Abstract
The type X collagen gene (Col10a1) is a specific molecular marker of hypertrophic chondrocytes during endochondral bone formation. Mutations in human COL10A1 and altered chondrocyte hypertrophy have been associated with multiple skeletal disorders. However, until recently, the cis-enhancer element that specifies Col10a1 expression in hypertrophic chondrocytes in vivo has remained unidentified. Previously, we and others have shown that the Col10a1 distal promoter (−4.4 to −3.8 kb) may harbor a critical enhancer that mediates its tissue specificity in transgenic mice studies. Here, we report further localization of the cis-enhancer element within this Col10a1 distal promoter by using a similar transgenic mouse approach. We identify a 150-bp Col10a1 promoter element (−4296 to −4147 bp) that is sufficient to direct its tissue-specific expression in vivo. In silico analysis identified several putative transcription factor binding sites including two potential activator protein-1 (AP-1) sites within its 5′- and 3′-ends (−4276 to −4243 and −4166 to −4152 bp), respectively. Interestingly, transgenic mice using a reporter construct deleted for these two AP-1 elements still showed tissue-specific reporter activity. EMSAs using oligonucleotide probes derived from this region and MCT cell nuclear extracts identified DNA/protein complexes that were enriched from cells stimulated to hypertrophy. Moreover, these elements mediated increased reporter activity on transfection into MCT cells. These data define a 90-bp cis-enhancer required for tissue-specific Col10a1 expression in vivo and putative DNA/protein complexes that contribute to the regulation of chondrocyte hypertrophy. This work will enable us to identify candidate transcription factors essential both for skeletal development and for the pathogenesis of skeletal disorders.
Key words: type X collagen gene, cis-enhancer element, transgenic mice, hypertrophic chondrocyte, activator protein-1
INTRODUCTION
The type X collagen gene (Col10a1) is a specific molecular marker of hypertrophic chondrocytes, a terminal stage of endochondral ossification. Altered COL10A1 expression and chondrocyte hypertrophy have been associated with a spectrum of skeletal disorders such as Schmid metaphyseal chondrodysplasia (SMCD), cleidocranial dysplasia (CCD), and osteoarthritis.(1–6) It has been reported that Col10a1-null mice have subtle growth plate compressions partially resembling SMCD.(7) Abnormal hypertrophic cartilage and Col10a1 expression in the cranial base have also been observed in certain mouse models, suggesting its potential role in craniofacial development.(8) Characterization of the tissue-specific cis-acting elements that control Col10a1 transcription during chondrocyte hypertrophy is, therefore, critical toward identification of transcription factors that specify this critical step during endochondral bone formation and the pathogenesis of skeletal dysplasia and osteoarthritis.
Multiple cis-elements and trans-acting factors have been reported to regulate Col10a1 expression both in vitro and in vivo. The immediate upstream sequence of the Col10a1 transcription start site (−120 to +1 bp) shows a high level of conservation across species and is likely the basal promoter.(9) In vitro transfection studies have identified multiple regulatory elements within human, murine, and chicken Col10a1 promoter regions that further regulate Col10a1 expression.(10,11) We have previously reported identification of a 4-kb murine Col10a1 promoter (−4018 to +185 bp) that can mediate reporter activity selectively in lower hypertrophic chondrocytes in transgenic mice. We have also shown that Runx2 contributes directly to transactivation of this Col10a1 promoter both in vitro and in vivo through putative Runx2 binding sites found in this promoter region.(12)
Recently, a 4.6-kb murine Col10a1 promoter fragment (−4410 to +634 bp) was shown to drive higher levels of tissue-specific expression in hypertrophic cartilage in vivo.(13) These data together with our previous studies suggest that the major cis-enhancer essential for tissue-specific Col10a1 expression in vivo is within the Col10a1 distal promoter (−4.4 to −3.8 kb), where it locates the conservative enhancer.(12–14) Interestingly, not all hypertrophic chondrocytes in the growth plate of these transgenic mice show reporter (LacZ) expression.(13) More recently, a bacterial artificial chromosome (BAC) construct that contains the complete Col10a1 gene and large flanking sequences was shown to control efficient and specific LacZ expression in all hypertrophic chondrocytes in transgenic mice.(15) This shows that additional cis-elements located elsewhere (i.e., further upstream of the Col10a1 promoter or intronic sequence) may be needed to contribute to high-level cell-specific Col10a1/reporter expression.
Here we report a systematic dissection of the Col10a1 promoter/intronic region by transgenic studies using various-sized Col10a1 elements ranging from 10 to 4.6 kb upstream of the LacZ as reporter. Our results confirmed the observation that the Col10a1 distal promoter (−4.4 to −3.8 kb) harbors a critical enhancer that mediates its tissue specificity.(13,14) We further report localization of a minimal 90-bp cis-enhancer within 150 bp (−4296 to −4147 bp) of this Col10a1 promoter that is sufficient to direct high-level hypertrophic chondrocyte-specific reporter expression in vivo. Putative transcription factors that may bind this Col10a1 cis-enhancer and together mediate its tissue specificity are also discussed.
MATERIALS AND METHODS
Generation of Col10a1 promoter reporter constructs
The Col10a1 regulatory elements used for generation of different reporter constructs in this study were derived from the Col10a1 BAC(12) by either PCR amplification or restriction enzyme digestion and subcloned into pBluescript II KS vector. The 8-kb (−4018 to +4220 bp) transgenic reporter construct (Tg-8kb) encompasses the original 4-kb Col10a1 promoter(12) and a large second intron (an 8.2-kb XhoI fragment) that drives LacZ as a reporter (Fig. 1A; Table 1). The10-kb (−6022 to +4220 bp) reporter construct (Tg-10kb) contains an additional 2-kb upstream sequence (generated by PCR) of the Col10a1 promotor compared with the above 8-kb construct. (The sequences of the primers are available on request). The 6-kb (−6022 to + 185) and 4.6 kb (−4580 to + 185 bp) reporter construct (Tg-6kb and Tg-4.6kb) each contains a 2-kb or a 0.6-kb upstream sequence respectively followed by the original 4-kb Col10a1 promoter/intronic element and the LacZ gene (Fig. 1A; Table 1).
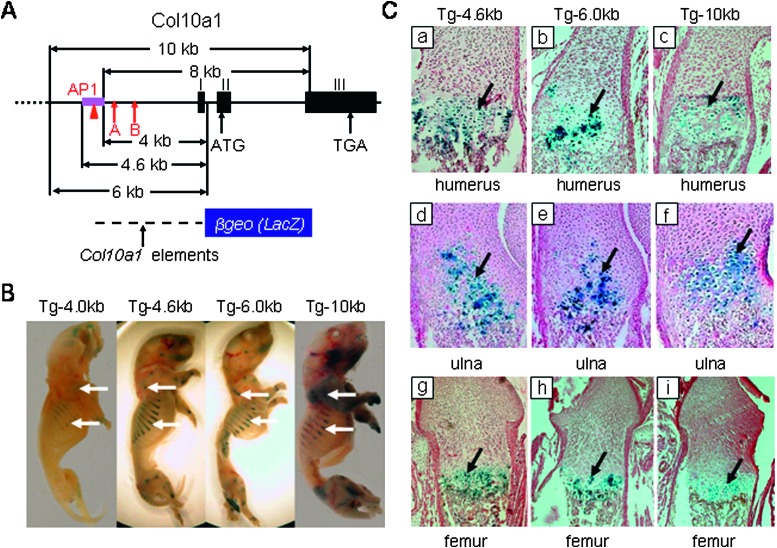
FIG. 1.
Transgenic mouse studies using Col10a1 promoter/intronic elements. (A) Analysis of mouse type X collagen gene: Col10a1 is composed of three exons numbered I, II, and III. Positions of the conserved Runx2 binding sites(12) (A and B, arrows) and the AP1 element (red triangle) are as indicated. The 4-, 4.6-, and 6-kb Col10a1 fragments used in transgenic studies are indicated by arrows and shown at bottom. The 8- and 10-kb Col10a1 elements with endogenous ATGs within exon II were mutated into ACGs. (B) Whole mount staining of Col10a1 4-, 4.6-, 6-, and 10-kb transgenic P1 mice. X-gal staining of postnatal day 1 (P1) transgenic mice showed that the 4.6-, 6-, and 10-kb Col10a1 regulatory element containing the conserved AP-1 element can direct higher level reporter expression (blue staining) of transgene compared with the staining pattern of 4-kb transgenic mice (Tg-4 kb, Tg-4.6 kb, Tg-6 kb, and Tg-10 kb, arrows).(12) The 8-kb Col10a1 promoter/intronic element directs weak but more specific reporter expression (Tg-8 kb) compared with Col10a1 4-kb transgenic mice(12) (data not shown). (C) Histological analysis of Col10a1-4.6-, 6-, and 10-kb transgenic P1 mice. Paraffin-embedded sections of representative P1 mice from X-gal–stained Col10a1 4.6-, 6-, and 10-kb transgenic mice were counterstained with nuclear fast red. Sagittal sections of distal humerus, proximal ulna, and femur from these transgenic mice show blue staining throughout the zone of hypertrophy (Cc, Cf, and Ci, Tg-10 kb; Cb, Ce, and Ch, Tg-6 kb; Ca, Cd, and Cg, Tg-4.6 kb). Similar results were also observed in ribs and all other long bone sections (data not shown). Staining is representative of data from at least three transgenic founder mice or lines. The wildtype littermate controls showed no staining (data not shown). Tg, transgenic mice.
Table 1.
Transgenic Studies of Col10a1
| Transgenic lines | Position to transcription start site | Expression pattern | Founders or lines analyzed |
| Tg-4kb | 4-kb promoter and first intron (−4018 to +185 bp) | Weak reporter expression in lower hypertrophic zone | Three founders and two lines at E15.5 and P1 stage |
| Tg-8kb | Original 4-kb element and second intron (−4018 to +4220 bp) | Stronger but still low level reporter expression in hypertrophic zone | Three founders and three lines at E15.5 and P1 stage |
| Tg-10kb | 2-kb upstream sequence and the 8-kb fragment (−6022 to +4220 bp) | High-level lacZ expression in hypertrophic zone with some staining in other tissue/cells | Two founders at P1, one line at E15.5 and P1 stage |
| Tg-6kb | 2-kb upstream sequence and the 4-kb element (−6022 to +185 bp) | High-level tissue-specific expression, no background | Three founders at P1, two lines at E15.5 and P1 stage |
| Tg-4.6kb | 600-bp upstream sequence and the 4-kb element (−4580 to +185 bp) | High-level tissue-specific expression, no background | Three founders and two lines at P1 stage |
| Tg-2x600bp | Two copies of the 600-bp (−4433 to −3790 bp) and ColX basal promoter | High-level tissue-specific expression, no background | Two founders, two lines at E15.5 and P1 stage |
| Tg-4x300bp | Four copies of the 300-bp (−4296 to −4009 bp) and ColX basal promoter | High-level tissue-specific expression, no background | Two founders at P1, one founder at E15.5 stage |
| Tg-4x150bp | Four copies of the 150-bp (−4296 to −4147 bp) and ColX basal promoter | High-level tissue-specific expression, no background | Two founders at P1, two lines at E15.5 and P1 stage |
| Tg-6xAP | Six copies of the two AP-1 elements (60 bp) and ColX basal promoter | No tissue-specific reporter expression was observed | Four founders at E15.5 |
| Tg-6×AP— | Six copies of the 150-bp without AP-1 sites (90 bp) and ColX basal promoter | High-level tissue-specific expression, no background | Two founders at E15.5, Two lines at E15.5 and P1 stage |
The Tg-1x600 and Tg-2x600 transgenic reporter construct was generated by inserting one or two copies of the conserved distal Col10a1 promoter element (−4433 to −3790 bp, an EcoRI restriction fragment) upstream of the Col10a1 basal promoter (−220 to +110 bp, a PCR derived fragment and the sequences of the primers are also available upon request) driving LacZ as reporter (Figs. 2A and 3A; Table 1). Transgenic reporter constructs Tg-1x300, Tg-4x300, and Tg-4x150 contain one or four copies of highly conserved 300 bp of Col10a1 promoter element (−4296 to −4009 bp) and four copies of the 150-bp element (−4296 to −4147 bp) upstream of the same Col10a1 basal promoter and LacZ gene (Figs. 2A and 3A; Table 1). The 300- and 150-bp Col10a1 promoter fragments were generated by PCR amplification with BamHI or BglII linkers added to the 5′ or 3′ end, respectively, such that multiple copies can be generated by enzyme digestion followed by linear ligation (Table 2). Two deletion mutant transgenic reporter constructs with or without the two putative activator protein-1 (AP-1) elements within 150 bp of the Col10a1 promoter region (5′AP 1: −4276 to −4243 bp and 3′ AP-1: −4166 to −4152 bp) were generated. The first reporter construct contains six copies of consecutive 5′ and 3′ AP-1 elements (60 bp, Tg-6xAP) upstream of the same Col10a1 basal promoter driving the LacZ gene (Figs. 4A and 4B). The second reporter construct was derived from the 150-bp Col10a1 regulatory fragment with the two AP-1 elements deleted (90 bp, Tg-6xAP −; Figs. 4A and 4B). These fragments were obtained by annealing the commercially synthesized oligos (IDT Technologies) with BamHI and BglII adapters for generating multiple copies (Table 2). The strategy of using BamHI and BglII linkers/adapters enables us to generate reporter constructs with multiple copies of the putative enhancers (Col10a1 distal promoter elements) and only in the forward direction. These constructs include Tg-1x300, Tg-4x300, Tg-4x150 Tg-6xAP, and Tg-6xAP − as described. The enhancer element (−4433 to −3790 bp, an EcoRI restriction fragment) of reporter construct Tg-1x600 is also in the forward direction. However, because of random ligation, both copies of the EcoRI fragment used in reporter construct Tg-2x600 are in the reverse direction by sequencing confirmation. The Col10a1 basal promoter used in all these reporter constructs is in the forward direction. Additional reporter constructs using elements derived from the Col10a1 distal promoter (−4.4 to −3.8 kb) are described in the following sections (see also Fig. 3A).
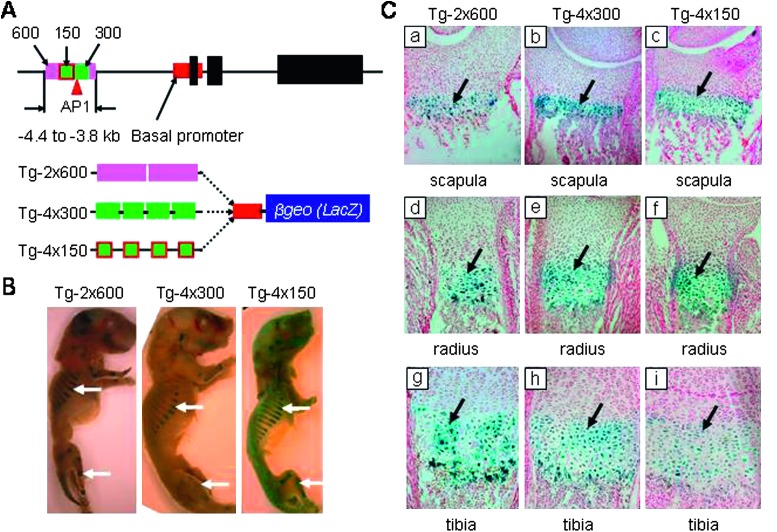
FIG. 2.
Distal Col10a1 promoter elements used in transgenic mouse studies. (A) Transgenic studies using 600, 300, and 150 bp of Col10a1 distal promoter elements The 600 bp of the putative enhancer element (−4.4 to −3.8 kb) and the highly conserved 300-bp fragment (−4.3 to −4.0 kb) are highlighted as purple and green bars, respectively (top). The 5′ portion of the 150-bp element within the 300-bp fragment is shown as a red square. The transgenic reporter constructs containing two copies of the 600-bp fragment (Tg-2x600) or four copies of the 300-bp (Tg-4x300) and the 5′ 150 bp of fragment (Tg-4x150) upstream of the Col10a1 basal promoter (red bar) driving the LacZ gene are shown at the bottom. (B) Whole mount staining of Tg-2x600, Tg-4x300, and Tg-4x150 transgenic P1 mice. Whole mount X-gal–stained postnatal day 1 (P1) transgenic mice of Tg-2x600, Tg-4x300, and Tg-4x150 showed a reporter expression pattern (blue staining) in the chondro-osseous junction of limbs and ribs (white arrows). (C) Histological analysis of Tg-2x600, Tg-4x300, and Tg-4x150 transgenic P1 mice. Paraffin-embedded sagittal sections of the scapula, radius, and tibia of representative P1 mice from X-gal–stained Tg-2x600 (Ca, Cd, and Cg), Tg-4x300 (Cb, Ce, and Ch), or Tg-4x150 (Cc, Cf, and Ci) transgenic lines show blue staining throughout the zone of hypertrophy (black arrows and data not shown). No staining was observed in wildtype littermates. Data were collected from at least three founders or F1 mice from each transgenic mouse line.
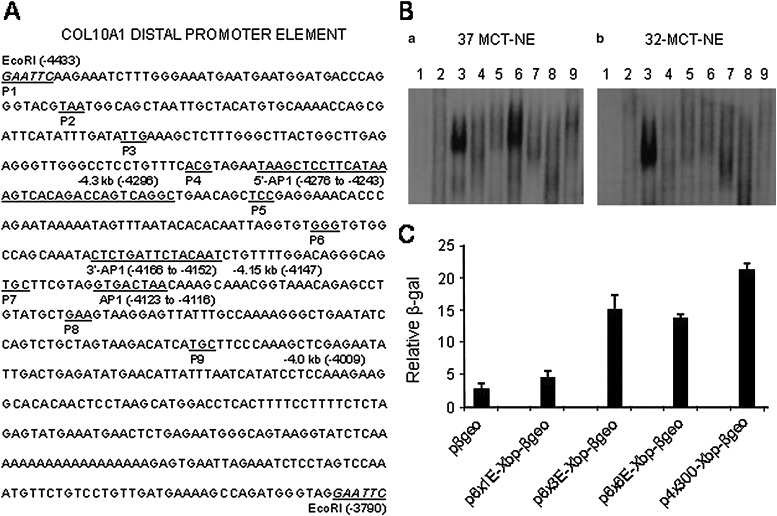
FIG. 3.
In vitro studies using cis-elements derived from the Col10a1 distal promoter. (A) Dissecting the Col10a1 distal promoter elements. This is a schematic map covering the entire putative enhancer sequence: the EcoRI fragment within the Col10a1 distal promoter ranging from −4433 to −3790 bp. The 600-, 300-, and 150-bp enhancer sequences used for generation of various reporter constructs are indicated. The newly identified 5′ and 3′ AP-1 binding sites and the original AP-1 site(13) that is outside of the 150-bp enhancer region (−4.3 to −4.15 kb) are underlined. A series of nine pairs of DNA oligos (∼50 base with a few bases of overlapping sequence between junctions) within this Col10a1 distal promoter (−4.4 to −3.8 kb) were commercially synthesized by IDT Technologies. These oligos were designed with BamHI (G-GATC-C) and BglII (A-GATC-T) adapters for generating multiple copies and for cloning of corresponding reporter constructs as shown in Table 2. (B) Putative enhancer element binding in MCT cell nuclear extracts. EMSAs were performed using hypertrophic MCT cell nuclear extracts and nine consecutive pairs of DNA oligos as probe. Two specific DNA/protein complexes formed with the third and sixth double-stranded oligos (a, lanes 3 and 6). These DNA/protein complexes diminished with competition when 100-fold of cold probe was used (data not shown). We also performed EMSA using proliferative MCT cell nuclear extracts (grown at 32°C) and these double-stranded oligos. Interestingly, formation of the DNA/protein complex can only be seen with the third DNA element (b, lanes 3 and 6). (C) Transfection studies using reporter plasmids derived from the putative cis-elements. Reporter plasmids were generated each with the first, third, and sixth element and were transfected into hypertrophic MCT cells. An RSV-luc luciferase expression plasmid was cotransfected as an internal control for transfection efficiency. The results showed that the reporter plasmid p4x300-Xbp-βgeo (Tg-4x300) that shows in vivo relevance (Fig. 2) showed significant reporter activity. Reporter plasmids with the third or sixth element, which form DNA/protein binding complexes, also show upregulated reporter activity (p6x3E-Xbp-βgeo, p6x6E-Xbp-βgeo), whereas the reporter plasmid derived from the first element (p6x1E-Xbp-βgeo) only shows basal promoter activity. Bars represent the average ratios of β-galactosidase to luciferase activity. The SDs are indicated by the error bars. pβgeo, pSA-βgeo-bpA as empty vector control; Xbp, Col10a1 basal promoter; E, element.
Table 2.
Primer and Linker/Adapter Sequences for Reporter Constructs
| Primer names | Primer sequences |
| 300 forward | 5′-AATGGATCCTCCTGTTTCACGTAG-3′ (GGATCC: BamHI linker) |
| 300 reverse | 5′-TATAGATCTCGAGCTTTGGGAAG-3′ (AGATCT: BgIII linker) |
| 150 forward | 5′-AATGGATCCTCCTGTTTCACGTAG-3′ (GGATCC: BamHI linker) |
| 150 reverse | 5′-AACAGATCTGTAGAATCAGAG-3′ (AGATCT: BgIII linker) |
| 6 × AP forward | 5′-GATCCGAATAAGCTCCTTCATAAAGTCACAGACCAGTCAGGCTGAACAAATACTCTGATTCTACAATCTGTTA-3′ (GATCC/A:BamHI/BgIII adapter) |
| 6 × AP reverse | 5′-GATCTAACAGATTGTAGAATCAGAGTATTTGTTCAGCCTGACTGGTCTGTGACTTTATGAAGGAGCTTATTCG-3′ (GATCT/G: BgIII/BamHI adapter) |
| 6 × AP− forward | 5′-GATCCGCCTCCTGTTTCACGTACAGCTCCGAGGAAACACCCAGAATAAAAATAGTTTAATACACACAATTAGGTGTGGGTGTGGCCAGCA-3′ (GATCC/A: BamHI/BgIII adapter) |
| 6 × AP− reverse | 5′-GATCTGCTGGCCACACCCACACCTAATTGTGTGTATTAAACTATTTTTATTCTGGGTGTTTCCTCGGAGCTGTACGTGAAACAGGAGGCG-3′ (GATCT/G: BgIII/BamHI adapter) |
FIG. 4.
Analysis of the putative AP-1 elements within the 150-bp Col10a1 promoter. (A) The two putative AP-1 sites within the 5′ (−4276 to −4243 bp) and 3′ (−4166 to −4152 bp) portion of the 150-bp sequence are underlined. (B) The red rectangle represents the 150-bp fragment; the 5′ and 3′ AP-1 elements are shown as black and blue dots, respectively. Transgenic reporter constructs containing six copies of consecutive 5′ and 3′ AP-1 elements (Tg-6xAP) or AP-1 deletion mutant (Tg-6xAP −) upstream of the same Col10a1 basal promoter (red bar) driving the LacZ gene are shown at the bottom (also see Table 2). (C) X-gal–stained E15.5 transgenic founder mouse embryo of Tg-6xAP − showed reporter expression pattern (blue staining) in the chondro-osseous junction of limbs and ribs (white arrows). No staining was observed in Tg-6xAP transgenic mouse embryos (left). (D) Sagittal section of X-gal–stained E15.5 mouse embryo from one Tg-6xAP − founder showed reporter activity (blue staining) exclusively throughout the hypertrophic zone in the fibula, radius, ulna, and other long bone and rib sections (black arrows and data not shown). a and c are lower magnifications compared with b and d. No staining pattern was observed in wildtype littermate controls.
EMSA
A series of nine pairs of DNA oligos (∼50 bases) derived from the conserved Col10a1 distal promoter (−4.4 to −3.8 kb) have been commercially synthesized by IDT Technologies (Fig. 3A). EMSAs using MCT cell nuclear extracts and these annealed DNA oligos as probes have been carried out as previously described.(12) Briefly, 10 fmol of a 32P-end–labeled double-stranded probe corresponding to these series of annealed DNA oligos was incubated with 3 μg of MCT cell nuclear extracts. Characteristics of MCT cells (mouse chondrocytes, a cell model of chondrocyte hypertrophy), preparation of the MCT cell nuclear extracts, and the conditions for the DNA-binding experiments were as described previously.(12,16–19) Complexes were electrophoresed on a 4% nondenaturing polyacrylamide gel and visualized by autoradiography.
Cell culture and transfection studies
MCT cells were grown at 32°C in standard DMEM with 8% FBS (Gibco BRL) and 8% CO2 as per the published protocol.(18) Three reporter constructs were selectively generated by inserting six copies of the first, third, and sixth cis-elements (corresponding to the annealed pairs of DNA oligos used in EMSA studies) upstream of the Col10a1 basal promoter on a pSAβgeobpA backbone. The reporter plasmids and the pSAβgeobpA control vector and the transgenic reporter construct (Tg-4x300) were transfected into hypertrophic MCT cells as previously described.(12) Briefly, MCT cells grown at 32°C were transfected for 6 h using Lipofectamine-plus (Gibco BRL), incubated for an additional 48 h at 37°C, and harvested for a β-galactosidase activity assay. A luciferase expression plasmid pRSVluc was added to all transfections and used as an internal control for normalizing the cell transfection efficiency.(20) Transfections were also performed in triplicate at three doses to ensure a linear dose response.
Generation and histochemical analysis of transgenic mice
The DNA fragments containing entire transgenic cassettes that include various Col10a1 promoter/intronic elements on a SAßgeobpA reporter plasmid backbone were released by NotI and SalI digestion. Purified DNA was redissolved and microinjected into fertilized mouse eggs and implanted into FVB pseudopregnant foster mothers(21) using the Axiovert 200 transgenic apparatus (Carl Zeiss). Transgenic founder mice were genotyped by PCR amplification using LacZ-specific primer pairs and analyzed by X-gal staining of mouse embryos at the stages of E15.5 and P1 for β-galactosidase expression.(22) After staining, mice were paraffin embedded, sectioned, and counterstained with nuclear fast red (Poly Scientific; R&D). Sections of all long bones and ribs were analyzed, and comparisons were made only among littermates at the same magnifications using the Axioplan 2 imaging system as described previously.(12,22) At least 30 sections of each growth plate were analyzed. The studies were approved by the animal care and oversight committees at Baylor College of Medicine and Rush University Medical Center.
RESULTS
Col10a1 distal promoter (−4.4 to −3.8 kb) harbors essential enhancer(s)
To study whether the conserved enhancer within the Col10a1 distal promoter and whether additional cis-elements located elsewhere are needed for high-level tissue-specific expression in vivo, we generated a series of transgenic mouse lines in addition to the original one using the 4-kb Col10a1 promoter for further promoter analysis (Figs. 1A and 1B, Tg-4 kb).(12) The 8-kb Col10a1 regulatory element that includes the 4-kb proximal promoter and the large second intron was shown to mediate weak reporter expression (LacZ) within the zone of hypertrophy as previously observed (Fig. 1A, Tg-8kb; data not shown).(12) The 10-kb Col10a1 fragment that adds an additional 2 kb upstream sequence can direct higher-level reporter expression throughout the hypertrophic zone of long bones and ribs (Figs. 1B and 1C, Tg-10kb; data not shown). Although some nonspecific reporter expression was also observed in resting chondrocytes of digits as shown by histological analysis (data not shown), these in vivo data suggest the presence of strong enhancer elements in the upstream 2 kb (−6.0 to −4.0 kb) of the Col10a1 distal promoter but not the intronic region. To further map this distal element, we generated two additional transgenic mouse lines using 6- and 4.6-kb Col10a1 promoters, each driving the LacZ reporter (Fig. 1A). High-level tissue-specific reporter expression was also observed in these transgenic mice (Figs. 1B and 1C, Tg-6kb and Tg-4.6kb). Compared with the original 4-kb Col10a1 promoter element (−4018 to +185 bp),(12) the 6- and 4.6-kb Col10a1 regulatory elements added an additional 2 and 0.6 kb of distal promoter, respectively. Although there is a slight sequence difference between our Tg-4.6kb reporter construct (−4580 to +185 bp) and the reported Col10a1 regulatory element (−4410 to +634 bp)(13) used for transgenic studies, both results suggest that the tissue-specific cis-enhancer for Col10a1/reporter expression is located in the Col10a1 distal promoter (−4.4 to −3.8 kb) where the conservative enhancer is located as previously reported (Fig. 1A; Table 1).(13,14)
One hundred fifty base pairs of the Col10a1 distal promoter (−4.3 to −4.15 kb) contain a major cis-enhancer
To study the contribution of the putative enhancer (−4433 to −3790 bp) within the Col10a1 distal promoter to its expression in hypertrophic chondrocytes in vivo, we generated a transgenic mouse line using only this enhancer upstream of the Col10a1 basal promoter (−220 to +110 bp) driving the LacZ gene. No tissue-specific reporter expression pattern was observed in transgenic founder mice (Tg-1x600, data not shown). However, when the reporter constructs were generated by using two copies of this putative enhancer and in the reverse direction as described above, the transgenic founder mice showed similar high-level tissue-specific reporter expression pattern compared with that of Tg-4.6kb, Tg-6kb, and Tg-10kb transgenic mice (Figs. 1B, 2A, and 2B, Tg-2x600). Histological analysis showed X-gal staining correlated with β-galactosidase reporter activity throughout the hypertrophic zone of the long bones and ribs (Fig. 2C, Tg-2x600; data not shown). This result showed that the 600-bp Col10a1 distal promoter (−4433 to −3790 bp) contains an essential enhancer that is responsible for its tissue specificity in vivo.
Comparative sequence analyses have shown that this 600-bp putative murine enhancer is 60-70% homologous to that of the corresponding human enhancer.(13,14) However, a detailed sequence analysis identified a highly conserved 300-bp fragment (−4.3 to −4.0 kb), which shows >80% identity to that of human. Therefore, we generated transgenic mice carrying a reporter construct containing this highly conserved 300-bp fragment upstream of the same Col10a1 basal promoter and the LacZ gene (Fig. 2A, Tg-1x300). These Tg-1x300 transgenic founder mice did not show reporter expression patterns in long bones and ribs after whole embryo staining (data not shown). However, when four copies of the highly conserved 300-bp fragment were used for generation of the transgenic reporter construct, high-level tissue-specific reporter expression was observed in Tg-4x300 transgenic founder mice, as shown by whole embryo staining and histological analysis (Figs. 2B and 2C, Tg-4x300). These data suggest that the highly conserved 300-bp Col10a1 regulatory element contains the major enhancer that mediates its tissue specificity.
To further refine this enhancer element, we divided this 300-bp enhancer into 5′ and 3′ portions with a 20-bp overlapping sequence for additional transgenic studies. We first generated a reporter construct in which the Lac Z reporter gene was driven by four copies of 3′ portion of this 300-bp fragment and the basal promoter (−4.17 to −4.0 kb, Tg-4x3'-150, data not shown). No reporter expression pattern (blue staining) was observed in any tissues of all four transgenic founder mice. In another reporter construct, when the 5′ portion (−4.3 to −4.15 kb; Fig. 3A) of this 300-bp fragment was used, a tissue-specific reporter expression pattern was observed in both the transgenic founders and the F1 mice (Figs. 2A–2C, Tg-4x150; data not shown). These results localize the major cis-enhancer element to a 150-bp Col10a1 distal promoter between −4296 and −4147 bp.
In vitro studies with cis-elements derived from the Col10a1 distal promoter
To determine whether cis-enhancer element (s) within the Col10a1 distal promoter (−4.4 to −3.8 kb; Fig. 3A) could bind putative transcription factors in the cell model of chondrocyte hypertrophy, we performed EMSA assays using proliferative or hypertrophic MCT cell nuclear extracts and a series of DNA 50-mers derived from this region. Two specific DNA/protein complexes formed with probes 3 and 6 (Fig. 3B) when the hypertrophic MCT cell nuclear extracts were used, whereas only probe 3 showed a DNA/protein complex when incubated with proliferative MCT cell nuclear extracts. Interestingly, probes 3 and 6 overlap with the 5′ and 3′ ends of this 150-bp enhancer (−4.3 kb to −4.15 kb) defined by in vivo studies (Fig. 3A).
To determine whether these probes and the corresponding DNA/protein complex correlated with detectable transactivation activities in this cell model, we performed transfection studies using hypertrophic MCT cells as previously described.(12) Reporter constructs containing six copies of either probe 1, 3, or 6 were generated. As a positive control, reporter construct p4x300-Xbp-βgeo or Tg-4x300, which shows specific reporter activity in hypertrophic chondrocytes of transgenic mice, was used (Figs. 2A and 3C). As expected, p4x300-Xbp-βgeo gave the highest β-galactosidase activity when transfected in hypertrophic MCT cells (Tg-4x300; Fig. 3C). Significant upregulation of reporter activity was also seen in reporter constructs containing probes 3 or 6 (p6x3/6/E-Xbp-βgeo; Fig. 3C) but not probe 1. These results suggest that putative transcription factors could bind cis-elements within this 150-bp region and mediate Col10a1/reporter expression.
Characterize the role of two AP-1 elements within the 150-bp Col10a1 promoter in vivo
To identify the putative transcription factor binding sites that may mediate tissue-specific Col10a1/reporter expression, we performed in silico cross-species analysis of this conserved 150-bp element using the rVISTA tool.(23) We identified several putative transcription factor binding sites including two potential novel AP-1 sites within the 5′ and 3′ ends as described (−4276 to −4243 and −4166 to −4152 bp; Fig. 4A). Previously, putative AP-1 binding sites within the Col10a1 distal promoter have been reported to contribute to its expression by in vitro experiments.(13) Given the importance of AP-1 family members in cell fate determination and specifically in chondrogenesis,(13,24–31) we further studied whether these newly identified AP-1 elements contribute to tissue-specific Col10a1 expression in vivo. We generated two deletion mutant reporter constructs with or without the two putative AP-1 elements for transgenic studies (Tg-6xAP and Tg-6xAP−; Fig. 4B). Interestingly, tissue-specific reporter activity was only observed in transgenic mouse embryos with the two AP-1 elements deleted (Tg-6xAP−; Figs. 4C and 4D) but not in the one with the putative AP-1 elements (Tg-6xAP; Fig. 4C; data not shown).
DISCUSSION
Tissue-specific gene expression typically requires binding of specific transcription factors to major enhancer(s) in addition to action of general transcription factors to the core promoter region.(32–34) These enhancers are cis-acting DNA regulatory elements that are known to stimulate transcription, independent of their position and orientation.(34,35) The type X collagen gene (Col10a1) is a specific molecular marker of hypertrophic chondrocytes, a cell type that undergoes terminal differentiation and apoptosis and may help mineralization and angiogenesis during long bone development.(36) However, until recently, the cis-enhancer element that mediates hypertrophic chondrocyte-specific Col10a1 expression in vivo has not been identified. Putative transcription factors that are suggested to be responsible for Col10a1 expression are therefore still illusive.(12,13,37)
In this study, our data together with others suggested that the cis-acting regulatory element responsible for high-level Col10a1/reporter expression is located within the Col10a1 distal promoter (−4.4 to −3.8 kb; Fig. 1, Tg-10kb, -6kb, -4.6kb).(13) A putative enhancer that mediates upregulated Col10a1 promoter activity in vitro was previously reported within this region.(13,14) In our further transgenic studies, whereas a single copy of this putative Col10a1 enhancer (Tg-1x600, Tg-1x300) did not confer tissue specificity, multiple copies of the enhancer (Tg-2x600, Tg-4x300) directed high-level reporter expression throughout hypertrophic zone of transgenic mouse embryos (Fig. 2). Interestingly, a putative enhancer used for generation of the Tg-2x600 transgenic reporter construct was found in the reverse direction. Although, we did not make a reporter construct by placing two copies of the 600-bp enhancer in the forward direction, our data suggest that this enhancer works independent of its orientation and functions better when presented in multiple tandem copies.(33,34) Moreover, we further performed promoter analysis and successfully refined mapping the major cis-enhancer to 150 bp (−4296 to −4147 bp) of the Col10a1 promoter that is sufficient to mediate its tissue specificity in vivo (Fig. 2, Tg-4x150).
Our in vitro DNA binding experiments showed that specific DNA/protein complexes formed when MCT cell nuclear extracts were incubated with annealed DNA oligos derived from the 5′ or 3′ end of the 150-bp Col10a1 promoter (Fig. 3B). Correspondingly, reporter constructs derived from the above correlated oligos (3 and 6) showed upregulated reporter activity when transfected into hypertrophic MCT cells (Fig. 3C). It has been well shown that MCT cells grew continuously at a temperature permissive for the activity of the SV40 large tumor antigen (32°C). However, these cells undergo growth arrest when cultured at a nonpermissive temperature (37°C) and show significant upregulation of Col10a1 expression.(12,18) This will make MCT cells a suitable model to study Col10a1 gene regulation in vitro. Formation of specific DNA/protein complex(es) and the upregulated promoter activity seen in corresponding reporter constructs suggest that certain transcription factors might bind these elements and specify Col10a1 expression. Hypertrophic MCT cells also express type I collagen, indicating their acquirement of osteoblastic characteristics.(18) However, given the in vivo relevance of this 150-bp refined enhancer, transcription factors that upregulate Col10a1/reporter expression are very likely to interact with cis-elements within this enhancer and together mediate reporter activity. Therefore, these in vitro data provide valuable information in finding the molecular determinants important for Col10a1 gene regulation by means of EMSA candidate approach and proteomics methods.
Multiple transcription factors and signaling pathways have been shown to contribute to regulation of chondrocyte maturation and type X collagen expression by in vitro transfection and mouse genetic studies. These include the transcription factors AP-1, specific protein-1, and Runx2, and the Ihh/PTH-related protein (PTHrP), BMP/Smad, Wnt/β-catenin, fibroblast growth factor (FGF), and TGFβ signaling pathways.(8,11–14,37–49) Previously, multiple AP-1 binding sites were identified within the Col10a1 distal promoter, one of which was shown to be functionally active (−4123 to −4116 bp) and specific for induction of reporter activity in hypertrophic chondrocytes by in vitro transfection studies.(13) Interestingly, these reported AP-1 binding sites implicated in Col10a1 expression are located outside and downstream of the 150-bp Col10a1 enhancer described here. Here, using the rVista 2.0 tool,(23) we report the identification of two potential AP-1 binding sites within this 150-bp enhancer that were not reported previously.(13) Transgenic studies using a reporter construct that deleted the two AP-1 elements from the 150-bp enhancer still gave tissue-specific reporter activity (Tg-6xAP−; Figs. 4C and 4D). Whereas another reporter construct that contained only the AP-1 elements and the LacZ as a reporter did not show any tissue specificity (Tg-6xAP, data not shown). These data do not rule out AP-1 involvement in regulating Col10a1 expression but do further restrict the major cis-enhancer element to 90 bp of the Col10a1 promoter. AP-1 family members are known to have multiple biological roles by functioning as both transcriptional activators and repressors.(25–29) The repressor activity of AP-1 family members on Col10a1/reporter expression has been shown previously by in vitro studies.(13,50) These AP-1 elements may serve as silencers and be part of the enhanceosome that mediates transcriptional repression to exclude gene activity in inappropriate tissues as previously described.(33,34)
Our previous results showed that the contribution of Runx2 regulating Col10a1 expression is mediated by conservative Runx2 binding sites within the proximal 4-kb Col10a1 promoter.(12) No Runx2 binding sites were identified within this 90-bp Col10a1 enhancer by preliminary in silico analysis.(23) It is possible that additional cis-elements located elsewhere or multiple transcription factors in addition to Runx2 and AP-1 may work together or independently to contribute to Col10a1 expression. It has been shown that, beside the core promoter region, enhancers, as well as silencers and insulators, may be present in the distant 5′ upstream or 3′ downstream region, in introns and even in interchromosomal DNA, which constitute auxiliary transcriptional regulatory elements.(51,52)
In summary, dissection of type X collagen gene regulation during chondrocyte hypertrophy has been attempted by both in vitro and in vivo studies. Recently, this has also drawn extensive attention from scientists all over the world, because chondrocyte maturation is not only a critical stage of skeletal development but also an essential player that is involved in the pathogenesis of skeletal dysplasia and osteoarthritis.(1–6) Published results to date point to a plethora of possible transcription factors and cis-enhancer elements(12,13,15,37) that contribute to regulation of Col10a1 expression. None, however, to date have been able to define a sufficiently refined element in vitro and in vivo to facilitate cloning of candidate transcription factors. In this study, we successfully showed that the major cis-enhancer element that is responsible for Col10a1 expression is within the 90-bp Col10a1 promoter as described above. Although it is not practical to characterize many of the putative binding factors based on the results of in silico sequence analysis (such as CdxA, CRE-BP, Nkx-2, and NF-κB), this refined Col10a1 cis-enhancer element will enable us to identify the candidate transcription factor(s) that mediate its tissue specificity by using candidate EMSA, proteomics, and/or one-hybrid screening approaches. Moreover, such a short element will have direct applications in targeting genes of interest selectively to hypertrophic chondrocytes by combining transgenic and/or Cre/LoxP recombination approaches.(53) The easy cloning strategy described in this study that involves replacement of the LacZ reporter only enhances these processes. The mouse models generated in this study will also help genetically to elucidate the potential transcriptional determinants for Col10a1 gene regulation during terminal chondrocyte differentiation.
ACKNOWLEDGMENTS
We thank Ming Ming Jiang and Sujatha Kakuru for technical support on injection and histological analysis. We thank Dr. Benoit de Crombrugghe for the MCT cells. This work was supported by National Institutes of Health Grants R03 DE16041 to Q.Z., AR44738 to B.L., ES11253 to B.L., and HD22657 to B.L., the Arthritis Foundation (Q.Z., B.L.), Astrazeneca (A.P.), Baylor College of Medicine Child Health Research Center (B.L.), and DFG Za96/10-2/3 (B.Z.).
Footnotes
The authors state that they have no conflicts of interest.
REFERENCES
- 1.Warman ML, Abbott M, Apte SS, Hefferon T, McIntosh I, Cohn DH, Hecht JT, Olsen BR, Francomano CA. A type X collagen mutation causes Schmid metaphyseal chondrodysplasia. Nat Genet. 1993;5:79–82. doi: 10.1038/ng0993-79. [DOI] [PubMed] [Google Scholar]
- 2.von der Mark K, Frischholz S, Aigner T, Beier F, Belke J, Erdmann S, Burkhardt H. Upregulation of type X collagen expression in osteoarthritic cartilage. Acta Orthop Scand Suppl. 1995;266:125–129. [PubMed] [Google Scholar]
- 3.Girkontaite I, Frischholz S, Lammi P, Wagner K, Swoboda B, Aigner T, Von der Mark K. Immunolocalization of type X collagen in normal fetal and adult osteoarthritic cartilage with monoclonal antibodies. Matrix Biol. 1996;15:231–238. doi: 10.1016/s0945-053x(96)90114-6. [DOI] [PubMed] [Google Scholar]
- 4.Drissi H, Zuscik M, Rosier R, O'Keefe R. Transcriptional regulation of chondrocyte maturation: Potential involvement of transcription factors in OA pathogenesis. Mol Aspects Med. 2005;26:169–179. doi: 10.1016/j.mam.2005.01.003. [DOI] [PubMed] [Google Scholar]
- 5.Tchetina EV, Squires G, Poole AR. Increased type II collagen degradation and very early focal cartilage degeneration is associated with upregulation of chondrocyte differentiation related genes in early human articular cartilage lesions. J Rheumatol. 2005;32:876–886. [PubMed] [Google Scholar]
- 6.Zheng Q, Sebald E, Zhou G, Chen Y, Wilcox W, Lee B, Krakow D. Dysregulation of chondrogenesis in human cleidocranial dysplasia. Am J Hum Genet. 2005;77:305–312. doi: 10.1086/432261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kwan KM, Pang MK, Zhou S, Cowan SK, Kong RY, Pfordte T, Olsen BR, Sillence DO, Tam PP, Cheah KS. Abnormal compartmentalization of cartilage matrix components in mice lacking collagen X: Implications for function. J Cell Biol. 1997;136:459–471. doi: 10.1083/jcb.136.2.459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Shum L, Wang X, Kane AA, Nuckolls GH. BMP4 promotes chondrocyte proliferation and hypertrophy in the endochondral cranial base. Int J Dev Biol. 2003;47:423–431. [PubMed] [Google Scholar]
- 9.Beier F, Lammi MJ, Bertling W, von der Mark K. Transcriptional regulation of the human type X collagen gene expression. Ann N Y Acad Sci. 1996;785:209–211. doi: 10.1111/j.1749-6632.1996.tb56263.x. [DOI] [PubMed] [Google Scholar]
- 10.Dourado G, LuValle P. Proximal DNA elements mediate repressor activity conferred by the distal portion of the chicken collagen X promoter. J Cell Biochem. 1998;70:507–516. doi: 10.1002/(sici)1097-4644(19980915)70:4<507::aid-jcb7>3.0.co;2-n. [DOI] [PubMed] [Google Scholar]
- 11.Long F, Linsenmayer TF. Tissue-specific regulation of the type X collagen gene: Analyses by in vivo footprinting and transfection with a proximal promoter region. J Biol Chem. 1995;270:31310–31314. doi: 10.1074/jbc.270.52.31310. [DOI] [PubMed] [Google Scholar]
- 12.Zheng Q, Zhou G, Morello R, Chen Y, Garcia-Rojas X, Lee B. Type X collagen gene regulation by Runx2 contributes directly to its hypertrophic chondrocyte-specific expression in vivo. J Cell Biol. 2003;162:833–842. doi: 10.1083/jcb.200211089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gebhard S, Pöschl E, Riemer S, Bauer E, Hattori T, Eberspaecher H, Zhang Z, Lefebvre V, de Crombrugghe B, von der Mark K. A highly conserved enhancer in mammalian type X collagen genes drives high levels of tissue-specific expression in hypertrophic cartilage in vitro and in vivo. Matrix Biol. 2004;23:309–322. doi: 10.1016/j.matbio.2004.05.010. [DOI] [PubMed] [Google Scholar]
- 14.Chambers D, Young DA, Howard C, Thomas JT, Boam DS, Grant ME, Wallis GA, Boot-Handford RP. An enhancer complex confers both high-level and cell-specific expression of the human type X collagen gene. FEBS Lett. 2002;531:505–508. doi: 10.1016/s0014-5793(02)03606-2. [DOI] [PubMed] [Google Scholar]
- 15.Gebhard S, Hattori T, Bauer E, Bösl MR, Schlund B, Pöschl E, Adam N, de Crombrugghe B, von der Mark K. BAC constructs in transgenic reporter mouse lines control efficient and specific LacZ expression in hypertrophic chondrocytes under the complete Col10a1 promoter. Histochem Cell Biol. 2007;127:183–194. doi: 10.1007/s00418-006-0236-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dignam JD, Lebovitz RM, Roeder RG. Accurate transcription initiation by RNA polymerase II in a soluble extract from isolated mammalian nuclei. Nucleic Acids Res. 1983;11:1475–1489. doi: 10.1093/nar/11.5.1475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dyer RB, Herzog NK. Isolation of intact nuclei for nuclear extract preparation from a fragile B-lymphocyte cell line. Biotechniques. 1995;19:192–195. [PubMed] [Google Scholar]
- 18.Lefebvre V, Garofalo S, de Crombrugghe B. Type X collagen gene expression in mouse chondrocytes immortalized by a temperature-sensitive simian virus 40 large tumor antigen. J Cell Biol. 1995;128:239–245. doi: 10.1083/jcb.128.1.239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhou G, Zheng Q, Engin F, Munivez E, Chen Y, Sebald E, Krakow D, Lee B. Dominance of SOX9 function over RUNX2 during skeletogenesis. Proc Natl Acad Sci USA. 2006;103:19004–19009. doi: 10.1073/pnas.0605170103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhou G, Chen Y, Zhou L, Thirunavukkarasu K, Hecht J, Chitayat D, Gelb BD, Pirinen S, Berry SA, Greenberg CR, Karsenty G, Lee B. CBFA1 mutation analysis and functional correlation with phenotypic variability in cleidocranial dysplasia. Hum Mol Genet. 1999;8:2311–2316. doi: 10.1093/hmg/8.12.2311. [DOI] [PubMed] [Google Scholar]
- 21.Hogan BLM, Costantini F, Lacey E. NY, USA: Cold Spring Harbor Laboratory Press, Cold Spring Harbor; 1986. Manipulating the Mouse Embryo. A Laboratory Manual. [Google Scholar]
- 22.Zhou G, Garofalo S, Mukhopadhyay K, Lefebvre V, Smith CN, Eberspaecher H, de Crombrugghe B. A 182-bp fragment of the mouse pro-alpha-1(II) collagen gene is sufficient to direct chondrocyte expression in transgenic mice. J Cell Sci. 1995;108:3677–3684. doi: 10.1242/jcs.108.12.3677. [DOI] [PubMed] [Google Scholar]
- 23.Loots GG, Ovcharenko I. rVISTA 2.0: Evolutionary analysis of transcription factor binding sites. Nucleic Acids Res. 2004;32:W217–W221. doi: 10.1093/nar/gkh383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jochum W, David JP, Elliott C, Wutz A, Plenk H, Jr, Matsuo K, Wagner EF. Increased bone formation and osteosclerosis in mice overexpressing the transcription factor Fra-1. Nat Med. 2000;6:980–984. doi: 10.1038/79676. [DOI] [PubMed] [Google Scholar]
- 25.Hess J, Porte D, Munz C, Angel P. AP-1 and Cbfa/runt physically interact and regulate parathyroid hormone-dependent MMP13 expression in osteoblasts through a new osteoblast-specific element 2/AP-1 composite element. J Biol Chem. 2001;276:20029–20038. doi: 10.1074/jbc.M010601200. [DOI] [PubMed] [Google Scholar]
- 26.D'Alonzo RC, Selvamurugan N, Karsenty G, Partridge NC. Physical interaction of the activator protein-1 factors c-Fos and c-Jun with Cbfa1 for collagenase-3 promoter activation. J Biol Chem. 2002;277:816–822. doi: 10.1074/jbc.M107082200. [DOI] [PubMed] [Google Scholar]
- 27.MacLean HE, Kim JI, Glimcher MJ, Wang J, Kronenberg HM, Glimcher LH. Absence of transcription factor c-maf causes abnormal terminal differentiation of hypertrophic chondrocytes during endochondral bone development. Dev Biol. 2003;262:51–63. doi: 10.1016/s0012-1606(03)00324-5. [DOI] [PubMed] [Google Scholar]
- 28.Hess J, Hartenstein B, Teurich S, Schmidt D, Schorpp-Kistner M, Angel P. Defective endochondral ossification in mice with strongly compromised expression of JunB. J Cell Sci. 2003;116:4587–4596. doi: 10.1242/jcs.00772. [DOI] [PubMed] [Google Scholar]
- 29.Hess J, Angel P, Schorpp-Kistner M. AP-1 subunits: Quarrel and harmony among siblings. J Cell Sci. 2004;117:5965–5973. doi: 10.1242/jcs.01589. [DOI] [PubMed] [Google Scholar]
- 30.Papachristou D, Pirttiniemi P, Kantomaa T, Agnantis N, Basdra EK. Fos- and Jun-related transcription factors are involved in the signal transduction pathway of mechanical loading in condylar chondrocytes. Eur J Orthod. 2006;28:20–26. doi: 10.1093/ejo/cji101. [DOI] [PubMed] [Google Scholar]
- 31.Goldring MB, Tsuchimochi K, Ijiri K. The control of chondrogenesis. J Cell Biochem. 2006;97:33–44. doi: 10.1002/jcb.20652. [DOI] [PubMed] [Google Scholar]
- 32.Tamura T, Konishi Y, Makino Y, Mikoshiba K. Mechanisms of transcriptional regulation and neural gene expression. Neurochem Int. 1996;29:573–581. doi: 10.1016/s0197-0186(96)00048-4. [DOI] [PubMed] [Google Scholar]
- 33.Harafuji N, Keys DN, Levine M. Genome-wide identification of tissue-specific enhancers in the Ciona tadpole. Proc Natl Acad Sci USA. 2002;99:6802–6805. doi: 10.1073/pnas.052024999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Arnosti DN, Kulkarni MM. Transcriptional enhancers: Intelligent enhanceosomes or flexible billboards? J Cell Biochem. 2005;94:890–898. doi: 10.1002/jcb.20352. [DOI] [PubMed] [Google Scholar]
- 35.Banerji J, Rusconi S, Schaffner W. Expression of a beta-globin gene is enhanced by remote SV40 DNA sequences. Cell. 1981;27:299–308. doi: 10.1016/0092-8674(81)90413-x. [DOI] [PubMed] [Google Scholar]
- 36.Zenmyo M, Komiya S, Kawabata R, Sasaguri Y, Inoue A, Morimatsu M. Morphological and biochemical evidence for apoptosis in the terminal hypertrophic chondrocytes of the growth plate. J Pathol. 1996;180:430–433. doi: 10.1002/(SICI)1096-9896(199612)180:4<430::AID-PATH691>3.0.CO;2-H. [DOI] [PubMed] [Google Scholar]
- 37.Dong Y, Drissi H, Chen M, Chen D, Zuscik MJ, Schwarz EM, O'Keefe RJ. Wnt-mediated regulation of chondrocyte maturation: Modulation by TGF-beta. J Cell Biochem. 2005;95:1057–1068. doi: 10.1002/jcb.20466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Long F, Sonenshein GE, Linsenmayer TF. Multiple transcriptional elements in the avian type X collagen gene. Identification of Sp1 family proteins as regulators for high level expression in hypertrophic chondrocytes. J Biol Chem. 1998;273:6542–6549. doi: 10.1074/jbc.273.11.6542. [DOI] [PubMed] [Google Scholar]
- 39.St-Jacques B, Hammerschmidt M, McMahon AP. Indian hedgehog signaling regulates proliferation and differentiation of chondrocytes and is essential for bone formation. Genes Dev. 1999;13:2072–2086. doi: 10.1101/gad.13.16.2072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chung UI, Schipani E, McMahon AP, Kronenberg HM. Indian hedgehog couples chondrogenesis to osteogenesis in endochondral bone development. J Clin Invest. 2001;107:295–304. doi: 10.1172/JCI11706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Minina E, Wenzel HM, Kreschel C, Karp S, Gaffield W, McMahon AP, Vortkamp A. BMP and Ihh/PTHrP signaling interact to coordinate chondrocyte proliferation and differentiation. Development. 2001;128:4523–4534. doi: 10.1242/dev.128.22.4523. [DOI] [PubMed] [Google Scholar]
- 42.Riemer S, Gebhard S, Beier F, Pöschl E, von der Mark K. Role of c-fos in the regulation of type X collagen gene expression by PTH and PTHrP: Localization of a PTH/PTHrP-responsive region in the human COL10A1 enhancer. J Cell Biochem. 2002;86:688–699. doi: 10.1002/jcb.10260. [DOI] [PubMed] [Google Scholar]
- 43.Adams SL, Pallante KM, Niu Z, Cohen AJ, Lu J, LeBoy PS. Stimulation of type-X collagen gene transcription by retinoids occurs in part through the BMP signaling pathway. J Bone Joint Surg Am. 2003;85(Suppl 3):29–33. doi: 10.2106/00004623-200300003-00006. [DOI] [PubMed] [Google Scholar]
- 44.Schipani E, Provot S. PTHrP, PTH, and the PTH/PTHrP receptor in endochondral bone development. Birth Defects Res C Embryo Today. 2003;69:352–362. doi: 10.1002/bdrc.10028. [DOI] [PubMed] [Google Scholar]
- 45.Ijiri K, Zerbini LF, Peng H, Correa RG, Lu B, Walsh N, Zhao Y, Taniguchi N, Huang XL, Otu H, Wang H, Wang JF, Komiya S, Ducy P, Rahman MU, Flavell RA, Gravallese EM, Oettgen P, Libermann TA, Goldring MB. A novel role for GADD45beta as a mediator of MMP-13 gene expression during chondrocyte terminal differentiation. J Biol Chem. 2005;280:38544–38555. doi: 10.1074/jbc.M504202200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Li TF, O'Keefe RJ, Chen D. TGF-beta signaling in chondrocytes. Front Biosci. 2005;10:681–688. doi: 10.2741/1563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Magee C, Nurminskaya M, Faverman L, Galera P, Linsenmayer TF. SP3/SP1 transcription activity regulates specific expression of collagen type X in hypertrophic chondrocytes. J Biol Chem. 2005;280:25331–25338. doi: 10.1074/jbc.M412549200. [DOI] [PubMed] [Google Scholar]
- 48.Dong YF, Soung do Y, Chang Y, Enomoto-Iwamoto M, Paris M, O'Keefe RJ, Schwarz EM, Drissi H. Transforming growth factor-beta and Wnt signals regulate chondrocyte differentiation through Twist1 in a stage-specific manner. Mol Endocrinol. 2007;21:2805–2820. doi: 10.1210/me.2007-0199. [DOI] [PubMed] [Google Scholar]
- 49.Tchetina EV, Kobayashi M, Yasuda T, Meijers T, Pidoux I, Poole AR. Chondrocyte hypertrophy can be induced by a cryptic sequence of type II collagen and is accompanied by the induction of MMP-13 and collagenase activity: Implications for development and arthritis. Matrix Biol. 2007;26:247–258. doi: 10.1016/j.matbio.2007.01.006. [DOI] [PubMed] [Google Scholar]
- 50.Thomas DP, Sunters A, Gentry A, Grigoriadis AE. Inhibition of chondrocyte differentiation in vitro by constitutive and inducible overexpression of the c-fos proto-oncogene. J Cell Sci. 2000;113:439–450. doi: 10.1242/jcs.113.3.439. [DOI] [PubMed] [Google Scholar]
- 51.GuhaThakurta D. Computational identification of transcriptional regulatory elements in DNA sequence. Nucleic Acids Res. 2006;34:3585–3598. doi: 10.1093/nar/gkl372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Apostolou E, Thanos D. Virus infection induces NF-kappaB-dependent interchromosomal associations mediating monoallelic IFN-beta gene expression. Cell. 2008;134:85–96. doi: 10.1016/j.cell.2008.05.052. [DOI] [PubMed] [Google Scholar]
- 53.Gebhard S, Hattori T, Bauer E, Schlund B, Bösl MR, de Crombrugghe B, von der Mark K. Specific expression of Cre recombinase in hypertrophic cartilage under the control of a BAC-Col10a1 promoter. Matrix Biol. 2008;27:693–699. doi: 10.1016/j.matbio.2008.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]