Abstract
We report that microtubule (MT) nucleation at the Golgi apparatus requires AKAP450, a centrosomal γ-TuRC-interacting protein that also forms a distinct network associated with the Golgi. Depletion of AKAP450 abolished MT nucleation at the Golgi, whereas depletion of the cis-Golgi protein GM130 led to the disorganisation of AKAP450 network and impairment of MT nucleation. Brefeldin-A treatment induced relocalisation of AKAP450 to ER exit sites and concomitant redistribution of MT nucleation capacity to the ER. AKAP450 specifically binds the cis-side of the Golgi in an MT-independent, GM130-dependent manner. Short AKAP450-dependent growing MTs are covered by CLASP2. Like for centrosome, dynein/dynactin complexes are necessary to anchor MTs growing from the Golgi. We further show that Golgi-associated AKAP450 has a role in cell migration rather than in cell polarisation of the centrosome–Golgi apparatus. We propose that the recruitment of AKAP450 on the Golgi membranes through GM130 allows centrosome-associated nucleating activity to extend to the Golgi, to control the assembly of subsets of MTs ensuring specific functions within the Golgi or for transporting specific cargos to the cell periphery.
Keywords: AKAP450, centrosome, GM130, Golgi apparatus, MTs
Introduction
In most mammalian cells, the Golgi apparatus (GA) is a single-copy organelle shaped like a ribbon and closely associated with the centrosome. Both the structure and positioning of the GA are highly dependent on the microtubule (MT) cytoskeleton. In the absence of MTs, the Golgi ribbon fragments, giving rise to discrete Golgi elements, which are dispersed throughout the cell. This suggests that MTs are required to link stacks into a single organelle and to ensure its pericentrosomal location (Thyberg and Moskalewski, 1999). The centrosome is the major site for MT nucleation and anchorage in proliferating and migrating cultured animal cells. In these cells, MT arrays are organised with MT minus ends anchored at the centrosome and plus ends extended toward the cell periphery. By contrast, in most differentiated animal cells as myotubes, neurons or epithelial cells, MTs are arranged into noncentrosomal nonradial arrays (Bartolini and Gundersen, 2006). Noncentrosomal MTs can be nucleated by the centrosome, released and then stabilised into nonradial arrays, can be assembled in the cytoplasm or can be nucleated at noncentrosomal sites (Luders and Stearns, 2007). Recent data in a range of cell types indicate that γ-tubulin-dependent formation of new MTs can take place in the GA (Efimov et al, 2007).
The ability of Golgi membranes to assemble and stabilise MTs was first observed in two hepatic cell lines in MT-repolymerisation experiments. In addition, purified rat liver Golgi membranes were shown to support MT nucleation in vitro (Chabin-Brion et al, 2001). More recently, the GA was identified as an MT nucleating centre in RPE1 cells by time-lapse imaging and laser ablation of the centrosome. MT nucleation at the GA was demonstrated to require γ-tubulin and CLASPs, MT plus end-binding proteins that are recruited to the GA by interacting with the TGN-associated protein GCC185. A role for these Golgi-emanating MTs in establishing cell polarisation, in post-Golgi trafficking and in cell migration has been proposed (Efimov et al, 2007).
Two important questions are how and where these MTs are nucleated. To date, two Golgi-associated γ-tubulin-interacting proteins have been described: GMAP210 (Rios et al, 2004) and AKAP450 (Takahashi et al, 2002). GMAP210 was proposed to anchor minus ends of centrosome-nucleated MTs. AKAP450, which interacts with γ-tubulin complexes through binding with γ-tubulin complex protein 2 (GCP2), was reported to provide MT nucleation sites to the centrosome.
In this work, we identify AKAP450, and the cis-Golgi-associated protein GM130, as important players in the nucleation of MTs at the GA. AKAP450, also known as AKAP350 or CG-NAP, is a multiply spliced AKAP family member localised to the centrosome and the GA (Schmidt et al, 1999; Takahashi et al, 1999; Witczak et al, 1999). The role of AKAP450 in centrosome dynamics has been studied (Takahashi et al, 2002; Keryer et al, 2003a, 2003b; Nishimura et al, 2005). Centrosome-targeting mechanism through the calmodulin-binding PACT domain is also established (Gillingham and Munro, 2000). However, few studies concerning the role of this protein in GA function are available (Larocca et al, 2004; Kim et al, 2007). GM130 has been proposed to be involved in protein transport and GA biogenesis (Nakamura et al, 1997; Lowe et al, 1998; Seemann et al, 2000; Puthenveedu et al, 2006; Marra et al, 2007) and more recently, in regulating centrosome morphology and function during interphase (Kodani and Sutterlin, 2008). We now report that, by anchoring AKAP450, GM130 also participates in MT nucleation at the GA.
Results
AKAP450 localisation and dynamics in normal cells
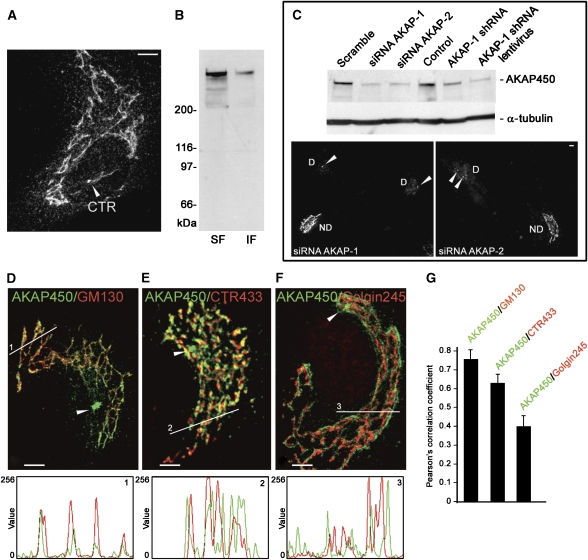
The polyclonal antibody A24 raised against peptides encoded by exons 24–27 of AKAP450 cDNA (Keryer et al, 2003b) revealed the centrosome and a network congruent with the GA in RPE1 cells (Figure 1A). By western blotting (WB), A24 antibody recognised a major high molecular weight band, in both detergent soluble and insoluble fractions (Figure 1B). To further test the specificity of A24, an siRNA approach was employed, using three complementary RNAi-based methods and targeting two different sequences, inside (AKAP-1) or outside (AKAP-2) the AKAP450 cDNA. After 72 h of AKAP-1 or AKAP-2 siRNA transfection, the band recognised by the A24 antibody was depleted by 80% as assayed by WB (Figure 1C). By immunofluorescence (IF), we observed that GA and cytoplasmic labellings had almost completely disappeared in siRNA-transfected cells further showing the specificity of the antibody (Figure 1C). siRNA knockdown was not efficient enough to completely deplete AKAP450 from the centrosome. In addition, about 10–15% of cells were untransfected in these experiments, which may account for the residual AKAP450 amount detected by WB. Similar results were obtained when the sequence targeted by siRNA AKAP-1 was used to construct knockdown pSUPER or lentiviral vectors containing the corresponding short hairpin (Figure 1C).
Figure 1.
AKAP450 is present into a network associated to the cis-face of the GA. (A) RPE1 cells stained with A24 antibody. (B) NP-40-soluble (SF) and insoluble (IF) fractions from RPE1 cells were analysed by WB with A24 antibody. (C) RPE1 cells were transfected with scramble or knockdown siRNAs (AKAP-1 or AKAP-2), short hairpin expressing pSUPER (AKAP-1 shRNA) or lentiviral (AKAP-1 shRNA lentivirus) vectors. After 72 h, a total lysate was prepared and analysed by WB for AKAP450 or α-tubulin as a loading control. Alternatively cells were fixed and processed by IF with A24 antibody. D, depleted cells; ND, nondepleted cells. Arrowheads indicate the centrosome (CTR). (D–F) Double-labelled cells for AKAP450 (green) and GM130 (D), CTR433 (E) or Golgin245 (F) were analysed by confocal microscopy. Merged images are shown. Arrowheads point to the centrosome. Bars, 10 μm. Fluorescence intensity profiles (at bottom) correspond to lines drawn in images D, E and F (lines 1, 2 and 3 as indicated). (G) Quantification of colocalisation by ICA based on the Pearson's correlation coefficient. The values are means of 10 lines drawn at random positions in 16 different cells, each line contained 500 pixels in average. Errors bars indicate ±s.e.m.
To precisely determine the distribution of AKAP450 network in the GA, we analysed by confocal microscopy RPE1 cells co-stained with A24 and different Golgi compartment markers. As shown in Figure 1D–F, AKAP450 overlapped more with the cis-Golgi protein GM130 than with medial CTR433 or trans-Golgin245 markers. Fluorescence intensities profiles along lines drawn over the Golgi area show the degree of correlation between two stainings. Image correlation analysis, which is a pixel-to pixel-comparison based on the Pearson's correlation coefficient, is shown in Figure 1G. Whereas AKAP450 showed a high degree of colocalisation with GM130 (r=0.7669) the correlation with Golgin245 (r=0.342) was significantly low. These results indicate that AKAP450 network is associated with the cis-side of the GA (see also Figure 2B and C).
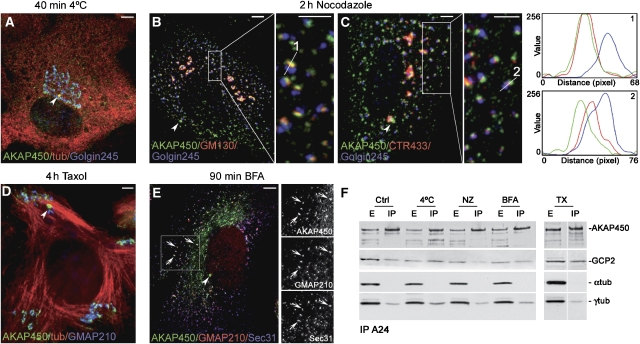
Figure 2.
AKAP450 association with membranes is preserved in the absence of MTs. (A) RPE1 cells were incubated at 4°C for 40 min and labelled for AKAP450 (green), tubulin (red) and Golgin245 (blue) or were treated with NZ for 2 h (B, C) and triple labelled for AKAP450 (green), GM130 (B, red) or CTR433 (C, red) and Golgin245 (blue). Boxes in B and C are enlarged on the right of each image. Fluorescence intensity profiles taken from the lines indicated in the respective enlarged boxes (1 and 2) are depicted at right. Note the nearly identical localisation of AKAP450 and GM130 in line 1. (D) Taxol-treated cells triple labelled for AKAP450 (green), tubulin (red) and GMAP210 (blue). (E) AKAP450 network is sensitive to BFA treatment. Merged image showing the association of AKAP450 (green) with ERES revealed with anti-GMAP210 (red) and anti-Sec31 antibodies (blue). Gray images of each single channel from the inset area are shown at right. Arrowheads point to the centrosome. Bars, 10 μm. (F) Co-immunoprecipitation of AKAP450, GCP2 and γ-tubulin from control or treated cells as indicated, using A24 antibody. Blots were also revealed for α-tubulin as a negative control. E, total extract; IP, immunoprecipitates.
AKAP450 network, but not AKAP450 binding to membranes, depends on MT cytoskeleton and GA integrity
To gain insights into AKAP450 network dynamics, RPE1 cells were subjected to different treatments. Depolymerisation of MTs either by incubation at 4°C that preserves Golgi localisation (Figure 2A) or by Nocodazole (NZ) treatment that leads to GA fragmentation and dispersion of Golgi ministacks (Figure 2B and C) perturbed AKAP450 network. Under these conditions, centrosome staining was maintained and a significant amount of AKAP450 appeared as punctuated structures dispersed throughout the cytoplasm. However, in contrast with data obtained in HeLa cells (Kim et al, 2007), a large fraction of AKAP450 remained associated with the intact GA in cold-treated cells or with NZ-induced Golgi ministacks (insets in Figure 2B and C), suggesting that the interaction of AKAP450 with Golgi membranes does not depend on MTs. The presence of AKAP450 in Golgi elements was correlated with the same series of markers as above. Again, a high correlation coefficient was obtained with GM130 (r=0.5995) as compared with r=0.4988 with CTR433 and only r=0.3226 with Golgin245. Intensity profiles of Golgi elements reveal almost identical localisation of AKAP450 and GM130.
Taxol treatment, which promotes the complete polymerisation of tubulin into noncentrosomal MT bundles, also preserved AKAP450 binding to the centrosome and to Golgi elements that appeared associated with MT minus ends located at one side of MT bundles (Figure 2D). Strikingly, AKAP450 network was also sensitive to Brefeldin-A (BFA) treatment and the protein partially codistributed with GMAP210 and Sec31 in ER exit sites (ERES, Figure 2E). Thus, AKAP450 behaves as the so-called cis-Golgi matrix proteins. These data show that the assembly of AKAP450 network, but not AKAP450 binding to membranes, requires MT cytoskeleton and GA integrity. We next wondered whether the previously reported interaction between AKAP450 and γ-TuRC complexes is maintained under conditions in which AKAP450 network is disrupted. Immunoprecipitation with A24 pulled down GCP2 and γ-tubulin, but not α-tubulin, regardless of MT or Golgi state (Figure 2F). Altogether, these results support the existence of MT nucleating complexes containing AKAP450 and γ-TuRC on the surface of Golgi membranes even in the absence of MTs or a stacked GA.
AKAP450 depletion inhibits MT nucleation from the GA and alters the MT array
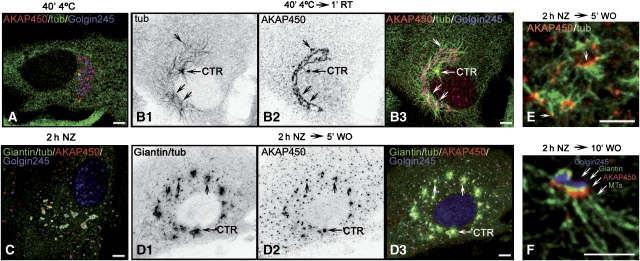
To determine whether AKAP450 is involved in MT nucleation at the GA, we first analysed the kinetics of MT nucleation after cold or NZ-induced depolymerisation in cells. In cold-treated cells, a large number of noncentrosomal MTs in addition to the centrosomal aster was observed only 1 min after rewarming (Figure 3B1). These MTs emanated from the GA (Figure 3B2 and B3). Strikingly, the lengths of MTs nucleated by the centrosome or the GA were similar, suggesting that nucleation started simultaneously on both structures. Identical results were obtained in NZ-recovering cells (Figure 3C and D). We carried out a four-labelling approach in NZ washout assays (see Materials and methods and legend of Figure 3). This approach allowed us to clearly visualise MT asters radiating from Golgi fragments that contained AKAP450 (arrows in Figure 3D1–D3). High magnifications revealed that most minus ends of growing MTs were anchored to AKAP450 containing structures (Figure 3E) and that Golgi-associated MT nucleation occurred at the cis-side of the GA where AKAP450 localises (Figure 3F).
Figure 3.
MT nucleation at the GA. (A, B) MTs formed after cold-induced depolymerisation (A) and rewarming (B1, arrows), colocalised with AKAP450 (B2, arrows) and with the GA labelled for Golgin245 (B3, merged image). (C, D) NZ-treated cells at 0 (C) or 5 min (D) after washout were quadruple labelled as follows: monoclonal anti-giantin and anti-tubulin antibodies were revealed with the same secondary anti-mouse antibody, whereas AKAP450 and Golgin245 were revealed by anti-rabbit and anti-human antibodies, respectively. In D1, both giantin and tubulin labellings are shown. In C and D3, giantin and tubulin are in green, AKAP450 in red and Golgin245 in blue. B1, B2, D1and D2 are inverted images. (E) A high magnification view of cells processed like in D shows that growing MTs contain AKAP450 at their minus ends. (F) A Golgi mini-stack quadruple labelled like in D showing that MT outgrowth occurs from the cis-face where AKAP450 is located. Bars, 10 μm.
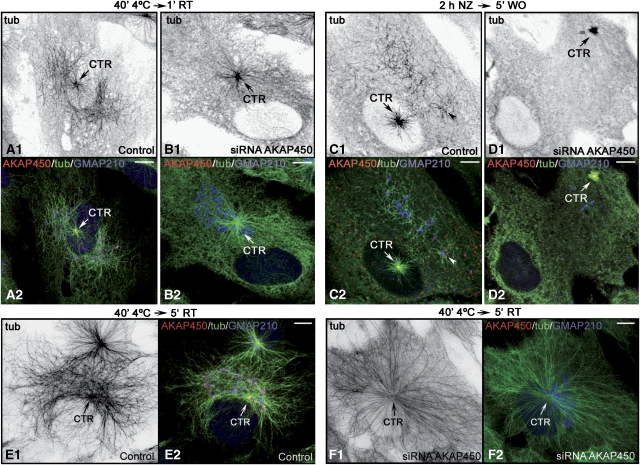
Then, we compared MT repolymerisation in scramble or siRNA AKAP-1 transfected cells recovering from cold (Figure 4A and B) or NZ (Figure 4C and D) treatments. Unlike control cells, AKAP450-depleted cells recovering from cold treatment did not show MTs growing from the GA (compare Figure 4A1 and B1). Centrosomal MT nucleation was not apparently altered but as mentioned earlier, siRNA did not completely deplete AKAP450 from the centrosome. Similar results were obtained after NZ washout experiments. Because MTs only originated from the centrosome, a radial symmetric MT array was formed in depleted cells, whereas in AKAP450-containing cells, a dense meshwork of MTs was also present at the Golgi area after 5 min of cold recovery (Figure 4E and F). We conclude that MT nucleation at the GA is a cis-Golgi-associated process that requires AKAP450.
Figure 4.
AKAP450 is required for MT nucleation at the GA. (A–D) Control (A, C) or AKAP450-depleted (B, D) cold-treated cells after 1 min of rewarming (A, B) or NZ-treated cells after 5 min washout (C, D) were labelled for AKAP450 (red), tubulin (green) and GMAP210 (blue). A1–D1 are inverted images of tubulin staining. A2–D2 are merged images. In E, F, AKAP450 nondepleted (E) or AKAP450-depleted (F) cold-treated cells after 5 min at room temperature are shown labelled for AKAP450 in red, tubulin in green and GMAP210 in blue. E1 and F1 are inverted images of tubulin staining. Merged images are shown in E2 and F2. Arrows indicate the centrosome. Bars, 10 μm.
AKAP450-dependent MTs are coated with CLASP2 at early stages of regrowth
Depletion of CLASPs has been reported to decrease the number of Golgi-based MTs. In addition, GFP-CLASP2 was found to entirely cover newly generated MTs at the initial stages of regrowth (Efimov et al, 2007). To investigate the relationship between AKAP450-nucleated MTs and CLASP2-stabilised MTs, we compared the distribution of both proteins in MT regrowth experiments. As shown in Figure 5A, both proteins colocalised at peripheral Golgi ministacks of NZ-treated cells and at the centrosome. Three minutes after NZ washout, most of the short MTs growing from AKAP450 appeared covered by CLASP2 (Figure 5B), suggesting that both proteins participate in the formation of the same subset of MTs at the GA.
Figure 5.
AKAP450-nucleated MTs are coated with CLASP2 at early stages of regrowth. (A, B) CLASP2 localisations before and after NZ washout. In NZ, both CLASP2 and AKAP450 associate with Golgi ministacks and with the centrosome (A). Three minutes after NZ removal, CLASP2 is detected at the newly formed MTs arising from AKAP450 containing elements (B, arrowheads). A1 and B1 are inverted images of tubulin staining; the same cells stained for AKAP450 (red) and CLAPS2 (green) are shown in A2 and B2. Boxes are enlarged at right. Bars, 10 μm.
In BFA-treated cells, MT nucleation occurs at ERES in an AKAP450-dependent manner
To gain further support for the role of AKAP450 in MT nucleation, we induced its redistribution to ERES by treating cells with BFA and then tested whether AKAP450-containing ERES were able to sustain MT assembly. Control or AKAP450-knockdown cells were incubated with BFA for 90 min and then with NZ for additional two hours. Double labelling with A24 and GMAP210 or Sec31 (not shown) revealed an almost complete relocalisation of AKAP450 to ERES under these conditions (Figure 6A1 and A2). Five minutes after NZ washout, MTs growing from AKAP450-containing ERES were clearly visible (Figure 6B1 and B2). At longer times, a perturbed and unfocused MT network was formed in which cytoplasmic MTs seem to be anchored to AKAP450-containing ERES (Figure 6C1 and C2). Upon AKAP450 silencing, centrosomal MT nucleation was apparently unaltered, whereas no MTs emanating from ERES were observed at any time after release (Figure 6D and E). These results indicate that AKAP450 is required for membrane-associated MT nucleation.
Figure 6.
In BFA-treated cells, AKAP450-dependent MT nucleation activity is redistributed to ERES. (A–C) Double BFA-NZ-treated cells fixed at 0 (A), 5 (B) or 10 min (C) after NZ washout stained for AKAP450, tubulin and GMAP210. A1–C1 are inverted images of tubulin staining. A2–C2 are merged images with AKAP450 in red, tubulin in green and GMAP210 in blue. Arrowheads in B and C indicate MTs emanating from ERES. (D–E) Similar experiments in AKAP450-depleted cells. Note that MTs only grow from the centrosome. Arrow indicates the centrosome (CTR). Bars, 10 μm.
GM130 anchors AKAP450 network to the cis-Golgi
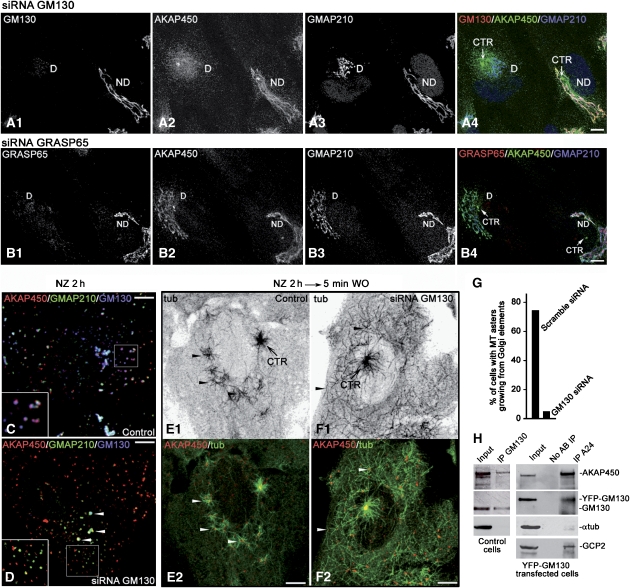
As AKAP450 behaved as a cis-Golgi matrix protein and colocalised with GMAP210 or GM130 under all conditions tested, we investigated by siRNA whether one or both of these proteins was involved in its recruitment to GA. Depletion of GMAP210 did not apparently affect AKAP450 binding to membranes (data not shown). By contrast, depletion of GM130 had a dramatic effect on AKAP450 network that appeared completely disassembled although the protein remained in a pericentrosomal localisation (Figure 7A1 and A2) where a slightly fragmented GA was localised (Figure 7A3). As depletion of GM130 displaces GRASP65 from the GA, we also assessed whether AKAP450 could interact with GM130 through GRASP65 by looking at the integrity of the AKAP450 network in cells lacking GRASP65. As shown in Figure 7B1 and B2, the AKAP450 network remained intact in the absence of GRASP65 (compare depleted cells in A2 and B2) and congruent with an intact GA (Figure 7B3), demonstrating that AKAP450 does not interact with GM130 through GRASP65. When GM130-depleted cells were treated with NZ, AKAP450 network dispersed throughout the cytoplasm as small aggregates that, in contrast to nondepleted cells, did not mostly colocalise with Golgi elements (Figure 7C and D, compare insets). Only in some elements containing residual GM130, AKAP450 remained associated to membranes (Figure 7D). Altogether, our data indicates that GM130 is necessary for the cis-GA localisation of AKAP450 network.
Figure 7.
Docking of AKAP450 network to cis-GA is mediated by GM130. (A, B) Depletion of GM130 (A1) disrupts AKAP450 network (A2) and slightly modified Golgi morphology as revealed by staining for GMAP210 (A3). On the contrary, GRASP65 knockdown (B1) did not affect neither AKAP450 network (B2) nor Golgi ribbon integrity (B3). Merged images are shown in A4 and B4. (C, D) Nocodazole treatment in control (C) and GM130-depleted (D) cells. Merged images are shown with AKAP450 in red, GMAP210 in green and GM130 in blue. (E, F) Control (E) or GM130-depleted cells (F) 5 min after NZ washout labelled for AKAP450 (red) and α-tubulin (green). In top panels (E1 and F1) inverted images of tubulin staining are shown. Arrowheads in (E) show MTs growing from distinct centres, whereas they indicate single MTs growing from dispersed AKAP450 aggregates in (F). The centrosome is indicated (CTR). Bottom panels (E2 and F2) are merged images with tubulin in green and AKAP450 in red. Bars, 10 μm. (G) The percentage of cells containing MT asters growing from Golgi elements in control and GM130-knockdown cells was determined (n=100 cells, two experiments). (H) RPE1 cell extracts were incubated with anti-GM130 antibody. Immunoprecipitates were analysed by WB for AKAP450, GM130 and α-tubulin as a negative control. Alternatively, immunoprecipitation experiments were carried out with A24 antibody on lysates from RPE1 cells transfected with a YFP-GM130 coding vector. Co-immunoprecipitating proteins were analysed by WB for AKAP450, YFP, α-tubulin as a negative control and GCP2 as a positive control. Antibody-free beads were used as controls (no AB IP).
To test whether AKAP450 is able to nucleate MTs when it is not associated with Golgi membranes, we analysed MT regrowth after NZ washout in cells where AKAP450 was displaced from the cis-GA by GM130 depletion. A striking difference between MT nucleation patterns of GM130-containing and GM130-depleted cells was evident (Figure 7E and F). In more than 70% of control cells, noncentrosomal MTs appeared as small asters growing from Golgi elements (Figure 7E1 and E2), whereas only 2% of GM130-knockdown cells exhibited these structures (Figure 7G). Unlike controls, MTs formed in GM130-depleted cells did not arise from distinct centres. Instead, single MTs were randomly distributed throughout the cytoplasm (Figure 7F1 and F2), indicating that membrane binding was not necessary for MT nucleation. Finally, we examined by co-immunoprecipitation experiments whether AKAP450 and GM130 form a complex. A24 pulled down AKAP450 but hardly endogenous GM130 (not shown), whereas anti-GM130 antibody coprecipitated both GM130 and AKAP450 (Figure 7H). Further, A24 coprecipitated endogenous AKAP450 with ectopically expressed YFP-tagged GM130. Thus, we conclude that by recruiting AKAP450 to the cis-GA, GM130 participates in MT assembly at the GA. Whether AKAP450–GM130 interaction is direct or not remains to be determined.
Dynein/dynactin complexes are necessary for MT formation at the GA
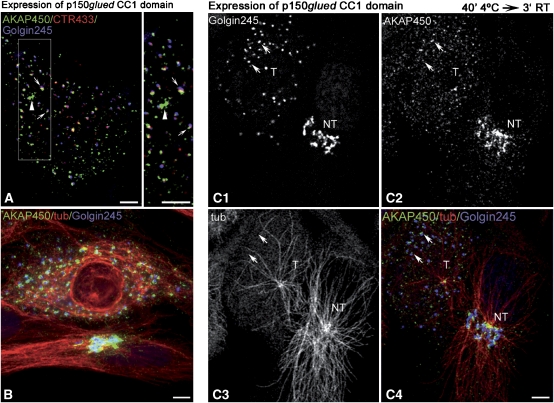
It is widely accepted that successful formation of MTs requires not only MT nucleation by γ-TuRC but also MT anchorage. Live observations have recently revealed that dynein inhibition enhanced the detachment of MTs from the centrosome (Burakov et al, 2008). Dynein/dynactin complexes are also essential for Golgi structure and localisation (Schroer, 2004). In addition, interaction of AKAP450 with p150glued has been reported recently and proposed to play a role in AKAP450 recruitment to minus ends of MTs (Kim et al, 2007). To evaluate whether the MT-anchoring activity of dynactin is necessary for MT formation at the GA, we transfected a dominant negative mutant of p150glued (CC1 domain) in RPE1 cells or overexpressed dynamitin (not shown). In p150glued CC1-expressing cells, the GA appeared fragmented and MT network profoundly disturbed (Figure 8A and B) as reported by others (Quintyne et al, 1999). AKAP450 binding either to centrosome or to Golgi elements was apparently unaffected (Figure 8A). Golgi fragments from cold-treated transfected cells were unable to support MT outgrowth after rewarming (Figure 8C1–C4). The number of centrosomal nucleated MTs was also significantly lower in transfected cells (Figure 8C3). These results suggest that, like for centrosomes, MT assembly at the GA requires MT anchoring activity of dynein/dynactin complexes.
Figure 8.
Dynein/dynactin complex is involved in MTs anchoring at the GA. (A, B) Merged images of RPE1 cells transfected with the CC1 domain of p150glued and stained for AKAP450 (green), Golgin245 (blue) and CTR433 (red, A) or tubulin (red, B). Box from A is enlarged at right to show AKAP450 association with the centrosome (arrowhead) and with stacked Golgi elements (arrows). (C) MT repolymerisation experiments in p150glued CC1 transfected cells. Three minutes after rewarming, cells were fixed and stained for Golgin245 (C1), AKAP450 (C2) and tubulin (C3). A transfected (T) and a nontransfected cell (NT) are shown. In C4, a merged image is shown with AKAP450 in green, tubulin in red and Golgin245 in blue. Arrows indicate some AKAP450-containing Golgi elements in a transfected cell that do not nucleate MTs. Bars, 10 μm.
Acetylated MTs are absent of the Golgi region in cells lacking AKAP450
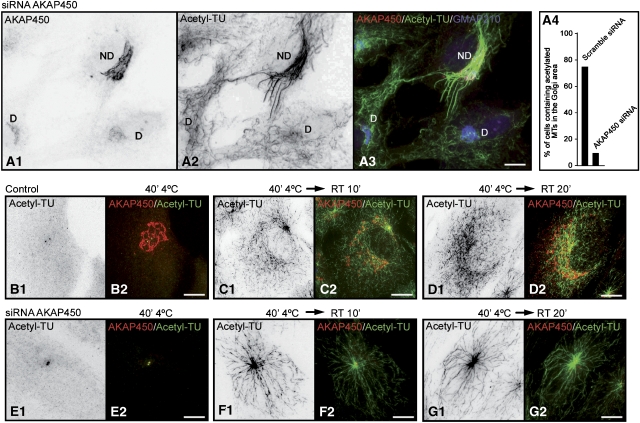
As Golgi-associated MTs are heavily acetylated in RPE1 cells, we investigated the effect of AKAP450 depletion on MT acetylation (Figure 9A1–A3). Loss of AKAP450 led to a substantial decrease in the density of acetylated MTs in the Golgi area in comparison to what it is observed in neighbour control cells. No differences were detected between depleted or nondepleted cells in the amount of acetylated MTs outside of the Golgi region. Quantification of these experiments indicated that more than 75% of nonsilenced cells contained acetylated MTs in the Golgi area (Figure 9A4). In contrast, only 8% of AKAP450-depleted cells contained Golgi-associated acetylated MTs.
Figure 9.
AKAP450-depletion decreased MT acetylation in the Golgi area. (A1–A3) AKAP450 depleted (D) and nondepleted (ND) cells were fixed and labelled for AKAP450 (A1), acetylated tubulin (A2) and GMAP210 (A3, merged image). A1 and A2 are inverted images. (A4) A graphic representation of the percentage of AKAP450 depleted or nondepleted cells containing acetylated MTs in the Golgi area (n=200 cells, two experiments). (B–G) Nondepleted (B–D) or depleted (E–G) RPE1 cells were cold treated for 40 min (B, E) and then rewarmed at RT for 10 min (C, F) or 20 min (D, G). After fixation, cells were labelled for acetylated tubulin (acetyl-TU) and AKAP450. In left panels of each figure, inverted images of acetylated MTs are shown. In right panels, merged images with acetylated MTs in green and AKAP450 labelling in red. Bars, 10 μm.
We also compared the timing of MT acetylation in control or AKAP450-depleted cells recovering from cold treatment, as it has been reported that MTs growing from the GA were more rapidly acetylated than those growing from centrosomes (Chabin-Brion et al, 2001). In RPE1 cells, however, acetylation proceeded at similar rates on centrosome- or Golgi-nucleated MTs (Figure 9B–D). Ten minutes after rewarming, acetylated tubulin was observed in radially organised MTs growing from the centrosome but also in nonoriented MTs colocalising with the GA (Figure 9C1 and C2). After 20 min of repolymerisation, a dense network of Golgi-associated acetylated MTs could be detected (Figure 9D1 and D2). In AKAP450-silenced cells, in contrast, acetylated tubulin was only detected in centrosome-anchored MTs (Figure 9E–G). We conclude that AKAP450 contributes significantly to maintaining the subset of acetylated MTs present in the Golgi area in normal cells.
AKAP450-depleted cells are defective in cell migration
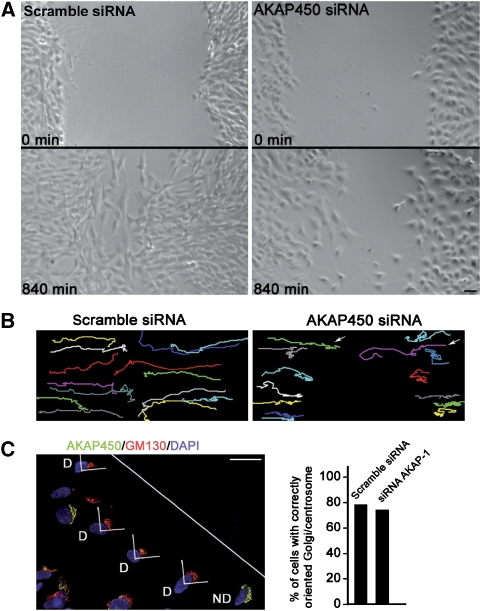
A role for Golgi-nucleated MTs in cell polarisation and motility has been proposed (Efimov et al, 2007). To evaluate the ability of cells to migrate in the absence of Golgi-based MTs, we tested whether AKAP450 depletion had an effect in cell migration. RPE1 cells transfected with scramble or AKAP-1 siRNAs were grown to confluence and then a wound-healing assay was used to initiate directional migration of cells toward the wound edge. Using time-lapse recording, we observed that AKAP450-knockdown cells appeared to advance into the wound at a slower rate that control cells (Figure 10A). By measuring the surface covered by cells in the first and the last frames of the movies from both control and depleted cells, we determined a decrease in cell migration of 60%. As in our RNAi experiments about 10–15% of cells were untransfected, the real cell migration inhibition is probably higher. Further confirmation was obtained by tracking randomly picked cells over 22 h of recording. Compared with scramble siRNA-transfected cells, 12 of 14 AKAP450-knockdown cells exhibited weaker motility (Figure 10B). The few cells displaying normal migration rates, probably corresponded to nondepleted cells. These results strongly support that AKAP450, and possibly Golgi-based MTs, are required for cell migration. To further know whether this defect in migration was due to the inability of AKAP450-depleted cells to reorientate their Golgi/centrosome, scramble or AKAP-1 siRNA-transfected cells were fixed 6 h after wounding and stained for Golgi/centrosome (Figure 10C). The Golgi/centrosome area was counted as oriented when the majority of the labelling was located within the 90°C angle facing the wound. Quantification of these experiments did not reveal significant differences in Golgi/centrosome reorientation between cells containing or lacking AKAP450 pointing to a specific defect in cell migration in the absence of AKAP450.
Figure 10.
AKAP450-depleted cells are defective in cell migration. (A) Living RPE1 cells transfected with scramble siRNA or with AKAP450 siRNA were imaged at 30-min intervals for 14 h. The first (0 min, top) and last (840 min, bottom) frames of the movie are presented. For estimation of the surface covered by cells in the first and the last frames of control and AKAP450-depleted cells, the MethaMorph software was used. (B) For tracking, scramble siRNA and AKAP450 siRNA-transfected cells were recorded during 22 h. Migration tracks of 14 randomly picked cells are represented in different colours. Arrows indicate two cells exhibiting normal migration. (C) Eight hours after wounding, siRNA AKAP-1 transfected cells were fixed and stained with anti-AKAP450 (green), anti-GM130 (red) and DAPI (blue). D, depleted cells; ND, nondepleted cells. The white line indicates the scratch orientation. Wound-edge cells having their Golgi/centrosome area in the quadrant that is in the front of the nucleus and facing the wound (as represented) were considered as correctly reoriented. At right, percentage of control or knockdown cells with Golgi/centrosome correctly reoriented. In knockdown experiments, only depleted cells (D) were counted. Results shown are means of two independent experiments for a total of at least 200 cells being scored in each condition. Bars, 50 μm.
Discussion
Our data indicate that the GA becomes a major site for MT nucleation by acting as a preferential γ-TuRC docking site according to a mechanism similar to that operating at the centrosome. We show that this is accomplished through the peripheral protein GM130 that recruits AKAP450, which in turn tethers γ-TuRCs, thus conferring MT nucleation capacity to the cis-Golgi compartment. Three lines of evidence reveal the essential role of AKAP450 in the process. First, depletion of AKAP450 abolished MT nucleation at the GA. Second, BFA treatment induced the redistribution of AKAP450 to ERES and concomitantly, the acquisition by ERES of AKAP450-dependent MT nucleation capacity, and third, dissociation of AKAP450 from the GA by GM130 knockdown dissociated MT nucleation ability from the GA. We further showed that AKAP450-dependent MT nucleation does not require Golgi binding, as it takes place in the cytoplasm when AKAP450 is displaced from the GA. Altogether, these results strongly support the notion that the cis-Golgi becomes an MT nucleating compartment through AKAP450 recruitment at this location.
We have observed in RPE1 cells that a fraction of AKAP450 dispersed throughout the cytoplasm in the absence of MTs. This is consistent with the proposal that AKAP450 is recruited to the GA in an MT-dependent manner through its interaction with p150glued (Kim et al, 2007). However, a significant fraction of AKAP450 remained associated with Golgi membranes. We also observed that AKAP450 binding to the cis-Golgi was preserved in cells expressing a dominant negative mutant of p150glued arguing against an essential role of dynein in AKAP450 recruitment to the GA. Remarkably, we further showed that the integrity of the AKAP450 network was also perturbed by BFA treatment that induced its redistribution to ERES. Therefore, an MT-independent mechanism should be involved in AKAP450 binding to membranes. In our view, AKAP450 could be delivered to MT minus ends in a dynein-dependent manner and then anchored to the cis-GA through a direct or indirect interaction with GM130. The network itself would form through homodimerisation or oligomerisation of AKAP450 mediated by its N-terminal domain (Takahashi et al, 1999). MT minus ends binding of AKAP450 might further contribute to the network stability. Our data also reveal a role for dynein/dynactin complexes in the retention of MT seeds at GA as has been proposed for the centrosome in interphase or for spindle poles during mitosis (Burakov et al, 2008). Thus, centrosome and GA seem to share the same components for MT nucleation and minus ends anchoring.
Noteworthy, MT nucleation at the GA has not been previously observed in other cell lines (Piehl et al, 2004; Rios et al, 2004). As these cells contain AKAP450, GM130 and p150glued, the reason for this different behaviour is not understood. One possibility is that these cells lack a still unidentified factor necessary for MTs formation. Another possibility is that different AKAP450 isoforms are expressed depending on the cell type: the human gene is composed of 50 exons that are alternatively spliced to produce multiple isoforms (Schmidt et al, 1999), most of which are still uncharacterised. The A24 antibody used in this study recognises exons 24–27-containing isoforms, whereas a monoclonal antibody recognising exon 28 only decorates the centrosome and some punctated structures in the AKAP450 network by IF (our unpublished data). A similar situation has been reported for p150glued (Dixit et al, 2008). For example, the absence of the MT binding domain of p150glued does not affect Golgi structure and localisation in HeLa cells, whereas a point mutation in this domain disrupts the GA in neurons (Dixit et al, 2008).
We have also shown that AKAP450-dependent MTs were coated by CLASP2 shortly after their nucleation. As both AKAP450 (this work) and CLASPs (Efimov et al, 2007) depletion completely abrogate MT formation at the GA, it seems clear that both proteins participate in the mechanism of MT assembly at the GA. Our results broadly support the recently proposed model according to which MT seeds generated at the cis-Golgi would be subsequently stabilised by TGN-associated MT plus end binding CLASP proteins (Efimov et al, 2007). Although no evidence has been provided for translocation of MTs seeds from cis-to-trans face of the GA to be stabilised and further elongated toward the cell periphery, our data do not exclude this possibility. However, MTs could as well elongate from the cis face of Golgi stacks, where they are nucleated, and their plus ends stabilised by TGN-associated CLASPs. Admittedly, this would generate short MTs within, and rather parallel to, the Golgi ribbon in addition of long cytoplasmic MTs. As a matter of fact, most cell types display a subpopulation of short, convoluted and detyrosylated or acetylated MTs that colocalise with the GA (Thyberg and Moskalewski, 1999; Rios and Bornens, 2003). Additional molecular players such as GMAP210 (Rios et al, 2004) or the centrosomal protein CAP350 (Hoppeler-Lebel et al, 2007) could also contribute to maintain and stabilise this subset of MTs. A direct experimental support for this possibility can also be found in very short-term regrowth experiments after cold treatment, where the GA is not dispersed like after NZ treatment (see Figures 3B and 4A). In that case, it is an intra-GA subset of MTs that is first assembled.
A striking feature of these Golgi-associated MTs is their high level of post-translationally modified tubulin. Whether these modified MTs perform specific functions in Golgi dynamics remains to be elucidated. They might control protein trafficking by promoting binding and transport of specific motors as has been reported for MT acetylation and kinesin-1 in neurons (Reed et al, 2006). Other Golgi-associated MTs growing out of the GA could be in this way loaded with specific cargos that could not be loaded on centrosome-associated MTs and that would be delivered to the leading edge of migrating cells. Interestingly, AKAP450-depleted cells lack acetylated MTs in the Golgi area and are defective in cell migration. In good agreement with our results, GM130-depleted cells were unable to form stable, acetylated MTs and to migrate in wound healing assays (Kodani and Sutterlin, 2008). These effects could be due to the impairment of AKAP450 recruitment to the pericentrosomal GA. However, GM130-depleted cells are unable to orientate the centrosome toward the leading edge (Kodani and Sutterlin, 2008) contrary to AKAP450-depleted cells (this work). GM130 depletion might cause specific defects in centrosome function in addition to the effect on AKAP450 association to the GA. AKAP450-knockdown cells, in which centrosomal AKAP450 was not depleted, display an apparently functional centrosome. Centrosome nucleating activity might be sufficient to promote reorientation of the centrosome and then of the GA, as GA maintains an association with the centrosome in AKAP450-depleted cells (see Figure 4).
Deciphering the role of AKAP450 on cell migration will deserve more work. AKAP450 is a huge protein that acts as a signalling platform by anchoring several enzymes including kinases, phosphatases and phosphodiesterases (Takahashi et al, 1999, 2000; Tasken et al, 2001; Sillibourne et al, 2002). It has also been reported to interact with calmodulin and with the CDC42-binding protein CIP4, as well as with other regulatory proteins (Keryer et al, 2003b; Larocca et al, 2004). Thus, there are other possible pathways in which AKAP450 could have a direct impact on cell motility.
In conclusion, we propose that the ability of the centrosomal scaffolding protein AKAP450 to be recruited to the cis-GA through GM130 results in the spatial extension of the region where MT nucleation activity is possible. As AKAP450 interacts with dynein/dynactin complexes, the capacity of MT minus end anchoring is also transferred to the GA by AKAP450. On the contrary, the stabilisation of the MT plus ends by CLASPs proteins could be a specific property of the GA. This strategy would allow the formation of a subset of modified MTs that could participate in Golgi dynamics during cell polarisation and migration. How enlargement of the centrosome, from the centriole pair to a broad pericentrosomal region limited by the GA, is regulated during cell cycle progression is another important question for the future.
Materials and methods
Cells, antibodies and treatments
Immortalised human pigment epithelial cells hTERT-RPE1 (Clontech) were grown in DMEM/F12 with 10% fetal bovine serum at 37°C in 5% CO2. Rabbit anti-GMAP210 (RM130), anti-AKAP450 (A24), anti-GCP2 and mouse CTR433 monoclonal antibodies have been produced in our laboratories. Autoimmune sera recognising GMAP210 and Golgin245 (unpublished data) have been also characterised (Infante et al, 1999). Anti-CLASP2 and anti-giantin antibodies were generous gifts of I Kaverina, N Galjart and HP Hauri. Monoclonal anti-α-tubulin, anti-γ-tubulin (clone GTU-88) and anti-acetylated tubulin (clone 611B) antibodies were purchased from Sigma and monoclonal anti-GM130, anti-AKAP450 and anti-Sec31 antibodies were from BD Biosciences. Polyclonal anti-GM130 was from Abcam, goat anti-GRASP65 from Santa Cruz and rabbit anti-GFP from ICL. Cells were treated with either 10 μM NZ for 2 h, 10 μM taxol (Paclitaxel) for 4 h or 5 μg/ml BFA for 90 min at 37°C (Sigma).
MT regrowth assay
MTs were depolymerised on ice for 40 min or with NZ (10 μM) for 2 h. Regrowth was induced by incubation in prewarmed medium (37°C). For NZ washout assays, cells were rinsed five times with ice-cold medium and then moved to a dish with warm (37°C) medium as described (Efimov et al, 2007). All the MT regrowth experiments were carried out at room temperature.
siRNAs, constructs, transfection and co-immunoprecipitation
To deplete AKAP450 expression, we used two siRNAs, AKAP-1 (5′-AACTTTGA AGTTAACTATCAA-3′) and AKAP-2 (5′-ATATGAACACAGCTTATGA-3′). To deplete GM130 expression, we used a mixture of two siRNAs, GM130A (5′-CAATGCTGCTACTCTACAATT-3′) and GM130B (5′-GGAGTCGG TTAGACAACTATT-3′). Duplex were from Sigma-Proligo or Dharmacon. To deplete GRASP65, we used a pool of three target-specific 20–25 nt siRNAs (Santa Cruz; Ref. sc-41228). Scramble siRNA (5′-CGUACGCGGAAUACUUCGATT-3′) was used as a control. Knockdown plasmid were constructed in pSUPER as described (Brummelkamp et al, 2002). Targeting sequence within AKAP450 cDNA corresponds to that of AKAP-1 siRNA. YFP-GM130 and p150-CC1 constructs were provided by V Malhotra and T Schorer, respectively. We used Oligofectamine (Invitrogen) for siRNA transfection, and Lipofectamine2000 (Invitrogen) for plasmid transfections. Extract preparations and co-immunoprecipitations from control or transfected cells were performed as described (Rios et al, 2004). Because of the high molecular weight of AKAP450, immunoprecipitates were analysed in parallel in 6 or 8% acrylamide-containing gels as indicated.
Lentiviral vectors
The lentiviral vector pLVTHM, as well as the second-generation lentivirus packaging (psPAX2) and envelope plasmids (pMD2.G), were kindly provided by Dr D Trono, Switzerland (Wiznerowicz and Trono, 2003). The map and the sequences of these plasmids are available at http://tronolab.epfl.ch/page58115.html. The H1-shRNA cassette from pSUPER-AKAP-1 was excised and cloned into the lentiviral pLVTHM vector within EcoRI-ClaI sites. The viral particles were produced by transient transfection of 293T cells and lentiviral particles titers were determined by flow cytometry of RPE-1 transducted cells. RPE-1 were seeded at 8 × 104 cells in a 6-well plate 24 h prior transduction, then lentiviral vectors were added and the culture medium was replaced 24 h after transduction.
IF and confocal microscopy
For IF experiments, RPE1 cells grown in coverslips were fixed in 100% methanol at −20°C for 6 min and then labelled with appropriate antibodies. Triple labelling experiments were always carried out by combining rabbit polyclonal and mouse monoclonal antibodies with one of available human autoimmune sera (anti-GMAP210 or anti-Golgin245) except in Figure 5 in which cells were first incubated with a mouse anti-AKAP450 and a rabbit anti-CLASP2 antibodies, washed and then incubated with corresponding secondary antibodies conjugated with Alexa fluor633 and TRITC. After several washes for 2 h, cells were re-incubated with monoclonal anti-α-tubulin, washed and incubated with secondary anti-mouse conjugated with Alexa fluor488. A similar strategy was used to perform the four-labelling approach as shown in Figure 3: fixed cells were first incubated with anti-giantin and anti-AKAP450 antibodies, washed and incubated with secondary anti-mouse and anti-rabbit antibodies conjugated with Alexa fluor488 and TRITC. After 4 h of washing, cells were re-incubated with anti-tubulin and anti-Golgin245 antibodies, washed and incubated with secondary anti-mouse and anti-human antibodies conjugated with Alexa fluor488 and Alexa fluor633 antibodies. Cells were examined under a motorised upright wide-field microscope (LEICA DM6000B). Confocal images were captured by a Confocal Leica TCS SP5 using a HCX PL APO Lambda blue 63 × 1.4 OIL objective at 22°C. Image analysis was carried out using the Leica and Adobe Photoshop software. For calculation of the Pearson's correlation coefficient, images were processed using MetaMorph Offline 7.1.7.0 using the command Linescan to view the intensity values along a given line in graphical format. All the values of Pearson's correlation coefficients were processed using SPSS for Windows 10.0.1.
Wound healing assay and Golgi/centrosome reorientation
RPE-1 cells were plated onto a 40-mm LabTek chambered coverglass dish and transfected 24 h later with Scramble or AKAP-1 siRNAs. Two days after transfection, individual wound were made with a micropipette tip. Cell migration was observed with a Leica DMI6000 inverted microscope equipped with a Hamamatsu ORCA-ER camera and using a LEICA N PLAN 10 × /0.25 objective. Processing, quantification and analysis were carried out using MetaMorph software. For centrosome/GA reorientation assays, 2 days after transfection serum was removed. After 24 h, scratches were made and serum was added. Cells were fixed 6 h after serum induction. Centrosome and GA were visualised by staining for AKAP450 and GM130. Golgi/centrosome located in front of the nucleus, within the quadrant facing the wound, were scored as correctly oriented.
Acknowledgments
The authors thank G Keryer, A Delouvee, W Kemmner, T Schroer, I Kaverina, N Galjart, HP Hauri, P Gleeson and V Malhotra for providing reagents, G Keryer for ongoing discussions, P Dominguez for confocal technical support and G Egea for reading of the manuscript. This study was supported by the MEC, Spain (to RMR) and the CNRS, France (to MB). SR was supported by a predoctoral fellowship from the University of Seville and JC by a grant from the MEC.
References
- Bartolini F, Gundersen GG (2006) Generation of noncentrosomal microtubule arrays. J Cell Sci 119: 4155–4163 [DOI] [PubMed] [Google Scholar]
- Brummelkamp TR, Bernards R, Agami R (2002) A system for stable expression of short interfering RNAs in mammalian cells. Science 296: 550–553 [DOI] [PubMed] [Google Scholar]
- Burakov A, Kovalenko O, Semenova I, Zhapparova O, Nadezhdina E, Rodionov V (2008) Cytoplasmic dynein is involved in the retention of microtubules at the centrosome in interphase cells. Traffic 9: 472–480 [DOI] [PubMed] [Google Scholar]
- Chabin-Brion K, Marceiller J, Perez F, Settegrana C, Drechou A, Durand G, Pous C (2001) The Golgi complex is a microtubule-organizing organelle. Mol Biol Cell 12: 2047–2060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dixit R, Levy JR, Tokito M, Ligon LA, Holzbaur EL (2008) Regulation of dynactin through the differential expression of p150glued isoforms. J Biol Chem 283: 33611–33619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Efimov A, Kharitonov A, Efimova N, Loncarek J, Miller PM, Andreyeva N, Gleeson P, Galjart N, Maia AR, McLeod IX, Yates JR III, Maiato H, Khodjakov A, Akhmanova A, Kaverina I (2007) Asymmetric CLASP-dependent nucleation of noncentrosomal microtubules at the trans-Golgi network. Dev Cell 12: 917–930 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gillingham AK, Munro S (2000) The PACT domain, a conserved centrosomal targeting motif in the coiled-coil proteins AKAP450 and pericentrin. EMBO Rep 1: 524–529 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoppeler-Lebel A, Celati C, Bellett G, Mogensen MM, Klein-Hitpass L, Bornens M, Tassin AM (2007) Centrosomal CAP350 protein stabilises microtubules associated with the Golgi complex. J Cell Sci 120: 3299–3308 [DOI] [PubMed] [Google Scholar]
- Infante C, Ramos-Morales F, Fedriani C, Bornens M, Rios RM (1999) GMAP-210, a cis-Golgi network-associated protein, is a minus end microtubule-binding protein. J Cell Biol 145: 83–98 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keryer G, Di Fiore B, Celati C, Lechtreck KF, Mogensen M, Delouvee A, Lavia P, Bornens M, Tassin AM (2003a) Part of Ran is associated with AKAP450 at the centrosome: involvement in microtubule-organizing activity. Mol Biol Cell 14: 4260–4271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keryer G, Witczak O, Delouvee A, Kemmner WA, Rouillard D, Tasken K, Bornens M (2003b) Dissociating the centrosomal matrix protein AKAP450 from centrioles impairs centriole duplication and cell cycle progression. Mol Biol Cell 14: 2436–2446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim HS, Takahashi M, Matsuo K, Ono Y (2007) Recruitment of CG-NAP to the Golgi apparatus through interaction with dynein-dynactin complex. Genes Cells 12: 421–434 [DOI] [PubMed] [Google Scholar]
- Kodani A, Sutterlin C (2008) The Golgi protein GM130 regulates centrosome morphology and function. Mol Biol Cell 19: 745–753 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larocca MC, Shanks RA, Tian L, Nelson DL, Stewart DM, Goldenring JR (2004) AKAP350 interaction with cdc42 interacting protein 4 at the Golgi apparatus. Mol Biol Cell 15: 2771–2781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowe M, Rabouille C, Nakamura N, Watson R, Jackman M, Jamsa E, Rahman D, Pappin DJ, Warren G (1998) Cdc2 kinase directly phosphorylates the cis-Golgi matrix protein GM130 and is required for Golgi fragmentation in mitosis. Cell 94: 783–793 [DOI] [PubMed] [Google Scholar]
- Luders J, Stearns T (2007) Microtubule-organizing centres: a re-evaluation. Nat Rev Mol Cell Biol 8: 161–167 [DOI] [PubMed] [Google Scholar]
- Marra P, Salvatore L, Mironov A Jr, Di Campli A, Di Tullio G, Trucco A, Beznoussenko G, Mironov A, De Matteis MA (2007) The biogenesis of the Golgi ribbon: the roles of membrane input from the ER and of GM130. Mol Biol Cell 18: 1595–1608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura N, Lowe M, Levine TP, Rabouille C, Warren G (1997) The vesicle docking protein p115 binds GM130, a cis-Golgi matrix protein, in a mitotically regulated manner. Cell 89: 445–455 [DOI] [PubMed] [Google Scholar]
- Nishimura T, Takahashi M, Kim HS, Mukai H, Ono Y (2005) Centrosome-targeting region of CG-NAP causes centrosome amplification by recruiting cyclin E-cdk2 complex. Genes Cells 10: 75–86 [DOI] [PubMed] [Google Scholar]
- Piehl M, Tulu US, Wadsworth P, Cassimeris L (2004) Centrosome maturation: measurement of microtubule nucleation throughout the cell cycle by using GFP-tagged EB1. Proc Natl Acad Sci USA 101: 1584–1588 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puthenveedu MA, Bachert C, Puri S, Lanni F, Linstedt AD (2006) GM130 and GRASP65-dependent lateral cisternal fusion allows uniform Golgi-enzyme distribution. Nat Cell Biol 8: 238–248 [DOI] [PubMed] [Google Scholar]
- Quintyne NJ, Gill SR, Eckley DM, Crego CL, Compton DA, Schroer TA (1999) Dynactin is required for microtubule anchoring at centrosomes. J Cell Biol 147: 321–334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reed NA, Cai D, Blasius TL, Jih GT, Meyhofer E, Gaertig J, Verhey KJ (2006) Microtubule acetylation promotes kinesin-1 binding and transport. Curr Biol 16: 2166–2172 [DOI] [PubMed] [Google Scholar]
- Rios RM, Bornens M (2003) The Golgi apparatus at the cell centre. Curr Opin Cell Biol 15: 60–66 [DOI] [PubMed] [Google Scholar]
- Rios RM, Sanchis A, Tassin AM, Fedriani C, Bornens M (2004) GMAP-210 recruits gamma-tubulin complexes to cis-Golgi membranes and is required for Golgi ribbon formation. Cell 118: 323–335 [DOI] [PubMed] [Google Scholar]
- Schmidt PH, Dransfield DT, Claudio JO, Hawley RG, Trotter KW, Milgram SL, Goldenring JR (1999) AKAP350, a multiply spliced protein kinase A-anchoring protein associated with centrosomes. J Biol Chem 274: 3055–3066 [DOI] [PubMed] [Google Scholar]
- Schroer TA (2004) Dynactin. Annu Rev Cell Dev Biol 20: 759–779 [DOI] [PubMed] [Google Scholar]
- Seemann J, Jokitalo EJ, Warren G (2000) The role of the tethering proteins p115 and GM130 in transport through the Golgi apparatus in vivo. Mol Biol Cell 11: 635–645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sillibourne JE, Milne DM, Takahashi M, Ono Y, Meek DW (2002) Centrosomal anchoring of the protein kinase CK1delta mediated by attachment to the large, coiled-coil scaffolding protein CG-NAP/AKAP450. J Mol Biol 322: 785–797 [DOI] [PubMed] [Google Scholar]
- Takahashi M, Mukai H, Oishi K, Isagawa T, Ono Y (2000) Association of immature hypophosphorylated protein kinase cepsilon with an anchoring protein CG-NAP. J Biol Chem 275: 34592–34596 [DOI] [PubMed] [Google Scholar]
- Takahashi M, Shibata H, Shimakawa M, Miyamoto M, Mukai H, Ono Y (1999) Characterization of a novel giant scaffolding protein, CG-NAP, that anchors multiple signaling enzymes to centrosome and the Golgi apparatus. J Biol Chem 274: 17267–17274 [DOI] [PubMed] [Google Scholar]
- Takahashi M, Yamagiwa A, Nishimura T, Mukai H, Ono Y (2002) Centrosomal proteins CG-NAP and kendrin provide microtubule nucleation sites by anchoring gamma-tubulin ring complex. Mol Biol Cell 13: 3235–3245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tasken KA, Collas P, Kemmner WA, Witczak O, Conti M, Tasken K (2001) Phosphodiesterase 4D and protein kinase a type II constitute a signaling unit in the centrosomal area. J Biol Chem 276: 21999–22002 [DOI] [PubMed] [Google Scholar]
- Thyberg J, Moskalewski S (1999) Role of microtubules in the organization of the Golgi complex. Exp Cell Res 246: 263–279 [DOI] [PubMed] [Google Scholar]
- Witczak O, Skalhegg BS, Keryer G, Bornens M, Tasken K, Jahnsen T, Orstavik S (1999) Cloning and characterization of a cDNA encoding an A-kinase anchoring protein located in the centrosome, AKAP450. EMBO J 18: 1858–1868 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiznerowicz M, Trono D (2003) Conditional suppression of cellular genes: lentivirus vector-mediated drug-inducible RNA interference. J Virol 77: 8957–8961 [DOI] [PMC free article] [PubMed] [Google Scholar]