Abstract
Fenofibrate, widely used for the treatment of dyslipidemia, activates the nuclear receptor, peroxisome proliferator-activated receptor α. However, liver toxicity, including liver cancer, occurs in rodents treated with fibrate drugs. Marked species differences occur in response to fibrate drugs, especially between rodents and humans, the latter of which are resistant to fibrate-induced cancer. Fenofibrate metabolism, which also shows species differences, has not been fully determined in humans and surrogate primates. In the present study, the metabolism of fenofibrate was investigated in cynomolgus monkeys by ultraperformance liquid chromatography-quadrupole time-of-flight mass spectrometry (UPLC-QTOFMS)-based metabolomics. Urine samples were collected before and after oral doses of fenofibrate. The samples were analyzed in both positive-ion and negative-ion modes by UPLC-QTOFMS, and after data deconvolution, the resulting data matrices were subjected to multivariate data analysis. Pattern recognition was performed on the retention time, mass/charge ratio, and other metabolite-related variables. Synthesized or purchased authentic compounds were used for metabolite identification and structure elucidation by liquid chromatographytandem mass spectrometry. Several metabolites were identified, including fenofibric acid, reduced fenofibric acid, fenofibric acid ester glucuronide, reduced fenofibric acid ester glucuronide, and compound X. Another two metabolites (compound B and compound AR), not previously reported in other species, were characterized in cynomolgus monkeys. More importantly, previously unknown metabolites, fenofibric acid taurine conjugate and reduced fenofibric acid taurine conjugate were identified, revealing a previously unrecognized conjugation pathway for fenofibrate.
Fibrates are agonists of the peroxisome proliferator-activated receptor α (PPARα), a nuclear receptor that modulates expression of genes involved in lipid, glucose, and amino acid homeostasis (Qi et al., 2000; Peters et al., 2005). In clinical studies, fibrates are widely used for the treatment of dyslipidemia, yet adverse affects have been reported such as myotoxicity (Blane, 1987; Clouâtre et al., 1999; Ritter and Nabulsi, 2001; Ghosh et al., 2004) as well as the generation of oxidative stress in rats (Nishimura et al., 2007). In addition, chronic administration of fibrates to rodents causes hepatocellular carcinoma. It is interesting to note that both humans and nonhuman primates are refractory to hepatocellular carcinoma caused by PPARα agonists (Reddy et al., 1980; Rao and Reddy, 1987; Ward et al., 1998; Klaunig et al., 2003; Hoivik et al., 2004), yet the mechanism behind the species-related difference of susceptibility to these drugs remains to be firmly established (Peters et al., 2000; Hoivik et al., 2004). Some metabolites of PPARα agonists have been reported to cause toxicity and complex drug-drug interactions, underscoring the need for detailed metabolic studies of PPARα agonists (Sallustio et al., 1997; Backman et al., 2002; Prueksaritanont et al., 2002; Shitara et al., 2004).
Fenofibrate metabolism has been documented in humans, rats, guinea pigs, and dogs (Weil et al., 1988, 1990; Cornu-Chagnon et al., 1995). However, fenofibrate metabolism has not been studied in nonhuman-primates (NHPs). Fenofibric acid (FA) and reduced fenofibric acid (RFA) have been identified in rat, guinea pig, dog, and human, and also in rat hepatocytes. In addition, fenofibric acid ester glucuronide (FAEG) and reduced fenofibric acid glucuronide (RFAEG) have been reported as metabolites in rat, guinea pig, and human but were not detected in dog (Weil et al., 1988). It is interesting to note that a structural analog of fenofibrate, clofibrate, also shows similar species differences (Cayen et al., 1977; Emudianughe et al., 1983). Dogs, cats, and ferrets form the taurine conjugate of clofibric acid, but humans, rodents, and rabbits do not. We were surprised to find that no taurine conjugates of fenofibrate have been reported in any species.
Metabolomics can be defined as the global profiling of small molecules in a biofluid. Metabolomics has been exceptionally useful for identifying unique chemical fingerprints associated with a particular physiologic state, drug treatment, and/or disease (Chen et al., 2007, 2008; Zhen et al., 2007; Ma et al., 2008). When combined with multivariate data analysis (MDA), ultraperformance liquid chromatography (UPLC)-quadrupole time-of-flight mass spectrometry (QTOFMS) is a highly sensitive and powerful means for uncovering drug metabolites.
In the present study, cynomolgus monkeys (Macaca fascicularis) were dosed orally with fenofibrate. Urine samples before and after fenofibrate administration were collected and profiled by UPLC-QTOFMS. After chromatogram deconvolution, fenofibrate metabolites were identified using projection to latent structures discriminant analysis (PLS-DA). Potentially novel metabolites were structurally elucidated by tandem mass spectrometry comparison of the urinary constituent with the synthesized or commercially available authentic compounds. In addition to previously reported metabolites, four new metabolites were characterized including fenofibric acid taurine conjugate (FAT) and reduced fenofibric acid taurine conjugate (RFAT), revealing a previously unknown conjugation pathway of fenofibrate in cynomolgus monkeys.
Materials and Methods
Chemicals and Reagent. Fenofibrate, FA, and 4-chloro-4′-hydroxybenzophenone (compound A) were purchased from Shangqiu Chemry Chemicals Co., Ltd. (Shangqiu, China). Taurine was purchased from Shanghai Jiachen Co., Ltd. (Shanghai, China). High-performance liquid chromatography-grade acetonitrile, methanol, and formic acid were purchased from Sigma-Aldrich (St. Louis, MO). Purified water was obtained from an Elix system (Millipore Corporation, Billerica, MA). Biochemical analysis kits for alanine aminotransferase, aspartate aminotransferase, creatinine, blood urea nitrogen, and lactate dehydrogenase analysis were purchased from Shanghai Kehua Bio-engineering Co., Ltd. (Shanghai, China). All other chemicals used for experiments were analytical grade.
Synthesis of FAT, RFAT, Compound X, RFA, Compound AR, and Compound B. FA was used as the starting material for synthesis of RFA, compound X, FAT, and RFAT. Compound A was used as the starting material for compound AR and 4-chloro-4′-isopropoxybenzophenone (compound B). Taurine was used as an intermediate for synthesis of FAT and RFAT. Synthesized compounds were subjected to 1H NMR (AV-400; Bruker, Bremen, Germany) for structure and purity confirmation.
To synthesize FAT, FA (318.5 mg, 1 mmol) and triethylamine (0.14 ml, 1.2 mmol) were dissolved in 5 ml of acetone at –5°C (ice bath). A solution of methylchloroformate (0.1 ml, 5 mM) in 1 ml of dichloromethane was added dropwise. The mixture was stirred at –5 to 0°C until the disappearance of starting material was noted by TLC (5–10 min). A solution of taurine (125 mg, 1 mmol) in 2 ml of aqueous sodium hydroxide (2 M) was then added over a period of 10 min. After the reaction was complete (noted by TLC, 3–4 h), the mixture was evaporated at ambient temperature using a rotary evaporator. The residue was purified by preparative TLC on a silica gel plate using a mobile phase composed of dichloromethane, methanol, and acetic acid (15:1:0.1).
For RFAT, the above purified product of FAT (160 mg, 0.5 mmol) was dissolved in 2 ml of 3:1 tetrahydrofuran-methanol solution at 0°C (ice bath). NaBH4 (95 mg, 2.5 mmol) was then added portionwise over 30 min. The mixture was stirred at 0 to 25°C until the disappearance of starting material was noted by TLC (2∼3 h). The reaction mixture was concentrated on a rotary evaporator. The residue was dissolved in 4 ml of water, and the solution was acidified to pH 6 with the slow addition of 6 M citric acid and then extracted with ethyl acetate three times. The ethyl acetate layers were dried and concentrated by rotary evaporation to yield the product.
The chemical structures of synthesized compounds, the synthetic roadmap, and detailed procedures for compound X, RFA, compound AR, and compound B are shown in Supplemental Fig. 1.
Animals and Treatments. All work was performed in accordance with Public Health Service policies, the Animal Welfare Act, and the Institutional Animal Care and Use Committee of Guangzhou Institute of Biomedicine and Health, Chinese Academy of Sciences Policy on the Humane Care and Use of Vertebrate Animals. The protocol and standard operating procedures were approved by the Institutional Animal Care and Use Committee of Guangzhou Institute of Biomedicine and Health. Three male and three female cynomolgus monkeys, 4 to 5 years old and weighing 3 to 6 kg, were obtained from South-China Primate Research and Development Center [SCXK (Guangdong Province) 2004-001020066025]. Monkeys were individually housed in suspended, stainless steel cages and were maintained under a standard 12-h light/dark cycle with free access to purified water and a pelleted diet. The certified primate diet was provided twice daily, and food was offered 2 to 3 h after dosing in the morning and before dosing in the afternoon.
A two-stage investigation was designed using a dose of 30 mg/kg/day followed by 2500 mg/kg/day. This approach was used to identify metabolic pathways that may be dose-dependent. A drug-free period of at least 4 weeks was allowed between the 30 and the 2500 mg/kg/day dose. Four days before the start of each dosing period, the vehicle consisting of 0.5% sodium carboxymethylcellulose was given to the animals according to their initial individual body weight. Animals were administered fenofibrate by oral gavage twice daily for 12 days from day 5. Animals were weighed every 3 days, and the individual doses were adjusted according to the new body weight measurement. To monitor potential toxicity, serum was collected for measuring markers of hepatotoxicity and nephrotoxicity using standard biochemical methods.
Urine Collections. All urine samples were collected into stainless steel pans in the morning. Those contaminated by feces and diet were discarded, and urine samples in the afternoon were collected instead. All of the samples collected were immediately placed in dry ice and transferred to –80°C until analysis.
UPLC-QTOFMS Analyses. Urine samples were diluted with an equal volume of acetonitrile and centrifuged at 16,000g at 4°C for 20 min to remove particulates and protein. Five microliters of each sample was chromatographed on a ACQUITY 1.7-μm BEH C18 column (50 mm × 2.1 mm; Waters, Milford, MA) using an ACQUITY UPLC system (Waters). Flow rate was maintained at 0.5 ml/min in a 10-min run with a gradient mobile phase (acetonitrile: 5–100% from 0.5 to 10 min). The eluent was introduced directly into the mass spectrometer by electrospray. Mass spectrometry was performed on a Waters Q-TOF Premier mass spectrometer operating in positive and negative ion modes. Sulfamethoxine was used as the lock mass (m/z of 311.0814) for accurate mass calibration. Data were acquired in the centroid mode from 50 to 850 m/z.
MDA and Identification of Metabolites. Centroided and integrated mass chromatographic data were processed by MarkerLynx mass spectrometry software (Waters) to generate a multivariate data matrix. The resulting data matrices were then exported into SIMCA-P+ 11 (Umetrics, Umea, Sweden) for MDA. Urine samples after more than 7 days of treatment for each dose were selected as observations. Pretreatment methods and pattern recognition methods were optimized for effective classification and signal extraction. PLS-DA was performed on Pareto-scaled data. Suspected metabolites were identified from the corresponding loadings plots. The 35Cl/37Cl isotope ratio was used as additional inclusion criteria for fenofibrate metabolites.
Structural Elucidation of Metabolites. After the metabolites were synthesized and characterized by NMR or the authentic compounds were purchased, mass spectra of both the standards and the putative urinary constituents were recorded on an ABI 3000 mass spectrometer (Applied Biosystems, Foster City, CA). For MS/MS fragmentation of target ions, collision energies ranging from 10 to 60 V were applied. Further confirmation of identity was sought by comparison of high-performance liquid chromatography (Shimadzu 10A; Shimadzu Corporation, Kyoto, Japan) retention times in multiple reaction monitoring (MRM) mode. For MRM identification in negative ion mode, the urine samples were diluted 1:20 with methanol, and for MRM identification in positive ion mode, samples were diluted 1:4. For FAT and RFAT, which were considered as unreported metabolites, two ion pairs as identified from synthesized compounds were used for LC-MS/MS comparison. For FAEG and RFAEG, identifications were made based on fragmentation profiles and two ion pairs detection in MRM, the main product ions of which apparently came from FA (317.1), RFA (319.1), and their glucuronide (175.4).
In MRM detection, the compounds were separated on a CapCell PAK C18 column (5 μm, 2.0 × 50 mm) at room temperature with the flow rate of 0.2 ml/min. The mass spectrometer was operated in negative ion mode using MRM with transitions m/z 317.0/230.9 for FA, 319.2/233.1 for RFA, and 233.1/215.0 for compound AR and the double ion pairs 426.3/192.2 and 426.1/124.2 for RFAT, 424.2/192.2 and 424.2/231.2 for FAT, 493.5/175.4 and 493.5/317.1 for FAEG, and 495.4/175.4 and 495.5/319.1 for RFAEG. Transition m/z 275.3/233.1 for compound B and 333.3/233.1 for compound X were used in positive ion mode. All of the raw data were processed using Analyst software 1.4.2.
Identified metabolites were checked by LC-MS/MS analysis of six blank urine samples and six fenofibrate-treated samples urine samples, which were desalted on an solid-phase extraction column (Supelco, Bellefonte, PA).
Exposure and Excretion of New Taurine Conjugates. Urine samples and plasma samples collected when a steady state of fenofibrate treatment was reached were subjected to LC-MS/MS analysis for exposure and excretion evaluation. Synthesized FAT and RFAT were used to establish calibration curves. Comparison was made with FA, which was the pharmacologically active metabolite of fenofibrate. Chromatographic conditions and analytical parameters were same as those indicated in structural elucidation of metabolites. Concentrations of the three metabolites in monkey urine samples and plasma samples were determined by validated methods for which bezafibrate (m/z 360.1/274.4) was used as internal standard. The results were transformed to micromolar concentrations and are expressed as means ± S.D. for both dose treatments.
Results
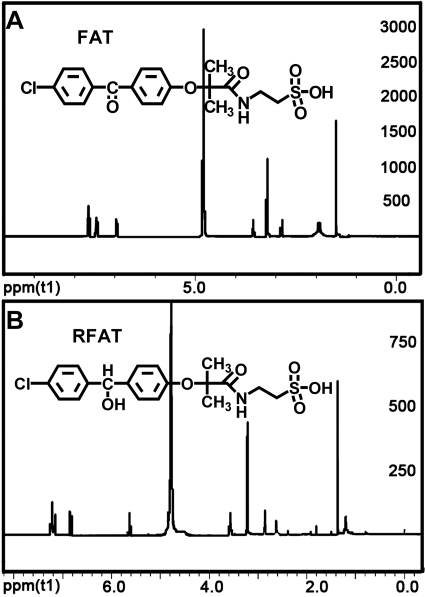
Synthesis of Fenofibrate Metabolites. The NMR spectra for RFA, compound AR, FAT, RFAT, compound B, and compound X were acquired. The purity of these chemicals used for MS validation was ≥95% according to 1H NMR analyses. NMR spectra for FAT and RFAT, which were abundant metabolites of a new metabolic pathway, are shown in Fig. 1, A and B (NMR spectra of compound X, RFA, compound AR, and compound B are shown in Supplemental Fig. 2).
Fig. 1.
A, NMR spectra of synthesized FAT. No obvious signal peaks from impurities were observed indicating the total amount of impurities was <5% (solvent: CD3OD). 1H NMR (400 MHz, MeOD) δ 1.530 (s, 6H), δ 2.825–2.858 (t, 2H), δ 3.530–3.575 (m, 2H), δ 6.939–6.960 (d, 2H), δ 7.432–7.453 (d, 2H), δ 7.629–7.66 7 (m, 4H). B, NMR spectra of synthesized RFAT. No obvious signal peaks from impurities were observed indicating the total amount of impurities was <5% (solvent: CD3OD). 1H NMR (400 MHz, MeOD) δ 1.362 (s, 6H), δ 2.843–2.876 (t, 2H), δ 3.542–3.575 (t, 2H), δ 5.623 (s, 1H), δ 6.814–6.835 (d, 2H), δ 7.148–7.253 (m, 6H).
Phenotypes of Cynomolgus Monkeys Treated with Fenofibrate. SPSS 11.5 for Windows was used for data analysis of phenotypes of fenofibrate treatment. A paired t test indicated that administration of both low and high doses of fenofibrate did not affect monkey body weights (p = 0.586 for low dose and p = 0.695 for high dose). Blood samples were collected and serum biochemical analysis was performed before and after fenofibrate administration. One-way analysis of variance revealed no significant changes in alanine aminotransferase, aspartate aminotransferase, creatinine, blood urea nitrogen, and lactate dehydrogenase for either dose treatment throughout the study, demonstrating that fenofibrate was well tolerated in NHPs (data for the high dose are shown in Supplemental Fig. 3; data for the low dose are not shown).
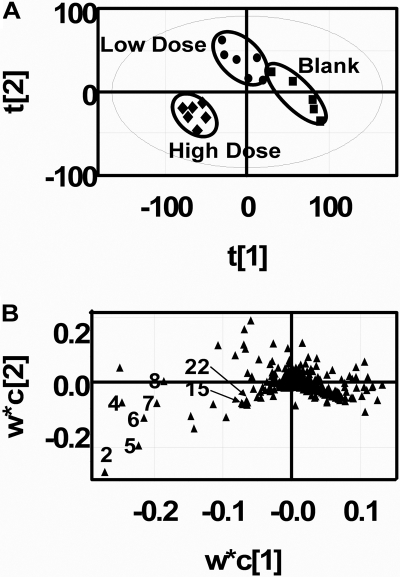
MDA and Selection of Signals. Supervised PLS-DA was used for pattern recognition, and the best scores plot and loadings plot were reached (Fig. 2) [R2X (cumulative) = 0.439′Q2 (cumulative) = 0.518]. The scores plot revealed three clusters corresponding to urine samples of the control, low-dose, and high-dose groups (Fig. 2A). The loadings plots revealed the ions that contributed most to the group separation along the first component (Fig. 2B). Suspected fenofibrate metabolites were identified from the loadings plots. In-source fragments and adducts were excluded after inspection of the raw chromatograms. After confirming the absence of suspected fenofibrate metabolites in the vehicle-only urine samples, eight variables were extracted as metabolite signals from the negative data matrix (Table 1). The positive data matrix was analyzed similarly, but no further new information was acquired.
Fig. 2.
Score plots and loading plots of negative data matrix with PLS-DA and Pareto scaling. Score plot A demonstrates the discriminate power of supervised PLS-DA (for first two-component R2X (cumulative) = 0.439′Q2 (cumulative) = 0.518). The low-dose cluster was closer to the blank cluster, and the high-dose cluster deviated significantly from the blank dose. The first component contributed most to the grouping power. The metabolite signals suspected and identified from the negative data matrix and their ranks are indicated as numerals in plot B.
TABLE 1.
Variables of identified metabolites extracted from negative and positive data matrices
Responses are expressed as mean ± S.D. (n = 6). Metabolites B and X were not listed in the top 50 variables contributing to grouping; thus their ranks are not provided. For RFAEG, a chiral separation effect caused two signals in UPLC-QTOFMS data as shown.
| Retention Time | m/z | Rank | Blank | Low Dose | High Dose | Proposed Metabolites |
|---|---|---|---|---|---|---|
| 5.22 min | 426.078- | 2 | 0.21 ± 0.21 | 71.08 ± 61.05 | 740.33 ± 275.79 | RFAT |
| 5.35 min | 495.104- | 15 | 0 | 10.87 ± 15.39 | 61.22 ± 43.01 | RFAEG |
| 5.45 min | 495.104- | 5 | 0.06 ± 0.09 | 74.75 ± 69.26 | 445.15 ± 48.59 | RFAEG |
| 5.46 min | 424.062- | 22 | 0.03 ± 0.05 | 7.44 ± 8.35 | 54.16 ± 49.21 | FAT |
| 5.74 min | 493.089- | 4 | 0.26 ± 0.14 | 247.24 ± 159.28 | 515.42 ± 98.12 | FAEG |
| 5.91 min | 319.074- | 6 | 0.008 ± 0.02 | 143.36 ± 106.44 | 404.34 ± 97.35 | RFA |
| 5.92 min | 233.036- | 7 | 0.04 ± 0.1 | 145.4 ± 84.12 | 320.11 ± 54.12 | AR |
| 6.32 min | 317.059- | 8 | 0 | 202.19 ± 132.97 | 319.86 ± 115.18 | FA |
| 6.90 min | 333.09+ | 0 | 0.66 ± 0.75 | 3.62 ± 3.38 | X | |
| N.D. | N.D. | N.D. | N.D. | N.D. | B |
-, negative mode; +, positive mode; N.D., not detected.
Compound X was reported as a fenofibrate metabolite in rat hepatocytes, and compound B was structurally derived from fenofibrate. Although MDA of both negative and positive ion data matrices indicated no information related to these two compounds (as shown in Table 1), we suspected that they were probably metabolites. Hence, synthesized compound X and B were used for identification by the LC-MS/MS analysis and fragmentation profile.
Identification and Structural Elucidation of Suspected Metabolites. FA, RFA, FAEG, RFAEG, and compound X were reported previously in humans, rats, and dogs. They were also found as metabolites in the cynomolgus urine samples in this study. For FA, RFA, and compound X, the identical MS/MS profiles from authentic or synthetic compounds and urine samples demonstrate identical fragmentation patterns. As further evidence for identity, their retention times by LC-MS/MS were determined to be almost identical (Supplemental Figs. 4–6). For FAEG and RFAEG, the double ion pairs detected in the urine samples by MRM in negative mode shared identical retention times, and their fragments came from the substrates FA (317.1), RFA (319.1), and the ligand of glucuronic acid (175.4) (Supplemental Fig. 7).
Compounds B and AR, which were structurally part of FA and RFA but not identified as metabolites in other species, were found in this study. Both compounds B and AR in cynomolgus urine samples chromatographed with the same retention times as the authentic or synthetic compounds. In their MS/MS profiles, the main fragments from the synthetic compounds were identical to those in urine samples (Supplemental Figs. 8 and 9).
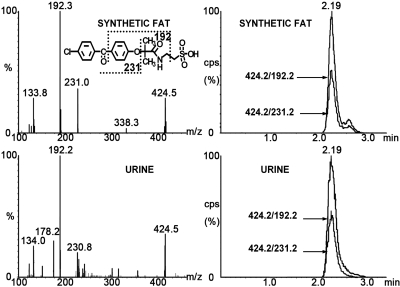
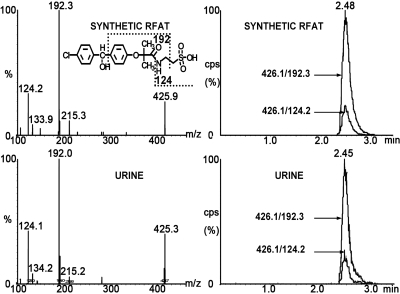
Two taurine conjugates of fenofibrate, FAT and RFAT, were found in cynomolgus monkeys and have not been reported in other species. The fragmentation profiles of synthetic FAT and RFAT were same as those found in urine samples. MRM detection in negative mode with double ion pairs showed that the urine samples shared retention times that were identical to those of the synthetic compounds (Figs. 3 and 4). Thus, they can be defined as new metabolites of fenofibrate in cynomolgus monkey.
Fig. 3.
Fragmentation profiles and LC-MS/MS chromatographs of synthetic FAT and cynomolgus urine samples detected in double ion pairs. The fragmentation profiles and retention time for both ion pairs were identical for synthetic compound and urine sample.
Fig. 4.
Fragmentation profiles and LC-MS/MS chromatographs of synthetic RFAT and cynomolgus urine samples detected in double ion pairs. The fragmentation profiles and retention time for both ion pairs were identical for synthetic compound and urine sample.
For further verification, the blank urine samples and fenofibrate-treated urine samples from the six individual monkeys were subjected to LC-MS/MS analysis in MRM in both negative and positive modes (data not shown). The results for absence of the above-identified metabolites in blank samples and presence in fenofibrate-treated urine samples confirmed the metabolites found by this metabolomics approach.
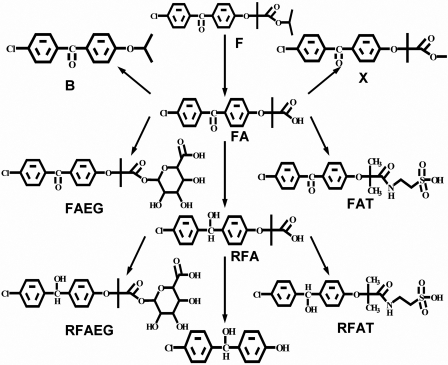
Combining the taurine conjugation pathway of fenofibrate, which was newly found in this study, the previously reported known metabolites (summarized in Table 2), together with the metabolic intermediate found in this study, we proposed the metabolic pathway of fenofibrate in cynomolgus monkey as shown in Fig. 5.
TABLE 2.
Summary of metabolites of fenofibrate in different species
Metabolites of fenofibrate in human, guinea pig, dog, rat, and rat hepatocytes were obtained from the indicated references in the text (Weil et al., 1988, 1990; Cornu-Chagnon et al., 1995).
|
Metabolites
|
Human
|
Monkey
|
Guinea Pig
|
Dog
|
Rat
|
|
|---|---|---|---|---|---|---|
| Rat | Rat Hepatocyte | |||||
| FA | Yes | Yes | Yes | Yes | Yes | Yes |
| FAEG | Yes | Yes | Yes | Yes | Yes | |
| RFA | Yes | Yes | Yes | Yes | Yes | Yes |
| RFAEG | Yes | Yes | Yes | Yes | ||
| AR | Yes | |||||
| Compound B | Yes | |||||
| Compound X | Yes | Yes | ||||
| FAT | Yes | |||||
| RFAT | Yes | |||||
Fig. 5.
Proposed metabolic map of fenofibrate in cynomolgus monkeys.
Exposure Level and Excretion of FAT and RFAT. Quantitation results for FAT, RFAT, and FA are shown in Table 3. FA was the most abundant metabolite of fenofibrate in plasma and exposure to high-dose treatment was approximately 7 times that of low-dose treatment. For these three metabolites, the concentration in urine was higher than that in plasma because of the kidney concentration effect. The ratio of taurine conjugates and FA increased from less than 1:10 in the low-dose group to more 1:4 in the high-dose group. Excretion of metabolites in urine showed dose dependence, but taurine conjugates in urine increased much more than FA when the high dose was given (20- and 60-fold for FAT and RFAT versus 3-fold for FA). During the high-dose treatment, FAT and RFAT molar concentrations were markedly higher than that of FA, which was different from that during the low-dose treatment. Thus, when the dose was higher, more FA would be transformed to FAT and RFAT and these taurine conjugates were more easily excreted in urine.
TABLE 3.
Trough concentration of FA, FAT, and RFAT in plasma at steady state and their excretion in urine expressed in micromolar concentration
Results are expresses as mean ± S.D. (n = 18).
|
FA
|
FAT
|
RFAT
|
||||
|---|---|---|---|---|---|---|
| Low | High | Low | High | Low | High | |
| Plasma | 4.1 ± 2.8 | 28.6 ± 27.4 | 0.0026 ± 0.0022 | 0.064 ± 0.14 | 0.37 ± 0.25 | 7.5 ± 3.6 |
| Urine | 32.9 ± 30.5 | 107.1 ± 83.4 | 0.47 ± 0.52 | 9.18 ± 7.2 | 16.52 ± 13.6 | 982.6 ± 809 |
Discussion
Fibrates are a class of hypolipidemic drugs that have been used extensively for more than 30 years. However, fenofibrate was reported to be a peroxisome proliferator and to produce hepatocarcinogenesis in some rodent species, but there is no evidence for hepatotoxic or carcinogenic effects of these compounds in humans or NHPs (Hoivik et al., 2004; Peters et al., 2005). In mice, PPARα was found to mediate the liver toxicity and cancer properties of fibrate drugs and related PPARα agonists (Ward et al., 1998; Peters et al., 2000). Hepatic cell proliferation was found to be involved in peroxisome proliferator-induced hepatocarcinogenesis (Yang et al., 2007). Fibrates also cause myotoxicity clinically, which was estimated to be a 5.5-fold increased risk compared with that for statins. Their coadministration with statins increased risk by an additional 2-fold versus that for fibrates alone (Graham et al., 2004). In muscle homogenates, fenofibrate induces mitochondrial dysfunction predominantly by inhibition of complex I of the respiratory chain (Brunmair et al., 2004). A fiber type-selective response caused by PPARα activation was consistent with increased fatty acid uptake and β-oxidation, which represent the major pathways responsible for the clinical benefits of fibrate drugs (De Souza et al., 2006). In rat skeletal muscle cultures, PPARα activation was found to mediate in part the toxic response to PPARα agonists (Johnson et al., 2005). The mechanistic investigation of species-related differences and myotoxicity of fibrates and whether metabolites may play a role is still unresolved.
The metabolism of fenofibrate was investigated in vivo in human volunteers, dog, rat, and guinea pig (Table 2). Both in vivo and in vitro studies revealed metabolic pathways of ester hydrolysis, reduction of the carbonyl group, and glucuronidation (Cornu-Chagnon et al., 1995). FA, RFA, FAEG, and RFAEG were identified as metabolites of fenofibrate. In the current study, UPLC-QTOFMS-based metabolomics coupled with highly sensitive LC-MS/MS was used, and four new metabolites hitherto unreported were identified. Among the newly found metabolites, FAT and RFAT indicate a previously unknown conjugation pathway for fenofibrate.
Compounds B and X contain the same scaffold of fenofibrate, and compound X has been reported as a metabolite in rat hepatocytes. However, they were not detected as significant variables by UPLC-QTOFMS because of their low abundance in monkey urine, yet were identified as metabolites by targeted LC-MS/MS. Their retention times were identical with those of authentic compounds (3.87 and 3.54 min) and longer than that of larger molecular weight ions; therefore, the possibility of in-source-fragmentation was excluded. Furthermore, these two compounds are unlikely to be impurities or products caused by pretreatment with methanol, because in another study, in which protein precipitation was performed by addition of acetonitrile, both compounds B and X were produced by incubation of rat hepatocytes with FA (data not shown). The abundance of compound AR was considerable and extracted by MDA from the negative data matrices. It is also structurally related to other metabolites, but its retention time was shorter than that of metabolites with larger molecular mass (2.11 min) and thus in-source-fragmentation was excluded, similar to compounds B and X.
Although species differences of taurine conjugation metabolites for clofibrate have already been studied (Cayen et al., 1977), the taurine conjugation pathway of fenofibrate, which is a structural analog of clofibrate, has not been reported. In this study, taurine conjugates of FA and RFA were identified as metabolites of fenofibrate by identical fragmentation profiles and retention times of double ion pairs. This result differed from that for clofibrate, for which taurine conjugation was not noted in humans (Cayen et al., 1977). Taurine conjugates were of higher abundance than other metabolites as detected by UPLC-QTOFMS. By quantitation of the primary metabolite FA and newly identified taurine metabolites, exposure to FA at the high dose was approximately 7 times that at the low dose (Table 3), which was similar to the 4 times reported previously (2500 mg/kg/day versus 200 mg/kg/day) (Hoivik et al., 2004). In addition, exposure to RFAT was more than one-fourth that of FA by molar concentration, indicating a potential warning sign of metabolite safety; however, the phenotype analysis showed that fenofibrate was well tolerated in cynomolgus monkeys (Supplemental Fig. 3) in the presence of high exposure to RFAT (Table 3). Thus, these new taurine metabolites probably are safe in this NHP. The higher concentrations of RFAT than FA in urine and its in vivo toxicity tolerance in this study are in accord with the function of phase II metabolism of xenobiotics.
To increase the survivability of drug candidates, metabolite safety issues have been receiving more attention in recent years (Baillie et al., 2002). Safety testing is required when systemic exposure of a metabolite is more than 10% of that of the parent compound according to guidance from the U.S. Food and Drug Administration. In the present study, RFAT and FAT, previously unreported metabolites, were found; systemic exposure of RFAT was approximately 25% of that of FA in the high-dose treatment group. FA, a carboxylic acid, is the pharmacologically active metabolite of fenofibrate, and although it was reported that 9 of the 29 drugs withdrawn between 1974 and 1993 were carboxylic acid-containing drugs (Bakke et al., 1995; Skonberg et al., 2008), no safety issues have ever been reported for FA. In addition, acyl-CoAs were reported to be intermediates in phase II metabolism, including taurine conjugation with an active thioester bond (Skonberg et al., 2008). Thus, the taurine conjugation pathway reported here warrants additional drug and drug metabolite safety studies.
To find metabolites or biomarkers, NMR and MS are popular metabolomics platforms. Both approaches have their own distinct advantages in sensitivity, selectivity, and reproducibility (Robertson, 2005). It is also noteworthy that mass defect filtering, a method that depends on the acquisition of high-resolution MS data, was recently used to identify drug metabolites (Zhu et al., 2006). Mass defect filtering discriminates metabolites from matrix ions by setting a filtering window based on the similarity of the mass defect values of a compound and putative metabolites. However, in drug safety and toxicology assessment, for which biomarkers are usually unknown and endogenous, metabolomics is a powerful platform that combines the advantages of UPLC-QTOF (or NMR) and MDA, especially when coupled with LC-MS/MS, as demonstrated by identification of compounds B and X in the current study.
In conclusion, nine metabolites of fenofibrate were identified by metabolomics from fenofibrate-treated cynomolgus monkey urine samples, of which compound AR, compound B, FAT, and RFAT have not previously been reported. Of these new metabolites, FAT and RFAT indicate a previously unreported conjugation pathway of fenofibrate in addition to the known glucuronidation pathway.
Supplementary Material
This work was supported in part by the National Institutes of Health National Cancer Institute [Grant Z01 BC005562-20] (Intramural Research Program); the National High-tech R&D Program [Grant 2006AA02Z339]; the Guangzhou Science and Technology Bureau [Grants 2006Z1-E4031, 2006P067]; and the Guangzhou Development District [Grant 2006Ss-P067].
Article, publication date, and citation information can be found at http://dmd.aspetjournals.org.
doi:10.1124/dmd.108.025817.
ABBREVIATIONS: PPARα, peroxisome proliferator-activated receptor α; NHP, nonhuman-primate; FA, fenofibric acid; RFA, reduced fenofibric acid; FAEG, fenofibric acid ester glucuronide; RFAEG, reduced fenofibric acid ester glucuronide; MDA, multivariate data analysis; UPLC, ultraperformance liquid chromatography; QTOFMS, quadrupole time-of-flight mass spectrometry; PLS-DA, projection to latent structures discriminant analysis; FAT, fenofibric acid taurine conjugate; RFAT, reduced fenofibric acid taurine conjugate; compound A, 4-chloro-4′-hydroxybenzophenone; compound AR, compound A reduced; compound X, 2-[4-(4-chloro-benzoyl)-phenoxy]-2-methylpropionic acid methyl ester; compound B, 4-chloro-4′-isopropoxybenzophenone; TLC, thin-layer chromatography; MS/MS, tandem mass spectrometry; MRM, multiple reaction monitoring; LC, liquid chromatography; MS, mass spectrometry.
The online version of this article (available at http://dmd.aspetjournals.org) contains supplemental material.
References
- Backman JT, Kyrklund C, Neuvonen M, and Neuvonen PJ (2002) Gemfibrozil greatly increases plasma concentrations of cerivastatin. Clin Pharmacol Ther 72 685–691. [DOI] [PubMed] [Google Scholar]
- Baillie TA, Cayen MN, Fouda H, Gerson RJ, Green JD, Grossman SJ, Klunk LJ, LeBlanc B, Perkins DG, and Shipley LA (2002) Drug metabolites in safety testing. Toxicol Appl Pharmacol 182 188–196. [DOI] [PubMed] [Google Scholar]
- Bakke OM, Manocchia M, de Abajo F, Kaitin KI, and Lasagna L (1995) Drug safety discontinuations in the United Kingdom, the United States, and Spain from 1974 through 1993: a regulatory perspective. Clin Pharmacol Ther 58 108–117. [DOI] [PubMed] [Google Scholar]
- Blane GF (1987) Comparative toxicity and safety profile of fenofibrate and other fibric acid derivatives. Am J Med 83 26–36. [DOI] [PubMed] [Google Scholar]
- Brunmair B, Lest A, Staniek K, Gras F, Scharf N, Roden M, Nohl H, Waldhäusl W, and Fürnsinn C (2004) Fenofibrate impairs rat mitochondrial function by inhibition of respiratory complex I. J Pharmacol Exp Ther 311 109–114. [DOI] [PubMed] [Google Scholar]
- Cayen MN, Ferdinandi ES, Greselin E, Robinson WT, and Dvornik D (1977) Clofibrate and clofibric acid: comparison of the metabolic disposition in rats and dogs. J Pharmacol Exp Ther 200 33–43. [PubMed] [Google Scholar]
- Chen C, Krausz KW, Idle JR, and Gonzalez FJ (2008) Identification of novel toxicity-associated metabolites by metabolomics and mass isotopomer analysis of acetaminophen metabolism in wild-type and Cyp2e1-null mice. J Biol Chem 283 4543–4559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen C, Ma X, Malfatti MA, Krausz KW, Kimura S, Felton JS, Idle JR, and Gonzalez FJ (2007) A comprehensive investigation of 2-amino-1-methyl-6-phenylimidazo[4,5-b]pyridine (PhIP) metabolism in the mouse using a multivariate data analysis approach. Chem Res Toxicol 20 531–542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clouâtre Y, Leblanc M, Ouimet D, and Pichette V (1999) Fenofibrate-induced rhabdomyolysis in two dialysis patients with hypothyroidism. Nephrol Dial Transplant 14 1047–1048. [DOI] [PubMed] [Google Scholar]
- Cornu-Chagnon MC, Dupont H, and Edgar A (1995) Fenofibrate: metabolism and species differences for peroxisome proliferation in cultured hepatocytes. Fundam Appl Toxicol 26 63–74. [DOI] [PubMed] [Google Scholar]
- De Souza AT, Cornwell PD, Dai X, Caguyong MJ, and Ulrich RG (2006) Agonists of the peroxisome proliferator-activated receptor alpha induce a fiber-type-selective transcriptional response in rat skeletal muscle. Toxicol Sci 92 578–586. [DOI] [PubMed] [Google Scholar]
- Emudianughe TS, Caldwell J, Sinclair KA, and Smith RL (1983) Species differences in the metabolic conjugation of clofibric acid and clofibrate in laboratory animals and man. Drug Metab Dispos 11 97–102. [PubMed] [Google Scholar]
- Ghosh B, Sengupta S, Bhattacharjee B, Majumder A, and Sarkar SB (2004) Fenofibrate-induced myopathy. Neurol India 52 268–269. [PubMed] [Google Scholar]
- Graham DJ, Staffa JA, Shatin D, Andrade SE, Schech SD, La Grenade L, Gurwitz JH, Chan KA, Goodman MJ, and Platt R (2004) Incidence of hospitalized rhabdomyolysis in patients treated with lipid-lowering drugs. JAMA 292 2585–2590. [DOI] [PubMed] [Google Scholar]
- Hoivik DJ, Qualls CW Jr, Mirabile RC, Cariello NF, Kimbrough CL, Colton HM, Anderson SP, Santostefano MJ, Morgan RJ, Dahl RR, et al. (2004) Fibrates induce hepatic peroxisome and mitochondrial proliferation without overt evidence of cellular proliferation and oxidative stress in cynomolgus monkeys. Carcinogenesis 25 1757–1769. [DOI] [PubMed] [Google Scholar]
- Johnson TE, Zhang X, Shi S, and Umbenhauer DR (2005) Statins and PPARα agonists induce myotoxicity in differentiated rat skeletal muscle cultures but do not exhibit synergy with co-treatment. Toxicol Appl Pharmacol 208 210–221. [DOI] [PubMed] [Google Scholar]
- Klaunig JE, Babich MA, Baetcke KP, Cook JC, Corton JC, David RM, DeLuca JG, Lai DY, McKee RH, Peters JM, et al. (2003) PPARα agonist-induced rodent tumors: modes of action and human relevance. Crit Rev Toxicol 33 655–780. [DOI] [PubMed] [Google Scholar]
- Ma X, Chen C, Krausz KW, Idle JR, and Gonzalez FJ (2008) A metabolomic perspective of melatonin metabolism in the mouse. Endocrinology 149 1869–1879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishimura J, Dewa Y, Muguruma M, Kuroiwa Y, Yasuno H, Shima T, Jin M, Takahashi M, Umemura T, and Mitsumori K (2007) Effect of fenofibrate on oxidative DNA damage and on gene expression related to cell proliferation and apoptosis in rats. Toxicol Sci 97 44–54. [DOI] [PubMed] [Google Scholar]
- Peters JM, Cheung C, and Gonzalez FJ (2005) Peroxisome proliferator-activated receptor-α and liver cancer: where do we stand? J Mol Med 83 774–785. [DOI] [PubMed] [Google Scholar]
- Peters JM, Rusyn I, Rose ML, Gonzalez FJ, and Thurman RG (2000) Peroxisome proliferator-activated receptor α is restricted to hepatic parenchymal cells, not Kupffer cells: implications for the mechanism of action of peroxisome proliferators in hepatocarcinogenesis. Carcinogenesis 21 823–826. [DOI] [PubMed] [Google Scholar]
- Prueksaritanont T, Zhao JJ, Ma B, Roadcap BA, Tang C, Qiu Y, Liu L, Lin JH, Pearson PG, and Baillie TA (2002) Mechanistic studies on metabolic interactions between gemfibrozil and statins. J Pharmacol Exp Ther 301 1042–1051. [DOI] [PubMed] [Google Scholar]
- Qi C, Zhu Y, and Reddy JK (2000) Peroxisome proliferator-activated receptors, coactivators, and downstream targets. Cell Biochem Biophys 32 187–204. [DOI] [PubMed] [Google Scholar]
- Rao MS and Reddy JK (1987) Peroxisome proliferation and hepatocarcinogenesis. Carcinogenesis 8 631–636. [DOI] [PubMed] [Google Scholar]
- Reddy JK, Azarnoff DL, and Hignite CE (1980). Hypolipidaemic hepatic peroxisome proliferators form a novel class of chemical carcinogens. Nature 283 397–398. [DOI] [PubMed] [Google Scholar]
- Ritter JL and Nabulsi S (2001) Fenofibrate-induced elevation in serum creatinine. Pharmacotherapy 21 1145–1149. [DOI] [PubMed] [Google Scholar]
- Robertson DG (2005) Metabonomics in toxicology: a review. Toxicol Sci 85 809–822. [DOI] [PubMed] [Google Scholar]
- Sallustio BC, Harkin LA, Mann MC, Krivickas SJ, and Burcham PC (1997) Genotoxicity of acyl glucuronide metabolites formed from clofibric acid and gemfibrozil: a novel role for phase-II-mediated bioactivation in the hepatocarcinogenicity of the parent aglycones? Toxicol Appl Pharmacol 147 459–464. [DOI] [PubMed] [Google Scholar]
- Shitara Y, Hirano M, Sato H, and Sugiyama Y (2004) Gemfibrozil and its glucuronide inhibit the organic anion transporting polypeptide 2 (OATP2/OATP1B1:SLC21A6)-mediated hepatic uptake and CYP2C8-mediated metabolism of cerivastatin: analysis of the mechanism of the clinically relevant drug-drug interaction between cerivastatin and gemfibrozil. J Pharmacol Exp Ther 311 228–236. [DOI] [PubMed] [Google Scholar]
- Skonberg C, Olsen J, Madsen KG, Hansen SH, and Grillo MP (2008) Metabolic activation of carboxylic acids. Expert Opin Drug Metab Toxicol 4 425–438. [DOI] [PubMed] [Google Scholar]
- Ward JM, Peters JM, Perella CM, and Gonzalez FJ (1998) Receptor and nonreceptor-mediated organ-specific toxicity of di(2-ethylhexyl)phthalate (DEHP) in peroxisome proliferator-activated receptor α-null mice. Toxicol Pathol 26 240–246. [DOI] [PubMed] [Google Scholar]
- Weil A, Caldwell J, and Strolin-Benedetti M (1988) The metabolism and disposition of fenofibrate in rat, guinea pig, and dog. Drug Metab Dispos 16 302–309. [PubMed] [Google Scholar]
- Weil A, Caldwell J, and Strolin-Benedetti M (1990) The metabolism and disposition of 14C-fenofibrate in human volunteers. Drug Metab Dispos 18 115–120. [PubMed] [Google Scholar]
- Yang Q, Ito S, and Gonzalez FJ (2007) Hepatocyte-restricted constitutive activation of PPAR α induces hepatoproliferation but not hepatocarcinogenesis. Carcinogenesis 28 1171–1177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhen Y, Krausz KW, Chen C, Idle JR, and Gonzalez FJ (2007) Metabolomic and genetic analysis of biomarkers for peroxisome proliferator-activated receptor α expression and activation. Mol Endocrinol 21 2136–2151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu M, Ma L, Zhang D, Ray K, Zhao W, Humphreys WG, Skiles G, Sanders M, and Zhang H (2006) Detection and characterization of metabolites in biological matrices using mass defect filtering of liquid chromatography/high resolution mass spectrometry data. Drug Metab Dispos 34 1722–1733. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.