in the presence and absence of cyanide [133]. Due to the fact that the transfer rate from heme-loaded PhuS (Kd ~ 0.2 μM) to pa-HO is 30-fold faster than that to apo-myoglobin (kH = 10−8 μM), Bhakta et al. also suggested that heme transfer is independent of heme binding affinity.
Most heme proteins participating in O2 activation have imidazolate of histidine or thiolate of cysteine as proximal ligands and a few have a neutral imidazole as the proximal ligand [134]. In contrast to periplasmic heme transport protein PhuT and ShuT, HO binds substrate heme at the specific position of its pocket to form a heme-HO complex with histidine as a heme-iron ligand, as proposed in pa-HO [135], nm-HO from N. meningitides [136], and cd-HO from C. diphtheriae [137].
The mechanism of heme oxidation by the HO has been proposed as follows [14, 134]: the reaction starts with the formation of the ferric heme-HO complex with histidine as an iron ligand. Ferric iron is then reduced to a ferrous iron by the electron donated from NADPH-cytochrome P450 reductase. Next, oxygen binds to the complex forming a metastable oxy-form. The iron-bound oxygen is converted to a hydroperoxide intermediate (Fe3+-OOH), by receiving another electron from the reductase and a proton from the distal pocket water. Lastly, the terminal oxygen of Fe3+-OOH attacks the α-meso-carbon of the porphyrin ring to form ferric α-meso-hydroxyheme, which is converted to the ferric verdoheme in the presence of oxygen. The mechanism of the verdoheme degradation to ferric biliverdin is not well understood.
Heme uptake in Gram-positive bacteria
Unlike that of the Gram-negative bacteria, the envelope of Gram-positive bacteria have a thick cell wall composed of the murein sacculus and the attached polysaccharides, teichoic acids, and cell wall proteins [138]. Studies of bacterial heme uptake systems have been mainly focusing on Gram-negative microbes, thus relatively less is known about the Gram-positive bacteria heme uptake pathway. Progress has recently been made in exploring the heme acquisition in certain Gram-positive bacteria. In Corynebacterium diphtheriae, the outer membrane receptors as well as the Ton system are missing; a HemT-like lipoprotein, HmuT, serves to function as a heme receptor [122, 139]. However, the ABC transporters are required for heme uptake not only in C. diphtheriae [122], but also in Streptococcus pyogenes, or group A streptococcus (GAS) [140–142], and S. aureus [21]. Cell surface-binding proteins (named Shr and Shp) and the ABC transporter lipoprotein (named HtsA) in human pathogen GAS have been identified recently [143, 144]. The Shr of S. pyogenes can bind heme and transfer it to the streptococcal heme-binding protein Shp [144]. Shp may then relay its heme to apo-HtsA to across the bacterial envelope. The HtsA (SiaA) from S. pyogenes has been characterized recently [145]. Resonance Raman (rR), magnetic circular dichroism (MCD), and nuclear magnetic resonance (NMR) spectroscopic studies suggest that the heme in SiaA is six-coordinate and low-spin, with methionine and histidine as axial ligands.
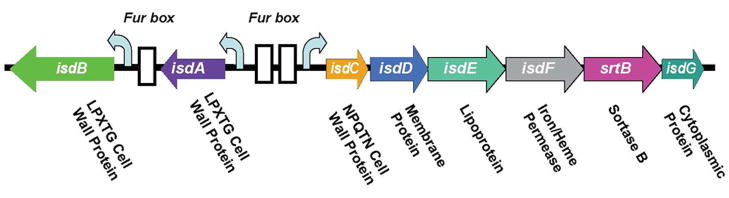
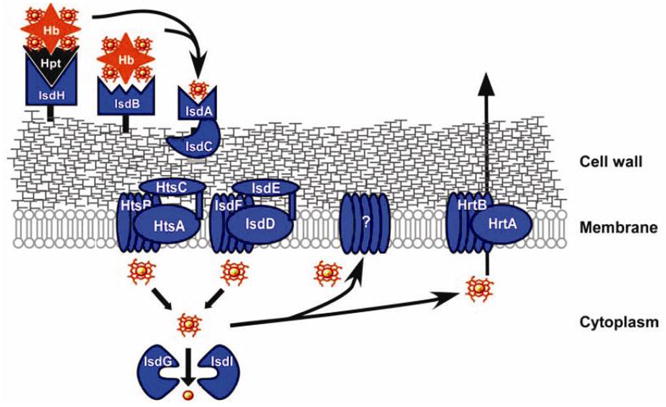
The Gram-positive bacterium S. aureus has developed a heme-acquisition system encoded by an isd (iron-regulated surface determinants) gene cluster (Figure 12) to acquire heme-bound iron during infection, via the secretion of hemolysins (which rupture the red blood cells to release hemoglobin) [146]. The process begins with binding of a hemoglobin or a haptoglobin-hemoglobin complex onto the bacterial surface receptors IsdB and IsdH (Figure 13). Heme is then removed from hemoglobin by IsdA and IsdB, transferred to the cell wall protein IsdC, and then passed through the membrane with the aid from translocation factors (IsdD, IsdE, and IsdF). A recent in vitro study [147] demonstrated that heme can be directly transferred from IsdA to IsdC via protein-protein interaction and that the process is rapid and affinity-driven.
Figure 12.
Organization of the isd locus of S. aureus encodes surface proteins, lipoprotein, membrane transporter, and cytoplasmic proteins (modified from [146]).
Figure 13.
Gram-positive bacteria S. aureus heme-acquisition system (adapted from [163]).
The crystal structure of the heme-IsdC complex was solved (Figure 14) [148]. IsdC has an architecture with a distinct binding pocket with the ligand located next to the hydrophobic core of the β-sandwich. The heme-iron is coordinated with a tyrosine surrounded by several non-polar side chains which cluster into a tightly packed proximal side. Meanwhile, the distal side is relatively exposed to a short helical peptide segment which acts as a lip and clasps onto almost half of the porphyrin plane. This structural feature was proposed to play a role in the mechanism of heme binding and release through conformational changes.
Figure 14.
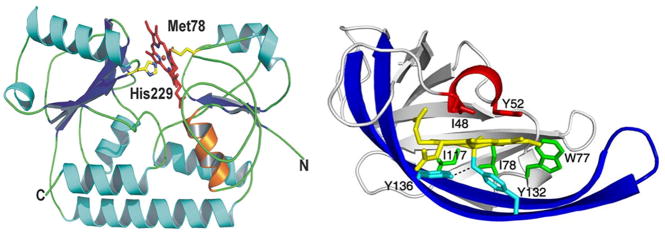
The overall structure of the IsdE-heme (A) and heme-IsdC complex (B). (A) Schematic representation of bi-lobed IsdE. Secondary structural elements of the protein are represented by strands, loops, and helices colored in blue, green, and cyan, respectively. Propionate stabilizing of helix-1 is represented in orange, and protein termini are labeled with N or C. Heme is shown as sticks within the binding pocket. Heme carbon and oxygen are shown as red, and nitrogen and iron are shown as blue and orange, respectively (adapted from [149]). (B) Schematic representation of IsdC highlights the lip region, shown in red, with the distal Ile48 and Tyr52 over the heme. The prominent β-hairpin structure is in dark blue, with Tyr136 hydrogen-bonded to the proximal Tyr132, both in cyan. Lastly, Trp77, Ile78, and Ile117, in green, pack along the equatorial plane of the porphyrin, effectively interlocking the heme into its binding pocket (adapted form [148]).
The crystal structure of the soluble portion of the IsdE lipoprotein in complex with heme (Figure 14) was reported recently [149]. The structure reveals a bi-lobed topology formed by an N- and C-terminal domain bridged by a single α-helix. The heme is bound in the groove of the domain interface, which is six-coordinated in a novel fashion for heme transporters by Met78 and His229. Both heme propionate side chains are secured by H-bonds to the IsdE main chain and side chain.
Once inside the cytoplasm, IsdG and IsdI, the cytoplasmic heme-iron binding proteins, may be involved in removing iron from heme. Heme may also be complexed with some unknown membrane-associated proteins as an enzyme cofactor. Another possibility is that excess heme or heme metabolites are pumped by HrtAB out of the cytoplasm to protect the bacteria from its toxicity [146].
IsdA and IsdB as well as IsdC are covalently anchored by sortases to the bacterial peptidoglycan. The LPXTG anchor structure is proposed to allow staphylococci to use IsdA and IsdB as receptors, whereas the NPQTN structure in IsdC may function in the transportation of heme across the cell wall. Magnetic circular dichroism (MCD) analysis of the recombinant IsdA from S. aureus suggests that it binds to a five-coordinate, high-spin ferric heme molecule, proximally coordinated by a tyrosyl residue [150]. Reduction of the IsdA heme yields a five-coordinate, high-spin ferrous heme with a neutral axial ligand, most likely the histidine [150]. The isd locus is also present in several other Gram-positive bacteria, such as Listeria monocytogenes, Bacillus halodurans, and Bacillus anthracis, suggesting that similar mechanisms of iron acquisition may exist [151].
Regulations on heme uptake
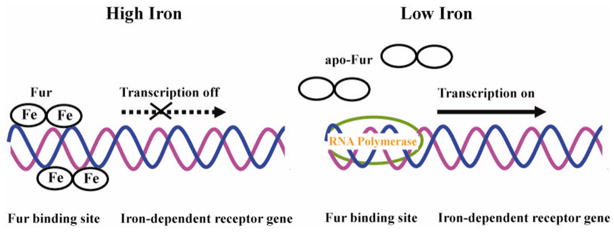
Iron is an essential nutrient for bacteria; however, overloaded iron could be harmful due to its toxic properties. Therefore, expression of iron/heme uptake proteins during different periods of time of infection is regulated in response to iron level in the environment. In many bacterial pathogens, such as V. cholerae [111], Y. pestis [130], Y. enterocolitica [64], and P. aeruginosa [84], iron/heme uptake regulation is negatively controlled by the global determinant ferric-uptake regulator protein (Fur) [152]. Fur, a homodimer consisting of 17-kDa subunits [153], are divided into two domains: N-terminal domain, which functions in DNA binding and the C-terminal domain, which is rich in His residues and is responsible for divalent iron binding and dimerization [153, 154]. As shown in Figure 15, under iron-rich conditions Fur binds to the divalent iron, which is capable of binding target DNA sequences designated as Fur box, a 19-bp consensus sequence, GATAATGATAATCATTATC. Two Fur dimers are suggested to bind at each Fur box on opposite faces of the DNA helix [155]. Hence, transcription of iron-dependent outer membrane receptor genes will be inhibited. On the other hand, when iron is scarcely available, divalent iron will be released from Fur and the RNA polymerase will gain access to the promoter region, and the genes will be transcribed.
Figure 15.
Schematic representation of Fur regulated gene expression. At high iron concentration, both iron-loaded Fur dimers bind Fur box on opposite faces of DNA. Transcription of the iron-dependent receptor gene will be turned off. When iron concentration decrease, iron is released from Fur and apo-Fur is displaced from Fur box. RNA polymerase binds at the promoter region initiating the transcription.
Another regulatory mechanism involves specific extracytoplasmic function (ECF) sigma factors. Figure 1 shows the has operon (hasRADEB) in S. marcescens. It codes for the receptor (HasR), the hemophore (HasA), two inner membrane hemophore secretion proteins (HasD and HasE) [26], and a TonB homolog (HasB), which is involved in heme uptake [103]. Two genes immediately upstream of the has operon encode HasI, an ECF sigma factor, and HasS, an anti-sigma factor. When enough heme is available in the media, the anti-sigma activity of HasS is turned off, which is positively regulated by HasI through a signaling cascade triggered by the heme-loaded HasA binding to HasR. When heme is in shortage, apo-HasA is displaced from HasR. As a result, HasS will be re-activated and the transcription of the has operon will be turned off.
To date, most of the heme transport systems are negatively regulated by the Fur repressor. Recently, positive regulation of bhu gene expression in the Gram-negative respiratory pathogen Bordetella pertussis and Bordetella bronchiseptica has been reported [156, 157]. The heme uptake regulator gene hurIR is immediately upstream of the heme uptake bhuRSTUV gene cluster and is predicted to encode ECF sigma factor, HurI, and cytoplasmic membrane regulator, HurR. In response to iron starvation conditions, Fur represses the hurI promoter activity. When enough iron is available in the media, Fur will derepress hurI allowing transcription of HurI and HurR and low levels of the bhu gene. However, HurI will remains inactive through its association with HurR until the outer membrane receptor, BhuR, binds heme. A receptor occupancy signal will be transduced through HurR, and HurI will be released and then it will associate with RNA polymerase to direct high levels of transcription of heme uptake genes. Fur repression will resume when intracellular iron stores are replenished.
Concluding remarks
Over the past few decades, it has been well established that heme is a major source of iron for bacteria. Many bacterial pathogens, including both Gram-negative and Gram-positive ones are armed with sophisticated mechanisms to steal heme from the host. Understanding their heme transport pathways is not only of scientific interest, but may also provide a basis for potential novel inhibitors/antibiotics design by targeting the heme transport processes. Structural and mechanistic studies of the proteins involved in the heme trafficking processes have been providing important insight to our understanding of this process at the molecular level. However, it is still challenging to understand the structures of the membrane proteins involved. Comparing with the heme-binding process, the mechanism of heme-releasing process is less understood. Although thermodynamics may play a significant role in certain cases [144, 147], it appears that heme transport processes are not simply driven by a sequential increase in heme-affinity from one protein to another in the heme-transport cascade. Specific protein-protein interactions with or without the coupled ATP hydrolysis, changes in heme-coordination and spin states may be critical to certain steps in the heme translocation processes. There is no doubt that further structural and mechanistic studies will be necessary to unravel the details of the heme acquisition processes in bacteria. This information will be important to guide the design of potential antimicrobial agents that target the heme-transport pathway.
Supplementary Material
Acknowledgments
This work was partially supported by the National Institutes of Health and University of Massachusetts Dartmouth. This publication/project was made possible by grant 1 R21 AT002743-02 from the National Center for Complementary and Alternative Medicine (NCCAM). Its contents are solely the responsibility of the authors and do not necessarily represent the official views of the NCCAM, or the National Institutes of Health. We thank Drs. T. Su, W. Dills, J. D. Smith and Ms Erin Sullivan (UMass Dartmouth, MA, USA) for helpful discussions.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Heinemann IU, Jahn M, Jahn D. Arch Biochem Biophys. 2008;474:238–251. doi: 10.1016/j.abb.2008.02.015. [DOI] [PubMed] [Google Scholar]
- 2.Rouault TA. Science. 2004;305:1577–1578. doi: 10.1126/science.1102975. [DOI] [PubMed] [Google Scholar]
- 3.Raymond KN, Dertz EA, Kim SS. Proc Natl Acad Sci U S A. 2003;100:3584–3588. doi: 10.1073/pnas.0630018100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Braun V. Int J Med Microbiol. 2001;291:67–79. doi: 10.1078/1438-4221-00103. [DOI] [PubMed] [Google Scholar]
- 5.Wandersman C, Delepelaire P. Annu Rev Microbiol. 2004;58:611–647. doi: 10.1146/annurev.micro.58.030603.123811. [DOI] [PubMed] [Google Scholar]
- 6.Skarr EP, Schneewind O. Microb Infect. 2004;6:390–397. doi: 10.1016/j.micinf.2003.12.008. [DOI] [PubMed] [Google Scholar]
- 7.Bullen JJ, Rogers HJ, Griffiths E. Curr Top Microbiol Immunol. 1978;80:1–35. doi: 10.1007/978-3-642-66956-9_1. [DOI] [PubMed] [Google Scholar]
- 8.Bullen JJ, Griffiths E. Iron and Infection: Molecular Physiological and Clinical Aspects. JohnWiley and Sons; NY: 1999. [Google Scholar]
- 9.Clarke TE, Tari LW, Vogel HJ. Curr Top Med Chem. 2001;1:7–30. doi: 10.2174/1568026013395623. [DOI] [PubMed] [Google Scholar]
- 10.Ratledge C, Dover LG. Annu Rev Microbiol. 2000;54:881–941. doi: 10.1146/annurev.micro.54.1.881. [DOI] [PubMed] [Google Scholar]
- 11.Chasteen ND, Harrison PM. J Struct Biol. 1999;126:182–194. doi: 10.1006/jsbi.1999.4118. [DOI] [PubMed] [Google Scholar]
- 12.Guo M, Harvey I, Yang W, Coghill L, Campopiano DJ, Parkinson JA, MacGillivray RT, Harris WR, Sadler PJ. J Biol Chem. 2003;278:2490–2502. doi: 10.1074/jbc.M208776200. [DOI] [PubMed] [Google Scholar]
- 13.Butler A. Nat Struct Biol. 2003;10:240–241. doi: 10.1038/nsb0403-240. [DOI] [PubMed] [Google Scholar]
- 14.Wilks A, Burkhard KA. Natural product reports. 2007;24:511–522. doi: 10.1039/b604193k. [DOI] [PubMed] [Google Scholar]
- 15.Reniere ML, Torres VJ, Skaar EP. Biometals. 2007;20:333–345. doi: 10.1007/s10534-006-9032-0. [DOI] [PubMed] [Google Scholar]
- 16.Cescau S, Cwerman H, Letoffe S, Delepelaire P, Wandersman C, Biville F. Biometals. 2007;20:603–613. doi: 10.1007/s10534-006-9050-y. [DOI] [PubMed] [Google Scholar]
- 17.Rajagopal A, Rao AU, Amigo J, Tian M, Upadhyay SK, Hall C, Uhm S, Mathew MK, Fleming MD, Paw BH, Krause M, Hamza I. Nature. 2008;453:1127–1131. doi: 10.1038/nature06934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sato E, Sagami I, Uchida T, Sato A, Kitagawa T, Igarashi J, Shimizu T. Biochemistry. 2004;43:14189–14198. doi: 10.1021/bi048742i. [DOI] [PubMed] [Google Scholar]
- 19.Poulos TL. Natural Product Reports. 2007;24:504–510. doi: 10.1039/b604195g. [DOI] [PubMed] [Google Scholar]
- 20.Paoli M, Marles-Wright J, Smith A. DNA Cell Biol. 2002;21:271–280. doi: 10.1089/104454902753759690. [DOI] [PubMed] [Google Scholar]
- 21.Skaar EP, Humayun M, Bae T, DeBord KL, Schneewind O. Science. 2004;305:1626–1628. doi: 10.1126/science.1099930. [DOI] [PubMed] [Google Scholar]
- 22.Ryter SW, Tyrrell RM. Free Radic Biol Med. 2000;28:289–309. doi: 10.1016/s0891-5849(99)00223-3. [DOI] [PubMed] [Google Scholar]
- 23.Vercellotti GM, Balla G, Balla J, Nath K, Eaton JW, Jacob HS. Artif Cells Blood Substit Immobil Biotechnol. 1994;22:207–213. doi: 10.3109/10731199409117415. [DOI] [PubMed] [Google Scholar]
- 24.Jeney V, Balla J, Yachie A, Varga Z, Vercellotti GM, Eaton JW, Balla G. Blood. 2002;100:879–887. doi: 10.1182/blood.v100.3.879. [DOI] [PubMed] [Google Scholar]
- 25.Zunszain PA, Ghuman J, Komatsu T, Tsuchida E, Curry S. BMC Struct Biol. 2003;3:6. doi: 10.1186/1472-6807-3-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ghigo JM, Letoffe S, Wandersman C. J Bacteriol. 1997;179:3572–3579. doi: 10.1128/jb.179.11.3572-3579.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bridges KR, Seligman PA. In: Blood: Principles and Practice of Hematology. Handlin RL, Lux SE, Stossel TP, editors. J.B. Lippincott Company; New York: 1995. pp. 1433–1472. [Google Scholar]
- 28.Keel SB, Doty RT, Yang Z, Quigley JG, Chen J, Knoblaugh S, Kingsley PD, De Domenico I, Vaughn MB, Kaplan J, Palis J, Abkowitz JL. Science. 2008;319:825–828. doi: 10.1126/science.1151133. [DOI] [PubMed] [Google Scholar]
- 29.Wejman JC, Hovsepian D, Wall JS, Hainfeld JF, Greer J. J Mol Biol. 1984;174:319–341. doi: 10.1016/0022-2836(84)90341-3. [DOI] [PubMed] [Google Scholar]
- 30.Evans RW, Crawley JB, Joannou CL, Sharma ND. In: Iron and Infection: Molecular, Physiological and Clinical Aspects. Bullen JJ, Griffiths E, editors. John Wiley and Sons; Chichester: 1999. pp. 27–86. [Google Scholar]
- 31.Kristiansen M, Graversen JH, Jacobsen C, Sonne O, Hoffman HJ, Law SK, Moestrup SK. Nature. 2001;409:198–201. doi: 10.1038/35051594. [DOI] [PubMed] [Google Scholar]
- 32.Jin H, Ren Z, Pozsgay JM, Elkins C, Whitby PW, Morton DJ, Stull TL. Infect Immun. 1996;64:3134–3141. doi: 10.1128/iai.64.8.3134-3141.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Stojiljkovic I, Larson J, Hwa V, Anic S, So M. J Bacteriol. 1996;178:4670–4678. doi: 10.1128/jb.178.15.4670-4678.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lewis LA, Gray E, Wang YP, Roe BA, Dyer DW. Mol Microbiol. 1997;23:737–749. doi: 10.1046/j.1365-2958.1997.2501619.x. [DOI] [PubMed] [Google Scholar]
- 35.Kahler CM, Blum E, Miller YK, Ryan D, Popovic T, Stephens DS. Infect Immun. 2001;69:1687–1696. doi: 10.1128/IAI.69.3.1687-1696.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Muller-Eberhard U. Methods Enzymol. 1988;163:536–565. doi: 10.1016/0076-6879(88)63049-7. [DOI] [PubMed] [Google Scholar]
- 37.Tolosano E, Altruda F. DNA Cell Biol. 2002;21:297–306. doi: 10.1089/104454902753759717. [DOI] [PubMed] [Google Scholar]
- 38.Hrkal Z, Vodrazka Z, Kalousek I. Eur J Biochem. 1974;43:73–78. doi: 10.1111/j.1432-1033.1974.tb03386.x. [DOI] [PubMed] [Google Scholar]
- 39.Dockal M, Carter DC, Ruker F. J Biol Chem. 1999;274:29303–29310. doi: 10.1074/jbc.274.41.29303. [DOI] [PubMed] [Google Scholar]
- 40.Curry S. Vox Sang. 2002;83(Suppl 1):315–319. doi: 10.1111/j.1423-0410.2002.tb05326.x. [DOI] [PubMed] [Google Scholar]
- 41.Wardell M, Wang Z, Ho JX, Robert J, Ruker F, Ruble J, Carter DC. Biochem Biophys Res Commun. 2002;291:813–819. doi: 10.1006/bbrc.2002.6540. [DOI] [PubMed] [Google Scholar]
- 42.Grinshtein N, Bamm VV, Tsemakhovich VA, Shaklai N. Biochemistry. 2003;42:6977–6985. doi: 10.1021/bi020647r. [DOI] [PubMed] [Google Scholar]
- 43.Stojiljkovic I, Hantke K. Embo J. 1992;11:4359–4367. doi: 10.1002/j.1460-2075.1992.tb05535.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hornung JM, Jones HA, Perry RD. Mol Microbiol. 1996;20:725–739. doi: 10.1111/j.1365-2958.1996.tb02512.x. [DOI] [PubMed] [Google Scholar]
- 45.Eakanunkul S, Lukat-Rodgers GS, Sumithran S, Ghosh A, Rodgers KR, Dawson JH, Wilks A. Biochemistry. 2005;44:13179–13191. doi: 10.1021/bi050422r. [DOI] [PubMed] [Google Scholar]
- 46.Burkhard KA, Wilks A. J Biol Chem. 2007;282:15126–15136. doi: 10.1074/jbc.M611121200. [DOI] [PubMed] [Google Scholar]
- 47.Lansky IB, Lukat-Rodgers GS, Block D, Rodgers KR, Ratliff M, Wilks A. J Biol Chem. 2006;281:13652–13662. doi: 10.1074/jbc.M600824200. [DOI] [PubMed] [Google Scholar]
- 48.Ho WW, Li H, Eakanunkul S, Tong Y, Wilks A, Guo M, Poulos TL. J Biol Chem. 2007;282:35796–35802. doi: 10.1074/jbc.M706761200. [DOI] [PubMed] [Google Scholar]
- 49.Tong Y, Guo M. J Biol Inorg Chem. 2007;12:735–750. doi: 10.1007/s00775-007-0226-x. [DOI] [PubMed] [Google Scholar]
- 50.Woloszczuk W, Sprinson DB, Ruis H. J Biol Chem. 1980;255:2624–2627. [PubMed] [Google Scholar]
- 51.Zou P, Borovok I, Ortiz de Orue Lucana D, Muller D, Schrempf H. Microbiology. 1999;145(Pt 3):549–559. doi: 10.1099/13500872-145-3-549. [DOI] [PubMed] [Google Scholar]
- 52.Vanderpool CK, Armstrong SK. J Bacteriol. 2001;183:4278–4287. doi: 10.1128/JB.183.14.4278-4287.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lewis LA, Dyer DW. J Bacteriol. 1995;177:1299–1306. doi: 10.1128/jb.177.5.1299-1306.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Cornelissen CN, Sparling PF. Mol Microbiol. 1994;14:843–850. doi: 10.1111/j.1365-2958.1994.tb01320.x. [DOI] [PubMed] [Google Scholar]
- 55.Lewis LA, Sung MH, Gipson M, Hartman K, Dyer DW. J Bacteriol. 1998;180:6043–6047. doi: 10.1128/jb.180.22.6043-6047.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Genco CA, Dixon DW. Mol Microbiol. 2001;39:1–11. doi: 10.1046/j.1365-2958.2001.02231.x. [DOI] [PubMed] [Google Scholar]
- 57.Letoffe S, Ghigo JM, Wandersman C. J Bacteriol. 1994;176:5372–5377. doi: 10.1128/jb.176.17.5372-5377.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Arnoux P, Haser R, Izadi N, Lecroisey A, Delepierre M, Wandersman C, Czjzek M. Nat Struct Biol. 1999;6:516–520. doi: 10.1038/9281. [DOI] [PubMed] [Google Scholar]
- 59.Arnoux P, Haser R, Izadi-Pruneyre N, Lecroisey A, Czjzek M. Proteins. 2000;41:202–210. doi: 10.1002/1097-0134(20001101)41:2<202::aid-prot50>3.0.co;2-8. [DOI] [PubMed] [Google Scholar]
- 60.Letoffe S, Omori K, Wandersman C. J Bacteriol. 2000;182:4401–4405. doi: 10.1128/jb.182.16.4401-4405.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Letoffe S, Redeker V, Wandersman C. Mol Microbiol. 1998;28:1223–1234. doi: 10.1046/j.1365-2958.1998.00885.x. [DOI] [PubMed] [Google Scholar]
- 62.Idei A, Kawai E, Akatsuka H, Omori K. J Bacteriol. 1999;181:7545–7551. doi: 10.1128/jb.181.24.7545-7551.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Rossi MS, Fetherston JD, Letoffe S, Carniel E, Perry RD, Ghigo JM. Infect Immun. 2001;69:6707–6717. doi: 10.1128/IAI.69.11.6707-6717.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Stojiljkovic I, Hantke K. Mol Microbiol. 1994;13:719–732. doi: 10.1111/j.1365-2958.1994.tb00465.x. [DOI] [PubMed] [Google Scholar]
- 65.Hanson MS, Pelzel SE, Latimer J, Muller-Eberhard U, Hansen EJ. Proc Natl Acad Sci U S A. 1992;89:1973–1977. doi: 10.1073/pnas.89.5.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Cope LD, Thomas SE, Hrkal Z, Hansen EJ. Infect Immun. 1998;66:4511–4516. doi: 10.1128/iai.66.9.4511-4516.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Deniau C, Gilli R, Izadi-Pruneyre N, Letoffe S, Delepierre M, Wandersman C, Briand C, Lecroisey A. Biochemistry. 2003;42:10627–10633. doi: 10.1021/bi030015k. [DOI] [PubMed] [Google Scholar]
- 68.Izadi N, Henry Y, Haladjian J, Goldberg ME, Wandersman C, Delepierre M, Lecroisey A. Biochemistry. 1997;36:7050–7057. doi: 10.1021/bi962577s. [DOI] [PubMed] [Google Scholar]
- 69.Letoffe S, Deniau C, Wolff N, Dassa E, Delepelaire P, Lecroisey A, Wandersman C. Mol Microbiol. 2001;41:439–450. doi: 10.1046/j.1365-2958.2001.02530.x. [DOI] [PubMed] [Google Scholar]
- 70.Caillet-Saguy C, Delepierre M, Lecroisey A, Bertini I, Piccioli M, Turano P. J Am Chem Soc. 2006;128:150–158. doi: 10.1021/ja054902h. [DOI] [PubMed] [Google Scholar]
- 71.Cwerman H, Wandersman C, Biville F. J Bacteriol. 2006;188:3357–3364. doi: 10.1128/JB.188.9.3357-3364.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Benevides-Matos N, Wandersman C, Biville F. J Bacteriol. 2008;190:21–27. doi: 10.1128/JB.01389-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Lefevre J, Delepelaire P, Delepierre M, Izadi-Pruneyre N. J Mol Biol. 2008;378:840–851. doi: 10.1016/j.jmb.2008.03.044. [DOI] [PubMed] [Google Scholar]
- 74.Letoffe S, Wecker K, Delepierre M, Delepelaire P, Wandersman C. J Bacteriol. 2005;187:4637–4645. doi: 10.1128/JB.187.13.4637-4645.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Bracken CS, Baer MT, Abdur-Rashid A, Helms W, Stojiljkovic I. J Bacteriol. 1999;181:6063–6072. doi: 10.1128/jb.181.19.6063-6072.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Letoffe S, Delepelaire P, Wandersman C. J Bacteriol. 2004;186:4067–4074. doi: 10.1128/JB.186.13.4067-4074.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Letoffe S, Nato F, Goldberg ME, Wandersman C. Mol Microbiol. 1999;33:546–555. doi: 10.1046/j.1365-2958.1999.01499.x. [DOI] [PubMed] [Google Scholar]
- 78.Izadi-Pruneyre N, Huche F, Lukat-Rodgers GS, Lecroisey A, Gilli R, Rodgers KR, Wandersman C, Delepelaire P. J Biol Chem. 2006;281:25541–25550. doi: 10.1074/jbc.M603698200. [DOI] [PubMed] [Google Scholar]
- 79.Letoffe S, Debarbieux L, Izadi N, Delepelaire P, Wandersman C. Mol Microbiol. 2003;50:77–88. doi: 10.1046/j.1365-2958.2003.03686.x. [DOI] [PubMed] [Google Scholar]
- 80.Cope LD, Yogev R, Muller-Eberhard U, Hansen EJ. J Bacteriol. 1995;177:2644–2653. doi: 10.1128/jb.177.10.2644-2653.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Cope LD, Thomas SE, Latimer JL, Slaughter CA, Muller-Eberhard U, Hansen EJ. Mol Microbiol. 1994;13:863–873. doi: 10.1111/j.1365-2958.1994.tb00478.x. [DOI] [PubMed] [Google Scholar]
- 82.Morton DJ, Seale TW, Madore LL, VanWagoner TM, Whitby PW, Stull TL. Microbiology. 2007;153:215–224. doi: 10.1099/mic.0.2006/000190-0. [DOI] [PubMed] [Google Scholar]
- 83.Cope LD, Love RP, Guinn SE, Gilep A, Usanov S, Estabrook RW, Hrkal Z, Hansen EJ. Infect Immun. 2001;69:2353–2363. doi: 10.1128/IAI.69.4.2353-2363.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Ochsner UA, Johnson Z, Vasil ML. Microbiology. 2000;146(Pt 1):185–198. doi: 10.1099/00221287-146-1-185. [DOI] [PubMed] [Google Scholar]
- 85.Stojiljkovic I, Perkins-Balding D. DNA Cell Biol. 2002;21:281–295. doi: 10.1089/104454902753759708. [DOI] [PubMed] [Google Scholar]
- 86.Friedman J, Lad L, Li H, Wilks A, Poulos TL. Biochemistry. 2004;43:5239–5245. doi: 10.1021/bi049687g. [DOI] [PubMed] [Google Scholar]
- 87.Wandersman C, Stojiljkovic I. Curr Opin Microbiol. 2000;3:215–220. doi: 10.1016/s1369-5274(00)00078-3. [DOI] [PubMed] [Google Scholar]
- 88.Simpson W, Olczak T, Genco CA. J Bacteriol. 2000;182:5737–5748. doi: 10.1128/jb.182.20.5737-5748.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Delanghe JR, Langlois MR. Clin Chim Acta. 2001;312:13–23. doi: 10.1016/s0009-8981(01)00586-1. [DOI] [PubMed] [Google Scholar]
- 90.Wong JC, Holland J, Parsons T, Smith A, Williams P. Infect Immun. 1994;62:48–59. doi: 10.1128/iai.62.1.48-59.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Stojiljkovic I, Hwa V, de Saint Martin L, O’Gaora P, Nassif X, Heffron F, So M. Mol Microbiol. 1995;15:531–541. doi: 10.1111/j.1365-2958.1995.tb02266.x. [DOI] [PubMed] [Google Scholar]
- 92.Ren Z, Jin H, Morton DJ, Stull TL. Infect Immun. 1998;66:4733–4741. doi: 10.1128/iai.66.10.4733-4741.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Morton DJ, Whitby PW, Jin H, Ren Z, Stull TL. Infect Immun. 1999;67:2729–2739. doi: 10.1128/iai.67.6.2729-2739.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Olczak T, Sroka A, Potempa J, Olczak M. Arch Microbiol. 2008;189:197–210. doi: 10.1007/s00203-007-0309-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Karunakaran T, Madden T, Kuramitsu H. J Bacteriol. 1997;179:1898–1908. doi: 10.1128/jb.179.6.1898-1908.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Olczak T, Dixon DW, Genco CA. J Bacteriol. 2001;183:5599–5608. doi: 10.1128/JB.183.19.5599-5608.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Olczak T, Simpsonv W, Liu X, Genco CA. FEMS Microbiol Rev. 2005;29:119–144. doi: 10.1016/j.femsre.2004.09.001. [DOI] [PubMed] [Google Scholar]
- 98.Liu X, Olczak T, Guo HC, Dixon DW, Genco CA. Infect Immun. 2006;74:1222–1232. doi: 10.1128/IAI.74.2.1222-1232.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Braun V. FEMS Microbiol Rev. 1995;16:295–307. doi: 10.1111/j.1574-6976.1995.tb00177.x. [DOI] [PubMed] [Google Scholar]
- 100.Letain TE, Postle K. Mol Microbiol. 1997;24:271–283. doi: 10.1046/j.1365-2958.1997.3331703.x. [DOI] [PubMed] [Google Scholar]
- 101.Higgs PI, Larsen RA, Postle K. Mol Microbiol. 2002;44:271–281. doi: 10.1046/j.1365-2958.2002.02880.x. [DOI] [PubMed] [Google Scholar]
- 102.Postle K, Skare JT. J Biol Chem. 1988;263:11000–11007. [PubMed] [Google Scholar]
- 103.Paquelin A, Ghigo JM, Bertin S, Wandersman C. Mol Microbiol. 2001;42:995–1005. doi: 10.1046/j.1365-2958.2001.02628.x. [DOI] [PubMed] [Google Scholar]
- 104.Pawelek PD, Croteau N, Ng-Thow-Hing C, Khursigara CM, Moiseeva N, Allaire M, Coulton JW. Science. 2006;312:1399–1402. doi: 10.1126/science.1128057. [DOI] [PubMed] [Google Scholar]
- 105.Postle K, Kadner RJ. Mol Microbiol. 2003;49:869–882. doi: 10.1046/j.1365-2958.2003.03629.x. [DOI] [PubMed] [Google Scholar]
- 106.Schoffler H, Braun V. Mol Gen Genet. 1989;217:378–383. doi: 10.1007/BF02464907. [DOI] [PubMed] [Google Scholar]
- 107.Bell PE, Nau CD, Brown JT, Konisky J, Kadner RJ. J Bacteriol. 1990;172:3826–3829. doi: 10.1128/jb.172.7.3826-3829.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Moeck GS, Letellier L. J Bacteriol. 2001;183:2755–2764. doi: 10.1128/JB.183.9.2755-2764.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Chang C, Mooser A, Pluckthun A, Wlodawer A. J Biol Chem. 2001;276:27535–27540. doi: 10.1074/jbc.M102778200. [DOI] [PubMed] [Google Scholar]
- 110.Zhao Q, Poole K. FEMS Microbiol Lett. 2000;184:127–132. doi: 10.1111/j.1574-6968.2000.tb09002.x. [DOI] [PubMed] [Google Scholar]
- 111.Occhino DA, Wyckoff EE, Henderson DP, Wrona TJ, Payne SM. Mol Microbiol. 1998;29:1493–1507. doi: 10.1046/j.1365-2958.1998.01034.x. [DOI] [PubMed] [Google Scholar]
- 112.Heidelberg JF, Eisen JA, Nelson WC, Clayton RA, Gwinn ML, Dodson RJ, Haft DH, Hickey EK, Peterson JD, Umayam L, Gill SR, Nelson KE, Read TD, Tettelin H, Richardson D, Ermolaeva MD, Vamathevan J, Bass S, Qin H, Dragoi I, Sellers P, McDonald L, Utterback T, Fleishmann RD, Nierman WC, White O, Salzberg SL, Smith HO, Colwell RR, Mekalanos JJ, Venter JC, Fraser CM. Nature. 2000;406:477–483. doi: 10.1038/35020000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Seliger SS, Mey AR, Valle AM, Payne SM. Mol Microbiol. 2001;39:801–812. doi: 10.1046/j.1365-2958.2001.02273.x. [DOI] [PubMed] [Google Scholar]
- 114.Higgins CF, Linton KJ. Nat Struct Mol Biol. 2004;11:918–926. doi: 10.1038/nsmb836. [DOI] [PubMed] [Google Scholar]
- 115.Hanson MS, Hansen EJ. Mol Microbiol. 1991;5:267–278. doi: 10.1111/j.1365-2958.1991.tb02107.x. [DOI] [PubMed] [Google Scholar]
- 116.Hanson MS, Slaughter C, Hansen EJ. Infect Immun. 1992;60:2257–2266. doi: 10.1128/iai.60.6.2257-2266.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Morton DJ, Madore LL, Smith A, Vanwagoner TM, Seale TW, Whitby PW, Stull TL. FEMS Microbiol Lett. 2005;253:193–199. doi: 10.1016/j.femsle.2005.09.016. [DOI] [PubMed] [Google Scholar]
- 118.Letoffe S, Delepelaire P, Wandersman C. Proc Natl Acad Sci U S A. 2006;103:12891–12896. doi: 10.1073/pnas.0605440103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Karpowich NK, Huang HH, Smith PC, Hunt JF. J Biol Chem. 2003;278:8429–8434. doi: 10.1074/jbc.M212239200. [DOI] [PubMed] [Google Scholar]
- 120.Kandt C, Xu Z, Tieleman DP. Biochemistry. 2006;45:13284–13292. doi: 10.1021/bi061280j. [DOI] [PubMed] [Google Scholar]
- 121.Clarke TE, Braun V, Winkelmann G, Tari LW, Vogel HJ. J Biol Chem. 2002;277:13966–13972. doi: 10.1074/jbc.M109385200. [DOI] [PubMed] [Google Scholar]
- 122.Burkhard KA, Wilks A. Biochemistry. 2008;47:7977–7979. doi: 10.1021/bi801005u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Chan AC, Lelj-Garolla B, Pedersen IRFKA, Mauk AG, Murphy ME. J Mol Biol. 2006;362:1108–1119. doi: 10.1016/j.jmb.2006.08.001. [DOI] [PubMed] [Google Scholar]
- 124.Wilks A. Antioxid Redox Signal. 2002;4:603–614. doi: 10.1089/15230860260220102. [DOI] [PubMed] [Google Scholar]
- 125.Maines MD. Annu Rev Pharmacol Toxicol. 1997;37:517–554. doi: 10.1146/annurev.pharmtox.37.1.517. [DOI] [PubMed] [Google Scholar]
- 126.Suits MD, Pal GP, Nakatsu K, Matte A, Cygler M, Jia Z. Proc Natl Acad Sci U S A. 2005;102:16955–16960. doi: 10.1073/pnas.0504289102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Schneider S, Sharp KH, Barker PD, Paoli M. J Biol Chem. 2006;281:32606–32610. doi: 10.1074/jbc.M607516200. [DOI] [PubMed] [Google Scholar]
- 128.Wilks A. Arch Biochem Biophys. 2001;387:137–142. doi: 10.1006/abbi.2000.2250. [DOI] [PubMed] [Google Scholar]
- 129.Wyckoff EE, Lopreato GF, Tipton KA, Payne SM. J Bacteriol. 2005;187:5658–5664. doi: 10.1128/JB.187.16.5658-5664.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Thompson JM, Jones HA, Perry RD. Infect Immun. 1999;67:3879–3892. doi: 10.1128/iai.67.8.3879-3892.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Wyckoff EE, Duncan D, Torres AG, Mills M, Maase K, Payne SM. Mol Microbiol. 1998;28:1139–1152. doi: 10.1046/j.1365-2958.1998.00873.x. [DOI] [PubMed] [Google Scholar]
- 132.Block DR, Lukat-Rodgers GS, Rodgers KR, Wilks A, Bhakta MN, Lansky IB. Biochemistry. 2007;46:14391–14402. doi: 10.1021/bi701509n. [DOI] [PubMed] [Google Scholar]
- 133.Bhakta MN, Wilks A. Biochemistry. 2006;45:11642–11649. doi: 10.1021/bi060980l. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Kikuchi G, Yoshida T, Noguchi M. Biochem Biophys Res Commun. 2005;338:558–567. doi: 10.1016/j.bbrc.2005.08.020. [DOI] [PubMed] [Google Scholar]
- 135.Ratliff M, Zhu W, Deshmukh R, Wilks A, Stojiljkovic I. J Bacteriol. 2001;183:6394–6403. doi: 10.1128/JB.183.21.6394-6403.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Zhu W, Wilks A, Stojiljkovic I. J Bacteriol. 2000;182:6783–6790. doi: 10.1128/jb.182.23.6783-6790.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Wilks A, Schmitt MP. J Biol Chem. 1998;273:837–841. doi: 10.1074/jbc.273.2.837. [DOI] [PubMed] [Google Scholar]
- 138.Navarre WW, Schneewind O. Microbiol Mol Biol Rev. 1999;63:174–229. doi: 10.1128/mmbr.63.1.174-229.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Schmitt MP, Drazek ES. J Bacteriol. 2001;183:1476–1481. doi: 10.1128/JB.183.4.1476-1481.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Bates CS, Montanez GE, Woods CR, Vincent RM, Eichenbaum Z. Infect Immun. 2003;71:1042–1055. doi: 10.1128/IAI.71.3.1042-1055.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Lei B, Smoot LM, Menning HM, Voyich JM, Kala SV, Deleo FR, Reid SD, Musser JM. Infect Immun. 2002;70:4494–4500. doi: 10.1128/IAI.70.8.4494-4500.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Lei B, Liu M, Voyich JM, Prater CI, Kala SV, DeLeo FR, Musser JM. Infect Immun. 2003;71:5962–5969. doi: 10.1128/IAI.71.10.5962-5969.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Liu M, Lei B. Infect Immun. 2005;73:5086–5092. doi: 10.1128/IAI.73.8.5086-5092.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Zhu H, Liu M, Lei B. BMC Microbiol. 2008;8:15. doi: 10.1186/1471-2180-8-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Sook BR, Block DR, Sumithran S, Montanez GE, Rodgers KR, Dawson JH, Eichenbaum Z, Dixon DW. Biochemistry. 2008;47:2678–2688. doi: 10.1021/bi701604y. [DOI] [PubMed] [Google Scholar]
- 146.Mazmanian SK, Skaar EP, Gaspar AH, Humayun M, Gornicki P, Jelenska J, Joachmiak A, Missiakas DM, Schneewind O. Science. 2003;299:906–909. doi: 10.1126/science.1081147. [DOI] [PubMed] [Google Scholar]
- 147.Liu M, Tanaka WN, Zhu H, Xie G, Dooley DM, Lei B. J Biol Chem. 2008;283:6668–6676. doi: 10.1074/jbc.M708372200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Sharp KH, Schneider S, Cockayne A, Paoli M. J Biol Chem. 2007;282:10625–10631. doi: 10.1074/jbc.M700234200. [DOI] [PubMed] [Google Scholar]
- 149.Grigg JC, Vermeiren CL, Heinrichs DE, Murphy ME. J Biol Chem. 2007;282:28815–28822. doi: 10.1074/jbc.M704602200. [DOI] [PubMed] [Google Scholar]
- 150.Vermeiren CL, Pluym M, Mack J, Heinrichs DE, Stillman MJ. Biochemistry. 2006;45:12867–12875. doi: 10.1021/bi0607711. [DOI] [PubMed] [Google Scholar]
- 151.Cabanes D, Dehoux P, Dussurget O, Frangeul L, Cossart P. Trends Microbiol. 2002;10:238–245. doi: 10.1016/s0966-842x(02)02342-9. [DOI] [PubMed] [Google Scholar]
- 152.Hantke K. Curr Opin Microbiol. 2001;4:172–177. doi: 10.1016/s1369-5274(00)00184-3. [DOI] [PubMed] [Google Scholar]
- 153.Coy M, Neilands JB. Biochemistry. 1991;30:8201–8210. doi: 10.1021/bi00247a016. [DOI] [PubMed] [Google Scholar]
- 154.Stojiljkovic I, Hantke K. Mol Gen Genet. 1995;247:199–205. doi: 10.1007/BF00705650. [DOI] [PubMed] [Google Scholar]
- 155.Lavrrar JL, Christoffersen CA, McIntosh MA. J Mol Biol. 2002;322:983–995. doi: 10.1016/s0022-2836(02)00849-5. [DOI] [PubMed] [Google Scholar]
- 156.Vanderpool CK, Armstrong SK. J Bacteriol. 2003;185:909–917. doi: 10.1128/JB.185.3.909-917.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Vanderpool CK, Armstrong SK. J Bacteriol. 2004;186:938–948. doi: 10.1128/JB.186.4.938-948.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Rossi MS, Paquelin A, Ghigo JM, Wandersman C. Mol Microbiol. 2003;48:1467–1480. doi: 10.1046/j.1365-2958.2003.03516.x. [DOI] [PubMed] [Google Scholar]
- 159.Koronakis V, Sharff A, Koronakis E, Luisi B, Hughes C. Nature. 2000;405:914–919. doi: 10.1038/35016007. [DOI] [PubMed] [Google Scholar]
- 160.Delepelaire P, Wandersman C. Embo J. 1998;17:936–944. doi: 10.1093/emboj/17.4.936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Letoffe S, Delepelaire P, Wandersman C. Embo J. 1996;15:5804–5811. [PMC free article] [PubMed] [Google Scholar]
- 162.DeLano WL. The PyMOL User’s Manual. DeLano Scientific; San Carlos: 2002. [Google Scholar]
- 163.Reniere ML, Torres VJ, Skaar EP. Biometals. 2007;20:333–345. doi: 10.1007/s10534-006-9032-0. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.