Abstract
High-throughput genomics methods are now being used to study a wide variety of viral diseases, in an effort to understand how host responses to infection can lead either to efficient elimination of the pathogen or the development of severe disease. This article reviews how gene expression studies are addressing important clinical issues related to hepatitis C virus infection, in which some 15–25% of infected individuals are able to clear the virus without treatment, while the remainder progress to chronic liver disease that can lead to cirrhosis and death. Similar methods are also being used in an effort to identify the mechanisms underlying the failure of some hepatitis C patients to respond to interferon-α/ribavirin therapy. By providing a detailed picture of virus-host interactions, high-throughput genomics could potentially lead to the identification of novel cellular targets for the treatment of hepatitis C.
I. Introduction: hepatitis C virus infection and liver disease
Hepatitis C virus (HCV), a blood-borne pathogen belonging to the Flaviviridae family, is a major cause of chronic hepatitis, progressive liver disease and hepatocellular carcinoma (Table 1) (Alter, 2007; Alter et al., 1992). Exposure to HCV has a variety of outcomes, ranging from spontaneous clearance of the virus to chronic infection and cirrhosis. The majority of patients develop persistent infection and approximately 20–30% of them will develop some form of liver disease. However, even among these patients, the clinical manifestations of HCV infection are highly variable, ranging from mild hepatitis to rapidly progressive fibrosis/cirrhosis. Current treatment is limited to combination PEG-IFN-α/ribavirin therapy which is associated with severe side effects, high cost and low efficacy in patients infected with HCV genotype 1. With nearly 200 million people infected worldwide and no available vaccine, HCV represents a significant health problem and is now the leading indicator for liver transplantation in the United States and Western Europe. Through the use of in vitro model systems, including subgenomic and genomic replicons as well as the recently developed HCV 2a infection system (Zhong et al., 2005) (Heller et al., 2005) (Lindenbach et al., 2005; Lindenbach et al., 2006), (Wakita et al., 2005) much has been learned about the molecular biology of HCV since its identification in 1989 as the cause of non-A, non-B hepatitis (Reviewed in (Lindenbach and Rice, 2005)).
Table 1.
Hepatitis C: the basics.
| Virus classification and structure | Hepatitis C virus (HCV) is the only member of the genus Hepacivirus in the family Flaviviridae. There are 6 major HCV genotypes; about 80% of patients in the US have genotype 1. The 9.6 kb single-stranded, positive-sense RNA genome encodes a single open reading frame flanked by 5′ and 3′ untranslated regions. |
| Infection Cycle | A number of hepatocyte surface molecules, including CD81 tetraspanin, scavenger receptor class B type I (SR-BI), Claudin-1, mannose-binding lectins DC-SIGN and L-SIGN have been identified as putative HCV receptors or co- receptors. Upon entry and uncoating of the virion, the plus-sense viral genome acts as messenger RNA. Translation is initiated at an internal ribosome entry site (IRES) located within the 5’ UTR. The 3,011 amino acid polyprotein is then co-and post-translationally cleaved into the viral structural and nonstructural proteins both by host signal peptidases and viral proteases. HCV structural proteins include the nucleocapsid core (C) and two envelope glycoproteins, E1 and E2. The non-structural proteins include a zinc-dependent metalloprotease encoded by the NS2-NS3 region, an NS3 serine protease-RNA helicase along with the NS4A peptide cofactor of NS3 protease activity, the NS4B phosphoprotein, and p7. The replicase complex includes the NS5A protein and the NS5B RNA-dependent RNA polymerase which is responsible for catalyzing viral RNA replication. |
| Epidemiology | HCV infects only humans. There are no approved vaccines. The virus is transmitted through blood transfusion, injecting drug use or other types of direct blood contact. Nearly 200 million people are infected worldwide. In the United States, there are estimated to be some 3.2 million infected persons, with about 20,000 new cases a year. Rates of HCV infection are higher among people infected with HIV. |
| Clinical course | A minority of patients develops mild, nonspecific symptoms during the months following infection, but most remain asymptomatic. Some 15–25% of patients are able to eliminate the virus without treatment. Chronic HCV infection is diagnosed on the basis of anti-HCV antibodies, PCR testing for circulating virus and the detection of abnormal liver function. The CDC estimates that for each 100 people with HCV, 75–80 will become chronically infected, 60–70 will develop liver disease, 5–20 will develop cirrhosis over a period of 20–30 years, and 1–5 will die of the consequences of viral infection. |
| Antiviral therapy | Antiviral therapy is initiated when progressive disease is indicated by biopsy and abnormal liver function tests. Current therapy consists of a combination of pegylated interferon alpha and ribavirin. Patients with genotypes 2 and 3 have a 60–80% response (disappearance of detectable HCV RNA in serum) to either a 24- or 48-week course of combination therapy, while the rate is 30–50% for genotype 1. |
Several areas of HCV research have been the focus of intense investigation for a number of years. This includes elucidating the immune responses underlying viral persistence/clearance, in particular identifying the parameters associated with protective immunity as well as the mechanisms by which HCV evades host immune responses. Another important area of research has been focused on studying the mechanisms of liver injury associated with HCV infection. Elucidating the exact cellular mechanisms underlying HCV pathogenesis has proven to be extremely challenging due to the lack of appropriate model systems. Currently, liver biopsies remain the “gold standard” to diagnose and monitor liver disease progression in patients, a procedure that is both invasive and expensive. A better understanding of how HCV infection induces liver damage will ultimately aid in the identification of biomarkers that could be used to both predict liver disease development and also to assess severity of liver disease in patients without the need to biopsy. Insight into the cellular pathways required for HCV replication and/or liver injury will also lead to the identification of novel therapeutic targets to be used in combination with current therapies. Finally, the host and viral factors responsible for the poor response rate to PEG-IFN-α/ribavirin therapy in patients remain undefined. Understanding the molecular mechanisms responsible for treatment failure as well as identifying markers that will accurately predict patient response would greatly improve patient care and perhaps eliminate unnecessary treatment.
This review will highlight how the use of high-throughput functional genomics is being utilized to study HCV infection. After briefly reviewing the types of model systems used for genomic analysis, this article will focus on the contribution genomics has made in addressing the following clinical questions:
Why do most patients fail to clear HCV infection following initial exposure?
Why does a subset of HCV patients develop severe liver disease?
Why do some patients fail to respond to IFN-ribavirin therapy?
Finally, this manuscript will also highlight how these insights could potentially lead to the development of novel therapeutics for treatment of chronic HCV infection.
II. Genomics studies of HCV infection
Microarray technology is a powerful tool that allows the generation of extensive amounts of data from limited quantities of biological material without the need for preconceived ideas about what is important. It also provides a very global perspective of the molecular events associated with a particular disease state, something which traditional research methods such as immunohistochemistry or confocal microscopy cannot achieve. This is particularly useful when studying HCV pathogenesis which likely involves a combination of direct HCV-mediated effects, virus-host interactions involving multiple cell types, and immune responses. Although analysis of such complex data presents certain challenges, there are now useful commercial and publicly available software which facilitate both analysis and presentation of gene expression data.
A. Human liver biopsies
Although there are animal models of HCV replication and infection, no actual model of HCV-associated liver disease exists. Furthermore, the extensive variation in clinical manifestations of chronic HCV infection indicates that host genetic factors influence disease severity and so utilizing model systems which lack this genetic heterogeneity are not necessarily ideal. Microarray studies aimed at investigating the host response to HCV infection and its relationship to disease or IFN therapy response are therefore mainly restricted to human clinical samples. Core needle liver biopsies have been extensively utilized for global transcriptional profiling of the host response to HCV infection from a variety of disease states including fibrosis, cirrhosis and hepatocellular carcinoma. While such samples are of obvious clinical relevance, they are of limited quantity and are comprised of a complex mixed cell population including hepatocytes, immune cells and sinusoidal endothelial cells.
B. Chimpanzees
Although HCV can cause persistent infections in chimpanzees, albeit at a lower rate than in humans, it is not associated with significant liver disease such as fibrosis/cirrhosis. It is currently unclear whether this is due to specific differences in host response or is simply an observational error, due to the small number of animals and lack of long-term follow-up. Despite these challenges, transcriptional profiling of hepatic tissue from HCV -infected chimpanzees has provided new insights into immune responses associated with either acute self-resolving or persistent infection.
C. The chimeric SCID-Alb/uPA mouse model
For many years, progress in the HCV field has been hindered by a lack of a small animal model in which to study both viral infection/replication and for antiviral screening. A significant breakthrough came with the development of the SCID-Alb/uPA mouse model (reviewed by Meulemans and Leroux-Roels, 2008). These animals are derived by transplantation of normal human hepatocytes into SCID mice carrying a plasminogen activator transgene (Alb-uPA) (Mercer et al., 2001; Meuleman et al., 2005; Hsu et al., 2003) (Lindenbach et al., 2006). Progressive depletion of the mouse hepatocytes as a result of the transgene provides an opportunity for the human hepatocytes to engraft and regenerate the mouse liver. The resulting human-mouse chimeric liver can then be infected with either HCV or HBV. While the absence of fibrosis in these animals makes the model unsuitable for the study of HCV-associated fibrogenesis, there are still significant advantages over in vitro systems in that it represents an in vivo infection in primary human hepatocytes, all HCV proteins are expressed at biologically relevant levels, and infectious virions are assembled and released from hepatocytes.
Perhaps the most valuable attribute of the model is the unique opportunity to characterize the host response to infection in multiple donor hepatocytes while at the same time having control over both viral inoculum and duration of infection. This will undoubtedly shed insight into genetic factors contributing to the variation in global gene expression profiles observed in microarray experiments from patient liver tissue. The model is also valuable for discerning direct virus-mediated transcriptional changes from immune-mediated effects, something which is extremely difficult to do with gene expression studies using patient biopsies.
III. Why do most patients fail to clear HCV infection?
To date, traditional immunological and molecular biology techniques have suggested that viral persistence is associated with impairment of cellular immune responses through a variety of mechanisms while spontaneous clearance is associated with a vigorous, multi-epitope T cell response (Reviewed in (Rehermann, 2007; Rehermann and Nascimbeni, 2005) (Shoukry et al., 2004)). The role of the humoral immune response and neutralizing antibodies in protective immunity remains unclear (Reviewed in (Keck et al., 2008), (Zeisel et al., 2007; Stamataki et al., 2008)). However, these studies typically utilized peripheral immune cells and it is not clear how the function of peripheral and liver-infiltrating immune cells parallel each other. Furthermore, they are not able to easily ascertain the role of hepatocyte innate immunity in HCV infection outcome. The following sections will outline the role microarray technology has played in characterizing persistent versus self-resolving HCV infections.
A. Intrahepatic gene expression profiles in chimpanzees
Chimpanzees have been especially useful for studying spontaneous viral clearance. Because patients are rarely identified during the very acute phase following exposure to HCV, such studies are difficult to conduct using human cohorts. Su et al examined the host transcriptional response in liver tissue from acutely-infected chimpanzees and found that HCV clearance was specifically associated with increased expression of interferon-γ-inducible genes (Su et al., 2002). Whether this is related to the direct antiviral activities of interferon-γ or simply representative of a more vigorous T-cell response is unclear. Increased expression of genes associated with T-cell recruitment, antigen processing, and presentation were also specifically associated with viral clearance. Interestingly, the induction of these genes is associated with higher levels of viremia during the acute phase of infection compared with that observed in an animal which developed a persistent infection. This suggests that a threshold level of viral replication is required to induce a successful adaptive immune response. A similar study of chimpanzees with acute HCV infection also demonstrated that viral clearance was associated with increased expression of genes associated with a Type 1 T cell response, including genes known to be induced by IFN-γ (Wieland et al., 2004). Bigger et al found that the expression patterns of interferon stimulated genes (ISGs) in a chimpanzee that cleared HCV were temporally distinct, and the authors speculated that different regulatory pathways or involvement of different cell types and interferon (Type I or II) function over time (Bigger et al., 2001). Similar to Su et al, immune cell markers were detected at later time points, possibly indicating infiltration of immune cells.
It is possible that induction of an interferon-α response early in infection plays an important role in limiting viral spread throughout the liver, similar to what was observed using the SCID-Alb/uPA mouse model of HCV infection (Walters et al., 2006a). Induction of a Type II interferon response, and associated adaptive immune response, may then eliminate the remaining HCV-infected hepatocytes. Interestingly, acute HBV infection in chimpanzees was not associated with induction of ISGs, suggesting that this virus does not induce an innate immune response at the site of infection (Wieland et al., 2004). This may be explained by the unique replication strategy employed by hepadnaviruses where viral genome replication occurs within the assembled capsid, thus preventing exposure of viral nucleic acid to dsRNA signaling mediators present in cytoplasm. Indeed, this strategy may have evolved in an attempt for the virus to circumvent these antiviral responses. Taken together, these results suggest that timely activation of both innate and adaptive immune responses are important in efficient elimination of HCV-infected hepatocytes in the absence of excessive liver injury. Failure of the host to quickly mount this protective response likely results in widespread HCV infection throughout the liver and subsequent chronic aberrant ineffective immune responses which fail to clear virus but instead mediate liver injury.
B. HCV and the innate antiviral response
Viral infections trigger a host cell antiviral response that results in the expression of Type 1 IFN (IFN-α/β) and subsequent induction of ISGs. Exposure to HCV typically results in a persistent infection, suggesting that the virus has developed mechanisms to either suppress the activation of innate antiviral pathways and/or to modulate the antiviral activity of effector proteins. In light of this, there has been a keen interest in studying the interaction between HCV and the innate antiviral signaling pathways.
Products of HCV replication, including double-stranded RNA (dsRNA) replication intermediates, can trigger the phosphorylation and activation of IFN-regulatory factor 3 (IRF3) and NF-κβ which mediate increased ISG production and establishment of an intracellular antiviral state (Sumpter, Jr. et al., 2005; Sumpter, Jr. et al., 2004). These cellular responses are initiated through both Toll-like receptor 3 (TLR-3) and retinoic acid inducible gene-1 (RIG-I)-dependent mechanisms of IRF3 activation. Consistent with the high rate of HCV persistence in exposed individuals, the HCV serine protease, NS3/4A, can block the phosphorylation and activation of the IRF3 transcription factor through disruption of both these pathways (Ferreon et al., 2005; li et al., 2005) (Foy et al., 2003; Foy et al., 2005) (Sumpter, Jr. et al., 2005). The TLR3-dependent activation of IRF3 is blocked by NS3/4A-mediated cleavage of Toll-IL1 receptor domain containing adaptor inducing IFN-β (TRIF), an adaptor protein which links TLR3 to kinases responsible for activating IRF3 and NF-κβ (li et al., 2005; Ferreon et al., 2005). NS3/4A also disrupts the RIG-I dependent intracellular pathway through cleavage of IPS-1 (also known as VISA, MAVS and Cardif), the recently identified RIG-I adapter protein (Breiman et al., 2005) (Foy et al., 2005) (Sumpter, Jr. et al., 2004) (Kawai et al., 2005) (Loo et al., 2006) (Meylan et al., 2005) (Seth et al., 2005) (Xu et al., 2005).
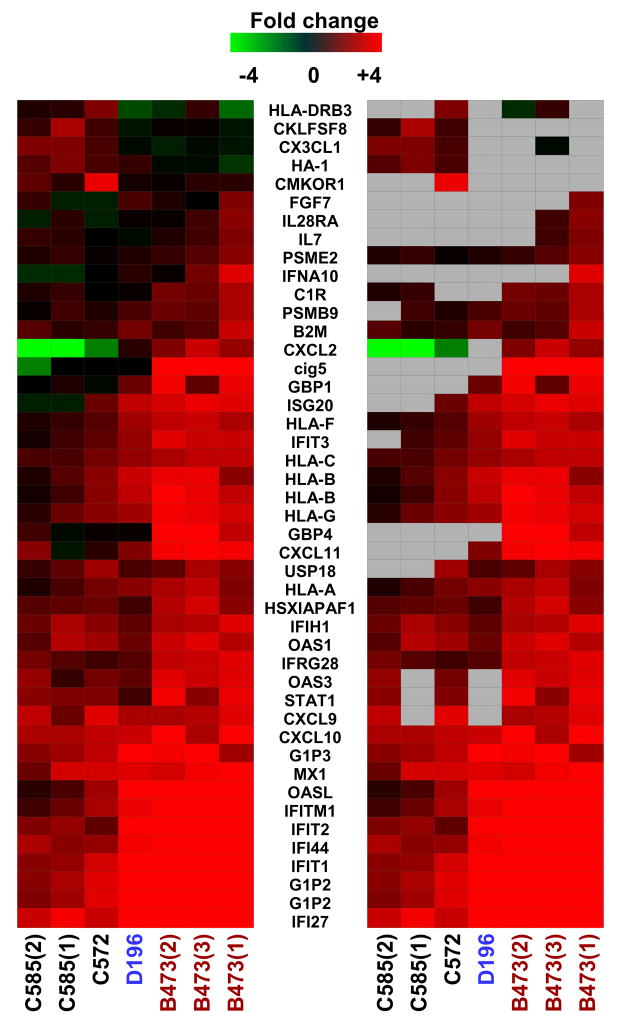
Based on the results of these extensive studies, it would be expected that induction of genes associated with the innate antiviral response would be significantly impaired in livers from chronically infected HCV patients. Intriguingly, this is in fact not the case as all gene expression studies of HCV-infected tissue demonstrate an active and prolonged stimulation of host innate dsRNA and Type I IFN pathways as indicated by induction of ISGs (Bigger et al., 2001; Bigger et al., 2004) (Su et al., 2002) (Smith et al., 2003; Smith et al., 2006) (Lederer et al., 2006) (Walters et al., 2006b; Walters et al., 2006a) (Bieche et al., 2005) (Lau et al., 2005) (Helbig et al., 2005). It is speculated that not all hepatocytes in a liver are infected and so it is feasible that the increased expression of ISGs occurs primarily in uninfected hepatocytes, induced in response to endogenous IFN released from adjacent HCV-infected hepatocytes. It has also been speculated that hepatocytes are not the main source of IFN in an infected liver but rather IFN is released from infiltrating immune cells such as plasmacytoid dendritic cells. However, it is important to note that induction of genes associated with IFN-signaling is observed in the SCID-Alb-uPA mouse model of infection in which human immune cells are absent (Figure 1). Furthermore, recent transcriptional profiling of HCV 2a-infected cultured hepatocytes revealed induction of numerous ISGs in cultures with 100% infection, indicating that ISG expression occurs in HCV-infected hepatocytes (Walters, unpublished data). It is possible that HCV-mediated antagonism of IRF-3 is incomplete or that induction of ISGs is occurring via a separate, yet identified pathway.
Figure 1.
Expression of genes associated with IFN-signaling in human liver tissue from HCV-infected uPA-SCID mice. A. Two-dimensional hierarchical clustering was done using Resolver System software with an agglomerative algorithm, complete link heuristic criteria, and Euclidean correlation metric. Each column represents gene expression data from an individual experiment (either individual HCV-infected mouse or individual liver sample). Genes were selected as at least 2-fold regulated (P value < 0.05) in at least 1 of 7 experiments. In the left panel, genes shown in red are up-regulated and genes shown in green are down-regulated in HCV-infected tissue relative to donor-matched uninfected tissue, while black indicates no change in gene expression. In the right panel, genes whose regulation showed P value > 0.05 are shown in gray.
A recent study attempted to partly address these questions by using microarray profiling and confocal microscopy to examine the relationship between IRF-3 activation and ISG expression in liver biopsy tissue from chronically infected patients (Lau et al., 2008). Comparison of signal intensities of ISGs in liver samples with and without IRF3-positive nuclei revealed a positive correlation between nuclear IRF-3 and higher expression levels of innate immune response genes, although this correlation was not always statistically significant. However, the study did not include an analysis of ISG expression, HCV infection and IRF-3 activation within individual hepatocytes. Such studies are obviously extremely technically challenging. One potential approach would be to utilize the chimeric SCID-Alb/uPA mouse model and a GFP-tagged HCV which would enable efficient separation of HCV-infected and uninfected cells. Microarray profiling could be then utilized to assess HCV replication and ISG expression levels in both HCV-GFP (+) and (−) hepatocytes.
IV. Why does a subset of HCV patients develop severe liver disease?
One of the most puzzling and poorly understood aspects of chronic HCV infection is why only a subset of patients develops significant liver disease while the majority remains asymptomatic despite active on-going viral replication. Although the reasons for the high variability in disease progression are not completely understood, several factors are considered to increase the risk of developing liver disease. These include alcohol consumption, co-infection with HIV-1, exposure to chemical agents, and disorders of iron metabolism. Inflammation associated with chronic HCV infection is thought to contribute to the liver injury, necrosis, and hepatic regeneration that underlie the progression to cirrhosis. Liver cell damage in chronic HCV infection is thought to be mediated primarily by apoptosis of HCV-infected hepatocytes (Bantel and Schulze-Osthoff, 2003; Kountouras et al., 2003). FAS-expressing hepatocytes are generally observed in areas of lymphocyte infiltration and it is speculated that HCV-specific T cells migrate into the liver, recognize viral antigen in infected hepatocytes, and induce apoptosis of FAS-expressing hepatocytes (Hiramatsu et al., 1994; Hayashi and Mita, 1999; Fukuzawa et al., 2001). However, liver disease progression is generally accelerated in immuno-compromised patients, including those co-infected by HIV or taking immuno-suppressive drugs (Garcia-Retortillo et al., 2002; Garcia-Samaniego et al., 1997) (Benhamou et al., 2001; Benhamou et al., 1999) (Soto et al., 1997; Graham et al., 2001) (Berenguer et al., 2000). Hepatocyte apoptosis has also been observed in the SCID-Alb/uPA mouse model of HCV infection, a model system which is unable to mount an adaptive HCV-specific immune response (Walters et al., 2006a). Finally, recent studies have indicated a direct HCV-mediated cytopathic effect in cultured hepatocytes through a variety of mechanisms, including ER stress and death receptor signaling (Zhu et al., 2007) (Sekine-Osajima et al., 2008).
In light of these studies, it is likely that liver injury results from a complex combination of viral and immune mediated effects. Therefore, traditional laboratory studies aimed at assessing the association of individual genes or proteins with disease progression have limitations with respect to elucidating cellular mechanisms of pathology. The lack of an appropriate model system also somewhat restricts studies to patient samples which are typically of limited quantity. Microarray technology is ideally suited to studying virus-host interactions on samples of limited quantity, such as that obtained by core needle biopsies, in that it allows the simultaneous measurement of thousands of individual genes, thus providing a very global perspective of the host response. It is perhaps because of these issues that the viral hepatitis field in particular has embraced the use of high-throughput genomics technology to study viral pathogenesis.
A. Gene expression profiling of the hepatic response to HCV infection
Studies aimed at identifying the host and viral factors that influence liver disease progression will ultimately improve patient care by helping to identify patients who are at risk of developing significant liver disease, thus providing an opportunity for early therapeutic intervention. Many studies have attempted to probe the complexities of the host response to HCV by performing global transcriptional profiling of liver tissue from HCV-infected individuals. The majority of these studies demonstrated an induction of genes associated with immune cell activation, consistent with the role cell mediated immunity is thought to play in liver injury (Smith et al., 2003) (Asselah et al., 2005; Bieche et al., 2005) (Shao et al., 2005) (Lau et al., 2005). Induction of genes associated with innate antiviral signaling pathways was also observed in the majority of studies, although the magnitude and intensity is highly variable (Smith et al., 2003; Bieche et al., 2005; Lau et al., 2005). Interestingly, one study found that three IFN-γ-inducible genes increased in parallel to necro-inflammation and stage of fibrosis which is again consistent with T cell mediated injury (Bieche et al., 2005).
Lau et al integrated intrahepatic gene expression profiles in HCV-infected patients at different stages of fibrosis with α-smooth muscle actin staining patterns and found that stellate cells are actually activated early in HCV-mediated injury. It was speculated that this was caused by oxidative stress from inflammation and lipid metabolism (Lau et al., 2005). A recent study suggests that stellate cells are also activated following phagocytosis of lymphocytes (Muhanna et al., 2008). The fact that stellate cell activation occurs even before histological evidence of fibrosis suggests the potential to serve as a predictive biomarker of liver disease development. Evidence of oxidative stress associated with disturbances in lipid metabolism was also observed in transcriptional profiling of livers from HCV-infected SCID-Alb/uPA mice (Walters et al., 2006a). HCV is known to be associated with disturbances in lipid metabolism pathways and so the oxidative stress may be a direct result of viral replication and not simply inflammation (Bigger et al., 2001) (Su et al., 2002) (Siagris et al., 2006) (Patel et al., 2005) (Hamamoto et al., 2005) (Petit et al., 2003) (Ye et al., 2003). Although these studies have been useful in identifying cellular pathways modulated by HCV infection, further in-depth analysis of the complex gene expression data and functional studies using in vitro or animal models are crucial to understanding the mechanisms by which virus-regulation mediates liver disease.
The majority of genomics studies have been cross-sectional in nature, making it difficult to assign clinical significance, with respect to ultimate patient outcome, to a particular gene expression pattern. Smith et al utilized serial liver biopsies obtained from liver transplant recipients with recurrent HCV to identify molecular processes influencing liver disease progression and to find potential gene markers of early fibrosis (Smith et al., 2006). This model system is ideal in that it allows a direct comparison of recurrent HCV in the presence and absence of disease. This will not only shed insight into molecular mechanisms of fibrosis development but also aspects of the host response which function to protect against liver disease. It also provides a unique opportunity to characterize the host response within an individual over time, an important consideration given the slow, progressive nature of HCV-associated liver disease.
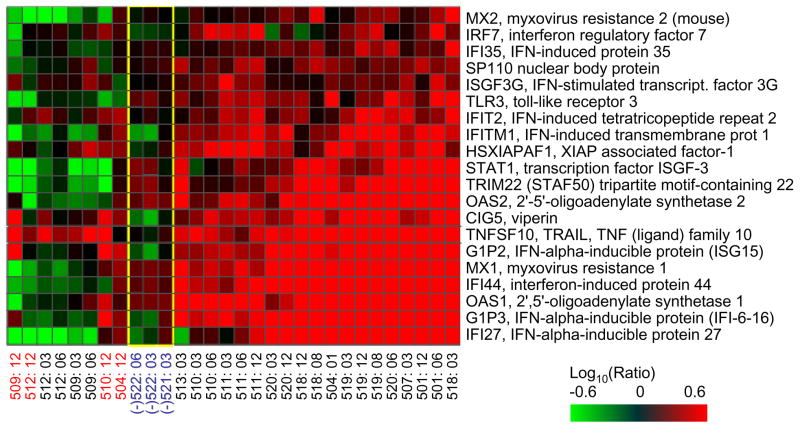
This longitudinal study involved gene expression profiling of liver tissue from 13 patients, four of which developed fibrosis within 15 months post-transplant. It was found that patients with rapidly progressive fibrosis accumulated higher numbers of differentially regulated genes, including induction of genes related to hepatic stellate cell activation, a finding similar that that of Lau et al (Lau et al., 2005). Interestingly, induction of genes associated with the IFN response was absent in two patients and suppressed in the 12 month biopsies in two other patients, all of which subsequently developed fibrosis (Figure 2). There was also a lack of induction of genes specific for immune cells, suggesting that fibrosis progression was associated with at least a partial impairment of the host immune response. It is possible that these results could be at least partially attributed to immunosuppressive therapy. The data suggests that protection from fibrosis development appears to be a fine balance between immune-mediated control of viral replication and immune-mediated liver injury, a scenario similar to what occurs during highly pathogenic influenza infection (Kash et al., 2006b; Kobasa et al., 2007). Significantly, the differences in expression of IFN-related and hepatic stellate cell-associated genes were present in patients prior to histological evidence of fibrosis and so have potential for use as predictive biomarkers. Although promising, the sample size of this study is relatively small and studies are on-going to expand these findings using a much larger cohort (Walters, unpublished data). Furthermore, while the study has been valuable in identifying differences in host response associated with different clinical outcomes, the data generated do not necessarily provide information as to why or how these differences occur.
Figure 2.
IFN-related gene expression patterns correlate with early progression to fibrosis in human patients. Expression profiles of 18 genes that encode components of IFN signaling and IFN-inducible proteins. Two-dimensional hierarchical clustering was performed with green and red bars showing decreased or increased levels of mRNA in a time point biopsy relative to the corresponding baseline biopsy (obtained from the same liver graft at the time of transplantation), Patients with early progression to fibrosis are indicated by red labels. HCV (−) patients are indicated by blue labels and the corresponding gene expression data are enclosed in the yellow box.
Many studies aimed at investigating HCV-associated liver disease focus on characterizing fibrotic tissue. However, as evidenced by the study by Smith et al, understanding the host response in HCV-infected individuals without disease may ultimately be even more valuable. While the chimpanzee model of HCV infection has been criticized for being an inappropriate model in which to study HCV pathogenesis, it may in fact be an ideal model in which to study the “protective” host response during a persistent infection.
B. Innate immunity and HCV pathogenesis
A finding common to the majority of these microarray studies is the increased expression of genes associated with innate antiviral signaling, albeit with extensive patient to patient variation in the magnitude of this response. However, it is important to note that transcriptional profiling of viral-infected cells, or tissue, enables the identification of only those ISGs which are induced in response to infection and may not necessarily reflect the complete transcriptional response to IFN, particularly as many viruses are well known to regulate activation of various aspects of these pathways. As such it is difficult to accurately assess viral regulation of innate signaling pathways. Experiments designed to examine the expression of ISGs in response to both infection and exogenous IFN treatment can provide significant insight into the magnitude of viral-mediated antagonism of host innate antiviral signaling pathways. For example, microarray profiling of in vitro IFN-treated and HCV-infected cultured hepatocytes demonstrated a remarkably similar regulation of ISGs, suggesting that HCV is not significantly impacting activation of innate signaling pathways (Walters, unpublished data). Similar experiments with the highly pathogenic acute filovirus infections using EBOV and MARV showed very different results. Infection with EBOV and MARV was associated with a significant antagonism of induction of genes associated with the antiviral response (Kash et al., 2006a).
For viruses such as HCV, which typically establish persistent infection characterized by minimal pathology (at least in the majority of individuals), extensive suppression of the innate antiviral response, such as what is seen with EBOV, would likely result in much higher levels of viral replication. This in turn may result in a more aggressive course of liver disease. In support of this, there is evidence to suggest that an attenuated IFN response can indeed result in increased or accelerated HCV-mediated pathogenesis. As discussed earlier, gene expression profiling of serial liver biopsies in liver transplant patients with recurrent HCV demonstrated that rapid fibrosis progression was associated with a lack of induction of genes associated with the IFN-mediated antiviral response, antigen presentation and cytotoxic response (Smith et al., 2006). This suggests that disease progression may result from an impaired immune response and subsequent lack of control of viral replication. A similar scenario was observed in a cohort of HCV and HCV/HIV-infected patients where a subset demonstrated impaired induction of several key genes involved in the IFN response (Walters et al., 2006b). Collectively, these studies suggest a direct pathogenic effect of HCV in the presence of high levels of virus.
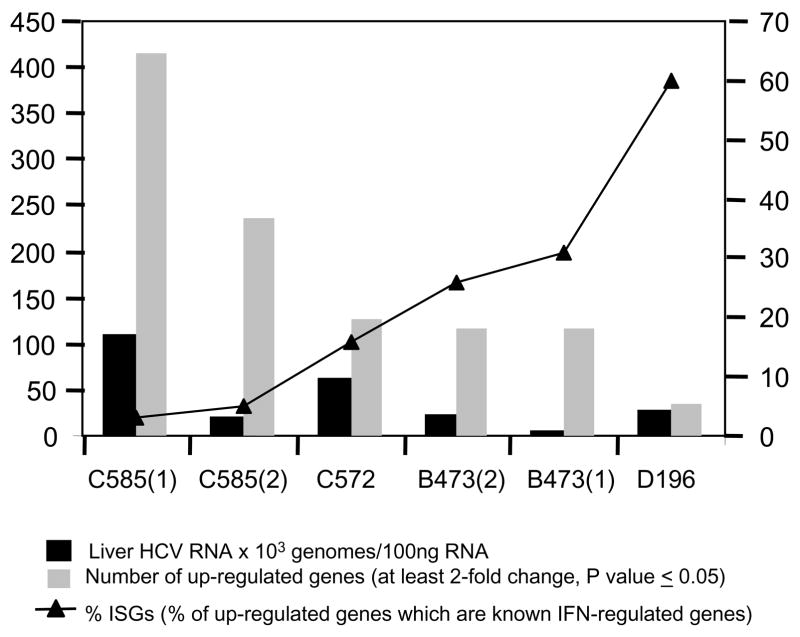
Further evidence that disease progression may result from an impaired immune response comes from transcriptional profiling of HCV-infected chimeric SCID-Alb/uPA mice which indicated that the nature of the host innate antiviral immune response during the acute phase of infection may determine the extent of viral-mediated effects on host gene expression. In this model, attenuated induction of ISGs associated with specific donor hepatocytes corresponded with higher levels of intrahepatic HCV replication. This in turn was associated with induction of lipid metabolism and oxidative stress genes which have the potential to cause cytopathic effects (Figure 3). Indeed, enhanced hepatocyte apoptosis was observed in HCV-infected animals. It is possible that viruses that establish chronic infections actually take advantage of host innate antiviral responses to limit their replication to a level that does not significantly impact the normal functions of the host cell. In patients who ultimately develop serious liver disease, the balance of IFN-mediated control of virus replication and virus-mediated suppression of innate antiviral signaling may shift toward the latter. Understanding the viral and host factors responsible for this variation is obviously critical.
Figure 3.
Association between intrahepatic HCV RNA levels, numbers of differentially expressed genes and IFN-stimulated genes in HCV-infected uPA-SCID mice.
C. Studies in chimeric SCID-Alb/uPA mice
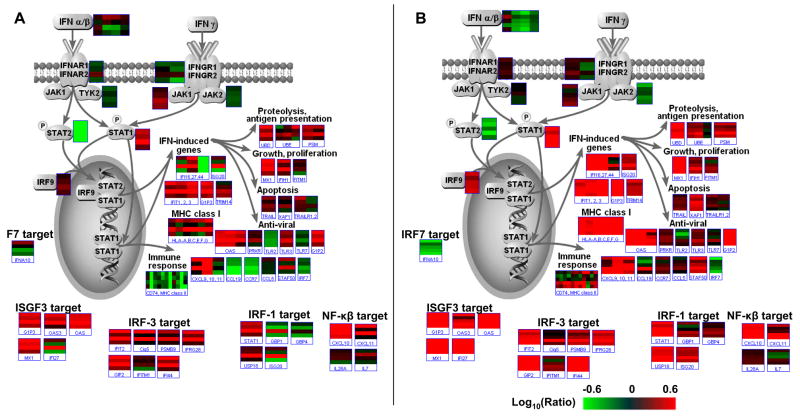
Microarray profiling has already demonstrated many similarities in the host response to HCV infection between the mouse model and chronically infected patients (Walters et al., 2006a). In particular, the activation of innate antiviral signaling pathways was shown to be remarkably similar, suggesting that the SCID-Alb/uPA mouse model may provide an excellent system with which to further investigate the interplay between HCV and the innate immune response (Figure 4). For example, genomic profiling of animals transplanted with hepatocytes from different donors and inoculated with the same virus source has revealed that host genetic factors influence the nature of the innate antiviral response. While all HCV-infected animals showed evidence of activation of innate antiviral response, differences in both the number and intensity of induction of ISGs were observed between mice containing hepatocytes from different donors (Walters et al., 2006a). These results were particularly intriguing as much emphasis has been placed on the ability of HCV to attenuate IFN signaling by multiple mechanisms (reviewed in (Gale, Jr. and Foy, 2005)). The results from the SCID-beige/Alb-uPA mice argue that host genetics also influence the effectiveness of the innate immune response. In light of the potential role of the magnitude of the innate IFN response in liver disease progression and therapy response, this model may prove to be extremely useful in understanding host genetic factors influence viral pathogenesis.
Figure 4.
Induction of IFN-signaling pathways in HCV-infected uPA-SCID mice (A) and patients (B). Pathway Builder (Protein Lounge) and the Pathway feature of Resolver were used to visualize the expression of genes from the IFN signaling pathway. The expression profiles of individual genes are shown in boxes and each bar within the box represents one experiment. Color schemes are as indicated in Figure 1. The reference for each microarray experiment was a pool of normal, uninfected human liver tissue.
V. Why do some patients fail to respond to IFN-ribavirin therapy?
Based on the importance of the innate antiviral response in viral clearance, it is not surprising that exogenous IFN therapy is currently utilized to treat a number of viral infections, including HCV. The primary goal of therapy is to obtain a sustained virological response, which is defined as absence of detectable HCV RNA in the serum using a sensitive qualitative HCV RNA assay 6 months following the completion of therapy (Wohnsland et al., 2007). Randomized controlled trials have demonstrated SVRs of 30 to 50% and 60 to 80% for patients infected with genotype 1 and 2/3, respectively (Thomson and Finch, 2005) (Lindsay, 2002) (Lau et al., 1998) (Manns et al., 2001). While these results are encouraging, there remains a large population of patients who fail to achieve a virological response to anti-HCV therapy, particularly patients infected with genotype 1. Since PEG-IFN-α/ribavirin is the only approved treatment for chronic HCV infection, there is a keen interest in elucidating the molecular mechanisms underlying treatment failure as well as identifying markers that can accurately predict patient response.
Much of the research in this area has focused on identifying the role of viral factors in IFN therapy resistance, including genotype, pre-treatment HCV RNA serum levels and viral quasispecies diversity (Reviewed in (Wohnsland et al., 2007)). In vitro studies have also demonstrated that viral proteins mediate IFN resistance through a variety of mechanisms, including inhibition of RIG-I and TLR3 via NS3/4A, disruption of the IFN-mediated Jak/STAT pathway via NS5A and core, as well as inhibition of PKR by the E2 and NS5A proteins (Reviewed in (Wohnsland et al., 2007)). However, virologic response rates have also been shown to be influenced by various host factors including age, weight, gender, race, liver enzymes and stage of fibrosis (Reviewed in (Gao et al., 2004)). A better understanding of the mechanism by which each of these factors contribute to therapy resistance may ultimately lead to individualized therapy protocols.
A. Gene expression signatures of patient response to IFN therapy
Microarray profiling of patient tissue obtained prior to and during therapy has played a key role in shedding light on gene expression signatures of patient response to IFN treatment. Two separate studies have compared liver transcriptional profiles of patients prior to initiation of therapy in an attempt to identify potential differences between eventual responders and non-responders. The results of both studies suggested that higher baseline levels of genes associated with innate antiviral signaling was associated with treatment failure. Microarray experiments performed on pre-treatment liver tissue from 31 chronic HCV patients who subsequently underwent IFN/ribavirin therapy identified a set of 18 genes whose expression differed significantly between all responders and all non-responders(Chen et al., 2005). These genes, of which many are IFN-regulated, were generally more highly induced in the livers of non-responders. Similarly, Feld et al demonstrated that non-responders had significantly higher intrahepatic pre-treatment expression levels of ISGs than those who achieved a sustained viral response(Feld et al., 2007). A lack of virological response to IFN-α treatment in HCV-infected chimpanzees is also thought to be due to already highly elevated hepatic ISG expression observed in these animals (Lanford et al., 2007).
While these results are intriguing, there are still many unanswered questions. First, it is important to note that neither group attempted to utilize these sets of genes to predict response in a separate cohort of patients. Such studies are crucial in determining the true predictive power of these potential biomarkers. Furthermore, differences in expression levels of these genes between responders and non-responders are often not striking. The implication of such minor differences with respect to antiviral function is unclear and the feasibility of using them for predicting patient response to therapy is questionable. Second, it remains to be determined if the higher basal expression level of the ISGs is the actual mechanism responsible for non-responsiveness to IFN therapy. It is unclear how higher expression levels of ISGs may result in resistance to IFN therapy but it may be related to de-sensitization of the IFN receptors. It is also possible that lack of response to exogenous IFN may be due to an already maximally induced ISG response in non-responders. Third, it is puzzling that patients have relatively high expression levels of ISGs within the liver yet fail to spontaneously clear infection. Collectively, these questions highlight the importance of studies aimed at elucidating the antiviral activity of individual ISGs, compartmentalization of ISG response within an HCV-infected liver as well as validation studies of potential markers in additional patient cohorts.
In addition to differences in expression levels of ISGs between responders and non-responders, transcriptional profiling has revealed that a potential mechanism of failed response may involve induction of genes associated with IFN-regulatory pathways. Feld et al found that response to IFN therapy was associated with a induction in the expression of ISGs, where treatment failure was associated with a greater change in the expression of genes associated with IFN-inhibitory pathways (Feld et al., 2007). Walsh et al found significantly increased intrahepatic expression of SOCS3 in patients who had failed IFN treatment (Walsh et al., 2006). SOCS proteins inhibit IFN signaling by inhibiting tyrosine phosphorylation and nuclear translocation of STAT (Walsh et al., 2006). Enhanced intrahepatic SOCS3 mRNA (and protein) is also speculated to contribute to the non-responsiveness of HCV-infected chimpanzees to IFN therapy (Huang et al., 2007a). Interestingly, it was found that SOCS3 increased significantly in HCV-infected but not naïve animals following IFN treatment. This suggests that HCV itself is modulating the expression of SOCS3, either directly or indirectly. However, this group also evaluated intrahepatic SOCS3 mRNA expression in a cohort of 21 patients prior to antiviral therapy and actually found higher levels in patients who went on to successfully respond to IFN(Huang et al., 2007a). In addition, no significant difference was observed between non-responders and responders in a separate cohort of 13 patients following IFN treatment. A potential criticism of this study is that the biopsies were obtained 24hrs following the administration of IFN-α. Genomic studies in chimpanzees have shown that the kinetics of the transcriptional response to IFN, in both liver and PBMC, is maximal between 4 and 8hrs and returns to baseline by 24hrs (Lanford et al., 2006). In addition, the pre-treatment and post-treatment biopsies used to calculate the level of induction of SOCS3 were actually taken from different patient cohorts and as such the comparison was indirect. Both of these drawbacks could explain why no significant difference in SOCS3 expression was observed between responders and non-responders following treatment. Clearly, additional studies are needed to more clearly define the relationship between treatment failure and induction of IFN inhibitory pathways.
D. Single nucleotide polymorphism analysis
Gene expression profiling of hepatic tissue from HCV-infected patients has demonstrated considerable variability in the activation of innate signaling pathways and subsequent induction of ISGs. While gene expression studies have suggested that disease progression is associated with an impaired induction of ISGs and that treatment failure is associated with higher pre-treatment expression of ISGs, it is currently unclear what factors are responsible for this variation. Studies using the SCID-Alb/uPA mouse model have suggested that this variation is influenced by host genetic factors. In light of the apparent importance of expression levels of these genes with respect to both disease progression and response to therapy, gaining an understanding of genetic factors that regulate this variation is crucial. For example, a particular SNP of the IFN-γ gene has been found to be functionally important for determining both spontaneous clearance and successful IFN treatment response in HCV-infected patients (Huang et al., 2007b). This SNP is in the IFN-γ gene promoter region and increases the binding affinity for the transcription factor HSF1, resulting in increased IFN-γ expression. Interestingly, these results are consistent with the gene expression studies in HCV-infected chimpanzees where viral clearance correlated with increased IFN-γ expression. As part of the Hepatitis C Antiviral Long-term Treatment against Cirrhosis (HALT-C) trial, patient genetic polymorphisms were assessed in individuals who had previously failed IFN therapy (Morgan et al., 2008). Among the non-Hispanic Caucasian patients re-treated with PEG-IFN-α/ribavirin, homozygosity for the previously identified ACC IL10 promoter diplotype was associated with sustained virological response. Similar types of analysis identifying SNPs associated with fibrosis progression may also shed light into the variation associated with ISG expression.
VI. Can high-throughput genomics identify cellular targets for therapy?
Antivirals aimed at exploiting cellular targets, rather than viral, provide the unique advantage in that the probability of a cellular gene developing a resistant phenotype is substantially less than what could be expected from a viral gene. This is particularly true for RNA viruses, including HCV, which typically lack efficient proofreading capabilities and so accumulate mutations at relatively high rates. Indeed, extensive efforts to develop drugs aimed at targeting HCV proteins such as the NS3 protease and NS5B polymerase have met with little success. This highlights the need for alternative therapeutic strategies to combat chronic HCV infection.
A common trend throughout the genomics studies involving HCV has been the role of the innate antiviral response in HCV clearance/persistence, disease progression and IFN treatment response. Microarray experiments have also been instrumental in revealing the true extent of the transcriptional response to IFN signaling. Transcriptional profiling of the response to a single dose of IFN-α in uninfected chimpanzee liver and PBMC samples revealed the altered expression (by at least two-fold) of nearly two thousand genes. (Lanford et al., 2006) This response is rapidly down-regulated, with maximal regulation occurring within 4 hours followed by a return to base-line levels by 24hrs following treatment. The response to IFN was also largely tissue-specific with unique transcriptional profiles observed in liver and PBMCs. Finally, an important observation of this study and others is the decreased expression of numerous genes following IFN treatment ((Kash et al., 2006a), Walters, unpublished observation). Interest in ISGs has primarily focused on genes which are induced in response to IFN but the extensive decreased expression of cellular genes, notably many associated with general metabolism, suggests that this also plays a significant role in the antiviral response and warrants further investigation.
It is now apparent that IFN signaling is much more complex than simply the activation of the PKR and RNaseL pathways. While microarray studies have shown that IFN signaling regulates the expression of thousands of genes that collectively establish an intracellular antiviral state, the function of the vast majority of these genes remains largely unknown. It is also quite likely that independent ISGs have varying antiviral activity against different families of viruses. This is an extremely important area of research which has only recently begun to be appreciated. Most recently, genomic studies aimed at examining the expression of IFN-regulated genes in liver tissue have led to the identification of genes that have anti-HCV activity. Such studies demonstrate the value of using transcriptional profiling to identify novel cellular targets for therapeutic intervention.
Among the 18 genes identified by Chen et al whose expression were found to distinguish responders from non-responders in IFN-treated HCV-infected patients was ubiquitin-specific protease 18 (USP18) (Chen et al., 2005). This gene, an ubiquitin-specific protease that cleaves ISG15 from its cellular targets, was found to be highly induced only in non-responders. Randall et al used siRNA to silence USP18 and subsequently measured the dose response of HCV replication to IFN-α in an in vitro HCV infection system (Randall et al., 2006). The knock-down of USP18 enhanced the ability of IFN to inhibit HCV replication in vitro. This enhanced antiviral activity was associated with increased protein ISGylation, prolonged STAT1 activation and increased expression of ISGs. Based on these results, modulation of USP18 was proposed as a strategy to improve response to IFN treatment. A similar study examining the expression of ISGs in the livers of nine individuals with chronic HCV identified the cellular gene viperin as being significantly induced in all patients (Helbig et al., 2005). In vitro studies demonstrated that viperin was induced by exposure to both IFN and poly (I:C) and that transient expression of vipirin significantly decreased replication of a HCV genomic replicon.
While these results are intriguing, it is puzzling that HCV persists in the livers of these patients despite high levels of a protein with apparent anti-HCV activity. This highlights the importance of studies aimed at identifying the source of the expression of these ISGs in the liver and their relationship to HCV replication levels. The chimeric SCID-Alb/uPA mouse model of infection may be the ideal model in which to study this. It has already been demonstrated that anti-HCV therapies, including response to IFN therapy, in chimeric mice parallel responses observed in patients (Kneteman et al., 2006). Genomics analysis of IFN-treated mice infected with both IFN-resistant and IFN-sensitive HCV in donor-matched animals could shed considerable insight into the host transcriptional response associated with a particular treatment outcome. The ability to infect chimeric mice with HCV containing mutations in specific viral genes will also help identify viral determinants of treatment response.
VII. Conclusion
This article has highlighted how high-throughput microarray technology has revealed insights into the relationship between the host response to HCV infection and liver disease. In particular, it has highlighted how the magnitude and extent of the host innate response may contribute to outcome of acute infection, severity of liver disease and IFN treatment failure. Future research should now include studies aimed at identifying the genetic and biochemical regulation of innate antiviral signaling activation.
Acknowledgments
Gene expression studies performed in the Katze laboratory were supported by funding provided by the Canadian Association for the Study of liver (CASL) and grants 5P01AI058113-050006, 5P30DA015625-07, 5R01HL080621-03, 5R21AI071892-02 from the National Institutes of Health. K.A.W. is supported by a CASL fellowship. We would like to thank Dr. Michael Bray, Dr. John Kash and Dr. Marcus Korth for their valuable scientific and editorial comments and manuscript preparation.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Reference List
- Alter MJ. Epidemiology of hepatitis C virus infection. World J Gastroenterol. 2007;13:2436–2441. doi: 10.3748/wjg.v13.i17.2436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alter MJH, Margolis HS, Krawczynski K, Judson F, Mares A, Alexander WJ, Hu PY, Miller JK, et al. The natural history of community-acquired hepatitis C in the United States. N Engl J Med. 1992;321:1494–1500. doi: 10.1056/NEJM199212313272702. [DOI] [PubMed] [Google Scholar]
- Asselah T, Bieche I, Laurendeau I, Paradis V, Vidaud D, Degott C, Martinot M, Bedossa P, Valla D, Vidaud M, Marcellin P. Liver gene expression signature of mild fibrosis in patients with chronic hepatitis C. Gastroenterology. 2005;129:2064–2075. doi: 10.1053/j.gastro.2005.09.010. [DOI] [PubMed] [Google Scholar]
- Bantel H, Schulze-Osthoff K. Apoptosis in hepatitis C virus infection. Cell Death Differ. 2003;10(Suppl 1):S48–S58. doi: 10.1038/sj.cdd.4401119. [DOI] [PubMed] [Google Scholar]
- Benhamou Y, Bochet M, Di MV, Charlotte F, Azria F, Coutellier A, Vidaud M, Bricaire F, Opolon P, Katlama C, Poynard T. Liver fibrosis progression in human immunodeficiency virus and hepatitis C virus coinfected patients. The Multivirc Group. Hepatology. 1999;30:1054–1058. doi: 10.1002/hep.510300409. [DOI] [PubMed] [Google Scholar]
- Benhamou Y, Di MV, Bochet M, Colombet G, Thibault V, Liou A, Katlama C, Poynard T. Factors affecting liver fibrosis in human immunodeficiency virus-and hepatitis C virus-coinfected patients: impact of protease inhibitor therapy. Hepatology. 2001;34:283–287. doi: 10.1053/jhep.2001.26517. [DOI] [PubMed] [Google Scholar]
- Berenguer M, Prieto M, Rayon JM, Mora J, Pastor M, Ortiz V, Carrasco D, San JF, Burgueno MD, Mir J, Berenguer J. Natural history of clinically compensated hepatitis C virus-related graft cirrhosis after liver transplantation. Hepatology. 2000;32:852–858. doi: 10.1053/jhep.2000.17924. [DOI] [PubMed] [Google Scholar]
- Bieche I, Asselah T, Laurendeau I, Vidaud D, Degot C, Paradis V, Bedossa P, Valla DC, Marcellin P, Vidaud M. Molecular profiling of early stage liver fibrosis in patients with chronic hepatitis C virus infection. Virology. 2005;332:130–144. doi: 10.1016/j.virol.2004.11.009. [DOI] [PubMed] [Google Scholar]
- Bigger CB, Brasky KM, Lanford RE. DNA microarray analysis of chimpanzee liver during acute resolving hepatitis C virus infection. J Virol. 2001;75:7059–7066. doi: 10.1128/JVI.75.15.7059-7066.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bigger CB, Guerra B, Brasky KM, Hubbard G, Beard MR, Luxon BA, Lemon SM, Lanford RE. Intrahepatic gene expression during chronic hepatitis C virus infection in chimpanzees. J Virol. 2004;78:13779–13792. doi: 10.1128/JVI.78.24.13779-13792.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breiman A, Grandvaux N, Lin R, Ottone C, Akira S, Yoneyama M, Fujita T, Hiscott J, Meurs EF. Inhibition of RIG-I-dependent signaling to the interferon pathway during hepatitis C virus expression and restoration of signaling by IKKepsilon. J Virol. 2005;79:3969–3978. doi: 10.1128/JVI.79.7.3969-3978.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L, Borozan I, Feld J, Sun J, Tannis LL, Coltescu C, Heathcote J, Edwards AM, McGilvray ID. Hepatic gene expression discriminates responders and nonresponders in treatment of chronic hepatitis C viral infection. Gastroenterology. 2005;128:1437–1444. doi: 10.1053/j.gastro.2005.01.059. [DOI] [PubMed] [Google Scholar]
- Feld JJ, Nanda S, Huang Y, Chen W, Cam M, Pusek SN, Schweigler LM, Theodore D, Zacks SL, Liang TJ, Fried MW. Hepatic gene expression during treatment with peginterferon and ribavirin: Identifying molecular pathways for treatment response. Hepatology. 2007;46:1548–1563. doi: 10.1002/hep.21853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferreon JC, Ferreon AC, li K, Lemon SM. Molecular determinants of TRIF proteolysis mediated by the hepatitis C virus NS3/4A protease. J Biol Chem. 2005;280:20483–20492. doi: 10.1074/jbc.M500422200. [DOI] [PubMed] [Google Scholar]
- Foy E, li K, Sumpter R, Jr, Loo YM, Johnson CL, Wang C, Fish PM, Yoneyama M, Fujita T, Lemon SM, Gale M., Jr Control of antiviral defenses through hepatitis C virus disruption of retinoic acid-inducible gene-I signaling. Proc Natl Acad Sci U S A. 2005;102:2986–2991. doi: 10.1073/pnas.0408707102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foy E, li K, Wang C, Sumpter RJ, Ikeda M, Lemon SM, Gale MJ. Regulation of interferon regulatory factor-3 by the hepatitis C virus serine protease. Science. 2003;300:1145–1148. doi: 10.1126/science.1082604. [DOI] [PubMed] [Google Scholar]
- Fukuzawa K, Takahashi K, Furuta K, Tagaya T, Ishikawa T, Wada K, Omoto Y, Koji T, Kakumu S. Expression of fas/fas ligand (fasL) and its involvement in infiltrating lymphocytes in hepatocellular carcinoma (HCC) J Gastroenterol. 2001;36:681–688. doi: 10.1007/s005350170031. [DOI] [PubMed] [Google Scholar]
- Gale M, Jr, Foy EM. Evasion of intracellular host defence by hepatitis C virus. Nature. 2005;436:939–945. doi: 10.1038/nature04078. [DOI] [PubMed] [Google Scholar]
- Gao B, Hong F, Radaeva S. Host factors and failure of interferon-alpha treatment in hepatitis C virus. Hepatology. 2004;39:880–890. doi: 10.1002/hep.20139. [DOI] [PubMed] [Google Scholar]
- Garcia-Retortillo M, Forns X, Feliu A, Moitinho E, Costa J, Navasa M, Rimola A, Rodes J. Hepatitis C virus kinetics during and immediately after liver transplantation. Hepatology. 2002;35:680–687. doi: 10.1053/jhep.2002.31773. [DOI] [PubMed] [Google Scholar]
- Garcia-Samaniego J, Soriano V, Castilla J, Bravo R, Moreno A, Carbo J, Iniguez A, Gonzalez J, Munoz F. Influence of hepatitis C virus genotypes and HIV infection on histological severity of chronic hepatitis C. The Hepatitis/HIV Spanish Study Group. Am J Gastroenterol. 1997;92:1130–1134. [PubMed] [Google Scholar]
- Graham CS, Baden LR, Yu E, Mrus JM, Carnie J, Heeren T, Koziel MJ. Influence of human immunodeficiency virus infection on the course of hepatitis C virus infection: a meta-analysis. Clin Infect Dis. 2001;33:562–569. doi: 10.1086/321909. [DOI] [PubMed] [Google Scholar]
- Hamamoto S, Uchida Y, Wada T, Moritani M, Sato S, Hamamoto N, Ishihara S, Watanabe M, Kinoshita Y. Changes in serum lipid concentrations in patients with chronic hepatitis C virus positive hepatitis responsive or non-responsive to interferon therapy. J Gastroenterol Hepatol. 2005;20:204–208. doi: 10.1111/j.1440-1746.2004.03526.x. [DOI] [PubMed] [Google Scholar]
- Hayashi N, Mita E. Involvement of Fas system-mediated apoptosis in pathogenesis of viral hepatitis. J Viral Hepat. 1999;6:357–365. doi: 10.1046/j.1365-2893.1999.00175.x. [DOI] [PubMed] [Google Scholar]
- Helbig KJ, Lau DT, Semendric L, Harley HA, Beard MR. Analysis of ISG expression in chronic hepatitis C identifies viperin as a potential antiviral effector. Hepatology. 2005;42:702–710. doi: 10.1002/hep.20844. [DOI] [PubMed] [Google Scholar]
- Heller T, Saito S, Auerbach J, Williams T, Moreen TR, Jazwinski A, Cruz B, Jeurkar N, Sapp R, Luo G, Liang TJ. An in vitro model of hepatitis C virion production. Proc Natl Acad Sci U S A. 2005;102:2579–2583. doi: 10.1073/pnas.0409666102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiramatsu N, Hayashi N, Katayama K, Mochizuki K, Kawanishi Y, Kasahara A, Fusamoto H, Kamada T. Immunohistochemical detection of Fas antigen in liver tissue of patients with chronic hepatitis C. Hepatology. 1994;19:1354–1359. [PubMed] [Google Scholar]
- Hsu EC, Hsi B, Hirota-Tsuchihara M, Ruland J, Iorio C, Sarangi F, Diao J, Migliaccio G, Tyrrell DL, Kneteman N, Richardson CD. Modified apoptotic molecule (BID) reduces hepatitis C virus infection in mice with chimeric human livers. Nat Biotechnol. 2003;21:519–525. doi: 10.1038/nbt817. [DOI] [PubMed] [Google Scholar]
- Huang Y, Feld JJ, Sapp RK, Nanda S, Lin JH, Blatt LM, Fried MW, Murthy K, Liang TJ. Defective hepatic response to interferon and activation of suppressor of cytokine signaling 3 in chronic hepatitis C. Gastroenterology. 2007a;132:733–744. doi: 10.1053/j.gastro.2006.11.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Y, Yang H, Borg BB, Su X, Rhodes SL, Yang K, Tong X, Tang G, Howell CD, Rosen HR, Thio CL, Thomas DL, Alter HJ, Sapp RK, Liang TJ. A functional SNP of interferon-gamma gene is important for interferon-alpha-induced and spontaneous recovery from hepatitis C virus infection. Proc Natl Acad Sci U S A. 2007b;104:985–990. doi: 10.1073/pnas.0609954104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kash JC, Mühlberger E, Carter V, Grosch O, Perwitasari O, Proll SC, Thomas MJ, Weber F, Klenk HD, Katze MG. Global suppression of the host antiviral response by Ebola- and Marburgviruses: increased antagonism of the type 1 interferon response is associated with enhanced virulence. J Virol. 2006a;80:3009–3020. doi: 10.1128/JVI.80.6.3009-3020.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kash JC, Tumpey TM, Proll SC, Carter V, Perwitasari O, Thomas MJ, Basler CF, Palese P, Taubenberger JK, García-Sastre A, Swayne DE, Katze MG. Genomic analysis of increased host immune and cell death responses induced by 1918 influenza virus. Nature. 2006b;443:578–581. doi: 10.1038/nature05181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawai T, Takahashi K, Sato S, Coban C, Kumar H, Kato H, Ishii KJ, Takeuchi O, Akira S. IPS-1, an adaptor triggering RIG-I- and Mda5-mediated type I interferon induction. Nat Immunol. 2005;6:981–988. doi: 10.1038/ni1243. [DOI] [PubMed] [Google Scholar]
- Keck ZY, Machida K, Lai MM, Ball JK, Patel AH, Foung SK. Therapeutic control of hepatitis C virus: the role of neutralizing monoclonal antibodies. Curr Top Microbiol Immunol. 2008;317:1–38. doi: 10.1007/978-3-540-72146-8_1. [DOI] [PubMed] [Google Scholar]
- Kneteman NM, Weiner AJ, O'Connell J, Collett M, Gao T, Aukerman L, Kovelsky R, Ni ZJ, Hashash A, Kline J, Hsi B, Schiller D, Douglas D, Tyrrell DL, Mercer DF. Anti-HCV therapies in chimeric scid-Alb/uPA mice parallel outcomes in human clinical application. Hepatology. 2006;43:1346–1353. doi: 10.1002/hep.21209. [DOI] [PubMed] [Google Scholar]
- Kobasa D, Jones SM, Shinya K, Kash JC, Copps J, Ebihara H, Hatta Y, Kim JH, Halfmann P, Hatta M, Feldmann F, Alimonti JB, Fernando L, Li Y, Katze MG, Feldmann H, Kawaoka Y. Aberrant innate immune response in lethal infection of macaques with the 1918 influenza virus. Nature. 2007;445:319–323. doi: 10.1038/nature05495. [DOI] [PubMed] [Google Scholar]
- Kountouras J, Zavos C, Chatzopoulos D. Apoptosis in hepatitis C. J Viral Hepat. 2003;10:335–342. doi: 10.1046/j.1365-2893.2003.00452.x. [DOI] [PubMed] [Google Scholar]
- Lanford RE, Guerra B, Bigger CB, Lee H, Chavez D, Brasky KM. Lack of response to exogenous interferon-alpha in the liver of chimpanzees chronically infected with hepatitis C virus. Hepatology. 2007;46:999–1008. doi: 10.1002/hep.21776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lanford RE, Guerra B, Lee H, Chavez D, Brasky KM, Bigger CB. Genomic response to interferon-alpha in chimpanzees: Implications of rapid downregulation for hepatitis C kinetics. Hepatology. 2006;43:961–972. doi: 10.1002/hep.21167. [DOI] [PubMed] [Google Scholar]
- Lau DT, Fish PM, Sinha M, Owen DM, Lemon SM, Gale M., Jr Interferon regulatory factor-3 activation, hepatic interferon-stimulated gene expression, and immune cell infiltration in hepatitis C virus patients. Hepatology. 2008 doi: 10.1002/hep.22076. [DOI] [PubMed] [Google Scholar]
- Lau DT, Kleiner DE, Ghany MG, Park Y, Schmid P, Hoofnagle JH. 10-Year follow-up after interferon-alpha therapy for chronic hepatitis C. Hepatology. 1998;28:1121–1127. doi: 10.1002/hep.510280430. [DOI] [PubMed] [Google Scholar]
- Lau DT, Luxon BA, Xiao SY, Beard MR, Lemon SM. Intrahepatic gene expression profiles and alpha-smooth muscle actin patterns in hepatitis C virus induced fibrosis. Hepatology. 2005;42:273–281. doi: 10.1002/hep.20767. [DOI] [PubMed] [Google Scholar]
- Lederer SL, Walters KA, Proll S, Paeper B, Robinzon S, Boix L, Fausto N, Bruix J, Katze MG. Distinct cellular responses differentiating alcohol- and hepatitis C virus-induced liver cirrhosis. Virol J. 2006;3:98. doi: 10.1186/1743-422X-3-98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- li K, Foy E, Ferreon JC, Nakamura M, Ferreon AC, Ikeda M, Ray SC, Gale M, Jr, Lemon SM. Immune evasion by hepatitis C virus NS3/4A protease-mediated cleavage of the Toll-like receptor 3 adaptor protein TRIF. Proc Natl Acad Sci USA. 2005;102:2992–2997. doi: 10.1073/pnas.0408824102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindenbach BD, Evans MJ, Syder AJ, Wolk B, Tellinghuisen TL, Liu CC, Maruyama T, Hynes RO, Burton DR, McKeating JA, Rice CM. Complete replication of hepatitis C virus in cell culture. Science. 2005;309:623–626. doi: 10.1126/science.1114016. [DOI] [PubMed] [Google Scholar]
- Lindenbach BD, Meuleman P, Ploss A, Vanwolleghem T, Syder AJ, McKeating JA, Lanford RE, Feinstone SM, Major ME, Leroux-Roels G, Rice CM. Cell culture-grown hepatitis C virus is infectious in vivo and can be recultured in vitro. Proc Natl Acad Sci U S A. 2006;103:3805–3809. doi: 10.1073/pnas.0511218103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindenbach BD, Rice CM. Unravelling hepatitis C virus replication from genome to function. Nature. 2005;436:933–938. doi: 10.1038/nature04077. [DOI] [PubMed] [Google Scholar]
- Lindsay KL. Introduction to therapy of hepatitis C. Hepatology. 2002;36:S114–S120. doi: 10.1053/jhep.2002.36226. [DOI] [PubMed] [Google Scholar]
- Loo YM, Owen DM, li K, Erickson AK, Johnson CL, Fish PM, Carney DS, Wang T, Ishida H, Yoneyama M, Fujita T, Saito T, Lee WM, Hagedorn CH, Lau DT, Weinman SA, Lemon SM, Gale M., Jr Viral and therapeutic control of IFN-beta promoter stimulator 1 during hepatitis C virus infection. Proc Natl Acad Sci USA. 2006;103:6001–6006. doi: 10.1073/pnas.0601523103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manns MP, McHutchison JC, Gordon SC, Rustgi VK, Shiffman M, Reindollar R, Goodman ZD, Koury K, Ling M, Albrecht JK. Peginterferon alfa-2b plus ribavirin compared with interferon alfa-2b plus ribavirin for initial treatment of chronic hepatitis C: a randomised trial. Lancet. 2001;358:958–965. doi: 10.1016/s0140-6736(01)06102-5. [DOI] [PubMed] [Google Scholar]
- Mercer DF, Schiller DE, Elliott JF, Douglas DN, Hao C, Rinfret A, Addison WR, Fischer KP, Churchill TA, Lakey JR, Tyrrell DL, Kneteman NM. Hepatitis C virus replication in mice with chimeric human livers. Nat Med. 2001;7:927–933. doi: 10.1038/90968. [DOI] [PubMed] [Google Scholar]
- Meuleman P, Libbrecht L, De VR, de HB, Gevaert K, Vandekerckhove J, Roskams T, Leroux-Roels G. Morphological and biochemical characterization of a human liver in a uPA-SCID mouse chimera. Hepatology. 2005;41:847–856. doi: 10.1002/hep.20657. [DOI] [PubMed] [Google Scholar]
- Meuleman P, Leroux-Roels G. The human liver-uPA-SCID mouse: a model for the evaluation of antiviral compounds against hepatitis B and C. Antiviral Res. 2008;80:231–8. doi: 10.1016/j.antiviral.2008.07.006. [DOI] [PubMed] [Google Scholar]
- Meylan E, Curran J, Hofmann K, Moradpour D, Binder M, Bartenschlager R, Tschopp J. Cardif is an adaptor protein in the RIG-I antiviral pathway and is targeted by hepatitis C virus. Nature. 2005;437:1167–1172. doi: 10.1038/nature04193. [DOI] [PubMed] [Google Scholar]
- Morgan TR, Lambrecht RW, Bonkovsky HL, Chung RT, Naishadham D, Sterling RK, Fontana RJ, Lee WM, Ghany MG, Wright EC, O'Brien TR. DNA polymorphisms and response to treatment in patients with chronic hepatitis C: Results from the HALT-C trial. J Hepatol. 2008;49:548–556. doi: 10.1016/j.jhep.2008.05.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muhanna N, Doron S, Wald O, Horani A, Eid A, Pappo O, Friedman SL, Safadi R. Activation of hepatic stellate cells after phagocytosis of lymphocytes: A novel pathway of fibrogenesis. Hepatology. 2008;48:963–977. doi: 10.1002/hep.22413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel K, Zekry A, McHutchison JG. Steatosis and chronic hepatitis C virus infection: mechanisms and significance. Clin Liver Dis. 2005;9:399–410. vi. doi: 10.1016/j.cld.2005.05.007. [DOI] [PubMed] [Google Scholar]
- Petit JM, Benichou M, Duvillard L, Jooste V, Bour JB, Minello A, Verges B, Brun JM, Gambert P, Hillon P. Hepatitis C virus-associated hypobetalipoproteinemia is correlated with plasma viral load, steatosis, and liver fibrosis. Am J Gastroenterol. 2003;98:1150–1154. doi: 10.1111/j.1572-0241.2003.07402.x. [DOI] [PubMed] [Google Scholar]
- Randall G, Chen L, Panis M, Fischer AK, Lindenbach BD, Sun J, Heathcote J, Rice CM, Edwards AM, McGilvray ID. Silencing of USP18 potentiates the antiviral activity of interferon against hepatitis C virus infection. Gastroenterology. 2006;131:1584–1591. doi: 10.1053/j.gastro.2006.08.043. [DOI] [PubMed] [Google Scholar]
- Rehermann B. Chronic infections with hepatotropic viruses: mechanisms of impairment of cellular immune responses. Semin Liver Dis. 2007;27:152–160. doi: 10.1055/s-2007-979468. [DOI] [PubMed] [Google Scholar]
- Rehermann B, Nascimbeni M. Immunology of hepatitis B virus and hepatitis C virus infection. Nat Rev Immunol. 2005;5:215–229. doi: 10.1038/nri1573. [DOI] [PubMed] [Google Scholar]
- Sekine-Osajima Y, Sakamoto N, Mishima K, Nakagawa M, Itsui Y, Tasaka M, Nishimura-Sakurai Y, Chen CH, Kanai T, Tsuchiya K, Wakita T, Enomoto N, Watanabe M. Development of plaque assays for hepatitis C virus-JFH1 strain and isolation of mutants with enhanced cytopathogenicity and replication capacity. Virology. 2008;371:71–85. doi: 10.1016/j.virol.2007.09.019. [DOI] [PubMed] [Google Scholar]
- Seth RB, Sun L, Ea CK, Chen ZJ. Identification and characterization of MAVS, a mitochondrial antiviral signaling protein that activates NF-kappaB and IRF 3. Cell. 2005;122:669–682. doi: 10.1016/j.cell.2005.08.012. [DOI] [PubMed] [Google Scholar]
- Shao RX, Hoshida Y, Otsuka M, Kato N, Tateishi R, Teratani T, Shiina S, Taniguchi H, Moriyama M, Kawabe T, Omata M. Hepatic gene expression profiles associated with fibrosis progression and hepatocarcinogenesis in hepatitis C patients. World J Gastroenterol. 2005;11:1995–1999. doi: 10.3748/wjg.v11.i13.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shoukry NH, Cawthon AG, Walker CM. Cell-mediated immunity and the outcome of hepatitis C virus infection. Annu Rev Microbiol. 2004;58:391–424. doi: 10.1146/annurev.micro.58.030603.123836. [DOI] [PubMed] [Google Scholar]
- Siagris D, Christofidou M, Theocharis GJ, Pagoni N, Papadimitriou C, Lekkou A, Thomopoulos K, Starakis I, Tsamandas AC, Labropoulou-Karatza C. Serum lipid pattern in chronic hepatitis C: histological and virological correlations. J Viral Hepat. 2006;13:56–61. doi: 10.1111/j.1365-2893.2005.00655.x. [DOI] [PubMed] [Google Scholar]
- Smith MW, Walters KA, Korth MJ, Fitzgibbon M, Proll SC, Thompson JC, Yeh MM, Shuhart MC, Furlong JC, Cox PP, Thomas DL, Phillips JD, Kushner JP, Fausto N, Carithers RL, Katze MG. Gene expression patterns that correlate with hepatitis C and early progression to fibrosis in liver transplant recipients. Gastroenterol. 2006;130:179–187. doi: 10.1053/j.gastro.2005.08.015. [DOI] [PubMed] [Google Scholar]
- Smith MW, Yue ZN, Korth MJ, Do HA, Boix L, Fausto N, Bruix J, Carithers RL, Jr, Katze MG. Hepatitis C virus and liver disease: global transcriptional profiling and identification of potential markers. Hepatology. 2003;38:1458–1467. doi: 10.1016/j.hep.2003.09.024. [DOI] [PubMed] [Google Scholar]
- Soto B, Sanchez-Quijano A, Rodrigo L, del Olmo JA, Garcia-Bengoechea M, Hernandez-Quero J, Rey C, Abad MA, Rodriguez M, Sales GM, Gonzalez F, Miron P, Caruz A, Relimpio F, Torronteras R, Leal M, Lissen E. Human immunodeficiency virus infection modifies the natural history of chronic parenterally-acquired hepatitis C with an unusually rapid progression to cirrhosis. J Hepatol. 1997;26:1–5. doi: 10.1016/s0168-8278(97)80001-3. [DOI] [PubMed] [Google Scholar]
- Stamataki Z, Grove J, Balfe P, McKeating JA. Hepatitis C virus entry and neutralization. Clin Liver Dis. 2008;12:693–712. x. doi: 10.1016/j.cld.2008.03.008. [DOI] [PubMed] [Google Scholar]
- Su AI, Pezacki JP, Wodicka L, Brideau AD, Supekova L, Thimme R, Wieland S, Bukh J, Purcell RH, Schultz PG, Chisari FV. Genomic analysis of the host response to hepatitis C virus infection. Proc Natl Acad Sci USA. 2002;99:15669–15674. doi: 10.1073/pnas.202608199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sumpter R, Jr, Loo YM, Foy E, li K, Yoneyama M, Fujita T, Lemon SM, Gale M., Jr Regulating intracellular antiviral defense and permissiveness to hepatitis C virus RNA replication through a cellular RNA helicase, RIG-I. J Virol. 2005;79:2689–2699. doi: 10.1128/JVI.79.5.2689-2699.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sumpter R, Jr, Wang C, Foy E, Loo YM, Gale M., Jr Viral evolution and interferon resistance of hepatitis C virus RNA replication in a cell culture model. J Virol. 2004;78:11591–11604. doi: 10.1128/JVI.78.21.11591-11604.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomson BJ, Finch RG. Hepatitis C virus infection. Clin Microbiol Infect. 2005;11:86–94. doi: 10.1111/j.1469-0691.2004.01061.x. [DOI] [PubMed] [Google Scholar]
- Wakita T, Pietschmann T, Kato T, Date T, Miyamoto M, Zhao Z, Murthy K, Habermann A, Krausslich HG, Mizokami M, Bartenschlager R, Liang TJ. Production of infectious hepatitis C virus in tissue culture from a cloned viral genome. Nat Med. 2005;11:791–796. doi: 10.1038/nm1268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walsh MJ, Jonsson JR, Richardson MM, Lipka GM, Purdie DM, Clouston AD, Powell EE. Non-response to antiviral therapy is associated with obesity and increased hepatic expression of suppressor of cytokine signalling 3 (SOCS-3) in patients with chronic hepatitis C, viral genotype 1. Gut. 2006;55:529–535. doi: 10.1136/gut.2005.069674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walters KA, Joyce MA, Thompson JC, Smith MW, Yeh MM, Proll S, Zhu LF, Gao TJ, Knetman NM, Tyrrell DL, Katze MG. Host-specific response to HCV infection in the chimeric SCID-beige/Alb-uPA mouse model: role of the innate antiviral immune response. PLoS Pathog. 2006a;2:591–601. doi: 10.1371/journal.ppat.0020059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walters KA, Smith MW, Pal S, Thompson JC, Thomas MJ, Yeh MM, Thomas DL, Fitzgibbon M, Proll S, Fausto N, Gretch DR, Carithers RL, Jr, Shuhart MC, Katze MG. Identification of a specific gene expression pattern associated with HCV-induced pathogenesis in HCV- and HCV/HIV-infected individuals. Virology. 2006b;350:453–464. doi: 10.1016/j.virol.2006.02.030. [DOI] [PubMed] [Google Scholar]
- Wieland S, Thimme R, Purcell RH, Chisari FV. Genomic analysis of the host response to hepatitis B virus infection. Proc Natl Acad Sci U S A. 2004;101:6669–6674. doi: 10.1073/pnas.0401771101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wohnsland A, Hofmann WP, Sarrazin C. Viral determinants of resistance to treatment in patients with hepatitis C. Clin Microbiol Rev. 2007;20:23–38. doi: 10.1128/CMR.00010-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu LG, Wang YY, Han KJ, Li LY, Zhai Z, Shu HB. VISA is an adapter protein required for virus-triggered IFN-beta signaling. Mol Cell. 2005;19:727–740. doi: 10.1016/j.molcel.2005.08.014. [DOI] [PubMed] [Google Scholar]
- Ye J, Wang C, Sumpter R, Jr, Brown MS, Goldstein JL, Gale M., Jr Disruption of hepatitis C virus RNA replication through inhibition of host protein geranylgeranylation. Proc Natl Acad Sci U S A. 2003;100:15865–15870. doi: 10.1073/pnas.2237238100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeisel MB, Fafi-Kremer S, Fofana I, Barth H, Stoll-Keller F, Doffoel M, Baumert TF. Neutralizing antibodies in hepatitis C virus infection. World J Gastroenterol. 2007;13:4824–4830. doi: 10.3748/wjg.v13.i36.4824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong J, Gastaminza P, Cheng G, Kapadia S, Kato T, Burton DR, Wieland SF, Uprichard SL, Wakita T, Chisari FV. Robust hepatitis C virus infection in vitro. Proc Natl Acad Sci U S A. 2005;102:9294–9299. doi: 10.1073/pnas.0503596102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu H, Dong H, Eksioglu E, Hemming A, Cao M, Crawford JM, Nelson DR, Liu C. Hepatitis C virus triggers apoptosis of a newly developed hepatoma cell line through antiviral defense system. Gastroenterology. 2007;133:1649–1659. doi: 10.1053/j.gastro.2007.09.017. [DOI] [PubMed] [Google Scholar]