Abstract
Proper regulation of NF-κB activity is critical to maintain and balance the inflammatory response. Inactivation of the NF-κB complex relies in part on the proteasome-mediated degradation of promoter-bound NF-κB, but the detailed molecular mechanism initiating this process remains elusive. Here, we show that the methylation of the RelA subunit of NF-κB has an important function in this process. Lysine methyltransferase Set9 physically associates with RelA in vitro and in vivo in response to TNF-α stimulation. Mutational and mass spectrometric analyses reveal that RelA is monomethylated by Set9 at lysine residues 314 and 315 in vitro and in vivo. Methylation of RelA inhibits NF-κB action by inducing the proteasome-mediated degradation of promoter-associated RelA. Depletion of Set9 by siRNA or mutation of the RelA methylation sites prolongs DNA binding of NF-κB and enhances TNF-α-induced expression of NF-κB target genes. Together, these findings unveil a novel mechanism by which methylation of RelA dictates the turnover of NF-κB and controls the NF-κB-mediated inflammatory response.
Keywords: degradation, methylation, RelA, Set9
Introduction
The transcription factor NF-κB has an important function in regulating immune and inflammatory responses, apoptosis, cell proliferation and differentiation and tumorigenesis (Baldwin, 1996; Ghosh et al, 1998). The prototypical NF-κB complex, consisting of a heterodimer of p50 and RelA, is sequestered in the cytoplasm by its association with its inhibitor IκBα (Baldwin, 1996; Ghosh et al, 1998; Karin, 1999). NF-κB is activated by a variety of stimuli, including various proinflammatory cytokines, T-cell receptor signals and viral and bacterial products. Stimulation by these agonists leads to the activation of the IκB kinase complex (IKK), which then phosphorylates IκBα, triggering its rapid ubiquitination and proteolytic degradation in the 26S proteasome (Karin and Ben-Neriah, 2000). The liberated NF-κB heterodimer rapidly translocates to the nucleus, in which it engages its cognate κB enhancer and stimulates gene expression through the transcriptional activation domain of RelA. NF-κB activates a variety of genes involved in different biological processes including inflammation, proliferation and cell survival (Ghosh and Karin, 2002).
Because of its pleiotropic effect and its important role in the inflammatory response, activation of NF-κB is tightly controlled at multiple levels. Activated NF-κB needs to be terminated after induction to limit the inflammation, as sustained NF-κB activity and inflammation lead to a variety of diseases including asthma, arthritis and septic shock (Ghosh and Karin, 2002). In fact, constitutively active NF-κB is frequently encountered in a wide variety of tumours and in some rheumatic conditions such as rheumatoid arthritis and systemic lupus erythematosus (Sethi et al, 2008). Cells use multiple mechanisms for the termination of NF-κB, including NF-κB-dependent resynthesis of IκBα and the deubiquitination of upstream signalling molecules by deubiquitinating enzymes A20 and CYLD (Hayden and Ghosh, 2008; Sun, 2008). A20 and CYLD are also target genes of NF-κB, which together with resynthesized IκBα form a negative feedback regulation of NF-κB (Hayden and Ghosh, 2008). In addition, recent studies also indicate that proteasome-mediated degradation of nuclear DNA-bound NF-κB provides another layer of termination independent of negative feedback regulation (Saccani et al, 2004; Natoli and Chiocca, 2008). These different levels of regulation define the tight control of NF-κB action and the inflammatory response.
The highly controlled degradation of proteins through the proteasome-dependent pathway represents a key mechanism for the regulation of a variety of signalling molecules, including transcription factors such as p53 and c-Jun (Muratani and Tansey, 2003; Dornan et al, 2004; Wertz et al, 2004). Termination of NF-κB also requires the polyubiquitination and degradation of the nuclear NF-κB (Natoli and Chiocca, 2008). Saccani et al reported that NF-κB response was terminated even in the absence of the inhibitor IκBα, and the inhibition of the proteasome function in this condition increased the expression of NF-κB target genes. These findings suggest that nuclear degradation of NF-κB contributes to the termination of NF-κB activity and prevents excessive induction of genes involved in inflammation (Saccani et al, 2004; Natoli and Chiocca, 2008). Once NF-κB binds to target gene promoters and activates gene transcription, a significant amount of RelA, estimated to be 25%, is ubiquitinated and degraded by the 26S proteasome in the nucleus (Saccani et al, 2004). In support of this, several E3 ligases, including SOCS1 and PDLIM2, have been identified to mediate the ubiquitination of NF-κB (Ryo et al, 2003; Maine et al, 2007; Tanaka et al, 2007). However, what signal triggers the ubiquitination and degradation of NF-κB is not clear.
Set9 (also known as Set7)-mediated lysine methylation has emerged recently as a key posttranslational modification that regulates the function of histone and non-histone proteins (Wang et al, 2001; Nishioka et al, 2002; Chuikov et al, 2004; Kouskouti et al, 2004; Huang et al, 2006; Huang and Berger, 2008). Set9 was originally identified as a methyltransferase that methylates lysine 4 of histone H3 (Wang et al, 2001; Nishioka et al, 2002); more recent studies indicate that Set9 is a methyltransferase preferentially for non-histone proteins, as studies from Set9 knock-out cells indicate no changes at the level of lysine methylation on histones (Ivanov et al, 2007; Kurash et al, 2008). In this regard, more non-histone proteins, especially transcription factors, have been identified as targets for Set9. p53 is the first transcription factor that was identified to be regulated by Set9-mediated methylation (Chuikov et al, 2004). Methylation of p53 by Set9 at lysine 372 stabilizes p53 and enhances its transcriptional activity, probably by enhancing the acetylation of p53, a modification that competes for the same lysine responsible for ubiquitination and subsequent degradation of p53 (Chuikov et al, 2004; Huang et al, 2006; Ivanov et al, 2007; Kurash et al, 2008).
In an effort to investigate the potential regulation of NF-κB signalling by lysine methylation, we find that the RelA subunit of NF-κB is monomethylated by Set9 at lysines 314 and 315. Methylation of RelA triggers the degradation of the activated form of the NF-κB complex and down-regulates NF-κB target gene expression.
Results
RelA is methylated by Set9 in vitro and in vivo in response to TNF-α stimulation
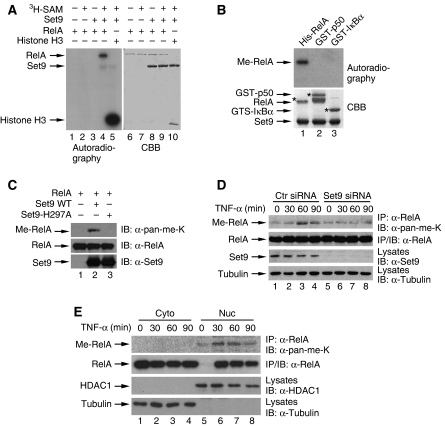
To investigate whether NF-κB is subjected to lysine methylation and the potential functional consequence of this modification, we first examined whether NF-κB could be a potential target of Set9 (Wang et al, 2001; Nishioka et al, 2002). In an in vitro methylation assay, when recombinant RelA was incubated with recombinant Set9 in the presence of 3H-S-adenosine-methionine (SAM), we found that recombinant RelA was methylated (Figure 1A, lane 4). Without either SAM or Set9, RelA was not methylated (Figure 1A, lanes 2 and 3), indicating that RelA is indeed methylated by Set9. As a positive control, histone H3 was also methylated by Set9 under the same condition as RelA, validating the in vitro methylation assay (Figure 1A, lane 5). However, when p50 and IκBα, two other key components in the NF-κB signalling pathway, were subjected to Set9-mediated methylation, no methylation was observed (Figure 1B).
Figure 1.
Set9 methylates RelA in vitro and in vivo. (A) Set9 methylates RelA in vitro. In vitro methylation was performed with 3H-S-adenosine-methionine (3H-SAM), recombinant Set9 and RelA or Histone H3 as indicated. Autoradiography and Coommassie Brilliant Blue staining (CBB) were used to show methylation and protein levels, respectively. (B) Set9 does not methylate p50 or IκBα. In vitro methylation of recombinant RelA, p50 or IκBα by Set9 were performed and detected as described in (A). (C) Enzymatic activity of Set9 is required for the methylation of RelA. In vitro methylation with RelA and recombinant wild-type (WT) Set9 or enzymatically inactive Set9-H297A mutant (Mut) of Set9 was performed as described in (A). Immunoblot (IB) analysis with anti-pan-methyl-lysine antibodies (α-pan-me-K) was used to detect the methylation of RelA. (D) TNF-α stimulates the methylation of RelA in vivo. U2OS cells were transfected with control or Set9 siRNA and stimulated with TNF-α as indicated. Immunoblot analysis of RelA immunoprecipitates from whole cell lysates was conducted with α-pan-me-K to detect methylation of RelA. (E) Only nuclear RelA is methylated upon TNF-α stimulation. U2OS cells were stimulated with TNF-α as indicated and fractionated. Methylation of RelA immunoprecipitates from cytoplasmic or nuclear extracts were assessed as described in (D).
An anti-pan-methyl-lysine antibody, which has been successfully used to detect the methylation of TAF10 and histone H3 (Su et al, 2003; Kouskouti et al, 2004), was able to detect the in vitro methylated RelA (Figure 1C). Methylation of RelA was recognized by this antibody when RelA was methylated by wild-type (WT) Set9 (Figure 1C, lane 2). However, when an enzymatically inactive form of Set9 (Set9-H297A) (Wang et al, 2001; Nishioka et al, 2002) was used for the same assay, no signal was detected, further confirming the methylation of RelA by Set9 (Figure 1C, lane 3).
We next examined whether endogenous RelA could be methylated in response to stimulation. Compared with unstimulated cells, TNF-α treatment of osteosarcoma U2OS cells led to increased methylation of RelA (Figure 1D). The methylated RelA appeared at 30 min and reached its maximal level around 60 min, then decreased at 90 min (Figure 1D, lanes 2–4), indicating a dynamic methylation status of RelA upon TNF-α stimulation. The signal recognized by the anti-pan-methyl-lysine antibodies in unstimulated cells probably represents a non-specific cross-reaction of the immunoprecipitates with the antibodies (Figure 1D, lane 1). To further examine whether TNF-α-induced methylation of RelA was mediated by Set9, we knocked down the expression of Set9 by siRNA. Depletion of Set9 by siRNA abolished TNF-α-induced methylation of RelA (Figure 1D). Fractionation experiments further demonstrate that only nuclear RelA undergoes signal-dependent methylation (Figure 1E). These results suggest that the activation-dependent RelA methylation requires endogenous Set9 and that nuclear RelA is likely an endogenous substrate for Set9.
RelA is methylated at lysines 314 and 315 in vitro and in vivo
Given that RelA is methylated in vitro and in vivo by Set9, we next aimed to identify the methylation site(s) of RelA. We purified bacterially expressed GST fusion proteins containing five different segments of RelA (Figure 2A) and used them as substrates for the in vitro methylation assay. Of these five fragments, only GST–RelA (271–364) was methylated by Set9 (Figure 2B, lane 5). There are six lysine residues on this fragment 271–364, so we further divided it into two different fragments: RelA (271–313) and RelA (311–347), with each fragment containing three lysine residues (Figure 2A). An in vitro methylation assay with these two fragments showed that the methylated lysine(s) were within the fragment RelA (311–347) (Figure 2C). To map the methylation site(s) and status, the in vitro methylated RelA (311–347) fragment was subjected to mass spectrometry analysis. Three different types of methylation were detected: single lysine monomethylation either at lysine 314 (K314me1, 30%) or lysine 315 (K315me1, 25%) or double lysine monomethylation at both lysines 314 and 315 (K314me1/K315me1, 35%) (Figure 2D). These data are consistent with the SET domain structure of Set9, which suggests that Set9 only monomethylates its substrate (Wilson et al, 2002; Kwon et al, 2003; Xiao et al, 2003).
Figure 2.
Set9 monomethylates RelA at lysines 314 and 315. (A) Schematic depiction of WT and deletion mutants of RelA. (B, C) In vitro methylation assay of deletion mutants of RelA shown in (A). (D) Mass spectrometry analysis of RelA fragment (a.a. 311–347) methylated by Set9. Top left: partial Fourier-Transform mass spectrum and monoisotopic masses. Both experimental (Exp) and theoretical (The) masses are shown. Bottom: fragmentation map depicting methylated lysines (circled) in all ECD fragment ions. Top right: percentage of fragments with different methylation states. (E) In vitro methylation of WT or point mutants of GST–RelA –(271–364) fragment. (F) In vitro methylation assay of full-length WT RelA and RelA-K314/315R mutant. (G) TNF-α stimulates the methylation of RelA at lysines 314 and 315. RelA or RelA-K314/315R reconstituted MEFs were stimulated with or without TNF-α (20 ng/ml), methylation of RelA was assessed as described in Figure 1D. ns represents a non-specific band.
To further confirm the methylation sites, we mutated lysines 314 and 315, individually or in combination, to arginine in fragment RelA (271–364), and compared the methylation of these mutants with WT RelA. Mutation of either lysine 314 or 315 impaired the methylation of RelA (Figure 2E, lanes 2 and 3). However, when both lysines were mutated, the methylation was completely abolished (Figure 2E, lane 4), Conversely, mutation of other lysines within this fragment, lysines 301, 303, 310 or 343, barely affected the methylation level (Figure 2E, lanes 5–8), confirming that lysines 314 and 315 but not the other lysines are methylated by Set9 in vitro. Further supporting this notion, mutation of both lysines 314 and 315 to arginine in full-length RelA (designated as RelA-K314/315R) also abolished the methylation of RelA (Figure 2F). The abolished methylation of RelA-K314/315R derives directly from the mutated lysines rather than from the possible binding defect of this mutant to Set9, as RelA-K314/315R binds to Set9 at a similar level as the WT RelA (Supplementary Figure S1).
To further investigate whether lysines 314 and 315 are methylated in vivo in response to stimuli, we reconstituted RelA-deficient mouse embryonic fibroblasts (MEFs) with WT RelA and RelA-K314/315R and examined the TNF-α-induced methylation of RelA. In the reconstituted MEFs, RelA and RelA-K314/315R express at a similar level as the endogenous RelA in WT MEFs (data not shown). In WT RelA reconstituted MEFs, TNF-α stimulation induced the methylation of RelA (Figure 2G, lane 2). However, TNF-α-induced methylation of RelA was abolished in RelA-K314/315R reconstituted MEFs (Figure 2G, lane 4). The background signal, which was also detected in RelA-deficient MEFs (data not shown), probably represents a non-specific protein captured in the co-immunoprecipitation (Figure 2G). All together, these results show that Set9 methylates RelA at the two lysine residues 314 and 315 in vivo in response to TNF-α. Hereafter, we refer to RelA-K314/315R mutant as the methylation-deficient mutant of RelA.
Set9 associates with RelA in vitro and in vivo
We next assessed the physical interaction between RelA and Set9 by in vivo co-immunoprecipitation assay. Immunoprecipitation of Flag-Set9 from transfected HEK293T cells co-purified T7-RelA (Figure 3A), indicating a physical interaction between RelA and Set9 in vivo. Reversed immunoprecipitation of T7-RelA also shows such an interaction (Figure 3B). In an in vitro GST pull-down assay, GST–Set9 pulled down a significant amount of RelA (Figure 3C), showing a direct interaction between Set9 and RelA. We next investigated whether there was a physical association between endogenous RelA and Set9. When HEK293T cells were stimulated with TNF-α, and endogenous Set9 was immunoprecipitated and examined for the associated RelA, we found that TNF-α stimulated the interaction between RelA and Set9 (Figure 3D), although levels of association varied at different time points. These data suggest that Set9 associates with RelA and induces its methylation in response to TNF-α stimulation.
Figure 3.
Set9 associates with RelA in vitro and in vivo. (A, B) Set9 interacts with RelA in vivo. HEK293T cells were transfected with T7-tagged RelA and Flag-tagged Set9 as indicated. At 40 h after transfection, T7-RelA or Flag-Set9 immunoprecipitates were prepared from whole-cell lysates and immunoblotted for Flag-Set9 or T7-RelA, respectively. (C) Set9 interacts with RelA in vitro. GST or GST–Set9 was incubated with recombinant RelA and precipitated with glutathione beads. The recovered materials were immunoblotted for RelA. (D) TNF-α stimulates the interaction between endogenous RelA and Set9. HEK293T cells were left untreated or stimulated with TNF-α (20 ng/ml) for indicated time periods. Set9 immunoprecipitates were prepared from whole-cell lysates and immunoblotted for RelA.
Set9-mediated methylation of RelA negatively regulates the transcriptional activation of NF-κB
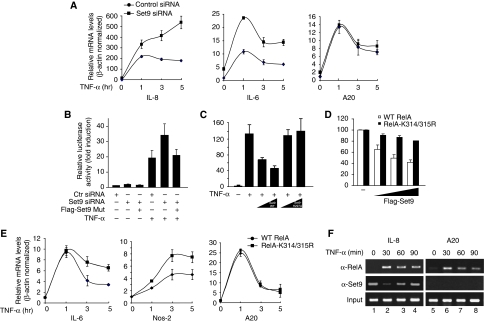
We next investigated the potential effects of Set9 on the RelA-dependent transcriptional activation of NF-κB. We used siRNA to knock-down the expression of Set9 in U2OS cells and evaluated TNF-α-induced activation of NF-κB target genes by quantitative real-time PCR. Depletion of Set9 (Supplementary Figure S2) enhanced TNF-α-induced transcription of both IL-8 and IL-6 genes compared with cells treated with control siRNA (Figure 4A, left and middle panels). Interestingly, depletion of Set9 barely affected TNF-α-induced expression of another NF-κB target gene, A20 (Figure 4A, right panel), indicating that Set9 is involved in the down-regulation of a subset of NF-κB targets and that the negative regulation of NF-κB by Set9 might be promoter specific. Similar results were obtained when Set9 was down-regulated in lung epithelial A549 cells using another siRNA targeting a different region of Set9 (Supplementary Figure S3), excluding a possible off-target effect of the siRNA in U2OS cells. Collectively, these results indicate that Set9 negatively regulates the expression of a subset of NF-κB target genes in different cell types.
Figure 4.
Set9 negatively regulates NF-κB activity. (A) Depletion of Set9 up-regulates transcription of a subset of NF-κB target genes. U2OS cells were transfected with control siRNA or Set9 siRNA. At 60 h after transfection, cells were left untreated or stimulated with TNF-α (20 ng/ml) for 1, 3 or 5 h. Total RNAs were isolated and relative levels of mRNA for IL-8, IL-6 and A20 were measured by quantitative real-time PCR. (B) Depletion of Set9 enhances TNF-α-induced activation of NF-κB. HEK293T cells were transfected with control siRNA or Set9 siRNA. After 36 h, cells were re-transfected with 5XκB luciferase reporter and Set9 siRNA resistant mutant and stimulated with TNF-α (2 ng/ml) for 5 h. Luciferase activity was measured and expressed as fold induction after normalization with Renilla luciferase. (C) Set9 inhibits TNF-α-induced activation of NF-κB. HEK293T cells were transfected with 5XκB luciferase reporter plasmid DNA and increasing amounts of plasmid DNA encoding WT Set9 or Set9-H279A. TNF-α-stimulated luciferase activity was measured as described in (B). (D) RelA-K314/315R displays more resistance than RelA WT to Set9-induced inhibition of RelA-mediated NF-κB activation. HEK293T cells were transfected with the expression vectors for RelA WT or RelA-K314/315R. After 24 h, cells were re-transfected with κB luciferase reporter plasmids together with increasing amounts of Set9. Luciferase activity was measured and reported as a percentage. (E) RelA-K314/315R displays enhanced and prolonged TNF-α-induced expression of NF-κB target genes in cells. RelA or RelA-K314/315R reconstituted MEFs were treated with TNF-α as indicated and the expression of IL-6, Nos-2 and A20 genes was assessed by real-time PCR as described in (A). (F) Set9 is differentially recruited to promoters of NF-κB target genes. U2OS cells were left untreated or stimulated with TNF-α (20 ng/ml) for 30, 60 or 90 min. ChIP assays were used to assess the binding of RelA and Set9 to the promoters of NF-κB targets IL-8 and A20. A representative PCR result from three independent experiments is shown.
To show that the enhanced expression of NF-κB target genes is due to the specific depletion of Set9 by the siRNA but not to a non-specific effect of the siRNA, we generated an siRNA resistant Set9 construct and examined its ability to rescue the effect of Set9 depletion. Consistent with the real-time PCR data (Figure 4A), depletion of Set9 by siRNA enhanced TNF-α-induced activation of NF-κB in a κB-luciferase assay (Figure 4B). However, when the siRNA resistant Set9 was reintroduced into the knock-down cells at a similar level to the endogenous Set9 (Supplementary Figure S4), TNF-α-induced NF-κB activation was reduced to a level comparable to that without siRNA treatment (Figure 4B). These data further support the conclusion that specific depletion of Set9 enhances the TNF-α-induced activation of NF-κB and that Set9-mediated methylation of RelA negatively regulates the transcriptional activation of NF-κB. Additionally, when Set9 was exogenously expressed in cells, TNF-α-induced NF-κB activity was reduced in a dose-dependent manner (Figure 4C). This Set9-mediated inhibition depends on the enzymatic activity of Set9, as Set9-H297A failed to inhibit the κB-luciferase activity (Figure 4C). Similar to the TNF-α-activated NF-κB, WT Set9, but not Set9-H297A, also inhibited RelA-mediated activation of κB-luciferase activity (Supplementary Figure S5). These data are consistent with the Set9 knock-down experiment, indicating that Set9 functions as a negative regulator for TNF-α-mediated activation of NF-κB.
To test whether the inhibitory effect of Set9 derives from the methylation of RelA at lysines 314 and 315, we examined the inhibitory effect of Set9 on WT RelA and RelA-K314/315R in a κB-luciferase assay. Transcriptional activation of WT RelA was significantly inhibited by Set9 (Figure 4D). However, transcriptional activity of RelA-K314/315R was only slightly affected by Set9 (Figure 4D). Furthermore, when we examined the TNF-α-induced expression of NF-κB target genes in MEFs reconstituted with WT RelA or RelA-K314/315R, we found that TNF-α-induced expression of IL-6 and Nos-2 was enhanced and prolonged in RelA-K314/315R reconstituted MEFs compared with the expression of the same genes in WT RelA reconstituted MEFs. In contrast, A20, whose expression was not regulated by Set9 (Figure 4A; Supplementary Figure S3), showed no significant expression difference between reconstituted WT and RelA-K314/315R MEFs. Collectively, these data, together with the real-time PCR results from Set9 knock-down cells (Figure 4A; Supplementary Figure S3), strongly indicate that methylation of RelA at lysines 314 and 315 by Set9 negatively regulates the expression of a subset of NF-κB target genes in response to TNF-α.
Set9 is differentially recruited to the promoters of NF-κB target genes
Having identified that Set9 negatively regulates the activation of NF-κB in a promoter-specific manner, we next used chromatin immunoprecipitation (ChIP) assays to examine the recruitment of Set9 to the promoters of NF-κB target genes. We stimulated U2OS cells with TNF-α for various time points and examined the binding of RelA and Set9 to the promoters of IL-8 and A20, whose expression is differentially regulated by Set9 (Figure 4A). As expected, RelA was recruited to the promoters of IL-8 after TNF-α stimulation. Interestingly, Set9 was found to reside on the promoter even before the stimulation (Figure 4F, left panel). After stimulation, promoter-bound Set9 decreased rapidly (∼30 min) and then increased gradually (Figure 4F), representing a dynamic binding of Set9 on the promoter in response to TNF-α stimulation. However, for the promoter of A20, Set9 was not recruited to the promoter before or after TNF-α stimulation even though RelA was still recruited to the promoter after TNF-α stimulation (Figure 4F, right panel). These ChIP data suggest that Set9 selectively occupies the promoters of NF-κB target genes and that preclearance of Set9 from the occupied promoters might be important for the initial recruitment and activation of NF-κB.
Methylation of RelA by Set9 down-regulates the stability of RelA
A role for lysine methylation in regulating the stability of transcription factors, including p53 and ERα, was reported recently (Chuikov et al, 2004; Huang et al, 2006; Kurash et al, 2008). Therefore, we examined whether Set9 also affected the stability of RelA. Co-expression of WT Set9 but not Set9-H297A in U2OS cells down-regulated the expression of RelA (Figure 5A), suggesting that Set9-mediated methylation of RelA might be involved in the down-regulation. Treatment with MG-132 gradually reversed the expression level of RelA and stimulated the accumulation of RelA ubiquiitnation in a time-dependent fashion (Figure 5B and C), suggesting that down-regulation of RelA is likely due to the Set9-induced ubiquitination and proteasome-mediated degradation of RelA. Further supporting these findings, we found that the co-expression of Set9 did not alter the RelA mRNA levels (Figure 5D).
Figure 5.
Set9 induces the degradation of RelA. (A) Set9, but not Set9-H297A, down-regulates the expression of RelA. U2OS cells were transfected with the expression vectors for T7-RelA and increasing amounts of WT Set9 or Set9-H279A. Whole cell lysates were immunoblotted as indicated. (B) Proteasome inhibitor MG-132 blocks Set9-induced down-regulation of RelA. U2OS cells were transfected with the expression vectors for T7-RelA and Flag-Set9. At 24 h after transfection, cells were treated with MG-132 (20 μM) for various time points, and whole cell lysates were immunoblotted as indicated. (C) Set9 induces polyubiquitination of RelA. U2OS cells were transfected and treated as described in (B). T7-RelA immunoprecipitates were prepared and immunoblotted for ubiquitin. (D) U2OS cells were transfected with the expression vectors for T7-RelA and WT Set9 or Set9-H279A. Quantitative real-time PCR with primers spanning the T7 tag and the N-terminus of RelA was performed to measure mRNA levels of T7-RelA. Data represent the average of three independent experiments. (E) Set9 shortens the half-life of RelA. U2OS cells were transfected with the expression vectors for T7-RelA or T7-RelA-K314/315R alone or together with Set9. At 24 h after transfection, cells were treated with cycloheximide (CHX), and immunoblotted for T7-RelA. A representative result from three independent experiments is shown. (F) Quantification of the results in (D). Data represent the average of three independent experiments.
To evaluate whether this degradation results from the methylation of RelA at lysines 314 and 315, we next compared the effect of Set9 on the half-life of WT RelA and RelA-K314/315R. Consistent with the notion that methylation of RelA promotes its degradation, the half-life of RelA was reduced from >12 to ∼3 h when RelA was co-expressed with Set9 (Figure 5E, upper two panels and Figure 5F). Conversely, RelA-K314/315R was more resistant to Set9-induced degradation, with a much longer half-life (>12 h) (Figure 5E, lower two panels and Figure 5F). Collectively, these findings show that methylation of RelA at lysines 314 and 315 by Set9 decreases the stability of RelA.
Methylation at lysines 314 and 315 promotes the degradation of DNA-bound RelA
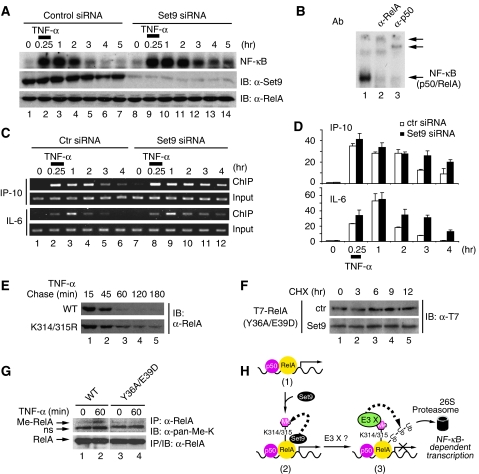
Proteasome-mediated degradation of DNA-bound RelA is essential for the prompt inactivation of NF-κB (Saccani et al, 2004; Maine et al, 2007). As Set9 negatively regulates NF-κB activity and down-regulates the stability of RelA, we next examined the effect of Set9-mediated methylation on the degradation of DNA-bound RelA. To exclude the interfering effect from re-synthesized IκBα, which binds to NF-κB and removes it from DNA (Baldwin, 1996; Ghosh et al, 1998), we compared the TNF-α-induced DNA-binding activity of NF-κB in IκBα-deficient cells that were transfected with control or Set9 siRNA. Pulse stimulation with TNF-α for 15 min induced the binding of the NF-κB complex, consisting of RelA and p50, to DNA (Figure 6A and B). However, the DNA-binding activity of NF-κB diminished gradually due to the degradation of RelA (Figure 6A, lanes 2–7) (Saccani et al, 2004). Importantly, when Set9 expression was down-regulated by siRNA, we observed a more sustained DNA-binding activity of NF-κB (Figure 6A, lanes 9–14). This prolonged DNA binding is similar to that observed when cells were treated with a proteasome inhibitor to prevent the degradation of RelA (Saccani et al, 2004). Together, these data suggest that proteasome-mediated degradation of RelA is impaired by the depletion of Set9 and that Set9-mediated methylation of RelA might be involved in the degradation of DNA-bound RelA.
Figure 6.
Set9 methylates and induces the degradation of DNA-bound RelA. (A) Set9 depletion prolongs the DNA-binding activity of NF-κB. IκBα-deficient MEFs were transfected with control or Set9 siRNA and left untreated or pulse stimulated with TNF-α for 15 min. Cells were harvested at the indicated time points. Whole cell lysates were prepared and DNA-binding activity of NF-κB was assessed by EMSA (top); protein levels of Set9 and RelA were measured by immunoblotting (middle and bottom). (B) Whole cell extracts from TNF-α-stimulated IκBα-deficient MEFs were used for supershift assay. Note that anti-RelA and anti-p50 antibodies (Ab) supershift the NF-κB complex. (C) Set9 depletion prolongs the binding of NF-κB to the native promoters of NF-κB target genes. IκBα-deficient MEFs were transfected and a pulse-chase experiment was performed as described in (A). The binding of RelA to the promoters of NF-κB targets IP-10 and IL-6 was assessed by ChIP. A representative PCR result from three independent experiments is shown. (D) Quantification of ChIP results in (C). Data represent the average of three independent experiments. (E) Mutation of lysines 314 and 315 stabilizes chromatin-associated RelA. WT RelA or RelA-K314/315R reconstituted MEFs were pulse stimulated with TNF-α for 15 min and harvested at indicated time points. Chromatin-associated proteins were extracted and immunoblotted for RelA. (F) Set9 has no effect on the half-life of DNA-binding-deficient mutant of RelA. Half-life of T7-RelA-Y36A/E39D in the absence or presence of Set9 was assessed as described in Figure 5E. (G) DNA-binding deficiency abolishes TNF-α-induced methylation of RelA. TNF-α-induced methylation of RelA from WT RelA or RelA-Y36A/E39D reconstituted MEFs was assessed as described in Figure 2G. (H) Model for the role of Set9-mediated methylation of RelA. Upon stimulation, Set9 methylates lysines 314 and 315 of the chromatin-bound RelA subunit of NF-κB. Methylation, probably by recruiting an unidentified ubiquitin E3 ligase (E3 X), triggers the ubiquitination and degradation of RelA by the 26S proteasome.
Next, we investigated the recruitment of endogenous RelA to the promoters of NF-κB target genes in control and Set9 knock-down cells by ChIPs. Consistent with the electrophoretic mobility shift assays (EMSA) experiments (Figure 6A), ChIP assays investigating the recruitment of RelA to the native promoters of IP-10 and IL-6 in IκBα-deficient cells show that the depletion of Set9 prolonged the binding of RelA to the promoters after pulse stimulation with TNF-α (Figure 6C and D). To further determine whether the sustained binding of RelA is due to the lack of methylation of RelA at lysines 314 and 315, we compared the stability of chromatin-associated WT RelA and RelA-K314/315R in reconstituted MEFs. When cells were pulse stimulated with TNF-α and the level of chromatin-associated RelA was examined, we found that WT RelA was degraded rapidly after pulse stimulation with TNF-α (Figure 6E). Conversely, chromatin-associated RelA-K314/315R displayed higher stability, consistent with the finding that RelA-K314/315R is more resistant to Set9-induced degradation than WT RelA (Figure 5E and F).
The DNA-binding property of RelA is important for the methylation of RelA by Set9
Having identified that Set9-mediated methylation of RelA is essential for the degradation of DNA-bound RelA, we next assessed whether the DNA-binding property of RelA is similarly important for Set9-mediated methylation and degradation. We generated a DNA-binding-deficient mutant of RelA (RelA-Y36A/E39D), which completely loses its DNA binding ability and is resistant to ubiquitination and proteasome-mediated degradation (Saccani et al, 2004). We first examined the ability of Set9 to induce the degradation of the RelA-Y36A/E39D mutant. A pulse-chase experiment measuring the half-life of this mutant in the presence and absence of exogenous Set9 revealed that the half-life of RelA-Y36A/E39D was not affected by Set9, indicating that Set9-mediated methylation and degradation only targets DNA-bound RelA (Figure 6F). To further assess whether binding of RelA to DNA is required for the methylation of RelA in vivo, we investigated TNF-α-induced methylation of RelA in WT RelA or RelA-Y36A/E39D reconstituted MEFs. When methylation of RelA was examined with anti-pan-methyl-lysine antibodies, we observed that TNF-α failed to stimulate the methylation of RelA-Y36A/E39D (Figure 6G). Taken together, these data suggest that Set9-mediated methylation occurs on DNA-bound RelA in vivo in response to TNF-α stimulation and that methylation of RelA at lysine residues 314 and 315 is both necessary and specific for the degradation of DNA-bound NF-κB.
Discussion
Proteasome-mediated degradation of promoter-bound NF-κB has emerged as a novel mechanism for the termination of nuclear NF-κB function (Natoli and Chiocca, 2008). Although each step within the NF-κB signalling pathway is tightly regulated, the regulation of the degradation of nuclear NF-κB is not yet clear. Our current studies reveal that the methylation of RelA has an important function in this regulation. The RelA subunit is subject to stimulus-coupled methylation mediated by Set9 at lysine residues 314 and 315. Methylation of these two lysines inhibits the transcription activity of NF-κB by inducing the degradation of promoter-associated NF-κB probably through an unidentified E3 ligase (Figure 6H). Set9-mediated lysine methylation of RelA thus serves as a ‘death' signal for the destruction of DNA-bound NF-κB (Figure 6H).
It is now well recognized that lysine methylation modifies not only histones but also non-histone proteins (Huang and Berger, 2008). Methylation of non-histone proteins plays an important role in regulating their functions. We find that Set9 methylates RelA in vitro and in vivo at lysines 314 and 315 (Figures 1 and 2). It has to be noted that lysines 314 and 315 are not conserved within Rel family proteins, although we also observed that c-Rel but not RelB was associated with and methylated by Set9 (Supplementary Figure S6). Methylation of RelA appears to be specific for Set9, as RelA is not methylated by other methyltransferases including Set8, SMYD2 and SMYD3 (data not shown). The ability of an anti-pan-methyl-lysine antibody to detect methylated RelA allows us to monitor the methylation status of endogenous RelA in response to various stimuli. TNF-α as well as LPS stimulates the methylation of nuclear RelA (Figure 1E; Supplementary Figure S7). Stimulus-coupled RelA methylation is dynamic, with increasing methylation shortly after stimulation reaching its maximum level around 60 min. This enhanced methylation of RelA is probably due to the increased association of RelA with Set9 after stimulation (Figure 3D). However, the nature of the diminished methylation signal at the later time point (90 min) (Figure 1D) is not clear. It may be caused by the degradation of methylated RelA or by a possible demethylation reaction by a demethylase. It is obvious that like other dynamic posttranslational modifications (Chen and Greene, 2004), methylation of RelA is also a tightly regulated event within cells.
Structural studies of Set9 suggest that Set9 targets lysine only for monomethylation (Wilson et al, 2002; Kwon et al, 2003; Xiao et al, 2003). Consistently, several Set9 substrates, including p53 and ERα, are found to be monomethylated at single lysine residues (Chuikov et al, 2004; Subramanian et al, 2008). Interestingly, our mass spectrometry data show that two lysine residues, 314 and 315, of RelA are monomethylated by Set9 either alone or together (Figure 2D). Mutagenesis studies further confirm the methylation of RelA at both lysine residues. Mutation of either lysine is not sufficient to eliminate the overall methylation signal, whereas mutation of both lysines completely abolishes the methylation of RelA in vitro and in vivo (Figure 2E–G). Currently, the mechanism underlying how Set9 mediates the methylation of two adjacent lysines in one substrate is not clear. Future structural studies of the interaction of the SET domain of Set9 and a RelA peptide harbouring lysines 314 and 315 might be able to provide insights into this mechanism.
Depletion of Set9 enhanced the TNF-α-induced expression of a subset of NF-κB target genes in both U2OS and A549 cells (Figure 4A; Supplementary Figure S3), indicating that Set9 negatively regulates NF-κB activation. This effect is likely due to the specific methylation of RelA by Set9, as Set9 does not methylate p50 or IκBα, the two key components in the NF-κB signalling pathway (Figure 1B). Further supporting this notion, RelA-deficient MEFs reconstituted with RelA-K314/315R displayed enhanced and prolonged TNF-α-induced expression of IL-6 and Nos-2 genes (Figure 4E). Unlike methylation of p53 and ERα, in which methylation enhances their activities by stabilizing both proteins, methylation of RelA inhibits NF-κB activity by inducing the degradation of RelA. It is likely that Set9-mediated methylation produces different biological outcomes in response to different stimuli and in different signalling pathways, functioning as a co-activator or a co-repressor. These distinct effects of Set9 might simply result from the binding of different factors to Set9 in a fashion similar to the lysine demethylase LSD1. LSD1 acts as an H3K4me2/me1 or H3K9me2/me2 demethylase to repress or activate gene transcription based on its association with its partner proteins CoREST or androgen receptor, respectively (Metzger et al, 2005; Shi et al, 2005; Wissmann et al, 2007).
NF-κB regulates the expression of >200 different kinds of genes with various biological functions in response to different stimuli (Ghosh et al, 1998). It is plausible that the regulation of NF-κB by Set9 could be cell-type specific and promoter specific. Supporting this, we found that TNFα-induced expression of A20 was not significantly affected in Set9 knock-down cells or RelA-K314/315R reconstituted MEFs (Figure 4A and E). The selective regulation of NF-κB target genes by Set9 is probably due to the differential recruitment of Set9 to the promoters (Figure 4F).
It is possible that Set9 might directly or indirectly modify histones to regulate NF-κB function, although genetic and structural analysis of Set9 suggests that it preferentially targets non-histone proteins for monomethylation, and our data show that Set9 directly targets RelA for methylation. In this regard, a recent report shows that Set9 up-regulates the expression of a subset of NF-κB target genes by targeting the tri-methylation of histone H3 K4 in monocytes (Li et al, 2008). In U2OS cells, we did not observe significant changes in the methylation status of histone proteins when the expression of Set9 was inhibited by siRNA (data not shown). However, in these cells, TNF-α-induced expression of several NF-κB target genes, including IL-6 and IL-8, is enhanced, and the methylation of RelA is abolished (Figures 4A and 1D). Future study will elucidate how Set9 differentially regulates NF-κB functions in different cellular contexts and in response to different stimuli.
In addition to methylation, RelA is also subjected to a variety of posttranslational modifications including acetylation and phosphorylation (Chen and Greene, 2004; Perkins, 2006). Recent studies indicate that phosphorylation also regulates the stability of RelA. Phosphorylation of RelA at serine 536 by IKK1 enhances the turnover of RelA in response to LPS in macrophages (Lawrence et al, 2005). It might be interesting to determine whether these modifications might function together or separately to regulate the degradation of different pools of NF-κB, although the phosphorylation status of RelA at serine 536 does not appear to affect its association with Set9 (Supplementary Figure S8). Intriguingly, many posttranslational modifications of RelA occur within a small region adjacent to the Rel homology domain of RelA (from a.a. 310 to 315). Within this region, lysine 310 is acetylated by p300/CBP (Chen et al, 2002), serine 311 is phosphorylated by PKCζ (Duran et al, 2003), and our current study shows that lysines 314 and 315 are methylated by Set9. It is likely that there is cross-talk amongst these modifications. Indeed, we find that RelA acetylated at lysine 310 is a poor substrate for Set9-mediated methylation at lysines 314 and 315 (Yang and Chen, unpublished data). Methylation and acetylation might be competitive and occur at different stages during NF-κB activation.
Proteasome-mediated degradation of promoter-bound NF-κB is critical for the postinduction inactivation of nuclear NF-κB independently of the resynthesis of IκBα (Saccani et al, 2004; Natoli and Chiocca, 2008). But the exact mechanism triggering the degradation is completely unknown. Our current studies show that methylation of RelA by Set9 has an important function in initiating this process. Methylation of RelA at lysines 314 and 315 is required for the degradation of promoter-bound RelA, and the methylation-deficient mutant of RelA (RelA-K314/315R) is resistant to Set9-induced degradation (Figure 5E and F). Furthermore, after TNF-α stimulation, chromatin-associated RelA-K314/315R is much more stable than WT RelA (Figure 6E). All these data highlight the importance of methylation in the degradation of promoter-bound RelA. Additionally, the DNA-binding property of RelA is required for its methylation. DNA-binding-defective mutant of RelA (RelA-Y36A/E39D), which is more stable than WT RelA (Figure 6F), cannot be methylated after TNF-α stimulation (Figure 6G). On the basis of these findings, we suspect that after activation of target genes by NF-κB, promoter-bound RelA is targeted by promoter-associated Set9 for methylation. Methylated RelA then recruits an E3 ligase for its ubiquitination and subsequent degradation (Figure 6H). Currently, the identity of the E3 ligase that mediates the ubiquitination of methylated RelA remains unknown. Recently, two NF-κB E3 ligases, SOCS1 and PDLIM2, have been identified, both of which have been shown to mediate the ubiquitination and degradation of NF-κB (Ryo et al, 2003; Maine et al, 2007; Tanaka et al, 2007). It may be interesting to see whether either is the E3 ligase that mediates the ubiquitination and degradation of promoter-bound methylated RelA. Our data indicate that Set9-mediated methylation of lysines 314 and 315 is a prerequisite for the degradation of promoter-bound RelA; without methylation, degradation of promoter-bound RelA is significantly impaired (Figure 6). It appears that the binding of the E3 ligase to promoter-bound RelA is methylation dependent. Methylated lysine can be recognized by various domains, including chromodomain, Tudor, PHD domains and WD40 repeats (Yang, 2005). It may be important to investigate whether proteins containing any of these domains possess E3 ligase activity and whether they mediate the polyubiquitination and degradation of methylated RelA.
In summary, our studies show that methylation of RelA at lysines 314 and 315 by Set9 negatively regulates the transcriptional activity of NF-κB by inducing the proteasome-mediated degradation of promoter-associated RelA (Figure 6H). Methylation of lysines 314 and 315 of RelA is required for the effective degradation of RelA, most likely by recruiting an as yet unidentified E3 ligase (Figure 6H). To our knowledge, this is the first study to reveal RelA as a substrate for Set9, and to show a role for Set9-mediated lysine methylation in the regulation of NF-κB. As protein methylation is a reversible modification mediated by methyltransferases and demethylases (Bannister and Kouzarides, 2005; Klose and Zhang, 2007), whether and how methylated RelA is subjected to demethylation and how Set9 activity is regulated during the inflammatory response merit further exploration.
Materials and methods
Reagents and plasmids
Pan-methyl-lysine antibody (ab7315) was purchased from Abcam; anti-RelA (C20 and F6), anti-Set9 (s4E5), anti-GST and anti-Flag were purchased from Santa Cruz; anti-Flag agarose beads (M2) and anti-tubulin were products from Sigma; anti-Set9 (07–314) and anti-T7 were purchased from Upstate and Novagen, respectively. Flag-Set9 and Flag-Set9-H279A plasmids were kindly provided by Drs Y Zhang and D Reinberg. A bacterial expression vector for His–Set9 was a gift from Dr R Trievel. Various GST–RelA fusion protein constructs were generated by PCR cloning of cDNA into pGEX4T1 vector. RelA point mutations and the siRNA resistant Set9 construct, which bears a three base-pair mutation on siRNA targeting region, were generated by site-directed mutagenesis (Stratagene). All PCR-derived sequences and point mutations were confirmed by DNA sequencing. Recombinant full-length RelA protein was purified from insect cells using the baculovirus system as described earlier (Chen et al, 2005). Various GST–RelA deletion proteins, GST–Set9 and His–Set9 proteins were expressed in and purified from the Escherichia coli strain BL21 (DE3).
In vitro methylation
In vitro methylation was performed as described earlier (Nishioka et al, 2002). Methylation was visualized either by autoradiography or by immunoblotting with anti-pan-methyl-lysine antibodies.
Mass spectrometry
A measure of 10 μg of GST–RelA (311–347) fusion protein was bound to glutathione beads and in vitro methylated by Set9. GST was then removed by thrombin digestion to elute GST-free RelA (311–347) fragments and purified by reverse phase liquid chromatography. Fractions of interest were recovered by drying to completion in a Speed-Vac. Purified samples were resuspended in 50 μl of ESI solution (50% acetonitrile, 49% H2O, 1% formic acid), and 10 μl was loaded into a nanospray robot (TriVersa Nanomate, Advion BioSciences, Ithaca, NY). Mass spectrometric analyses were carried out using a linear ion trap Fourier-Transform mass spectrometer operating at 12 Tesla. Different peptide species were isolated by the ion trap, transferred to the ion cyclotron resonance (ICR) cell and fragmented by electron capture dissociation (ECD) (Zubarev et al, 2000). Collected data were analysed using Xcalibur (Thermo Scientific) and manually interpreted, producing sets of intact mass data and fragment ion peak lists, which were loaded onto the ProSightPTM web server (www.prosightptm.scs.uiuc.edu) for comparing with the peptide sequence in absolute mass mode. The peptide ion relative ratio and fragmentation ion relative ratio values were calculated as described earlier (Pesavento et al, 2006).
Transfection, immunoprecipitation, immunoblotting analyses and EMSA
Transfection, immunoprecipitation and immunoblotting analyses and EMSA were performed as described earlier (Chen et al, 2001).
Fractionation
Fractionation of nuclear and cytoplasmic proteins was conducted as described earlier (Chen et al, 2001). For preparation of chromatin-associated proteins, 2 × 106 cells were stimulated with TNF-α for 15 min and then chased for various time periods. After treatment, cells were immediately washed with cold 1 × PBS buffer. Chromatin fraction was extracted as described earlier (Wysocka et al, 2001) and incubated in extraction buffer (20 mM HEPES pH 7.9, 1 mM EDTA, 420 mM NaCl, 0.5% NP-40, protease inhibitor cocktail, 1 mM PMSF) on ice for 30 min. Chromatin-associated proteins were recovered by centrifuging at 4°C at 14 000 r.p.m. for 20 min.
RNA interference
siRNAs were purchased from Ambion and transfected with Lipofectamine 2000 (Invitrogen) according to the manufacturer's instructions. Two different sets of human Set9 siRNA used to transfect human cell lines included siRNA1: 5′-GCCUUGUAGGAGAAGUAAAtt-3′ (sense) and 5′-UUUACUUCUCCUACAAGGCtt-3′ (anti-sense), siRNA2: 5′-GGGUUUAUGUUGCUGAAUCtt-3′ (sense) and 5′-GAUUCAGCAACAUAAACCCtt-3′ (anti-sense). One set of mouse Set9 siRNA used to transfect MEFs was: 5′-GGUAGCAGUUGGACCUAAUtt-3′ (sense) and 5′-AUUAGGUCCAACUGCUACCtt-3′ (anti-sense).
Chromatin immunoprecipitation
ChIP assays using anti-RelA (C20, Santa Cruz) or anti-Set9 (Upstate) were performed as described earlier (Chen et al, 2005). The sequence of ChIP primers will be provided upon request.
Real-time PCR
Real-time PCR was performed as described earlier (Chen et al, 2005). The primers used were purchased from Qiagen. To test the effect of Set9 on the mRNA level of co-expressed RelA, U2OS cells were transfected with T7-RelA alone or with Flag-Set9 or Flag-Set9-H297A. After 36 h, total RNA was extracted for quantitative real-time PCR assay. All real-time PCR data were normalized to β-actin.
Supplementary Material
Supplementary Figures S1–S8
Acknowledgments
We thank Y Zhang, D Reinberg and R Trievel for Set9 expression vectors; A Beg and A Hoffmann for providing the IκBα-deficient MEFs; WC Greene for reagents; members in the Chen lab for discussion. This work is supported in part by ICR provided by the University of Illinois at Urbana-Champaign and a Biomedical Research Grant from the American Lung Association. LFC is the recipient of an Arthritis Foundation Investigator Award.
References
- Baldwin AS Jr (1996) The NF-κB and IκB proteins: new discoveries and insights. Annu Rev Immunol 14: 649–683 [DOI] [PubMed] [Google Scholar]
- Bannister AJ, Kouzarides T (2005) Reversing histone methylation. Nature 436: 1103–1106 [DOI] [PubMed] [Google Scholar]
- Chen LF, Fischle W, Verdin E, Greene WC (2001) Duration of nuclear NF-κB action regulated by reversible acetylation. Science 293: 1653–1657 [DOI] [PubMed] [Google Scholar]
- Chen LF, Greene WC (2004) Shaping the nuclear action of NF-κB. Nat Rev Mol Cell Biol 5: 392–401 [DOI] [PubMed] [Google Scholar]
- Chen LF, Mu Y, Greene WC (2002) Acetylation of RelA at discrete sites regulates distinct nuclear functions of NF-κB. EMBO J 21: 6539–6548 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen LF, Williams SA, Mu Y, Nakano H, Duerr JM, Buckbinder L, Greene WC (2005) NF-kB RelA phosphorylation regulates RelA acetylation. Mol Cell Biol 25: 7966–7975 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chuikov S, Kurash JK, Wilson JR, Xiao B, Justin N, Ivanov GS, McKinney K, Tempst P, Prives C, Gamblin SJ, Barlev NA, Reinberg D (2004) Regulation of p53 activity through lysine methylation. Nature 432: 353–360 [DOI] [PubMed] [Google Scholar]
- Dornan D, Wertz I, Shimizu H, Arnott D, Frantz GD, Dowd P, O'Rourke K, Koeppen H, Dixit VM (2004) The ubiquitin ligase COP1 is a critical negative regulator of p53. Nature 429: 86–92 [DOI] [PubMed] [Google Scholar]
- Duran A, Diaz-Meco MT, Moscat J (2003) Essential role of RelA Ser311 phosphorylation by zetaPKC in NF-κB transcriptional activation. EMBO J 22: 3910–3918 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghosh S, Karin M (2002) Missing pieces in the NF-κB puzzle. Cell 109(Suppl): S81–S96 [DOI] [PubMed] [Google Scholar]
- Ghosh S, May MJ, Kopp EB (1998) NF-κB and Rel proteins: evolutionarily conserved mediators of immune responses. Annu Rev Immunol 16: 225–260 [DOI] [PubMed] [Google Scholar]
- Hayden MS, Ghosh S (2008) Shared principles in NF-κB signaling. Cell 132: 344–362 [DOI] [PubMed] [Google Scholar]
- Huang J, Berger SL (2008) The emerging field of dynamic lysine methylation of non-histone proteins. Curr Opin Genet Dev 18: 152–158 [DOI] [PubMed] [Google Scholar]
- Huang J, Perez-Burgos L, Placek BJ, Sengupta R, Richter M, Dorsey JA, Kubicek S, Opravil S, Jenuwein T, Berger SL (2006) Repression of p53 activity by Smyd2-mediated methylation. Nature 444: 629–632 [DOI] [PubMed] [Google Scholar]
- Ivanov GS, Ivanova T, Kurash J, Ivanov A, Chuikov S, Gizatullin F, Herrera-Medina EM, Rauscher F III, Reinberg D, Barlev NA (2007) Methylation-acetylation interplay activates p53 in response to DNA damage. Mol Cell Biol 27: 6756–6769 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karin M (1999) How NF-κB is activated: the role of the IκB kinase (IKK) complex. Oncogene 18: 6867–6874 [DOI] [PubMed] [Google Scholar]
- Karin M, Ben-Neriah Y (2000) Phosphorylation meets ubiquitination: the control of NF-κB activity. Annu Rev Immunol 18: 621–663 [DOI] [PubMed] [Google Scholar]
- Klose RJ, Zhang Y (2007) Regulation of histone methylation by demethylimination and demethylation. Nat Rev Mol Cell Biol 8: 307–318 [DOI] [PubMed] [Google Scholar]
- Kouskouti A, Scheer E, Staub A, Tora L, Talianidis I (2004) Gene-specific modulation of TAF10 function by SET9-mediated methylation. Mol Cell 14: 175–182 [DOI] [PubMed] [Google Scholar]
- Kurash JK, Lei H, Shen Q, Marston WL, Granda BW, Fan H, Wall D, Li E, Gaudet F (2008) Methylation of p53 by Set7/9 mediates p53 acetylation and activity in vivo. Mol Cell 29: 392–400 [DOI] [PubMed] [Google Scholar]
- Kwon T, Chang JH, Kwak E, Lee CW, Joachimiak A, Kim YC, Lee J, Cho Y (2003) Mechanism of histone lysine methyl transfer revealed by the structure of SET7/9-AdoMet. EMBO J 22: 292–303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawrence T, Bebien M, Liu GY, Nizet V, Karin M (2005) IKKalpha limits macrophage NF-κB activation and contributes to the resolution of inflammation. Nature 434: 1138–1143 [DOI] [PubMed] [Google Scholar]
- Li Y, Reddy MA, Miao F, Shanmugam N, Yee JK, Hawkins D, Ren B, Natarajan R (2008) Role of the histone H3 lysine 4 methyltransferase, SET7/9, in the regulation of NF-κB-dependent inflammatory genes: relevance to diabetes and inflammation. J Biol Chem 283: 26771–26781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maine GN, Mao X, Komarck CM, Burstein E (2007) COMMD1 promotes the ubiquitination of NF-κB subunits through a cullin-containing ubiquitin ligase. EMBO J 26: 436–447 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Metzger E, Wissmann M, Yin N, Muller JM, Schneider R, Peters AH, Gunther T, Buettner R, Schule R (2005) LSD1 demethylates repressive histone marks to promote androgen-receptor-dependent transcription. Nature 437: 436–439 [DOI] [PubMed] [Google Scholar]
- Muratani M, Tansey WP (2003) How the ubiquitin-proteasome system controls transcription. Nat Rev Mol Cell Biol 4: 192–201 [DOI] [PubMed] [Google Scholar]
- Natoli G, Chiocca S (2008) Nuclear ubiquitin ligases, NF-κB degradation, and the control of inflammation. Sci Signal 1: pe1. [DOI] [PubMed] [Google Scholar]
- Nishioka K, Chuikov S, Sarma K, Erdjument-Bromage H, Allis CD, Tempst P, Reinberg D (2002) Set9, a novel histone H3 methyltransferase that facilitates transcription by precluding histone tail modifications required for heterochromatin formation. Genes Dev 16: 479–489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perkins ND (2006) Post-translational modifications regulating the activity and function of the nuclear factor kappa B pathway. Oncogene 25: 6717–6730 [DOI] [PubMed] [Google Scholar]
- Pesavento JJ, Mizzen CA, Kelleher NL (2006) Quantitative analysis of modified proteins and their positional isomers by tandem mass spectrometry: human histone H4. Anal Chem 78: 4271–4280 [DOI] [PubMed] [Google Scholar]
- Ryo A, Suizu F, Yoshida Y, Perrem K, Liou YC, Wulf G, Rottapel R, Yamaoka S, Lu KP (2003) Regulation of NF-κB signaling by Pin1-dependent prolyl isomerization and ubiquitin-mediated proteolysis of p65/RelA. Mol Cell 12: 1413–1426 [DOI] [PubMed] [Google Scholar]
- Saccani S, Marazzi I, Beg AA, Natoli G (2004) Degradation of promoter-bound p65/RelA is essential for the prompt termination of the nuclear factor κB response. J Exp Med 200: 107–113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sethi G, Sung B, Aggarwal BB (2008) Nuclear factor-κB activation: from bench to bedside. Exp Biol Med (Maywood) 233: 21–31 [DOI] [PubMed] [Google Scholar]
- Shi YJ, Matson C, Lan F, Iwase S, Baba T, Shi Y (2005) Regulation of LSD1 histone demethylase activity by its associated factors. Mol Cell 19: 857–864 [DOI] [PubMed] [Google Scholar]
- Su IH, Basavaraj A, Krutchinsky AN, Hobert O, Ullrich A, Chait BT, Tarakhovsky A (2003) Ezh2 controls B cell development through histone H3 methylation and Igh rearrangement. Nat Immunol 4: 124–131 [DOI] [PubMed] [Google Scholar]
- Subramanian K, Jia D, Kapoor-Vazirani P, Powell DR, Collins RE, Sharma D, Peng J, Cheng X, Vertino PM (2008) Regulation of estrogen receptor alpha by the SET7 lysine methyltransferase. Mol Cell 30: 336–347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun SC (2008) Deubiquitylation and regulation of the immune response. Nat Rev Immunol 8: 501–511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka T, Grusby MJ, Kaisho T (2007) PDLIM2-mediated termination of transcription factor NF-κB activation by intranuclear sequestration and degradation of the p65 subunit. Nat Immunol 8: 584–591 [DOI] [PubMed] [Google Scholar]
- Wang H, Cao R, Xia L, Erdjument-Bromage H, Borchers C, Tempst P, Zhang Y (2001) Purification and functional characterization of a histone H3-lysine 4-specific methyltransferase. Mol Cell 8: 1207–1217 [DOI] [PubMed] [Google Scholar]
- Wertz IE, O'Rourke KM, Zhang Z, Dornan D, Arnott D, Deshaies RJ, Dixit VM (2004) Human De-etiolated-1 regulates c-Jun by assembling a CUL4A ubiquitin ligase. Science 303: 1371–1374 [DOI] [PubMed] [Google Scholar]
- Wilson JR, Jing C, Walker PA, Martin SR, Howell SA, Blackburn GM, Gamblin SJ, Xiao B (2002) Crystal structure and functional analysis of the histone methyltransferase SET7/9. Cell 111: 105–115 [DOI] [PubMed] [Google Scholar]
- Wissmann M, Yin N, Muller JM, Greschik H, Fodor BD, Jenuwein T, Vogler C, Schneider R, Gunther T, Buettner R, Metzger E, Schule R (2007) Cooperative demethylation by JMJD2C and LSD1 promotes androgen receptor-dependent gene expression. Nat Cell Biol 9: 347–353 [DOI] [PubMed] [Google Scholar]
- Wysocka J, Reilly PT, Herr W (2001) Loss of HCF-1-chromatin association precedes temperature-induced growth arrest of tsBN67 cells. Mol Cell Biol 21: 3820–3829 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao B, Jing C, Wilson JR, Walker PA, Vasisht N, Kelly G, Howell S, Taylor IA, Blackburn GM, Gamblin SJ (2003) Structure and catalytic mechanism of the human histone methyltransferase SET7/9. Nature 421: 652–656 [DOI] [PubMed] [Google Scholar]
- Yang XJ (2005) Multisite protein modification and intramolecular signaling. Oncogene 24: 1653–1662 [DOI] [PubMed] [Google Scholar]
- Zubarev RA, Horn DM, Fridriksson EK, Kelleher NL, Kruger NA, Lewis MA, Carpenter BK, McLafferty FW (2000) Electron capture dissociation for structural characterization of multiply charged protein cations. Anal Chem 72: 563–573 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figures S1–S8