Summary
Gonadotropin-releasing hormone (GnRH) regulates biosythesis in the pituitary gonadotrope via a complex signaling and gene network. Small non-coding microRNAs (miRNA) can play important roles in gene expression. We investigated the microtranscriptome in the mouse LβT2 gonadotrope cell line using microarray, single molecule coincidence detection assays, hairpin real time PCR and LNA (locked nucleic acid) primer-extension PCR. Expression of nearly 200 miRNAs were detected by array and a panel of 101 hairpin real-time PCR assays. Within this broad family of expressed miRNAs, GnRH induced upregulation of two miRNA products of the same primary transcript, miR-132 and miR-212, a result confirmed by single molecule, hairpin and LNA assays. Induction peaked 6 hours after GnRH exposure and showed no significant frequency sensitivity. Bioinformatics analysis was used to predict potential targets of each of these GnRH-regulated miRNAs. These findings suggest the importance of the microtranscriptome in gene control in the gonadotrope and implicate miR-132 and miR-212 in the regulation of GnRH-stimulated biosynthetic response.
Keywords: mouse, cell line, gonadotrope, microRNA, reproduction, pituitary
1. Introduction
Gonadotropin-releasing hormone (GnRH) mediates the hypothalamic control of gonadotropin gene induction and biosynthesis in the pituitary gonadotrope. GnRH binds to a high affinity heptahelical G-protein coupled receptor on the gonadotrope membrane and modulates a variety of signaling cascades, including inositol phosphate signaling, calcium mobilization, protein kinase C activation, and various phosphorylation cascades including the mitogen activated protein kinases ERK, p38 MK, and JNK (Ruf et al., 2003, Ruf and Sealfon, 2004). These intracellular signaling changes modulate a layered gene network consisting of dozens of immediate early genes and secondary genes. The initial wave of GnRH-activated genes encodes transcription factors that converge to regulate the gonadotropin genes, as well as regulatory proteins that feed back to the signaling pathway. Understanding the mechanisms by which this complex information transfer system integrates data about GnRH frequency and other extracellular signals to control reproductive timing and competency requires clarifying both the topology (connections) and the global dynamics (regulatory changes over time) of the components of the network.
Genomics studies have characterized the overall changes in mRNA expression in the gonadotrope induced by GnRH (Lawson et al., 2007, Wurmbach et al., 2001, Yuen et al., 2002). However, little is known about the expression and regulation of an important, more recently recognized class of genes, those encoding microRNAs (miRNA). miRNAs are small, approximately 22 nucleotide gene products that are increasingly recognized to serve, like transcription factors, as the basis for a combinatorial code that contributes to the regulation of expression of specific genes and proteins (Hobert, 2008). miRNAs hybridize with complementary 3′-UTR mRNA sequences leading to their recruitment into specialized protein complexes that mediate mRNA degradation, sequestration or translational repression (Williams, 2008). Given the complex orchestration of biosynthetic regulation in the gonadotrope that is necessary for normal reproductive physiology, it is important to study the role of miRNAs in these regulatory processes.
Because the study of miRNAs and the technologies for their measurement are relatively new, it is valuable to compare results using different methodologies. In order to obtain a reliable assessment of miRNA expression and regulation, we utilized four different approaches to meaure miRNAs in the LβT2 gonadotrope cell line. We identify expression of a large number of miRNAs in these cells and demonstrate using multiple assay technologies that GnRH induces the selective regulation of two miRNAs that are the product of the same gene.
2. Materials and Methods
2.1 Cell culture and RNA sample preparation
LβT2 gonadotrope cells (Turgeon et al., 1996) obtained from Pamela Mellon (University of California, San Diego) were maintained at 37 °C in 5% CO2 in humidified air in DMEM (Mediatech, Herndon, VA) containing 10% fetal bovine serum (Gemini, Calabasas, CA). 40–50 million cells were seeded in 15 cm dishes and medium was replaced 24 hours later with DMEM containing 25 mM HEPES (Mediatech) and glutamine and 10% charcoal stripped fetal bovine serum. On the next day, the cells were treated with 100 nM GnRH (Bachem, Torrance, CA) or vehicle and were returned to the CO2 incubator for 3 hours before harvesting for microarray and real time PCR experiments. For pulsing experiments, cells were incubated with 100 nM GnRH or vehicle for 10 minutes, washed once with DMEM, and incubated further with DMEM for 20, 50 or 110 minutes (for 30 min, 1 hour or 2 hours pulse frequencies). Cycles of GnRH or vehicle treatment were repeated and 6 replicate samples from each treatment were collected after 6 hours and 18 hours. Total RNA was isolated using mirVana miRNA Isolation Kit (ABI/Ambion, Austin, TX) according to the manufacturer’s protocol.
2.2 miRNA microarray printing, sample labeling and processing
LβT2 cells were treated with 100 nM GnRH or vehicle for 3 hours and total RNA was isolated using mirVana miRNA Isolation Kit. Small RNAs (<40 nucleotides) were size-selected by running through a flashPAGE fractionator (ABI/Ambion) with a starting quantity of 100 μg of total RNA. The small RNAs were precipitated overnight with 10 μg of glycogen (ABI/Ambion) as carrier and resuspended in 15 μl of DEPC-water for microarray analysis. A total of 350 oligonucleotides (mirVana miRNA probe set 1564V1, ABI/Ambion) complimentary to known miRNA sequences were printed in triplicate on SuperEpoxy glass slides (ArrayIt, Sunnyvale, CA) using a BioRobotics microGRID II arrayer (Digilab, Ann Arbor, MI). Two identical arrays were printed on each slide. Four control biosynthesized oligos (Control 1–4) were supplied and included in the array for labeling and hybridization quality control. Control 1 oligo which recognized the spiked-in control miRNA served as a positive control; whereas Control 2–4 oligos were derived from prokaryotic sequences and served as negative controls. Several anti-sense probes containing complementary sequence of known miRNAs were also included in the library and printed for additional hybridization quality control. A dye-swap assay was performed. The paired dye-swapped targets were hybridized onto the two arrays on a microarray slide. Three microliters of the PAGE-purified small RNA solution (equivalent to 20 μg of total RNA) spiked with miRNA positive control were labeled with either Cy-3 or Cy-5 (Amersham/GE Healthcare, Piscataway, NJ) using mirVana miRNA Labeling Kit. Hybridization and post-processing were carried out following the manufacturer’s instructions (mirVana miRNA Probe Set Kit). A raised-edge coverslip (Lifter-Slips, Erie Scientific) was used to improve hybridization efficiency. The fluorescence image was captured using a ScanArray HT scanner (PerkinElmer, Waltham, Massachusetts) and analyzed using ImaGene 6.0 (BioDiscovery, El Seguno, CA). The images were initially examined manually and the spots with non-specific brightness were flagged. Empty spot (signal < 16), negative spot (signal < 0) or spot whose signal intensity is less than 2 standard deviation of background intensity for both channels was flagged automatically by the program. The success of sample labeling and hybridization were evaluated with the control spots on the microarray where Control 1 spots showed strong signal intensity, and Control 2–4 spots as well as the mouse anti-sense spots were flagged as well as by consistency of results obtained with dye swapping. The actual signal intensity for each spot was corrected by subtracting its local median background intensity and then was log based 2 transformed. The global signal intensity was normalized by using a robust locally linear normalization function (Lowess) comparing the results obtained with each of the two fluorescence channels within each subgrid. All flagged spots were excluded from further analysis. Median signal intensity of each miRNA probe was calculated from the triplicates on an array. Non-flagged spots represented the expression of the corresponding miRNAs.
2.3 Single molecule miRNA detection analysis
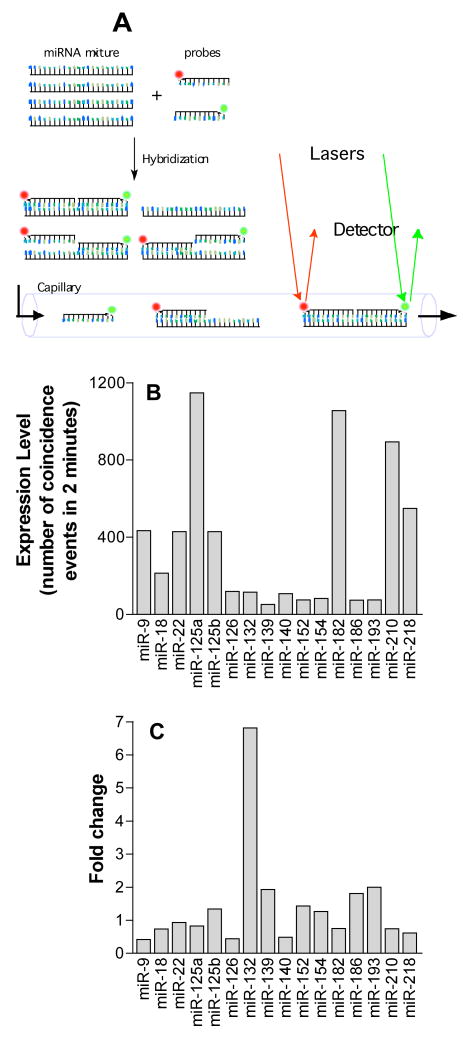
A published protocol developed by US Genomics (USG, Boston, MA) was followed as described (Chan et al., 2004, Neely et al., 2006). Briefly, a pair of LNA-DNA chimeric oligonucleotide probes, labeled with either Oyster 556 or Oyster 656, was designed for each target miRNA. In each of the miRNA assays, 1 nM probes were hybridized with 2 μg total RNA from LβT2 cells in the presence of 0.5 μl of RNase inhibitor (USB Corporation, Cleveland, OH) and 1X USG hybridization buffer for 2 hours at 55 °C. A final concentration of 1.3 nM of DNA quencher probes, containing A-quenchers (Exiqon, Woburn, MA) and synthetic miRNA templates (Integrated DNA Technologies, Coralville, IA) were added and the quenching reaction was allowed to proceed for 1 hour at 40 °C. The samples were diluted 10-fold in USG dilution buffer A before assay using a Trilogy 2020 Single Molecule Analyzer (US Genomics). Macromolecules were streamed through a microfluidic channel with a confocal laser-induced fluorescence detector (Fig. 1A). Single molecules of target miRNA were distinguished from background by counting the spatial coincidence events of the two fluorescent signals for each of the probes.
Figure 1.
A. Schematic of single molecule detection assay. A pair of LNA-DNA chimeric oligonucleotide probes complementary to specific miRNA targets were labeled with either Oyster 556 (green) or Oyster 656 (red). The labeled probes were hybridized with miRNA samples from LβT2 gonadotropes, and then were passed through a microfluidic capillary for confocal laser-induced fluorescence detection. Coincidence events of the green and red signals, which represent a single miRNA target molecule, were counted. B. Expression level of 16 miRNAs in control LβT2 cells determined by single molecule detection assay. C. Change in miRNA levels in LβT2 cells following 3 hour GnRH treatment.
2.4 Hairpin miRNA real-time PCR analysis
Quantitative real time PCR was performed in an ABI Prism 7900HT using TaqMan microRNA Human Panel (Early Access Kit) (P/N 4365381, Applied Biosystems, Forster city, CA) according to the manufacturer’s protocol. Briefly, 10 ng of total RNA was mixed with 1 U MultiScribe Reverse Transcriptase, 0.25 U RNase Inhibitor, 3 μl hairpin-looped miRNA-specific RT primer, 1 mM dNTPs and 1X Reverse Transcription Buf fer in a total volume of 15 μl. The RT primer contains DNA sequence complementary to the 3′ end of the miRNA to be assayed, and additional DNA sequence at the 5′ side of the primer, which forms a hairpin loop. This additional sequence provides an additional priming site for PCR amplification of the first strand cDNA from the miRNA template. The mixture was incubated at 16 °C for 30 minutes, 42 °C for another 30 minutes, and the reaction was stopped by heating to 85 °C for 5 minutes. Real time PCR reaction was set up in 20 μl volume with 1.33 μl first strand cDNA, 1X TaqMan MicroRNA Assay Mix and 1X TaqMan Universal PCR Master Mix. After activation of the AmpliTaq Gold DNA polymerase at 95 C for 10 minutes, 40 cycles of two-step PCR were run (95 °C for 15 seconds and 60 °C for 60 seconds). Data were collected and analyzed with SDS v.2.2.2 software (Applied Biosystems). The TaqMan microRNA Human Panel contains 157 human miRNA assays as well as several control assays. 101 of the mouse and human miRNA sequences included in the assay panel are identical and therefore 101 assays were determined to be suitable for mouse miRNA. Separate assays of several microRNAs were using the following synthesized primers and detection sequences. Let-7a: let-7a/RT (5′-GTCGTATCCAGTGCAGGGTCCGAGGTATTCGCACTGGATACGACAACTA-3′), let-7a/F (5′-GCCGCTGAGGTAGTAGGTTGTA-3′), let-7a/R (5′-GTGCAGGGTCCGAGGT-3′), let-7a/probe (5′-FAM-TGGATACGACAACTATAC-MGB-3′) miR-132: miR-132/RT (5′-GTCGTATCCAGTGCAGGGTCCGAGGTATTCGCACTGGATACGACCGACCA-3′) miR-132/F (5′-GCCGCTAACAGTCTACAGCCAT-3′) miR-132/R (5′-GTGCAGGGTCCGA GGT-3′) miR-132/probe (5′-FAM-TGGATACGACCGACCAT-MGB-3′) miR-212: miR-212/RT (5′-GTCGTATCCAGTGCAGGGTCCGAGGTAGGCGCACTGGATACGACGGCCGT -3′) miR-212/F (5′-GCCGCTAACAGTCTCCAGTCA-3′) miR-212/R (5′-GTGCAGGGTCCGAGGT-3′) miR-212/probe (5′-FAM-TGGATACGACGGCCGTG-MGB-3′). RNA oligonucleotides for miR-132 (5′-UAACAGUCUACAGCCAUGGUCG-3′) and miR-212 (5′-UAACAGUCUCCAGUCACGGCCA-3′) were used to generate standard curves for the miR-132 and miR-212 assays (Fig. S2).
2.5 Locked nucleic acid (LNA) microRNA analysis
Reactions were performed using the miRCURY locked nucleic acid (LNA) microRNA PCR system (Exiqon) according to the manufacturer’s recommendation. Briefly, 10 ng of total RNA was reverse transcribed into first strand cDNA in a reaction volume of 10 μl by incubating with 10 U Transcriptor reverse transcriptase, 20 U RNase inhibitor, 2 μl miR-specific reverse primer, 1X RT reaction buffer and 0.5 mM dNTP mix at 50 °C for 30 minutes. The first strand cDNA was then diluted to 50 μl. Real time PCR reaction was set up in 20 μl volume with 4 μl diluted first strand cDNA, 1X SYBR Green master mix, 1 μl LNA PCR primer and 1 μl Universal PCR primer. After an initial step of 95 °C for 10 minutes to activate the DNA polymerase, 40 two-step PCR cycles (95 °C for 20 seconds and 60 °C for 60 seconds) were performed in an ABI Prism 7900HT Real Time PCR machine. A dissociation run was performed after the PCR to ensure that a single product was amplified. SDS v.2.2.2 software (Applied Biosystems) was used for data collection and analysis.
2.6 Bioinformatic Target Prediction for miR-132 and miR-212
In order to identify potential targets for the two GnRH-regulated miRNAs identified, miRNA target prediction was undertaken using five different software tools: miRDB, v2.0 (http://mirdb.org/miRDB/) (Wang, 2008), miRanda, September 2008 Release (http://www.microrna.org/microrna/home.do) (John et al. 2004), miRBase, v5 (http://microrna.sanger.ac.uk/targets/v5/) (Griffiths-Jones et al., 2006), PicTar (http://pictar.mdc-berlin.de/) (Krek et al., 2005) and Target Scan, Release 4.2 (http://www.targetscan.org/index.html) (Lewis et al., 2003). A threshold score was selected for each prediction software in order to obtain between 100 and 300 targets for each miRNA evaluated (score ≥ 60 for miRDB, score ≥ 160 for miRanda, score ≥ 17 for miRBase, score ≥ 0 for PicTar, and score ≤ 0 for Target Scan). A list of potential targets was generated according to: 1. identification on at least two different analyses. 2. showing expression by Affymetric GeneChip U74A and U74Av2 microarray assay in LβT2 cells. This list was ordered in terms of how many different software analyses identified each target as well as by the rank within each software analysis.
3. Results
3.1 Characterization of global microtranscriptome in LβT2 cells
Two different approaches were used to characterize the global expression of miRNAs in gonadotrope cells, an oligonucleotide based microarray and a panel of hairpin real-time PCR assays. The oligonucleotide microarray assayed 350 miRNA targets and detected putative expression of a total of 175 (50%) of the miRNAs sequences tested (Table S1). Although the PCR panel was designed against human sequence, only the 101 assays which are identical in mouse were used for this study (see methods). Using this mouse PCR assay panel of 101 targets, 89 (88%) were found to be expressed (Table S1). The discrepancy between the percent of tested miRNAs detected by the two methodologies may reflect the lower sensitivity of the array assay for low level miRNAs, as well as inclusion of 147 miRNA targets on the array that may be spurious as they are not included in the Sanger miRNA mouse registry. If only the 203 microarray sequences included in the Sanger registry are considered, the array detects 139 or 68%, which is closer to the level of detection observed by the hairpin PCR panel. Overall, these results indicate that a large fraction of identified miRNAs are expressed in the gonadotrope.
3.2 Selective regulation of a specific miRNA gene by GnRH determined by single molecule miRNA assays
Constitutive expression of miRNAs are important mechanisms for developmental control of the expression levels of specific genes in differentiation of cell types (Kanellopoulou and Monticelli, 2008, Stefani and Slack, 2008). While the role of miRNAs in controlling gene expression is well established, the mechanisms controlling the regulation of miRNAs are less well studied. It has recently been found that some miRNAs are regulated by various types of cell signaling events (Coller et al., 2007, Kulshreshtha et al., 2007). In order to further understand the potential role of miRNAs in gonadotrope physiology, we next investigated the regulation of the microtranscriptome by GnRH.
The effects of GnRH on the expression of miRNAs were first studied using a single-molecule coincidence assay for miRNAs. This assay is based on hybridizing RNA isolated from LβT2 cells with two short locked nucleic acid (LNA) sequences, each labeled with a different fluorophore for each miRNA tested. The level of each miRNA is then assayed by using a microfluidics device to measure the number of events for which spatial coincidence of the two fluorophores is observed (Fig. 1A,B). The approach, which was originally developed for single molecule gene sequencing, provides single molecule miRNA detection (Neely et al., 2006). Using this assay, 16 miRNAs were tested in RNA samples from vehicle and GnRH-treated LβT2 cells. The results showed a selective regulation of miR-132 (Fig. 1C).
3.3. GnRH Regulation of miR-132 and miR-212
In order to validate and extend the results obtained with single molecule detection assays, we studied global regulation of the microtranscriptome using the miRNA array. When comparing miRNA expression on arrays hybridized with samples from GnRH- and vehicle-exposed LβT2 cells, statistically significant regulation was not observed for any miRNA assayed on the array (data not shown). While miR-132 was detected as expressed in LβT2 cells by the array (Table S1), it was not found to be significantly regulated by GnRH. Thus the results obtained using the global miRNA array contrast with the results obtained using single molecule detection (Fig. 1). These contradictory results obtained using two emerging technologies underscore the need to be cautious in interpreting results obtained relying on only one assay methodology.
In order to resolve the question of whether miR-132 was regulated by GnRH, additional studies were performed using the hairpin PCR panel for 101 miRNAs. When samples from cells treated for 0, 1, 3 and 6 and 24 hours were assayed with the panel, specific induction of miR-132 was observed at all time points (Table S2). These results support the finding by single-molecule detection assays that miR-132 was induced by GnRH.
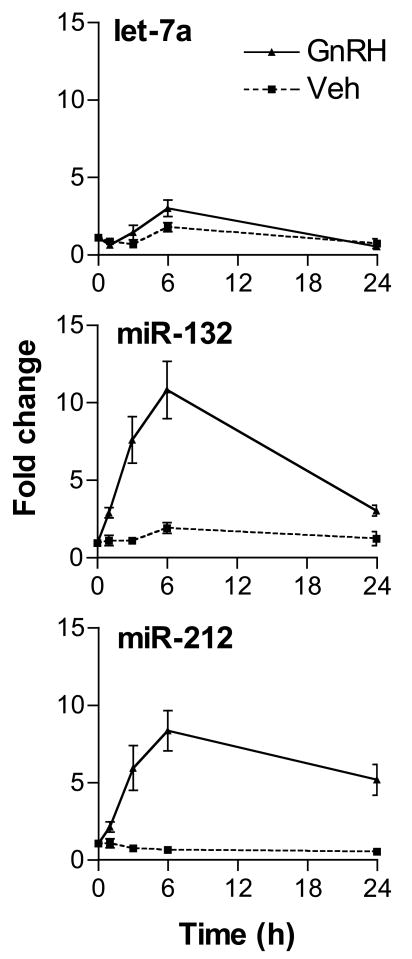
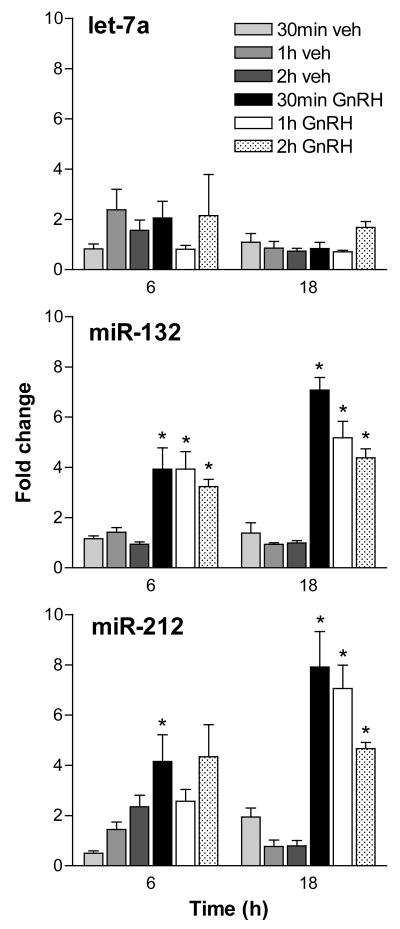
miR-132 is completely conserved throughout vertebrate evolution (Fig. 2). Notably, miR-132 maps to the same gene locus as a second highly conserved miRNA, miR-212 (Fig. 2). miR-212 was not included in the hairpin PCR panel. In order to study to coordinate regulation of both miRNAs, primers and probes for hairpin assay detection of both miRNAs and of let-7a as a control were synthesized and used to study their regulation by GnRH. Both miR-132 and miR-212 were found to be induced by GnRH in these experiments. Increased levels were observed by 1 hour, peaked by 6 hours and remained elevated after 24 hours exposure (Fig. 3). Notably, the time courses of both regulated microRNAs appear similar, as would be expected for products of the same gene locus. The regulation of both transcripts by pulsatile GnRH was also explored. Both transcripts were found to be regulated by pulses of GnRH administered over 6 and 18 hours, with no evident correlation of the level achieved and the interpulse intervals (Fig. 4).
Figure 2.
Evolutionary conservation of miR-132 and miR-212. The 22-nucleotide long miR-132 is completely conserved in vertebrates. The 21-nucleotide long miR-212 is conserved from fish to amphibians, and from rodents to humans. The human/rodent and the amphibian/fish miR-212 sequences differ only in 3 positions (at position 10, 17 and 21). Asterisks indicate base identity.
Figure 3.
Time course of miRNA induction by GnRH in LβT2 gonadotropes. Cells were treated with either 100 nM GnRH or vehicle and total RNA samples were isolated after 0, 1, 3, 6 and 24 hours of treatment. Expression levels of let-7a, miR-132 and miR-212 were determined by hairpin TaqMan real time PCR assays. Both miR-132 and miR-212 have similar trajectories. Increased levels were observed after 1 hour of GnRH treatment. Induction peaked at 6 hours and the miR-132 and miR-212 levels remained elevated after 24 hours. No significant induction was found in let-7a. This experiment is representative of three independent studies. Error bars denote standard error of the mean of three replicate samples.
Figure 4.
Induction of miR-132 and miR-212 by pulsatile GnRH treatment. Significant induction of miR-132 and miR-212 by 10-minute pulses of GnRH was observed after 6 hours of treatment. The induction was more pronounced after 18 hours of GnRH pulses. No clear pattern of correlation was found in samples treated with different pulse intervals of 30 minutes, 1 hour or 2 hours. No significant induction was found in let-7a. Fold-change is determined in comparison to untreated cells. Error bars denote standard error of the mean of six replicate samples. This experiment utilized 6 independent tissue culture samples treated and processed independently. Statistical analysis between vehicle- and GnRH-treated samples at corresponding pulse frequencies was performed using Student’s t-test, with the Bonferroni correction for multiple hypotheses. Asterisks denote p<0.05.
In order to further test the regulation of miR-132 and miR-212 by GnRH, the levels of both transcripts were assayed using the additional technique of LNA-based primer-extension real-time PCR. The LNA-based PCR assay was utilized because it has been reported to have increased sensitivity and specificity in comparison with the hairpin-based TaqMan assay (Raymond et al., 2005). The time course results for regulation of miR-212 and miR-132 by GnRH obtained with the LNA assays were found to be essentially identical to those observed using the hairpin TaqMan assays (compare Figs. 3 and S1). These results suggest that the lack of regulation of miR-132 observed using the array results from the lower sensitivity of the assay and indicate that both continuous and pulsatile GnRH induce a pronounced induction of both miRNA products of this single gene.
3.4 Bioinformatic analysis of potential targets for miR132 and miR-212
A number of algorithms have been developed to predict the mRNA targets of microRNAs. As described in the methods, we utilized five different software suites to identify potential targets of miR-132 and miR-212. The complete results of these analyses are shown in Table S3. A total of 117 potential miR-132 targets and 71 potential miR-212 targets were predicted by at least two of the analysis programs. 19 mRNAs that were found to be expressed in LβT2 cells by expression microarray experiment were predicted to be a target of miR-132 by at least three of the prediction programs. The validity of these analyses was supported by identifying Grit, a known target of miR-132 (Vo et al., 2005), as a target of both miR-132 and miR-212. 9 mRNAs expressed in LβT2 cells were predicted to be targets of miR-212 by at least three prediction programs. Two targets of miR-132 were predicted by all five analyses, Hn1 and Tjap1. Interestingly, the only target of miR-212 predicted by all five analyses, calumenin, was also predicted to be a target of miR-132 by four of the algorithms.
Discussion
This study reveals expression of a large number of miRNAs in LβT2 cells. In addition, using multiple assays, we demonstrate selective regulation of two miRNA products of a single gene, miR-132 and miR-212. These results suggest that miRNAs in general play an important role in the control of gene expression in the gonadotrope and that the induction of the two specific miRNAs miR-132 and miR-212, may contribute to the regulation of biosynthesis by GnRH.
In our experience, microRNAs are more accurately assayed using the hairpin TaqMan assay or the LNA probe assay in comparison with the array or the single molecule detection assay. miR-132 was not found to be regulated using a microarray assay of 350 miRNAs. Our results indicate that this lack of regulation was due to the relative lack of quantitative accuracy and sensitivity of this assay. In support of this conclusion is the demonstration of regulation of miR-132 by GnRH using three independent assay platforms: single molecule miRNA detection, hairpin miRNA TaqMan assay and LNA-based primer extension PCR assay. Further support for the regulation of miR-132 comes from the similar regulation observed for miR-212, a miRNA product of the same gene locus, using both the TaqMan assay and the LNA assay.
There are a large number of miRNAs expressed in the gonadotrope. Approximately 80% of all miRNAs assayed were found to be expressed. Given the breadth of this microtranscriptome, it is notable that miR-132 alone, among the 101 miRNAs tested by TaqMan assays, was the only miRNA showing significant regulation by GnRH.
The miR-132/miR-212 gene has been found to be regulated in several cell types and miR-132 is emerging as an important regulatory locus in several biological circuits. miR-132 is enriched in neurons and is transcriptionally regulated by the basic leucine zipper transcription factor cAMP-response element binding protein (CREB) (Vo et al., 2005). Brain-derived neurotropic factor (BDNF) induces a rapid and long-lasting miR-132 response in cortical neurons, which peaks at 4 hours of stimulation and remains elevated after 24 hours. miR-132 stimulates neurite outgrowth by translational repression of p250GAP, a member of the Rac/Rho family of GTPase activating proteins (Vo et al., 2005). It also inhibits translation of methyl CpG-binding protein 2 (MeCP2) which leads to decreased level of BDNF (Klein et al., 2007). This negative feedback provides homeostatic control of the MeCP2 level. Using DNA microarray expression profiling, miR-132 was found to be upregulated by lipopolysaccharide (LPS) in human acute monocytic leukemia cell line THP-1 (Taganov et al., 2006). In the suprachiasmatic nucleus of the mouse hypothalamus, which functions as the master circadian clock, miR-132 is light-inducible and exhibits circadian rhythm of expression, with peak level observed during the subjective day (Cheng et al., 2007). Circadian induction of miR-132 requires the MAP kinase cascade and targets regulatory factor X4 (RFX4), a member of the winged subfamily of helix-turn-helix transcription factors (Cheng et al., 2007). miR-132 is a positive regulator of CLOCK- and BMAL-dependent period1 (per1) transcription. Expression levels of miR-132 have also been studied in the hippocampal CA1 region of fetal, adult and age-matched Alzheimer brains, in which they were found to show no significant changes (Lukiw, 2007). In cancer, dysregulation of miRNAs is very common. Alteration of normal gene expression pattern in the cell by dysregulated miRNAs might contribute to carcinogenesis or tumor progression. miR-132 was found to be upregulated 3.54 fold in tongue squamous cell carcinoma when compared with normal tissue (Wong et al., 2008). Expression of miR-212 has been reported to be induced by ethanol (Tang et al., 2008). This induction was associated with a concomitant decrease in the protein level of zonula occludens 1 (ZO1) which is a major component of tight junctions that regulates intestinal permeability. Overexpression of miR-212 is correlated with hyperpermeability of the monolayer barrier. miR-212 levels were higher in colon biopsy samples in patients with alcoholic liver disease (Tang et al., 2008).
We identify many potential targets of miR-132 and miR-212 using bioinformatics approaches. Further studies will be needed to confirm the targets of miR-132 and miR-212 in the gonodotrope and to investigate the precise contributions of these miRNAs in mediating the effects of GnRH. The selectivity of the regulatory effects of GnRH on this miRNA gene locus identify miR-132 and miR-212 as potentially important regulators of protein and gene expression in the gonadotrope.
Supplementary Material
Table S1. Expression profile of miRNA in LβT2 gonadotropes determined by miRNA oligonucleotide microarray and hairpin TaqMan real time PCR panel. Expression level is represented by the fluorescence signal (log base 2) from the microarray, and threshold cycle value (Ct) from the real time PCR experiment. A presence call is made when the net fluorescence intensity (signal intensity subtracts local background intensity) is greater than or equal to 2 standard deviation of background intensity in the microarray, or when the threshold cycle (Ct) value is less than 30.0 in the hairpin TaqMan panel. ND, not determined; NaN, expression value not determined due to low slgnal-to-noise ratio; ✓, expressed; X, not expressed.
Table S2. Time course of miRNA expression determined by TaqMan hairpin real-time PCR assay panel. LβT2 cells were treated with 100 nM GnRH for 0, 1, 3, 6 and 24 hours, and the expression of 101 miRNAs were determined using TaqMan microRNA Assay Panel. Threshold cycle (Ct) values, adjusted using a global normalization procedure, are shown.
Table S3. miR-132 and miR-212 targets predicted by bioinformatics analysis. The software suite that predicted each target is indicated with a check mark (✓). mRNA transcripts that are predicted to be the targets of both miR-132 and miR-212 have their gene symbols highlighted in red. Transcripts that are found to be expressed in LβT2 cells from an Affymetrix GeneChip expression microarray experiment are denoted with a Present Call (P), and the mean signal intensity signal standard deviation and the number of separate samples and chips used in the experiment are indicated for these transcripts. Blank entries denote transcripts that were not assayed by the arrays. Although Tjap1 and Grit were not assayed in the microarray experiment, both were found to be expressed in LβT2 cells using quantitative PCR.
Figure S1. Regulation of miR-132 and miR-212 determined by LNA primer-extension real-time PCR assays. LβT2 cells were treated with either 100 nM GnRH or vehicle and total RNA samples were isolated after 0, 1, 3, 6 and 24 hours of treatment. Expression levels of let-7a, miR-132 and miR-212 were determined by LNA PCR assays. Note the similarity in the trajectories compared to that obtained with hairpin TaqMan PCR in Figure 3. Error bars denote standard error of the mean of three replicate samples.
Fig. S2. Standard curves for miR-132 and miR-212 hairpin TaqMan assays. Known amounts of synthetic RNA oligonucleotides identical to the sequence of miR-132 (A) or miR-212 (B) were used as starting material for hairpin TaqMan assays. Note the linearity and high sensitivity of the assays.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Chan EY, Goncalves NM, Haeusler RA, Hatch AJ, Larson JW, Maletta AM, Yantz GR, Carstea ED, Fuchs M, Wong GG, Gullans SR, Gilmanshin R. DNA mapping using microfluidic stretching and single-molecule detection of fluorescent site-specific tags. Genome Res. 2004;14:1137–46. doi: 10.1101/gr.1635204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng HY, Papp JW, Varlamova O, Dziema H, Russell B, Curfman JP, Nakazawa T, Shimizu K, Okamura H, Impey S, Obrietan K. microRNA modulation of circadian-clock period and entrainment. Neuron. 2007;54:813–29. doi: 10.1016/j.neuron.2007.05.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coller HA, Forman JJ, Legesse-Miller A. “Myc’ed messages”: myc induces transcription of E2F1 while inhibiting its translation via a microRNA polycistron. PLoS Genet. 2007;3:e146. doi: 10.1371/journal.pgen.0030146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffiths-Jones S, Grocock RJ, van Dongen S, Bateman A, Enright AJ. miRBase: microRNA sequences, targets and gene nomenclature. Nucleic Acids Res. 2006;34:D140–4. doi: 10.1093/nar/gkj112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hobert O. Gene regulation by transcription factors and microRNAs. Science. 2008;319:1785–6. doi: 10.1126/science.1151651. [DOI] [PubMed] [Google Scholar]
- John B, Enright AJ, Aravin A, Tuschl T, Sander C, Marks DS. Human MicroRNA targets. PLoS Biol. 2004;2:e363. doi: 10.1371/journal.pbio.0020363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanellopoulou C, Monticelli S. A role for microRNAs in the development of the immune system and in the pathogenesis of cancer. Semin Cancer Biol. 2008;18:79–88. doi: 10.1016/j.semcancer.2008.01.002. [DOI] [PubMed] [Google Scholar]
- Klein ME, Lioy DT, Ma L, Impey S, Mandel G, Goodman RH. Homeostatic regulation of MeCP2 expression by a CREB-induced microRNA. Nat Neurosci. 2007;10:1513–4. doi: 10.1038/nn2010. [DOI] [PubMed] [Google Scholar]
- Krek A, Grun D, Poy MN, Wolf R, Rosenberg L, Epstein EJ, MacMenamin P, da Piedade I, Gunsalus KC, Stoffel M, Rajewsky N. Combinatorial microRNA target predictions. Nat Genet. 2005;37:495–500. doi: 10.1038/ng1536. [DOI] [PubMed] [Google Scholar]
- Kulshreshtha R, Ferracin M, Negrini M, Calin GA, Davuluri RV, Ivan M. Regulation of microRNA expression: the hypoxic component. Cell Cycle. 2007;6:1426–31. [PubMed] [Google Scholar]
- Lawson MA, Tsutsumi R, Zhang H, Talukdar I, Butler BK, Santos SJ, Mellon PL, Webster NJ. Pulse sensitivity of the luteinizing hormone beta promoter is determined by a negative feedback loop Involving early growth response-1 and Ngfi-A binding protein 1 and 2. Mol Endocrinol. 2007;21:1175–91. doi: 10.1210/me.2006-0392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis BP, Shih IH, Jones-Rhoades MW, Bartel DP, Burge CB. Prediction of mammalian microRNA targets. Cell. 2003;115:787–98. doi: 10.1016/s0092-8674(03)01018-3. [DOI] [PubMed] [Google Scholar]
- Lukiw WJ. Micro-RNA speciation in fetal, adult and Alzheimer’s disease hippocampus. Neuroreport. 2007;18:297–300. doi: 10.1097/WNR.0b013e3280148e8b. [DOI] [PubMed] [Google Scholar]
- Neely LA, Patel S, Garver J, Gallo M, Hackett M, McLaughlin S, Nadel M, Harris J, Gullans S, Rooke J. A single-molecule method for the quantitation of microRNA gene expression. Nat Methods. 2006;3:41–6. doi: 10.1038/nmeth825. [DOI] [PubMed] [Google Scholar]
- Raymond CK, Roberts BS, Garrett-Engele P, Lim LP, Johnson JM. Simple, quantitative primer-extension PCR assay for direct monitoring of microRNAs and short-interfering RNAs. Rna. 2005;11:1737–44. doi: 10.1261/rna.2148705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruf F, Fink MY, Sealfon SC. Structure of the GnRH receptor-stimulated signaling network: insights from genomics. Front Neuroendocrinol. 2003;24:181–99. doi: 10.1016/s0091-3022(03)00027-x. [DOI] [PubMed] [Google Scholar]
- Ruf F, Sealfon SC. Genomics view of gonadotrope signaling circuits. Trends Endocrinol Metab. 2004;15:331–8. doi: 10.1016/j.tem.2004.07.008. [DOI] [PubMed] [Google Scholar]
- Stefani G, Slack FJ. Small non-coding RNAs in animal development. Nat Rev Mol Cell Biol. 2008;9:219–30. doi: 10.1038/nrm2347. [DOI] [PubMed] [Google Scholar]
- Taganov KD, Boldin MP, Chang KJ, Baltimore D. NF-kappaB-dependent induction of microRNA miR-146, an inhibitor targeted to signaling proteins of innate immune responses. Proc Natl Acad Sci U S A. 2006;103:12481–6. doi: 10.1073/pnas.0605298103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang Y, Banan A, Forsyth CB, Fields JZ, Lau CK, Zhang LJ, Keshavarzian A. Effect of alcohol on miR-212 expression in intestinal epithelial cells and its potential role in alcoholic liver disease. Alcohol Clin Exp Res. 2008;32:355–64. doi: 10.1111/j.1530-0277.2007.00584.x. [DOI] [PubMed] [Google Scholar]
- Turgeon JL, Kimura Y, Waring DW, Mellon PL. Steroid and pulsatile gonadotropin-releasing hormone (GnRH) regulation of luteinizing hormone and GnRH receptor in a novel gonadotrope cell line. Mol Endocrinol. 1996;10:439–50. doi: 10.1210/mend.10.4.8721988. [DOI] [PubMed] [Google Scholar]
- Vo N, Klein ME, Varlamova O, Keller DM, Yamamoto T, Goodman RH, Impey S. A cAMP-response element binding protein-induced microRNA regulates neuronal morphogenesis. Proc Natl Acad Sci U S A. 2005;102:16426–31. doi: 10.1073/pnas.0508448102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X. miRDB: a microRNA target prediction and functional annotation database with a wiki interface. RNA. 2008;14:1012–7. doi: 10.1261/rna.965408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams AE. Functional aspects of animal microRNAs. Cell Mol Life Sci. 2008;65:545–62. doi: 10.1007/s00018-007-7355-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong TS, Liu XB, Wong BY, Ng RW, Yuen AP, Wei WI. Mature miR-184 as Potential Oncogenic microRNA of Squamous Cell Carcinoma of Tongue. Clin Cancer Res. 2008;14:2588–92. doi: 10.1158/1078-0432.CCR-07-0666. [DOI] [PubMed] [Google Scholar]
- Wurmbach E, Yuen T, Ebersole BJ, Sealfon SC. Gonadotropin-releasing hormone receptor-coupled gene network organization. J Biol Chem. 2001;276:47195–201. doi: 10.1074/jbc.M108716200. [DOI] [PubMed] [Google Scholar]
- Yuen T, Wurmbach E, Ebersole BJ, Ruf F, Pfeffer RL, Sealfon SC. Coupling of GnRH concentration and the GnRH receptor-activated gene program. Mol Endocrinol. 2002;16:1145–53. doi: 10.1210/mend.16.6.0853. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1. Expression profile of miRNA in LβT2 gonadotropes determined by miRNA oligonucleotide microarray and hairpin TaqMan real time PCR panel. Expression level is represented by the fluorescence signal (log base 2) from the microarray, and threshold cycle value (Ct) from the real time PCR experiment. A presence call is made when the net fluorescence intensity (signal intensity subtracts local background intensity) is greater than or equal to 2 standard deviation of background intensity in the microarray, or when the threshold cycle (Ct) value is less than 30.0 in the hairpin TaqMan panel. ND, not determined; NaN, expression value not determined due to low slgnal-to-noise ratio; ✓, expressed; X, not expressed.
Table S2. Time course of miRNA expression determined by TaqMan hairpin real-time PCR assay panel. LβT2 cells were treated with 100 nM GnRH for 0, 1, 3, 6 and 24 hours, and the expression of 101 miRNAs were determined using TaqMan microRNA Assay Panel. Threshold cycle (Ct) values, adjusted using a global normalization procedure, are shown.
Table S3. miR-132 and miR-212 targets predicted by bioinformatics analysis. The software suite that predicted each target is indicated with a check mark (✓). mRNA transcripts that are predicted to be the targets of both miR-132 and miR-212 have their gene symbols highlighted in red. Transcripts that are found to be expressed in LβT2 cells from an Affymetrix GeneChip expression microarray experiment are denoted with a Present Call (P), and the mean signal intensity signal standard deviation and the number of separate samples and chips used in the experiment are indicated for these transcripts. Blank entries denote transcripts that were not assayed by the arrays. Although Tjap1 and Grit were not assayed in the microarray experiment, both were found to be expressed in LβT2 cells using quantitative PCR.
Figure S1. Regulation of miR-132 and miR-212 determined by LNA primer-extension real-time PCR assays. LβT2 cells were treated with either 100 nM GnRH or vehicle and total RNA samples were isolated after 0, 1, 3, 6 and 24 hours of treatment. Expression levels of let-7a, miR-132 and miR-212 were determined by LNA PCR assays. Note the similarity in the trajectories compared to that obtained with hairpin TaqMan PCR in Figure 3. Error bars denote standard error of the mean of three replicate samples.
Fig. S2. Standard curves for miR-132 and miR-212 hairpin TaqMan assays. Known amounts of synthetic RNA oligonucleotides identical to the sequence of miR-132 (A) or miR-212 (B) were used as starting material for hairpin TaqMan assays. Note the linearity and high sensitivity of the assays.