Abstract
The interaction between the integrin α6β4 and plectin is essential for the assembly and stability of hemidesmosomes, which are junctional adhesion complexes that anchor epithelial cells to the basement membrane. We describe the crystal structure at 2.75 Å resolution of the primary α6β4–plectin complex, formed by the first pair of fibronectin type III domains and the N-terminal region of the connecting segment of β4 and the actin-binding domain of plectin. Two missense mutations in β4 (R1225H and R1281W) linked to nonlethal forms of epidermolysis bullosa prevent essential intermolecular contacts. We also present two structures at 1.75 and 2.05 Å resolution of the β4 moiety in the absence of plectin, which reveal a major rearrangement of the connecting segment of β4 on binding to plectin. This conformational switch is correlated with the way α6β4 promotes stable adhesion or cell migration and suggests an allosteric control of the integrin.
Keywords: cell adhesion, epidermolysis bullosa, hemidesmosome, integrin, plakin
Introduction
Integrity of epithelial tissues requires the attachment of basal cells to the underlying basement membrane. In skin and other complex and stratified epithelia, multiprotein junctional complexes, named hemidesmosomes, mediate stable adhesion and provide resistance to mechanical stress by linking the extracellular matrix to the cytokeratin filament system in the cell. Hemidesmosomes contain three transmembrane components: the α6β4 integrin, the type XVII collagen BP180 and the integrin-associated tetraspanin CD151. In addition, two members of the plakin family of cytolinkers: plectin and BP230 (BPAG1e) are present at the cytoplasmic region of hemidesmosomes (Jones et al, 1998; Nievers et al, 1999).
The α6β4 integrin is a receptor for laminins, with preference for binding to laminin-332 (laminin-5), which is a major component of the epidermal basement membrane (Tsuruta et al, 2008). α6β4 is linked to the cytokeratin network by plectin and BP230, on which there are binding sites for α6β4, BP180 and intermediate filaments. The α6β4–plectin interaction is crucial for hemidesmosome stability, and it has been proposed that it acts as an initiation step in the assembly of hemidesmosomes facilitating the recruitment of BP180 and BP230 (Schaapveld et al, 1998). In fact, simple epithelia contain more rudimentary adhesion complexes, named type II hemidesmosomes, composed only of α6β4 and plectin (Uematsu et al, 1994).
Most of the intracellular interactions of α6β4, including binding to plectin, occur through the cytoplasmic moiety of the β4 subunit, which is unusually large (∼1000 residues) and shares no similarity with that of other integrin β subunits. The β4 cytodomain has a modular organisation with four fibronectin type III (FnIII) domains arranged in two pairs of tandem repeats separated by a region named the connecting segment (CS), upstream of the first FnIII a calx-β domain is present, whereas downstream of the fourth FnIII extends an 85-residue long C-terminal tail (Figure 1A). The N-terminal region of plectin interacts with the β4 subunit (Rezniczek et al, 1998). This region of plectin contains an actin-binding domain (ABD) build up of two calponin homology domains (CH1 and CH2). Adjacent to the ABD extends a region conserved among members of the plakin family named the plakin domain, which is composed of a tandem array of spectrin repeats and an SH3 domain (Sonnenberg et al, 2007).
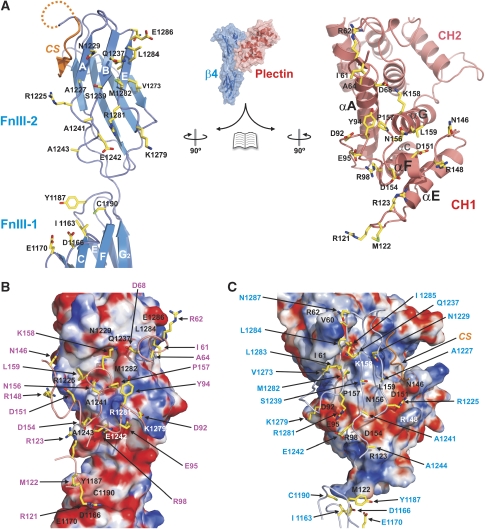
Figure 1.
Structure of the integrin β4–plectin complex. (A) Schematic representation of the domain organisation of plectin and the cytoplasmic domains of integrin α6β4. The regions of both proteins corresponding to the primary interaction site, whose crystal structure is presented herein, are highlight by a yellow box. (B) Ribbon representation in two orthogonal views of the integrin β4 (residues 1127–1318 in blue and 1324–1330 in orange) bound to plectin (residues 60–290 in red), with secondary structure elements labelled. Molecular figures were prepared using PyMOL (Delano, 2002).
The interaction between plectin and the cytoplasmic domain of the integrin β4 subunit occurs at multiple sites. The primary contact is established between the ABD of plectin and the first pair of FnIII domains (FnIII-1,2) and a small region of the CS of β4 (Geerts et al, 1999). This region of β4 is required for targeting plectin into hemidesmosomes (Niessen et al, 1997). Moreover, two missense mutations in the β4 gene (ITGB4) linked to nonlethal forms of epidermolysis bullosa, introduce point substitutions (R1225H and R1281W) in the second FnIII domain that prevents binding to plectin (Pulkkinen et al, 1998; Koster et al, 2001; Nakano et al, 2001); further illustrating the essential role in vivo of this primary contact. A second interaction is established between the plakin domain of plectin and a discontinuous site in β4 that includes the C-terminal area of the CS and the C-terminal tail downstream of the fourth FnIII domain (Koster et al, 2004).
Structural knowledge of the primary α6β4–plectin interaction sites includes the individual crystal structures of the FnIII-1,2 domains of β4 (de Pereda et al, 1999) and the ABD of plectin (Garcia-Alvarez et al, 2003; Sevcik et al, 2004). Using these structures and docking methods, we have previously constructed a theoretical model of the primary α6β4–plectin complex, and the contact interfaces derived from this model have been experimentally validated (Litjens et al, 2005). Despite these advances, the structural basis of the integrin α6β4 linkage to the cytoskeleton remained poorly understood. To decipher the molecular architecture of hemidesmosomes, we have determined the structure of the primary α6β4–plectin complex by X-ray crystallography. Combining structure-based mutagenesis and biophysical data, we have identified critical elements of the β4–plectin interaction. Furthermore, we present structures of the primary plectin-binding site on free β4, which reveals that there is a conformational switch in β4 linked to the binding to plectin. These results provide a molecular basis for diseases that target the hemidesmosome stability and have clear implications for models of hemidesmosome assembly and dynamics.
Results and discussion
Overall structure of the integrin β4–plectin complex
Crystals of the integrin β4–plectin complex were obtained using a fragment of human β4, residues 1126–1370, corresponding to the FnIII-1,2 domains and the N-terminal region of the CS, and a fragment of human plectin, residues 1–293, which contains the N-terminal tail and the ABD. The structure was solved by molecular replacement, and it was refined against data to a resolution of 2.75 Å (Table I). The asymmetric unit of the crystal contains two copies of the β4–plectin complex with one-to-one stoichiometry and a third unbound β4 molecule. Because of the moderate affinity of their interaction (see below), the complexes could not be purified before crystallisation; therefore, crystals were obtained using an equimolar mixture of the two proteins at a concentration about one order of magnitude higher than the equilibrium dissociation constant (Kd). Under these conditions, a small proportion of β4 and plectin is expected to be unbound, which explains the appearance of the free β4 molecule in the crystals.
Table 1.
Summary of crystallographic analysis
| β4–plectin complex | β4 (1126–1355) | β4 (1126–1370) | |
|---|---|---|---|
| Data collection | |||
| Space group | P3221 | P1 | C2 |
| Cell dimensions | |||
| a, b, c (Å) | 107.3, 107.3, 204.0 | 43.1, 49.5, 74.4 | 137.5, 42.2, 56.9 |
| α, β, γ (°) | 90, 90, 120 | 107.2, 96.1, 06.6 | 90, 109.9, 90 |
| Wavelength (Å) | 1.0723 | 1.5418 | 1.5418 |
| Resolution (Å) | 2.75 (2.90–2.75)a | 1.75 (1.81–1.75)a | 2.04 (2.11–2.04)a |
| Unique reflections | 35983 | 50859 | 19567 |
| Redundancy | 9.6 (9.9)a | 2.3 (2.2)a | 3.9 (3.6)a |
| Completeness (%) | 99.8 (100)a | 91.5 (84.8)a | 97.2 (85.9)a |
| Rmeas (%)b | 8.1 (60.7)a | 5.4 (36.0)a | 6.4 (28.9)a |
| 〈I/σI〉 | 20.4 (4.6)a | 16.3 (3.8)a | 16.6 (5.6)a |
| Refinement statistics | |||
| Resolution range (Å) | 85–2.75 | 22–1.75 | 36–2.04 |
| Unique reflections, work/free | 34 178/1805 | 48 246/2589 | 18 568/998 |
| R work/R free (%) | 20.4/25.7 | 15.5/18.2 | 17.6/21.5 |
| No. of molecules in the a.u.c | 2 × plectin/3 × β4 | 2 | 1 |
| No. of atoms | |||
| Protein | 8528 | 6832 | 3375 |
| Water | 20 | 502 | 142 |
| PEG | 15 | 56 | 18 |
| Ion | 1 | 2 | 1 |
| B-factors | |||
| Wilson plot | 63.6 | 25.7 | 31.6 |
| Proteind | 66.9/82.2/90.4/87.5/88.8 | 26.2/25.5 | 26.2 |
| Water | 55.6 | 36.8 | 32.6 |
| PEG | 71.9 | 56.4 | 49.4 |
| Ion | 43.4 | 25.6 | 26.2 |
| R.m.s. deviations | |||
| Bond lengths (Å) | 0.006 | 0.005 | 0.012 |
| Bond angles (°) | 0.907 | 0.878 | 1.130 |
| Ramachandran plot regions | |||
| Most favoured | 807 | 311 | 157 |
| Additionally allowed | 114 | 41 | 19 |
| Generously allowed | 2 | 0 | 0 |
| Disallowed | 0 | 0 | 0 |
| aNumbers in parenthesis correspond to the outer resolution shell. | |||
| bMultiplicity independent R factor. | |||
| cAsymmetric unit. | |||
| dValue per protein chain. | |||
The two copies of the β4–plectin complex have a similar structure. Each heterodimer has a ‘Y' shape in which the ABD branches at about 45° from the rod-like structure of the FnIII-1,2 domains of β4 (Figure 1B). The main contacts occur between the CH1 domain of plectin and the FnIII-2 domain of β4. Despite the overall similarities between the two copies of the β4–plectin complex, the binding interface is better defined in one of them, which has a lower average B-factor for the CH1 domain of plectin (74.8 versus 93.7 Å2) and the FnIII-2 of β4 (72.5 versus 105.2 Å2). When the β4 molecules from each complex were superimposed on top of each other, a difference in the orientation of the ABD in the poorly defined copy was revealed, that is, the ABD has rotated around an axis running parallel to the long axis of the β4 molecule by about 8°. The description of the structure of the heterodimer is based on what we observed in the well-defined copy.
The plectin-binding interface of integrin β4
The structure of the FnIII-1,2 domains of β4 in its complex with the plectin ABD is almost identical to that of the FnIII-1,2 domains, previously described by us, which did not contain the CS (PDB code 1QG3; r.m.s.d. of 0.79 Å for 191 Cα atoms) (de Pereda et al, 1999). The formation of the complex between β4 and plectin buries ∼1150 Å2 of the solvent accessible surface of each molecule, which corresponds to ∼10% of their surface. The core of the interface in the β4 subunit is formed by the ABE β-sheet and the BC loop of the FnIII-2 domain (Figure 2). This area accounts for ∼60% of the total surface buried in the complex. The interface of β4 is centred around a basic strip formed by R1225, K1279 and R1281, which faces an acidic area on the CH1 surface. R1225 and R1281 establish salt bridges with D151 and E95, respectively. A third salt bridge occurs between E1242 of β4 and R98 of plectin. Single point mutations R1225H or R1281W in β4, and E95S or R98Q in plectin abrogate the interaction of the ABD with β4 (Koster et al, 2001; Litjens et al, 2005); thus, the three intermolecular salt bridges are an essential part of the interaction. K1279 is located at the periphery of the interface and does not engage in any ionic interaction, which is in agreement that the K1279W substitution does not compromise the interaction with plectin (Koster et al, 2001). Direct contact between the backbones of both proteins occurs only at the level of the BC loop of β4 and the FG loop of plectin, where the carbonyl of D154 of plectin makes an H-bond with the nitrogen of E1242 of β4. The only large hydrophobic residue in the FnIII-2 domain that contributes to the interface is M1282 in the β-strand E, whose side chain contacts P157 and the aliphatic chain of K158 of plectin.
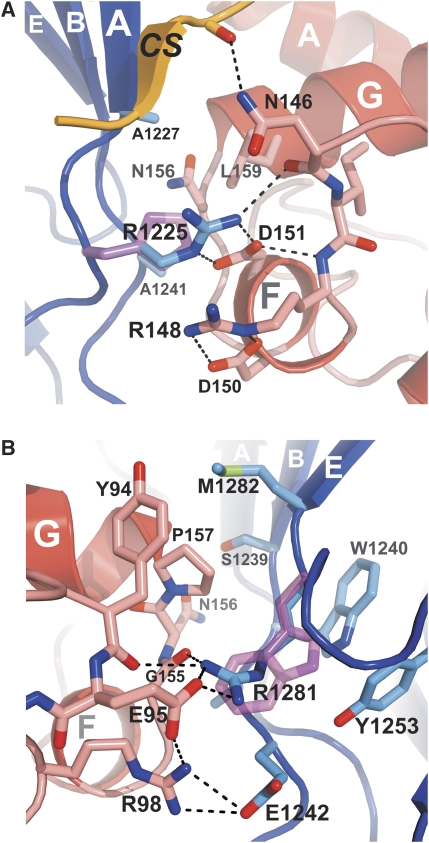
Figure 2.
Integrin β4–plectin interface. (A) Open-book view of the complex. Ribbon representation of the β4 (left) and plectin (right) structure with residues that constitute the interface shown as sticks. (B, C) Surface representation coloured by electrostatic potential (from -12 kT/e in red to 12 kT/e in blue) of β4 (B) and plectin (C) in the same orientation as in (A). Residues that form part of the interfaces are labelled on each surface. The backbone of the companion partner is partially shown as a wire, with the side chains that participate in the interaction shown as sticks and labelled with arrows.
A seven-residue long fragment extends adjacent to the β-strand A of the FnIII-2 domain of β4 and contributes to the binding interface. This fragment is not connected with any molecule in the crystal as concluded from the electron density. Yet, it is located near the C-terminus of the FnIII-2 and far from the N- or C-terminus of other molecules. Thus, this stretch was assigned to be part of the CS, which forms a short antiparallel β-strand. The CS extends the ABE β-sheet of the FnIII-2 and widens the plectin-binding interface by contacting the EF loop and the N-terminus of helix G in the CH1 of plectin.
The contribution of the FnIII-1 domain of β4 to the binding interface is much smaller than that of the FnIII-2 and the CS. The EF and CC′ loops of the FnIII-1 domain face the CE loop of the CH1 of plectin. This surface of β4 contains a hydrophobic crevice formed by I1163, Y1187 and C1190, and an acidic patch containing D1166 and E1170. The side chain of M122 in plectin is oriented toward the hydrophobic crevice of β4. Disruption of this hydrophobic area in the β4 double mutant Y1187R/C1190R inhibits binding to plectin (Litjens et al, 2005). R121 of plectin is located near the acidic CC′ loop of β4, but no electron density was observed for its side chain, suggesting that R121 does not engage in stable contacts. Replacement of the CC′ loop of the FnIII-1 domain by the equivalent loop of the FnIII-2 does not compromise the interaction with plectin (Litjens et al, 2005), which is in agreement with the lack of specific contacts between the CC′ loop of β4 and plectin. In summary, the plectin-binding interface of β4 is composed of three distinct elements: (1) the FnIII-2 domain at the centre of the interface, and additional contacts from (2) the FnIII-1 domain and (3) the CS.
Role of β4 regions to the interaction with plectin
To better understand the contribution of each of the above β4 elements to the interaction with plectin, we have determined the affinity of various β4 constructs for the ABD, using a fluorescence-based assay (Supplementary data and Supplementary Figure S1). The Kd of the interaction between the fragments used for crystallisation, β4 1126–1370 and plectin 1–293, is ∼49 μM (Table II). The affinity of the β4 fragment 1126–1322, which lacks the CS, for the ABD is approximately six-fold reduced, which is in agreement with the requirement of the 1329–1355 region for the recruitment of plectin into hemidesmosomes in vivo (Niessen et al, 1997). Electron density was observed only for a short region of the CS, which is located at the interface (see above). To more precisely define the sequence of the CS involved in the binding to the ABD of plectin, we have determined the affinity of additional β4 truncation mutants. The β4 fragment 1126–1355 has a slightly higher affinity for plectin than the 1126–1370 fragment, suggesting that residues 1356–1370 do not contribute to the interaction. A shorter β4 fragment, residues 1126–1348, has a three-fold lower affinity for the ABD than the 1126–1355 fragment. Deletion of additional regions of the CS as in the β4 fragments 1126–1343, 1126–1338 and 1126–1333 did not further reduce the affinity for the ABD. Taking together, these data reveal that the N-terminal region of the CS harbours two ABD-binding subsites that correspond to the sequences 1323PMSIPIIPDIP1333 and 1349YSDDVLR1355, respectively. It is reasonable to assume that one of these two subsites corresponds to the electron density located at the binding interface, which is located near to where the fluorescence probe is attached to a Cys engineered at position 162 of plectin. Thus, the region of the CS that occupies this position is expected to be a major determinant for the changes in the fluorescence of the AEDANS on complex formation. Although binding of the β4 fragments 1126–1370, 1126–1355, 1126–1348, 1126–1343, 1126–1338, 1126–1333 induces a similar change in the fluorescence emission spectrum of the AEDANS, as judged by the ratio between the fluorescence intensity of the fully bound and free states (Table II), the 1126–1322 fragment induces a much more moderate change in the fluorescence intensity. Thus, the first ABD-binding site of the CS, residues 1323–1333, is located at the binding surface near helix G of the CH1 of plectin. On the basis of the H-bond pattern of the polypeptide backbone, we have chosen residues 1324–1330 to be the fragment that forms a short antiparallel β-strand with the strand A of the FnIII-2 (Supplementary Figure S2A). The side chains of P1227 and I1329 face the CH1 of plectin and the carbonyl of P1227 makes an H-bond with the carboxamide group of N146 of plectin. On the other side, I1328 and P1330 pack against the FnIII-2 of β4. Simultaneous substitution of P1324A/P1227A in β4 1126–1355 does not alter its affinity for the ABD, suggesting that an Ala in position 1327 provides sufficient hydrophobic contacts to maintain the interaction. Notably, the double mutant P1330A/P1333A has the same affinity for the ABD as the wild-type protein, which is in agreement with a minimal effect of the P1330A/P1333A mutations on the interaction between β4 1115–1355 and plectin 1–339 in yeast two-hybrid experiments (Koster et al, 2004).
Table 2.
Affinity of the integrin β4–plectin interaction
| Kd (μM) | Ibound/ Ifreea | |
|---|---|---|
| Binding of integrin β4 fragments to plectin 1C (1–293) | ||
| β4 fragment | ||
| 1126–1370 (FnIII-1,2+CS) | 49±7 | 1.67±0.03 |
| 1126–1355 (FnIII-1,2+CS) | 31±4 | 1.67±0.04 |
| 1126–1348 (FnIII-1,2+CS) | 95±3 | 1.66±0.01 |
| 1126–1343 (FnIII-1,2+CS) | 108±8 | 1.57±0.01 |
| 1126–1338 (FnIII-1,2+CS) | 86±11 | 1.55±0.05 |
| 1126–1333 (FnIII-1,2+CS) | 90±3 | 1.59±0.02 |
| 1126–1322 (FnIII-1,2) | 288±92 | 1.20±0.04 |
| 1218–1355 (FnIII-2+CS) | 190±36 | 1.68±0.08 |
| 1126–1355 (FnIII-1,2+CS) P1323A/P1327A | 31±4 | 1.67±0.01 |
| 1126–1355 (FnIII-1,2+CS) P1330A/P1333A | 30±7 | 1.69±0.04 |
| 1126–1355 (FnIII-1,2+CS) R1225H | >5000b | ND |
| 1126–1355 (FnIII-1,2+CS) R1225A | >10 000b | ND |
| 1126–1355 (FnIII-1,2+CS) R1225K | 2600±170 | ND |
| 1126–1355 (FnIII-1,2+CS) R1281W | >10 000b | ND |
| 1126–1355 (FnIII-1,2+CS) R1281A | >10 000b | ND |
| 1126–1355 (FnIII-1,2+CS) R1281K | 1300±80 | ND |
| 1126–1355 (FnIII-1,2+CS) K1272C/S1345C reduced | 36±5 | 1.81±0.03 |
| 1126–1355 (FnIII-1,2+CS) K1272C/S1345C oxidized | 226±31 | 1.43±0.03 |
| Binding of plectin fragments to b4 (1126–1355) | ||
| Plectin fragment | ||
| 1C-ABD (1–293) | 31±4 | 1.67±0.04 |
| 1C(Δ1–58)-ABD (59–293) | 22±2 | 1.89±0.01 |
| 1A-ABD (1–266) | 28±5 | 1.51±0.02 |
| ABD (68–293)c | 16±4 | 2.08±0.12 |
| ND, not determined. | ||
| Data are presented as mean±standard deviation. | ||
| aRatio between the fluorescence intensities in the free and fully bound states (λex 340 nm, λem 475 nm). | ||
| bMinimal value compatible with the data. | ||
| cNumbers according to the 1C variant, correspond to 41–266 in the 1A isoform. | ||
Finally, deletion of the FnIII-1 domain of β4, as in the fragment 1218–1355, induces an approximately six-fold reduction in the affinity for the ABD compared with β4 1126–1355. The moderate contribution of the first FnIII domain to the affinity of binding is probably due to the fact that there are only few contacts with plectin. In summary, both the FnIII-1 and the CS contribute to the binding site and support the stability of the integrin β4–plectin ABD complex.
The β4-binding interface of plectin
The ABD of plectin binds to β4 through its CH1 domain, while there is no direct contact between the CH2 domain and β4. Nonetheless, the CH2 is required for binding to β4 (Geerts et al, 1999). There are extensive intramolecular contacts between the CH1 and CH2 of the ABD. Thus, it is likely that the CH2 favours binding to β4 by stabilising and offering the correct presentation of the CH1.
The β4-binding surface of the CH1 domain of plectin is centred on the FG loop, which is completely buried in the complex. Additional contacts involve the AC and CE loops, helix F and the N-terminal ends of helices E and G. The interface is characterised by a belt of charged residues (E95, R98 and D151), which establish essential intramolecular salt bridges (see above). The ABD of dystonin binds to the same region of β4 as the ABD of plectin, whereas the ABDs of dystrophin, α-actinin, utrophin and filamins A and B do not (Litjens et al, 2003). The plectin residues that form the β4-binding interface are conserved in dystonin, suggesting that the ABD of these proteins interacts with β4 in the same way.
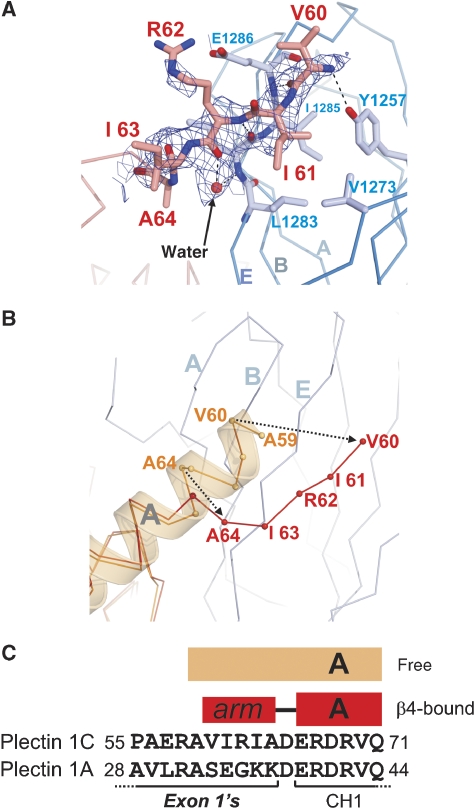
Plectin residues 60–64, which are located upstream of the ABD, form a second interaction site for β4 (Figure 3A). These residues adopt an extended conformation that projects away from the CH1 domain. Hence, it is named the N-terminal arm. The arm packs in antiparallel orientation against strand E of the FnIII-2 of β4 extending the ABE β-sheet on the opposite side from the CS. The plectin arm contacts β4 mainly through interactions between the polypeptide backbones, with the exception of I61 whose side chain makes hydrophobic contacts with V1273 and L1283. No electron density was observed for plectin residues 1–59. The affinity for β4 of the plectin fragment 59–293 is very similar to that of the 1–293 fragment (Table II). Thus, the plectin N-terminal region upstream the arm does not have a noticeable contribution to the interaction.
Figure 3.
Structure of the N-terminal arm of plectin. (A) Representation of the N-terminal arm of plectin (red) bound to the FnIII-2 of β4 (blue). An omit map (2mFobs-DFcalc, contoured at 1σ) is shown in blue; the map was calculated after simulated annealing refinement of a model in which the plectin arm was omitted. (B) Cα-trace of the N-terminal region of ABD in the absence of plectin (orange, PDB 1MB8) superimposed on the structure of the ABD (red) bound to β4 (blue). (C) Comparison of the sequences of plectin 1C and 1A upstream of the ABD. Only the initial residues of the CH1, which is common to the two isoforms, are shown. The secondary structure elements in the free (orange) and β4-bound (red) structures of the ABD are shown on top.
The PLEC-1 gene contains multiple alternative first exons that code for the N-terminal region upstream of the ABD. In humans, this results in four isoforms of which 1A and 1C are present in keratinocytes (Andra et al, 2003) and bind to β4 (Litjens et al, 2003). The complex in our crystals contains the plectin 1C isoform. The N-terminal arm of isoform 1A has no sequence similarity with that of plectin 1C (residues 60–64). Nonetheless, as the arm binds to β4 mainly through the polypeptide backbone, it is likely that the alternative region of the two isoforms interacts with β4 in a similar fashion. The plectin 1A fragment 1–266 has the same affinity for β4 as the analogous fragment of the 1C isoform (residues 1–293). Thus, the N-terminal region encoded by exon-1A does not enhance the interaction with the FnIII-1,2 domains of β4.
Notably, the isolated ABD (residues 68–293 in the 1C variant) has a slightly higher affinity for β4 than when the sequences encoded by exon-1A or exon-1C are also present in the fragment. This is in agreement with the observation that in vivo a plectin construct lacking the exon-1 encoded region is more strongly co-localised with β4 than the naturally occurring variants 1A or 1C (Litjens et al, 2003). In the structure of the free ABD (Garcia-Alvarez et al, 2003), the residues 59–64 are part of the helix A of the CH1 domain, while this region is extended in the N-terminal arm in the β4–plectin complex (Figure 3B and C). Therefore, the slight negative effect of the sequence upstream of the ABD on the binding to β4 may be due to an unfavourable energetic balance of the helix-to-strand transition of this segment, which would not be compensated by the interaction of the N-terminal arm with β4.
In summary, the selective targeting of the plectin 1A and 1C isoforms to hemidesmosomes does not rely on the interaction of the exon-1 coded regions with the primary binding site in β4. Yet, we cannot exclude that the N-terminal regions of plectin specific for the isoforms may harbour binding sites for other regions of β4 or other hemidesmosomal components.
Missense mutations of β4 linked to epidermolysis bullosa disrupt the binding interface
Two missense mutations in the ITGB4 gene that result in single amino acid substitutions in the integrin β4 subunit, R1225H and R1281W, have been detected in patients with nonlethal forms of epidermolysis bullosa with pyloric atresia (Nakano et al, 2001; Pulkkinen et al, 1998). These mutations abolish the interaction of β4 with the ABD of plectin as shown in yeast-two-hybrid assays and prevent the recruitment of plectin into hemidesmosomes in vivo (Koster et al, 2001). Both residues are located in the FnIII-2 domain and form part of the plectin-binding surface. R1225 forms an ionic pair with D151 and an H-bond with N146 in plectin. These contacts are likely to be further stabilised by the stacking of the guanidinium groups of R1225 of β4 and R148 of plectin (Figure 4A; Supplementary Figure S2B). R1281 contributes to a network of intermolecular polar contacts; its side chain forms a salt bridge with E95 and H-bonds with the carbonyl groups of G155 and Y94 of plectin. In addition, E95 of plectin establishes an intramolecular salt bridge with R98, which in turn makes an ionic pair with D1242 of β4 (Figure 4B).
Figure 4.
Structural basis of missense mutations of β4 linked to epidermolysis bullosa. (A) Close-up showing the intermolecular contacts between R1225 of β4 (blue) and plectin (red). A hypothetical conformation of the His side chain in the R1225H mutant is shown to illustrate the effect of this substitution, which prevents the interactions with N146, R148 and D151 in plectin. (B) Detailed view of the contacts established by R1281 of β4, and nearby residues (coloured as in A). A possible structure of the Trp side chain in the disease-linked mutation R1281W is shown. The imidazole ring does not maintain the H-bonds established by R1281 and most likely it causes steric clashes with W1240 of β4 and P157 of plectin.
To precisely assess the role of R1225 and R1281 in the binding to plectin and the effect of the disease-linked mutations, we have determined the affinity for the plectin ABD of the β4 fragment 1126–1355, in which R1225 was substituted by His, Ala or Lys, and R1281 by Trp, Ala or Lys (Table II). All these fragments with a single point mutation had the same far-UV circular dichroism spectrum and showed similar guanidinium hydrochloride-induced denaturation profile as the wild type (data not shown). Thus, these single amino acid changes do not alter the fold of the FnIII-2. Substitution R1225H severely reduces the affinity for plectin, increasing the Kd more than a 1000-fold. The His side chain is shorter than that of Arg, and it is unlikely that the imidazole group is protonated at neutral pH, resulting in the loss of the salt bridge with D151 in the mutant protein. As R1225 is located at the periphery of the binding interface, it is reasonable to assume that the His side chain becomes oriented in such a way that there is no steric hindrance between it and plectin. Thus, the deleterious result of the R1225H mutation appears to be mainly due to the prevention of specific contacts, rather than a distortion of the binding interface. This is supported by the similar reduction in affinity of the R1225A mutant. Furthermore, the substitution R1225K already induces a 100-fold increase in the Kd; the Lys is long enough to establish a salt bridge with D151 but not the H-bond with N146.
The substitution R1281W in β4 increases the Kd for plectin by at least 1000-fold. The affinity of the R1281A mutant is reduced to a similar level, which reveals the essential role of the intermolecular contacts made by the side chain of R1281. In addition, formation of the complex buries R1281 in such a way that the aliphatic portion of its side chain makes van der Waals contact with W1240 and Y1253 in β4 and P157 in plectin. Therefore, the R1281W mutation is likely to create additional steric hindrance at the binding site. The substitution R1281K induces an ∼50-fold increase in the Kd. This moderate reduction in affinity can be attributed to the loss of the salt bridge with E95 of plectin, while leaving the H-bonds with the carbonyls of G155 and Y94 intact.
In summary, disruption of the β4–plectin-binding interface is directly linked to the development of epidermolysis bullosa. R1225 and R1281 establish polar contacts that are essential for correct binding and constitute hot spots of the interaction. However, a major difference in the mechanism of action of these two disease-causing mutations is that the R1281W substitution is involved in steric hindrance.
Structure of free β4
In addition to the β4–plectin complexes, the asymmetric unit of the aforementioned crystal contains a β4 molecule not part of a complex, that is, free β4 (see above). The crystals were obtained using the β4 fragment 1126–1370. However, M1348 was found to be the most C-terminal residue with electron density in the unbound fragment; therefore, we refer to this β4 molecule as the ‘1126–1348 structure'. To have a more complete description of β4 in the unbound state, we have produced two additional crystals of the β4 fragments 1126–1355 and 1126–1370 in the absence of plectin and determined their structure to a resolution of 1.75 and 2.04 Å, respectively (Table I; Supplementary Data and Supplementary Figure S3). The models of these two crystal structures include the residues from 1126 to 1339, while no electron density was observed of amino acids downstream of S1339 in either structure. The structures of the two fragments are almost identical (r.m.s.d. of all Cα atoms between 0.30 and 0.45 Å), and we will describe the structure with the highest resolution, which we refer to as the ‘1126–1339 structure' to distinguish it from the structure of the free β4 molecule present in the crystal of the complex.
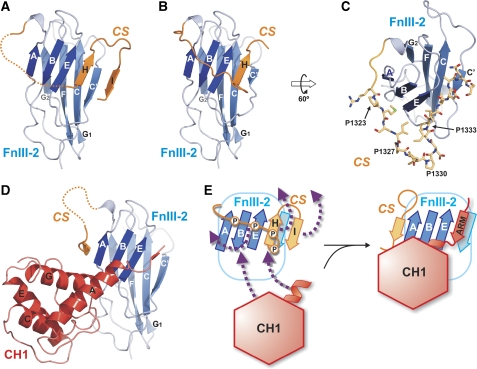
Residues 1333–1337 of the CS form an additional β-strand H that packs in a parallel fashion against strand E of FnIII-2 and enlarges the ABE sheet (Figure 5). In the structure of the 1126–1348 fragment, residues 1344–1348 form a second new β-strand I that packs with parallel H-bonding against strand C′ (Supplementary Figure S2C). In the 1126–1339 structure, the strand I is replaced by a contact in the crystal. Upstream of strand H, the CS contains a Pro-rich sequence (1323PMSIPIIPDI1332), which in the 1126–1339 structure forms a loop that extends over the ABE β-sheet and is highly exposed to the solvent. This region is stabilised by a contact in the crystal in which the 1324MSIPI1328 sequence forms a β-strand that makes antiparallel H-bonding to the strand C′ from the FnIII-2 of a neighbouring molecule in the crystal lattice. The only intramolecular contacts in the Pro-rich loop involve the side chains of M1324, I1326 and I1332, which are oriented toward a hydrophobic patch on the surface of the FnIII-2 that contains V1231 and L1284. No electron density was observed of the Pro-rich loop in the 1126–1348 structure, suggesting that despite being anchored at both ends to the FnIII-2, this loop has a large conformational mobility when there are no additional contacts.
Figure 5.
Conformational change of the connecting segment of β4 between the free and plectin-bound structures. (A) Ribbon representation of the 1126–1348 free β4 structure, with the FnIII-2 in blue and the CS in orange (the FnIII-1 has been omitted in all panels for clarity). The Pro-rich loop (residues 1324–1338), for which no electron density was observed, is shown as a dotted line. (B) Representation of the 1126–1339 free β4 structure in the same orientation and colouring as in (A). (C) Detailed view of the CS (shown as sticks, orange) in the 1126–1339 free β4 structure. P1323, P1327 and P1330 are highly exposed to the solvent, whereas P1333 contact the FnIII-2 right before the β-strand H. (D) Structure of the CS in the complex of β4 (blue) with plectin (red). The view is in the same orientation as in panels A and B, and the CH2 was omitted for clarity. (E) Schematic representation of the FnIII-2 and the CS of β4 in the free (left) and plectin-bound (right) structures. The arrows illustrate the structural reorganisation of the CS induced by plectin binding.
The Pro-rich loop has features that suggest that it may act as a docking site for protein–protein interactions. The presence of multiple H-bond donor and acceptor groups within this loop make it prone to engagement with other proteins through a β-zipper contact, such as the one observed in the 1126–1339 structure. In addition, P1327, P1330 and P1333 are in a PXXP pattern (where X is any other amino acid), which has a tendency to adopt a polyproline type II (PPII) helix. PPII helices are structural motifs recognised by multiple domains such as SH3, WW and EVH1 (Zarrinpar et al, 2003). Given the conformational variability of the Pro-rich region observed in the crystal structures, it is possible that it may adopt a PPII helical structure and serve as a protein interaction site.
The three crystal forms that contain the free β4 fragments were obtained under a wide range of conditions (see Materials and methods). Therefore, the structure of the CS observed in the absence of plectin probably represents a highly stable conformation. In summary, when β4 is not bound to plectin, the structure of the N-terminal region of its CS is organised in such a way that it makes extensive contacts with the FnIII-2, whereas the Pro-rich sequence is exposed in a highly mobile loop likely to act as a protein interaction site.
Conformational switch of β4
Comparison of the structures of free and plectin-bound β4 reveals a major conformational change of the CS between the two states; whereas there are minimal structural differences within the FnIII-1,2 domains (Figure 5D and E). Binding to plectin induces the disorganisation of strands H and I of the CS, and relocation of the Pro-rich sequence (amino acids 1323–1333) to the opposite edge of the ABE β-sheet of the FnIII-2. This conformational change of the CS is correlated with a small increase in the thermal stability of the β4 1126–1355 fragment on binding to plectin (Garcia-Alvarez et al, 2003). The surface of β4 around strand E, which engages with strand H in the free state, is occupied by the N-terminal arm of plectin in the complex. The CS is required for optimal binding, but the N-terminal arm of plectin is dispensable (see above). Therefore, rearrangement of the CS is induced by the interaction with the CH1 of plectin, for which binding of the N-terminal arm of plectin is not required.
The structural rearrangement of the CS linked to the binding to plectin suggests that the β4–plectin interaction could be regulated by a conformational control of β4. To address this issue we have created a β4 mutant, which can be locked in the conformation of the free β4 molecule. In this mutant of the 1126–1355 fragment, the four Cys occurring in the wild-type sequence were replaced either by Ala or Ser (see Material and methods). In addition, residues K1272 and S1345 were replaced by Cys. R1272C is located in the strand C′ of the FnIII-2, whereas S1345C is located in the strand I of the CS (Supplementary Figure S4). The engineered thiol groups are predicted to be adjacent in the structure of the unbound state. Thus, formation of a disulphide bond by mild oxidation locks the CS in the conformation of the free β4 molecule, whereas when reduced, the CS is able to relocate on binding to plectin. The affinity of the K1272C/S1345C mutant under reduced conditions (CS unlocked) is similar to that of the wild-type protein (Table II). Therefore, the six point mutations do not alter the interaction with plectin. On the other hand, when the K1272C/S1345C disulphide is formed (CS locked) there is a six-fold reduction in the affinity for plectin, compared with the unlocked protein. This reduction is similar to that observed after a complete ablation of the CS. In summary, the conformational switch of β4 involving the CS is necessary for binding to the ABD of plectin, suggesting that the plectin–β4 interaction may be allosterically inhibited by factors that stabilise the unbound conformation of β4.
Concluding remarks
The structure of the β4–plectin complex provides the first detailed insight in the molecular recognition mechanisms responsible for hemidesmosome assembly and stability. Moreover, a direct link between the disruption of the binding sites and the development of blistering diseases has been established. In addition, our data provide evidence of a conformational change linked to the function and subcellular localisation of α6β4 and suggests an allosteric control of this integrin.
It is likely that, before hemidesmosome assembly, the cytoplasmic moiety of β4 adopts an inactive or closed conformation held by an intramolecular interaction between the CS and the C-terminal tail (Rezniczek et al, 1998; Schaapveld et al, 1998; Koster et al, 2003). In cells that contain type I hemidesmosomes, such as those of the basal layer of the epidermis, α6β4 clusters at the basal cell surface where it binds to plectin as seen in the structure of the complex in the crystal. The β4–plectin interaction is an essential step in the assembly of hemidesmosomes, and it is required for the recruitment of BP180 and BP230 (Koster et al, 2004). Thus, plectin binding may unclasp the β4 cytoplasmic domain, leading to the exposure of binding sites for BP180 and BP230. The observed reorganisation of the CS caused by binding to the ABD provides a possible mechanism responsible for the destabilisation of the intramolecular contact with the C-terminal tail during hemidesmosome assembly.
Disruption of the β4–plectin association results in hemidesmosome disassembly (Geerts et al, 1999; Koster et al, 2001) and in the subcellular redistribution of the integrin α6β4 (Rabinovitz et al, 1999; Santoro et al, 2003). α6β4 stimulates cell migration, invasion and survival by activating signalling pathways; and these signalling functions are also coupled to the disruption of hemidesmosomes (Lipscomb and Mercurio, 2005; Wilhelmsen et al, 2006). On mobilisation of α6β4 from hemidesmosomes, the CS of β4 is likely to adopt the conformation observed in the structure of the unbound β4. How this rearrangement of the CS could be related to the signalling role of α6β4 is not known. Whether the part of the CS that becomes reorganised by binding to plectin is directly implicated in the mechanisms of α6β4 signalling has not been established. Another possibility is that the conformational change could favour the recruitment of signalling molecules by other regions of the β4 cytoplasmic domain. Finally, stabilisation of β4 in the original conformation of the unbound state (e.g. by binding of an adaptor protein, and/or posttranslational modifications) would hinder the re-association with plectin, and thus, prevent hemidesmosome assembly, keeping α6β4 available for signalling functions.
Materials and methods
Protein preparation
The cDNA of β4 and plectin constructs used were amplified by polymerase chain reaction and cloned into a modified pET15b vector (Novagen), which codes for an 8 × His-tag and a site recognised by the tobacco etch virus (TEV) protease at the N-terminus of the target protein. Amino acid substitutions were created by site directed mutagenesis using the QuikChange method (Stratagene, La Jolla, CA). Proteins were expressed in Escherichia coli strain BL21 (DE3) and were purified by affinity chromatography as described (Garcia-Alvarez et al, 2003). The His-tag was cleaved by digestion with the TEV protease and was removed by a second affinity chromatography. Proteins were finally dialysed against 20 mM Tris–HCl (pH 7.5), 150 mM NaCl.
Crystallisation and data collection
All crystals were grown at room temperature by vapour diffusion methods. Crystals of the complex of the β4 fragment 1126–1370 (Uniprot P16144-2), and the plectin 1C fragment 1–293 (Uniprot Q15149-2) were obtained by mixing equal volumes of a solution containing 500 μM of each protein and mother liquor consisting of 0.1 M Na acetate (pH 5.8), 0.2 M CaCl2, 10% (w/v) polyethylene glycol (PEG) 6000. Before freezing in liquid nitrogen, crystals were transferred into 0.1 M Na acetate (pH 5.8), 0.2 M CaCl2, 9% (w/v) PEG 6000, 25% (v/v) glycerol. Data were collected at 100 K in the beamline ID23.1 at the ESRF (Grenoble, France).
Crystals of the 1126–1355 fragment of β4 were grown by mixing a protein solution at 22 mg/ml with an equal volume of 0.1 M Tris–HCl (pH 7.5), 24% (v/v) PEG 200, 300 mM NaCl. Crystals were transferred into solutions as the mother liquor but with up to 35% (v/v) PEG 200 and were frozen in liquid nitrogen. Data were collected in house at 120 K.
Crystals of the 1126–1370 fragment of β4 were obtained by mixing equal volumes of protein at 48 mg/ml and mother liquor consisting of 0.1 M cacodylate (pH 6.5), 18.75% (v/v) PEG 600, 6.25% (v/v) glycerol, 0.7 M NaCl. Crystals were transferred to 0.1 M cacodylate (pH 6.5), 30% (v/v) PEG 600, 10% (v/v) glycerol, 0.7 M NaCl and were frozen in liquid nitrogen. Data were collected in house at 110 K. Data of all crystals were indexed with XDS and reduced with XSCALE (Kabsch, 1993).
Structure solution and refinement
Crystals of the β4 (1126–1370)–plectin (1–293) complex belong to space group P3221. The structure of the complex was solved by molecular replacement using PHASER (McCoy et al, 2005) and the structures of the ABD of plectin (PDB 1MB8) (Garcia-Alvarez et al, 2003) and the FnIII-1,2 of β4 (PDB 1QG3) (de Pereda et al, 1999) as search models. Two copies of the ABD and three β4 molecules were located. The structure was refined against data to 2.75 Å resolution using PHENIX REFINE (Afonine et al, 2005), alternated with model building with COOT (Emsley and Cowtan, 2004). At early stages, simulated annealing was carried out, whereas at later stages, restrained refinement in combination with TLS refinement (12 TLS groups) was used. Noncrystallographic symmetry (NCS) restraints were used; two-fold NCS was included for the plectin molecules, whereas three-fold NCS restraints were used for the β4 molecules with the exception of minor NCS groups, which were only conserved in the two plectin-bound β4 molecules. Distance restraints derived from the intramolecular main chain H-bonds of the high-resolution structures of β4 and plectin were included for the refinement. The refinement converged at Rwork and Rfree values of 20.4 and 25.7%, respectively (Table I). The final model contains three integrin β4 molecules, two plectin molecules, 20 solvent molecules, one calcium ion and three PEG molecules.
Crystals of the 1126–1355 fragment of β4 belong to space group P1 and they contain two molecules in the asymmetric unit (55% solvent content). The structure was solved by molecular replacement using PHASER (McCoy et al, 2005) and the structure of the FnIII-1,2 domains of β4, residues 1126–1320 (PDB 1QG3) (de Pereda et al, 1999), as search model. Refinement was done with PHENIX REFINE (Afonine et al, 2005) against data to 1.75 Å. After initial refinement by simulated annealing, additional density in 2Fo-Fc and Fo-Fc maps was readily assigned to residues downstream of 1320, not present in the search model. Manual model building with COOT (Emsley and Cowtan, 2004) was alternated with restrained refinement combined with the refinement of six TLS groups in each molecule. Two peaks in the anomalous difference maps located near residues 1139–1142 of each molecule were modelled as chloride ions. The final model contains residues 1126–1339 of each molecule, 502 waters, seven molecules of PEG fragments and two chloride ions (Table I).
Crystals of the 1126–1370 fragment of β4 belong to space group C2 and they contain a single β4 molecule in the asymmetric unit (56% solvent content.) The structure was solved by molecular replacement using PHASER (McCoy et al, 2005) and the structure of the β4 (1126–1355) protein as search model. The structure was refined against data to 2.04 Å resolution in a similar way as for the 1126–1355 fragment. The final structure contains residues 1126–1339, 142 waters, three PEG fragments and one chloride ion (Table I).
Fluorescence-based binding assay
Plectin fragments carrying the G162C substitution were labelled with 1,5-I-AEDANS as described (Haran et al, 1992). The labelling ratio was 0.95±0.03 molecules of AEDANS per molecule of plectin (average±standard deviation of four-labelling reactions). AEDANS-labelled plectin proteins at 4 μM in 20 mM Tris–HCl (pH 7.5), 150 mM NaCl were titrated with concentrated solutions of β4 proteins. The fluorescence of the AEDANS was measured with a FluoroMax-3 spectrofluorometer (HORIBA-Jobin-Yvon) at 25°C, exciting at 340 nm (0.5 nm bandwidth) and collecting the emission at 475 nm (5 nm bandwidth). The apparent dissociation Kd and the fluorescence intensity of the free (IF) and fully bound (IB) states were obtained by fitting a 1:1 binding model. A detailed description of the binding assay and its analysis is included in the Supplementary Data.
Design of the β4 mutant protein lockable in the free conformation
A β4 1126–1355 fragment carrying the substitutions C1190A, C1197A, C1213S, C1256S, K1272C and S1345C was expressed and purified as for the wild-type proteins. The disulphide bond between K1272C and S1345C was formed by incubation at room temperature for 5 h with 0.1 mM CuSO4, 0.3 mM 1,10-phenanthrolin in 20 mM Tris–HCl (pH 8.0), 150 mM NaCl. Alternatively, formation of the disulphide bond was prevented by keeping the protein in the presence of 0.2 mM DTT.
Accession numbers
The atomic coordinates and structure factors of the structure of the β4–plectin complex, the β4 1126–1355 and the β4 1126–1370 have been deposited in the Protein Data Bank under the codes of 3F7P, 3F7Q and 3F7R, respectively.
Supplementary Material
Supplementary Information
Acknowledgments
We thank German Rivas and Anastassis Perrakis for critical comments. We acknowledge the European Synchrotron Radiation Facility for provision of synchrotron radiation facilities, and we thank Stéphanie Monaco for assistance at beamline ID23-1. We thank Guillermo Montoya (Centro Nacional de Investigaciones Oncológicas), Juan A Hermoso, José M Mancheño (Instituto de Química Física Rocasolano) and Antonio Romero (Centro de Investigaciones Biológicas) for generous access to X-ray equipment and assistance during data collection. This work was supported by the Spanish Ministry of Science and Innovation and the European Regional Development Fund (grants SAF2003-02509 and BFU2006-01929/BMC to JMdP, and BFU2006-03905/BMC to MPL).
References
- Afonine PV, Grosse-Kunstleve RW, Adams PD (2005) The Phenix refinement framework. CCP4 Newsl 42, contribution 8. [Google Scholar]
- Andra K, Kornacker I, Jorgl A, Zorer M, Spazierer D, Fuchs P, Fischer I, Wiche G (2003) Plectin-isoform-specific rescue of hemidesmosomal defects in plectin (−/−) keratinocytes. J Invest Dermatol 120: 189–197 [DOI] [PubMed] [Google Scholar]
- de Pereda JM, Wiche G, Liddington RC (1999) Crystal structure of a tandem pair of fibronectin type III domains from the cytoplasmic tail of integrin alpha6beta4. EMBO J 18: 4087–4095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delano WL (2002) The PyMOL Molecular Graphics System. Palo Alto, CA, USA: DeLano Scientific [Google Scholar]
- Emsley P, Cowtan K (2004) Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr 60: 2126–2132 [DOI] [PubMed] [Google Scholar]
- Garcia-Alvarez B, Bobkov A, Sonnenberg A, de Pereda JM (2003) Structural and functional analysis of the actin binding domain of plectin suggests alternative mechanisms for binding to F-actin and to integrin a6b4. Structure 11: 615–625 [DOI] [PubMed] [Google Scholar]
- Geerts D, Fontao L, Nievers MG, Schaapveld RQ, Purkis PE, Wheeler GN, Lane EB, Leigh IM, Sonnenberg A (1999) Binding of integrin alpha6beta4 to plectin prevents plectin association with F-actin but does not interfere with intermediate filament binding. J Cell Biol 147: 417–434 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haran G, Haas E, Szpikowska BK, Mas MT (1992) Domain motions in phosphoglycerate kinase: determination of interdomain distance distributions by site-specific labeling and time-resolved fluorescence energy transfer. Proc Natl Acad Sci USA 89: 11764–11768 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones JC, Hopkinson SB, Goldfinger LE (1998) Structure and assembly of hemidesmosomes. Bioessays 20: 488–494 [DOI] [PubMed] [Google Scholar]
- Kabsch W (1993) Automatic processing of rotation diffraction data from crystals of initially unknown symmetry and cell constants. J Appl Cryst 26: 795–800 [Google Scholar]
- Koster J, Geerts D, Favre B, Borradori L, Sonnenberg A (2003) Analysis of the interactions between BP180, BP230, plectin and the integrin alpha6beta4 important for hemidesmosome assembly. J Cell Sci 116: 387–399 [DOI] [PubMed] [Google Scholar]
- Koster J, Kuikman I, Kreft M, Sonnenberg A (2001) Two different mutations in the cytoplasmic domain of the integrin beta 4 subunit in nonlethal forms of epidermolysis bullosa prevent interaction of beta 4 with plectin. J Invest Dermatol 117: 1405–1411 [DOI] [PubMed] [Google Scholar]
- Koster J, van Wilpe S, Kuikman I, Litjens SH, Sonnenberg A (2004) Role of binding of plectin to the integrin beta4 subunit in the assembly of hemidesmosomes. Mol Biol Cell 15: 1211–1223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lipscomb EA, Mercurio AM (2005) Mobilization and activation of a signaling competent alpha6beta4integrin underlies its contribution to carcinoma progression. Cancer Metastasis Rev 24: 413–423 [DOI] [PubMed] [Google Scholar]
- Litjens SH, Koster J, Kuikman I, van Wilpe S, de Pereda JM, Sonnenberg A (2003) Specificity of binding of the plectin actin-binding domain to beta4 integrin. Mol Biol Cell 14: 4039–4050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Litjens SH, Wilhelmsen K, de Pereda JM, Perrakis A, Sonnenberg A (2005) Modeling and experimental validation of the binary complex of the plectin actin-binding domain and the first pair of fibronectin type III (FNIII) domains of the beta4 integrin. J Biol Chem 280: 22270–22277 [DOI] [PubMed] [Google Scholar]
- McCoy AJ, Grosse-Kunstleve RW, Storoni LC, Read RJ (2005) Likelihood-enhanced fast translation functions. Acta Crystallogr D Biol Crystallogr 61: 458–464 [DOI] [PubMed] [Google Scholar]
- Nakano A, Pulkkinen L, Murrell D, Rico J, Lucky AW, Garzon M, Stevens CA, Robertson S, Pfendner E, Uitto J (2001) Epidermolysis bullosa with congenital pyloric atresia: novel mutations in the beta 4 integrin gene (ITGB4) and genotype/phenotype correlations. Pediatr Res 49: 618–626 [DOI] [PubMed] [Google Scholar]
- Niessen CM, Hulsman EH, Oomen LC, Kuikman I, Sonnenberg A (1997) A minimal region on the integrin beta4 subunit that is critical to its localization in hemidesmosomes regulates the distribution of HD1/plectin in COS-7 cells. J Cell Sci 110: 1705–1716 [DOI] [PubMed] [Google Scholar]
- Nievers MG, Schaapveld RQ, Sonnenberg A (1999) Biology and function of hemidesmosomes. Matrix Biol 18: 5–17 [DOI] [PubMed] [Google Scholar]
- Pulkkinen L, Rouan F, Bruckner-Tuderman L, Wallerstein R, Garzon M, Brown T, Smith L, Carter W, Uitto J (1998) Novel ITGB4 mutations in lethal and nonlethal variants of epidermolysis bullosa with pyloric atresia: missense versus nonsense. Am J Hum Genet 63: 1376–1387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rabinovitz I, Toker A, Mercurio AM (1999) Protein kinase C-dependent mobilization of the alpha6beta4 integrin from hemidesmosomes and its association with actin-rich cell protrusions drive the chemotactic migration of carcinoma cells. J Cell Biol 146: 1147–1160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rezniczek GA, de Pereda JM, Reipert S, Wiche G (1998) Linking integrin alpha6beta4-based cell adhesion to the intermediate filament cytoskeleton: direct interaction between the beta4 subunit and plectin at multiple molecular sites. J Cell Biol 141: 209–225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santoro MM, Gaudino G, Marchisio PC (2003) The MSP receptor regulates alpha6beta4 and alpha3beta1 integrins via 14-3-3 proteins in keratinocyte migration. Dev Cell 5: 257–271 [DOI] [PubMed] [Google Scholar]
- Schaapveld RQ, Borradori L, Geerts D, van Leusden MR, Kuikman I, Nievers MG, Niessen CM, Steenbergen RD, Snijders PJ, Sonnenberg A (1998) Hemidesmosome formation is initiated by the beta4 integrin subunit, requires complex formation of beta4 and HD1/plectin, and involves a direct interaction between beta4 and the bullous pemphigoid antigen 180. J Cell Biol 142: 271–284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sevcik J, Urbanikova L, Kost'an J, Janda L, Wiche G (2004) Actin-binding domain of mouse plectin. Crystal structure and binding to vimentin. Eur J Biochem 271: 1873–1884 [DOI] [PubMed] [Google Scholar]
- Sonnenberg A, Rojas AM, de Pereda JM (2007) The structure of a tandem pair of spectrin repeats of plectin reveals a modular organization of the plakin domain. J Mol Biol 368: 1379–1391 [DOI] [PubMed] [Google Scholar]
- Tsuruta D, Kobayashi H, Imanishi H, Sugawara K, Ishii M, Jones JC (2008) Laminin-332-integrin interaction: a target for cancer therapy? Curr Med Chem 15: 1968–1975 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uematsu J, Nishizawa Y, Sonnenberg A, Owaribe K (1994) Demonstration of type II hemidesmosomes in a mammary gland epithelial cell line, BMGE-H. J Biochem 115: 469–476 [DOI] [PubMed] [Google Scholar]
- Wilhelmsen K, Litjens SH, Sonnenberg A (2006) Multiple functions of the integrin alpha6beta4 in epidermal homeostasis and tumorigenesis. Mol Cell Biol 26: 2877–2886 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zarrinpar A, Bhattacharyya RP, Lim WA (2003) The structure and function of proline recognition domains. Sci STKE 2003: RE8. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Information