Abstract
Integration of genetic and metabolic profiling holds promise for providing insight into human disease. Coronary artery disease (CAD) is strongly heritable, but the heritability of metabolomic profiles has not been evaluated in humans. We performed quantitative mass spectrometry-based metabolic profiling in 117 individuals within eight multiplex families from the GENECARD study of premature CAD. Heritabilities were calculated using variance components. We found high heritabilities for amino acids (arginine, ornithine, alanine, proline, leucine/isoleucine, valine, glutamate/glutamine, phenylalanine and glycine; h2=0.33–0.80, P=0.005–1.9 × 10−16), free fatty acids (arachidonic, palmitic, linoleic; h2=0.48–0.59, P=0.002–0.00005) and acylcarnitines (h2=0.23–0.79, P=0.05–0.0000002). Principal components analysis was used to identify metabolite clusters. Reflecting individual metabolites, several components were heritable, including components comprised of ketones, β-hydroxybutyrate and C2-acylcarnitine (h2=0.61); short- and medium-chain acylcarnitines (h2=0.39); amino acids (h2=0.44); long-chain acylcarnitines (h2=0.39) and branched-chain amino acids (h2=0.27). We report a novel finding of high heritabilities of metabolites in premature CAD, establishing a possible genetic basis for these profiles. These results have implications for understanding CAD pathophysiology and genetics.
Keywords: acylcarnitines, amino acids, heritability, cardiovascular disease, metabolomics
Introduction
Coronary artery disease (CAD) is the leading cause of death in industrialized countries and, in concert with the epidemic of obesity and diabetes, is rapidly becoming the leading cause of death in developing countries (World Health Organization, 2002). The genetic predilection of CAD is well established; family history has been shown to be an independent risk factor for CAD (Shea et al, 1984), especially in early-onset forms (Rissanen, 1979). Despite this, the genetic architecture of CAD remains largely unknown.
Given the complex nature of CAD, evaluation of the disease with more comprehensive analytical tools may provide needed insights into biological pathways converging on this phenotype. High-throughput molecular profiling methods have become increasingly prevalent in basic and clinical research. Further, many commonly accepted risk factors for CAD are metabolic. Metabolomics, the study of small-molecule metabolites, may be useful for understanding metabolic imbalances and for diagnosis of human disease. Studies using unbiased metabolomic profiling have uncovered differences in profiles in myocardial ischemia (Sabatine et al, 2005), and have been suggested to predict the presence and severity of CAD (Brindle et al, 2002). However, another group has refuted the initial CAD findings (Kirschenlohr et al, 2006). In both studies, the majority of the analytes within the profile remained unidentified, making clinical and mechanistic interpretation difficult, and precluding correlation of the profiles with underlying genetics. More recently, targeted metabolite profiling studies have demonstrated heritability of metabolites in plants (Keurentjes et al, 2006) and mice (Ferrara et al, 2008), but this has never been explored in humans.
Therefore, in this study, we have performed quantitative profiling of 66 metabolites, including acylcarnitine species (by-products of mitochondrial fatty acid, carbohydrate and amino-acid oxidation), amino acids and free fatty acids, in plasma samples from eight large multiplex families heavily burdened with premature CAD. Our main goal was to perform a pilot study to test whether metabolites are heritable in families burdened with CAD, and to investigate the role of metabolomic profiles in etiologic heterogeneity in CAD pathogenesis.
Results
Heritability analysis
Metabolic profiling was performed on 117 individuals within eight multiplex Caucasian families (Supplementary information) from the GENECARD study of premature CAD. Of note, the majority of family members sampled for this study were as-yet-unaffected offspring of the original affected sibling pair, but who, as members of these families, were at high risk for development of premature CAD. As expected, there was a high burden of CAD risk factors, although the prevalence differed between families (Supplementary information).
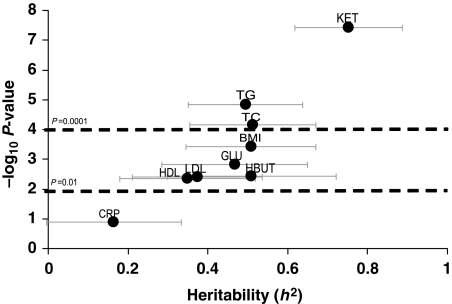
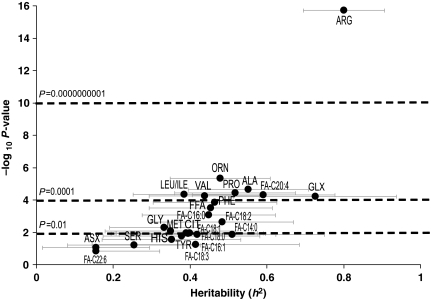
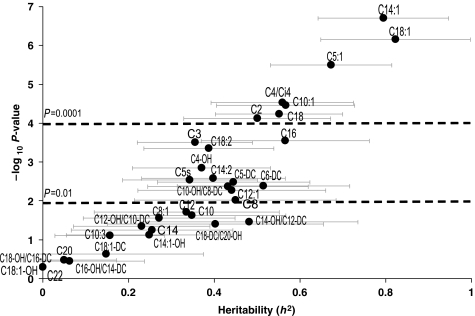
Consistent with prior reports (Beekman et al, 2002), we found high heritabilities for conventional risk factors such as lipids and body mass index (BMI) (Figure 1). Total ketones (h2=0.75, P=3.8 × 10−8) had the highest heritability among the metabolites analyzed by non-mass spectrometry-based methods, with similarly high heritability of the individual ketone β-hydroxybutyrate (HBUT; h2=0.51, P=0.004). Among analytes measured by mass spectrometry, several amino acids had high heritability (Figure 2; Supplementary information). Arginine (ARG) had the highest score (h2=0.80, P=1.9 × 10−16), with strong heritabilities also for glutamine/glutamate (GLX; h2=0.73, P=0.00006), alanine (ALA; h2=0.55, P=0.00002), proline (PRO; h2=0.52, P=0.00004), ornithine (ORN; h2=0.48, P=0.000005), phenylalanine (PHE; h2=0.46, P=0.0001) and the branched-chain amino acids leucine/isoleucine (LEU/ILE; h2=0.39, P=0.00005) and valine (VAL; h2=0.44, P=0.00006). Of the free fatty acids (Figure 2), FA-C20:4 (arachidonic acid, a key component in inflammatory pathways) was the most heritable (h2=0.59, P=0.00005), as well as FA-C18:2 (linoleic acid, precursor to arachidonic acid, h2=0.48, P=0.002). Many acylcarnitines also had high heritabilities (Figure 3; Supplementary information), the highest being the C18 acylcarnitines (C18, C18:1, and C18:2; h2=0.39–0.82, P=0.0000007–0.004); C14:1 (h2=0.79, P=0.0000002); C5:1 (h2=0.67, P=0.000003); the C10s (C10-OH:C8-DC, C10 and C10:1; h2=0.35–0.57, P=0.00003–0.02); C16 (h2=0.57, P=0.0002); C4:Ci4 (h2=0.56, P=0.00003); short chain dicarboxylacylcarnitines (C5-DC, C6-DC; h2=0.45–0.51, P=0.003–0.004) and C2 acylcarnitine (h2=0.50, P=0.00008). Interestingly, estimates for the genetic component of the variability of each metabolite often exceeded the proportion of variance explained by clinical covariates (Supplementary information).
Figure 1.
Heritabilities of conventional metabolites. Heritabilities of the conventional metabolites are displayed in graphical form. The Y-axis is the negative log10 of the P-value for the heritability estimate (X-axis). Error bars around heritability point estimates are in light gray.
Figure 2.
Heritabilities of amino acids and free fatty acids. Heritabilities of amino acids and free fatty acids are displayed in graphical form. The Y-axis is the negative log10 of the P-value for the heritability estimate (X-axis). Error bars around heritability point estimates are in light gray.
Figure 3.
Heritabilities of acylcarnitines. Heritabilities of the acylcarnitines are displayed in graphical form. The Y-axis is the negative log10 of the P-value for the heritability estimate (X-axis). Error bars around heritability point estimates are in light gray.
Metabolomic profiles within families
Given these strong findings, we sought to understand quantitative differences in metabolites between families. Multivariate linear models were used to test for the differences in metabolites between families. Of the amino acids, glutamate, ornithine, arginine, proline, histidine, phenylalanine, alanine and methionine (all P<0.0001), leucine/isoleucine (P<0.0001) and valine (P=0.003) best differentiated families. Of the acylcarnitines, the C18 (C18, C18:1 and C18:2) and the C14 acylcarnitines (C14 and C14:1) (all P<0.0001), along with C5:1 (P<0.0001) and C2 (P<0.0001) acylcarnitines best differentiated families. Many free fatty acids differentiated families, the strongest being arachidonic and palmitic acid (both P<0.0001). Of the conventional metabolites, ketones (P<0.0001) and β-hydroxybutyrate (P=0.0001) best differentiated families.
Principal components analysis (PCA)
Given the correlation of metabolites in biological pathways, we performed PCA to understand which clusters of metabolites were correlated and to identify factors that were most heritable. Fifteen factors were identified, demonstrating biologically consistent relationships (Table I). Factors accounting for the largest amount of variance within the data set were factor 1 (short- and medium-chain acylcarnitines); factor 2 (long-chain free fatty acids); factor 3 (long-chain acylcarnitines and amino acids (arginine, glutamate/glutamine and ornithine) possibly reporting on mitochondrial function); factor 4 (ketones, β-hydroxybutyrate, C2 and C4-OH (β-hydroxybutryl) acylcarnitines; all markers of terminal steps of fatty acid oxidation) and factor 5 (amino acids, including branched-chain amino acids and C3 and C5 acylcarnitines (by-products of branched-chain amino-acid catabolism)). As expected, given the results for individual metabolites, many factors were heritable.
Table 1.
Principal components analysis in GENECARD
| Factors | Metabolites within factora | Overall description of factor | Eigenvalue | Total var | Cum var | Heritability (SD) | P-value |
|---|---|---|---|---|---|---|---|
| 1 | C2, C6-DC, C8, C8:1, C10, C10:1, C10:3, C10-OH:C8-DC, C12, C12:1, C14, C14:1, C14:2, C14:1-OH, C14-OH:C12-DC | Short- and medium-chain acylcarnitines | 11.88 | 0.20 | 0.20 | 0.39 (0.16) | 0.0006 |
| 2 | Total FFA, FA-C14:0, FA-C16:0, FA-C16:1, FA-C18:0, FA-C18:1, FA-C18:2, FA-C18:3 | Free fatty acids | 7.55 | 0.13 | 0.32 | 0.35 (0.20) | 0.02 |
| 3 | ARG, GLX, ORN, C16, C18, C18:1, C18:2 | Amino acids, long-chain acylcarnitines (markers of overall mitochondrial function) | 5.89 | 0.10 | 0.42 | 0.40 (0.18) | 0.002 |
| 4 | C2, C4-OH, C14:1, C14:2, C14:1-OH, Ket, Hbut | FFA oxidation by-products | 3.51 | 0.06 | 0.48 | 0.61 (0.17) | 0.00004 |
| 5 | ALA, LEU/ILE, MET, PRO, TYR, VAL, PHE, C5, C3, C20 | Metabolites involved in amino-acid catabolism | 2.98 | 0.05 | 0.53 | 0.27 (0.15) | 0.01 |
| 6 | CIT, C5-DC, C8:1, C10:3 | Various | 2.36 | 0.04 | 0.57 | 0.51 (0.17) | 0.0008 |
| 7 | SER, GLY, CIT, MET | Amino acids | 2.04 | 0.03 | 0.60 | 0.44 (0.28) | 0.09 |
| 8 | C14-OH:C12-DC, C18:1-OH, C22 | Various | 1.89 | 0.03 | 0.64 | 0.40 (0.18) | 0.003 |
| 9 | C12-OH:C10-DC, C14, C14:1-OH, C20 | Various | 1.86 | 0.03 | 0.67 | 0.51 (0.17) | 0.0003 |
| 10 | C3, C4:Ci4, C22 | Various | 1.67 | 0.03 | 0.69 | 0.46 (0.19) | 0.002 |
| 11 | ASX, HIS | Amino acids | 1.48 | 0.02 | 0.72 | 0.33 (0.17) | 0.005 |
| 12 | FAC22:6, FAC20:4, C20 | Long-chain free fatty acids | 1.37 | 0.02 | 0.74 | 0.36 (0.17) | 0.007 |
| 13 | C16-OH:C14-DC | Various | 1.24 | 0.02 | 0.76 | 0.46 (0.20) | 0.002 |
| 14 | PRO, ALA, C18:1-DC | Various | 1.18 | 0.02 | 0.78 | 0.54 (0.16) | 0.0001 |
| 15 | C18-DC:C20-OH | Various | 1.06 | 0.02 | 0.80 | 0.45 (0.19) | 0.006 |
FFA, free fatty acids; tot var, total variance; cum var, cumulative variance.
Results of PCA in the data set are presented, including the key metabolites within each factor (i.e. those with a factor load ⩾∣0.4∣); an overall biochemical description of the key metabolites within each factor; and the eigenvalue, total and cumulative variance, heritability and P-value for the heritability point estimate for each factor.
Factor load ⩾∣0.4∣.
Discussion
In this study, we have applied a comprehensive set of analytical tools to gain a better understanding of the biochemical and physiologic underpinnings of cardiovascular disease, and how metabolomic profiles may relate to the known genetic component of CAD risk. We performed targeted, quantitative metabolic profiling in multiplex families burdened with premature CAD, the majority representing offspring of the affected generation that had not yet developed CAD, but in whom we hypothesized similar metabolic profiles as their affected family members, if such profiles were heritable. In fact, we found high heritabilities for many metabolites, many with higher heritabilities than for conventional risk factors. These high heritabilities suggest a strong correlation between genotype and phenotype, implying a strong genetic component to clustering of these metabolic signatures in families burdened with CAD. This study represents the first evaluation of the heritability of metabolite profiles in humans, extending upon prior studies in plants (Keurentjes et al, 2006) and mice (Ferrara et al, 2008).
As expected given the strong heritabilities, several individual metabolites distinguished families, the most prominent being, among the amino acids, arginine, ornithine and glutamate/glutamine; and among the lipid-derived metabolites, the long-chain acylcarnitines C18:0, C18:1 and C18:2. These findings suggest fundamental differences in mitochondrial function in these families, consistent with prior studies showing relationships between impaired mitochondrial function and insulin resistance (Petersen et al, 2003; An et al, 2004; Koves et al, 2005, 2008; Muoio and Newgard, 2008). Further studies are necessary to clarify the underlying mechanisms reflective of these differences.
We recognize that our study has some limitations. Given our sample size, some results had large standard errors despite strong statistical significance. However, it should be noted that the sample size of this study exceeds the limited number of previous human studies and was adequate to detect significant heritabilities. Given our studies were hypothesis generating, we did not adjust for multiple comparisons. However, with a Bonferroni correction at the level of the factors, nine factors remain significant (P<0.003). We did not account for dietary pattern (known influence on metabolites), renal function, or medications (unknown influence). To help minimize these ‘non-genetic' effects, we incorporated a household effect and included married-in individuals, partially controlling for shared nutritional and other environmental effects. The measures of household effects suggest minimal influence on heritability estimates with high heritabilities despite adjustment. Therefore, we believe our results reflect both underlying genetic and environmental effects, similar to traditional cholesterol parameters. Accordingly, we found a significant household effect for low-density lipoprotein (LDL)-cholesterol (proportion of variance due to household 0.11, P=0.02), but with a significant heritability despite adjustment for this environmental effect (h2=0.37, P=0.004).
Similarly, results could reflect differences in essential versus non-essential metabolites. However, we found similar heritabilities for the essential (h2=0.40, P=0.0004) and non-essential (h2=0.63, P=0.00002) amino acids when analyzed as groups, and for the essential (h2=0.50, P=0.003) compared with the non-essential (h2=0.33, P=0.03) fatty acids. Although underpowered for such analyses, we also examined the relationship of age with heritabilities related to these groups. Age was a significant covariate on heritability estimates for both essential (valine) and non-essential (proline, ornithine and citrulline) amino acids (Supplementary information). For the free fatty acids, age was a covariate only for non-essential fatty acids (palmitoleic, oleic and stearic acid). We also examined correlations of metabolites with age and found that both essential (tyrosine and linoleic acid) and non-essential (glutamine, ornithine, citrulline and oleic acid) metabolites were significantly correlated with age (data not shown). Therefore, there does not seem to be a consistent variation of metabolites with age, nor with heritability estimates, based on essential/non-essential groups. This may indicate that fundamental and genetically controlled metabolic processes (e.g. mitochondrial or microsomal catabolic pathways) are influencing the levels of both essential and non-essential metabolites that utilize these common elements of the metabolic machinery. Larger, population-based studies should delve deeper into these questions.
Other factors that could impact heritability estimates include variability in sample collection or processing. We used a standardized protocol to limit this type of variability, intra-individual variation was low in a set of repeated assays, and family members were collected at different locations and times. Finally, it is important to note that our heritability estimates, by design, are for families burdened with premature CAD, and therefore are not generalizable to a population without a genetic burden for CAD.
A major strength of the study is the use of a very accurate, targeted, quantitative approach to metabolomic profiling, allowing us to dissect biological mechanisms underlying CAD pathophysiology. Previous studies involving application of metabolomics to cardiovascular disease involved primarily non-targeted profiling and small sample sizes, and used modeling that is difficult to understand clinically and hard to integrate into our knowledge of pathophysiology. Another strength of our study is that we performed metabolite profiling within a family-based design. Using this approach, one can potentially distinguish shared environmental versus genetic influences, and develop hypotheses regarding metabolic and genetic risk in a sample of high-risk, as-of-yet unaffected offspring.
Our results have implications for future studies of CAD. Explicitly accounting for the heterogeneity introduced by the distinct metabolomic signatures, as we have done previously with conventional metabolites (Shah et al, 2006), may be important for mapping of CAD genes and for understanding the underlying genetic architecture of these metabolite profiles (so called ‘mQTLs') (Ferrara et al, 2008). In addition to furthering the understanding of CAD pathophysiology, these results may have significant implications for risk prediction. Future studies should include refining results in other cohorts, mapping the genes underlying these heritabilities and understanding the predictive capabilities of metabolites in non-familial cohorts.
Materials and methods
Study population
The GENECARD study enrolled 920 families to perform affected sibling pair linkage for the identification of genes for early-onset CAD (before the age of 51 years for men and 56 years for women) (Hauser et al, 2003). Families with at least two siblings each of whom met the criteria for early-onset CAD (before the age of 51 years for men and 56 years for women) were recruited. Unaffected family members were defined as no clinical evidence of CAD and age greater than 55 years for men (greater than 60 years for women). From this cohort, we selected eight representative families we believed would be particularly informative, based on the availability of a relatively large number of family members and a heavy burden of CAD in the proband and surrounding generations (Supplementary information). These families were recontacted; the affected sibling pair and family members not previously enrolled were ascertained regardless of CAD, focused on offspring of the affected sibling pair. This ascertainment strategy was based on the hypothesis that if abnormalities in metabolic profiles preceded development of CAD in these families, that significant concordance of metabolite levels within families would be evident even in the absence of overt CAD in the offspring. Sample collections within a given family were carried out at several different times and at different locations, by a single experienced phlebotomist. Blood samples were promptly processed after collection through peripheral venous phlebotomy (within minutes), frozen as soon as possible thereafter (at most within 12 h with the majority of samples being frozen within 1–2 h of collection), and stored as plasma samples in EDTA-treated tubes at −80°C. Samples were collected as often as possible in a fasting state; however, the consistency of this could not be determined. Institutional Review Boards approved study protocols; informed consent was obtained from each subject.
Biochemical measurements
Frozen plasma samples were used to quantitatively measure targeted metabolites, including 37 acylcarnitine species, 15 amino acids, 9 free fatty acids and conventional analytes, ketones and C-reactive protein (CRP). Sample preparation and coefficients of variation have been reported (Haqq et al, 2005). The laboratory was blinded to family identifiers and case–control status. Assay ranges are 0.05–40 μM (acylcarnitines); 5–1000 μM (amino acids) and 1–1000 mmol/l (fatty acids). For simplicity, the clinical shorthand of metabolites is used (Supplementary information). Intra-individual variability was assessed in samples from five individuals for which repeat profiling was performed on the same sample on five separate days. Coefficients of variation and correlation confirmed minimal inter-assay variability (Supplementary information).
Conventional metabolite analysis
Standard clinical chemistry methods were used for conventional metabolites, including glucose, total cholesterol, high-density lipoprotein (HDL)- and LDL-cholesterol, and triglycerides with reagents from Roche Diagnostics (Indianapolis, IN); and free fatty acids (total) and ketones (total and 3-hydroxybutyrate) with reagents from Wako (Richmond, VA). All measurements were performed using a Hitachi 911 clinical chemistry analyzer.
Acylcarnitines and amino acids
Proteins were first removed by precipitation with methanol. Aliquoted supernatants were dried, and then esterified with hot acidic methanol (acylcarnitines) or n-butanol (amino acids). Acylcarnitines and amino acids were analyzed by tandem MS with a Quattro Micro instrument (Waters Corporation, Milford, MA). In total, 37 acylcarnitine species and 15 amino acids in plasma were assayed by our previously described methods (Millington et al, 1990; An et al, 2004; Wu et al, 2004). Leucine/isoleucine (LEU/ILE) are reported as a single analyte because they are not resolved by our MS/MS method, and include contributions from allo-isoleucine and hydroxyproline. Under normal circumstances, these isobaric amino acids contribute little to the signal attributed to LEU/ILE (Chace et al, 1995). In addition, the acidic conditions used to form butyl esters results in partial hydrolysis of glutamine to glutamic acid and of asparagine to aspartate. Accordingly, values that are reported as GLU/GLN or ASP/ASN are not meant to signify the molar sum of glutamate and glutamine, or of aspartate and asparagine, but rather measure the amount of glutamate or aspartate plus the contribution of the partial hydrolysis reactions of glutamine and asparagine, respectively.
Free fatty acids
Free fatty acids were gently methylated using iodomethane and purified by solid-phase extraction (Patterson et al, 1999). Derivatized fatty acids were analyzed by capillary gas chromatography/mass spectrometry (GC/MS) using a Trace DSQ instrument (Thermo Electron Corporation, Austin, TX). Owing to sample volume considerations, free fatty acid measurements were performed only in 80 of the 117 individuals (five out of eight families).
All mass spectrometric analyses used stable isotope dilution. Quantification of the foregoing ‘targeted' intermediary metabolites was facilitated by the addition of mixtures of known quantities of stable isotope internal standards to samples from Isotec (St Louis, MO), Cambridge Isotope Laboratories (Andover, MA) and CDN Isotopes (Pointe-Claire, Quebec, CN) (Supplementary information).
Heritability analysis
Heritabilities were calculated using the Sequential Oligogenic Linkage Analysis Routines (SOLAR) software version 4.0.7 (Almasy and Blangero, 1998), which uses maximum-likelihood methods to estimate variance components, allowing incorporation of fixed effects for known covariates and variance components for genetic effects. This approach appropriately accounts for correlation between all family members and allows incorporation of extended pedigrees such as that are present in the current study. The total variation is partitioned into components for additive genetic variance and environmental variance, as well as a residual (unexplained) variability. The program uses the pedigree covariance matrix
where Ω is the covariance matrix, Φ is the matrix of kinship values, σg2 is the additive genetic variance, I represents the identity matrix, and σe2 is the random environmental variance (Almasy and Blangero, 1998). This model allows for complex pedigree data (i.e. beyond parent–offspring pairs) and hence, the resulting heritability estimates are more accurate than those obtained using only nuclear family members. For the current study, all sampled individuals from the pedigree were entered into the variance component models, including unaffected offspring, cousins and married-in family members. Incorporation of married-in family members (i.e. genetically unrelated but with shared environment) allows for better estimation of the environmental component of intrafamilial clustering of traits.
Values considered outliers were excluded from heritability analyses, defined as values falling outside of the mean±4 s.d. (1–2 outliers for each of 24 of the metabolites). Metabolite measurements below the lower limits of quantification (LOQ) were given a value of LOQ/2. Four metabolites having >25% of samples below LOQ were not further analyzed (C6, C5-OH:C3-DC, C4DC and C10:2 acylcarnitines). All measurements were natural log-transformed prior to analysis, resulting in most metabolites approximating a normal distribution, an important consideration for variance components analysis. Eighteen metabolites did not meet this criterion, and therefore, linear regression models adjusted for body mass index (BMI), age, sex, CAD, diabetes mellitus (DM (yes/no), hypertension (yes/no) and dyslipidemia (yes/no) were constructed for each of these metabolites, and the residuals were used for heritability estimates. Given the occasional low trait standard deviations for metabolites (<0.5), all log-transformed metabolites were multiplied by a factor of 4.7 prior to analysis.
Polygenic heritability models were then constructed. For the normally distributed metabolites (the majority of metabolites), polygenic heritability models were calculated using the log-transformed values, adjusting for age, sex, BMI, DM, dyslipidemia, hypertension and CAD. The proband and family members were not selected based on any metabolite values; however, the potential for ascertainment bias exists. Therefore, analyses were corrected based on which of the family members (proband) was the index member for ascertainment of the family for early-onset CAD. To account for factors such as diet (which are shared in households but are presumably not genetic), an additional variance component parameter corresponding to the fraction of variance associated with the effect of a common household (included in the model by a marker for residential address) was added to each model. All residual kurtoses for the final polygenic model were within normal range (i.e. <0.8), except for two amino acids (serine and phenylalanine), 11 acylcarnitines (C5, C10, C10:1, C10:3, C12:1, C14, C14-OH:C12-DC, C16-OH:C14-DC, C18:1-OH, C18:1-DC and C18-DC:C20-OH) and 3 free fatty acids (FAC14:0, FAC16:1 and FAC18:1). For these metabolites, removal of 1–4 of the most extreme values was necessary, which then resulted in a normal residual kurtosis. Two acylcarnitines required removal of a larger number of outliers to achieve a normal residual kurtosis (C16-OH:C14-DC and C12-OH:C10-DC), and hence, these results should be interpreted accordingly. For the 18 non-normally distributed metabolites, standardized residuals from adjusted regression models were used to estimate heritabilities using SOLAR, but as the normalized deviates were already adjusted for relevant covariates heritability models using these residuals were not further adjusted. Estimates of the proportion of variance explained by clinical covariates are reported for these non-normally distributed metabolites as estimated using the adjusted polygenic model constructed from the log-transformed crude values.
For understanding quantitative differences in metabolites between families, multivariate generalized linear models adjusted for sex, age, BMI, CAD, DM, dyslipidemia and hypertension were used to compare mean metabolite levels between families.
Unsupervised PCA
Given that many metabolites reside in overlapping pathways, correlation of metabolites is expected. To understand the correlation, we used PCA to reduce the large number of correlated variables (Supplementary information) into clusters of fewer uncorrelated factors using raw metabolite values without removal of outliers. The factor with the highest ‘eigenvalue' accounts for the largest amount of the variability within the data set. Standardized residuals calculated for each metabolite from linear regression models adjusted for age, sex, BMI, DM and CAD were used as inputs for PCA. PCA using residuals is recommended when, as in this case, the units for each variable vary significantly in magnitude (Johnson and Wichern, 1988). Factors with an eigenvalue ⩾1.0 were identified based on the commonly used Kaiser criterion (Kaiser, 1960). Varimax rotation was then performed to produce interpretable factors. Metabolites with a factor load ⩾∣0.4∣ are reported as composing a given factor, as is commonly used as an arbitrary threshold (Lawlor et al, 2004). Scoring coefficients were then used to compute factor scores for each individual (consisting of a weighted sum of the values of the standardized metabolites within that factor, weighted on the factor loading calculated for each individual metabolite). These factor scores were then used to calculate heritabilities for each factor with SOLAR as detailed above, using a polygenic model not further adjusted for covariates. Removal of 1–4 of the most extreme values for several of the factors was necessary to achieve a normal residual kurtosis.
As all analyses were exploratory in nature and given collinearity of the metabolites, nominal two-sided P-values unadjusted for multiple comparisons are presented; however, results interpreted in the context of a conservative Bonferroni correction are reported in the Discussion. Nominal statistical significance was defined as P-value⩽0.05. Statistical analyses used SAS version 9.1 (SAS Institute, Cary, NC), other than for heritability estimates that used SOLAR (Almasy and Blangero, 1998).
Supplementary Material
Supplementary Materials This file contains: Supplement 1. Pedigrees of the eight multiplex GENECARD families. Supplement 2. Clinical characteristics of GENECARD families. Supplement 3. Heritabilities, clinical covariates, and household effects for individual metabolites. Supplement 4. Nomenclature and Intra-Individual Variability of Metabolites. Supplement 5. Internal spiked standards for acylcarnitine and amino acid measurements. Supplement 6. Correlation matrix for metabolites. Supplementary Information
Dataset Supplementary Data
Acknowledgments
We acknowledge Lauren C Naliboff, Stephanie Decker and Sarah Nelson for their excellent technical assistance with this study. We are particularly grateful for the time and effort devoted by study participants. This study was supported by the American Heart Association (Fellow-to-Faculty Award, SH Shah) and the National Institutes of Health (NIH R01 HL073389, ER Hauser).
Footnotes
The authors declare that they have no conflict of interest.
References
- Almasy L, Blangero J (1998) Multipoint quantitative-trait linkage analysis in general pedigrees. Am J Hum Genet 62: 1198–1211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- An J, Muoio DM, Shiota M, Fujimoto Y, Cline GW, Shulman GI, Koves TR, Stevens R, Millington D, Newgard CB (2004) Hepatic expression of malonyl-CoA decarboxylase reverses muscle, liver and whole-animal insulin resistance. Nat Med 10: 268–274 [DOI] [PubMed] [Google Scholar]
- Beekman M, Heijmans BT, Martin NG, Pedersen NL, Whitfield JB, DeFaire U, van Baal GC, Snieder H, Vogler GP, Slagboom PE, Boomsma DI (2002) Heritabilities of apolipoprotein and lipid levels in three countries. Twin Res 5: 87–97 [DOI] [PubMed] [Google Scholar]
- Brindle JT, Antti H, Holmes E, Tranter G, Nicholson JK, Bethell HW, Clarke S, Schofield PM, McKilligin E, Mosedale DE, Grainger DJ (2002) Rapid and noninvasive diagnosis of the presence and severity of coronary heart disease using 1H-NMR-based metabonomics. Nat Med 8: 1439–1444 [DOI] [PubMed] [Google Scholar]
- Chace DH, Hillman SL, Millington DS, Kahler SG, Roe CR, Naylor EW (1995) Rapid diagnosis of maple syrup urine disease in blood spots from newborns by tandem mass spectrometry. Clin Chem 41: 62–68 [PubMed] [Google Scholar]
- Ferrara CT, Wang P, Neto EC, Stevens RD, Bain JR, Wenner BR, Ilkayeva OR, Keller MP, Blasiole DA, Kendziorski C, Yandell BS, Newgard CB, Attie AD (2008) Genetic networks of liver metabolism revealed by integration of metabolic and transcriptional profiling. PLoS Genet 4: e1000034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haqq AM, Lien LF, Boan J, Arlotto M, Slentz CA, Muehlbauer MJ, Rochon J, Gallup D, McMahon RL, Bain JR, Stevens R, Millington D, Butler MD, Newgard CB, Svetkey LP (2005) The Study of the Effects of Diet on Metabolism and Nutrition (STEDMAN) weight loss project: rationale and design. Contemp Clin Trials 26: 616–625 [DOI] [PubMed] [Google Scholar]
- Hauser ER, Mooser V, Crossman DC, Haines JL, Jones CH, Winkelmann BR, Schmidt S, Scott WK, Roses AD, Pericak-Vance MA, Granger CB, Kraus WE (2003) Design of the Genetics of Early Onset Cardiovascular Disease (GENECARD) study. Am Heart J 145: 602–613 [DOI] [PubMed] [Google Scholar]
- Johnson RA, Wichern DW (1988) Applied Multivariate Statistical Analysis. Englewood Cliffs, New Jersey: Prentice Hall [Google Scholar]
- Kaiser HF (1960) The application of electronic computers to factor analysis. Educ Psychol Meas 20: 141–151 [Google Scholar]
- Keurentjes JJ, Fu J, de Vos CH, Lommen A, Hall RD, Bino RJ, van der Plas LH, Jansen RC, Vreugdenhil D, Koornneef M (2006) The genetics of plant metabolism. Nat Genet 38: 842–849 [DOI] [PubMed] [Google Scholar]
- Kirschenlohr HL, Griffin JL, Clarke SC, Rhydwen R, Grace AA, Schofield PM, Brindle KM, Metcalfe JC (2006) Proton NMR analysis of plasma is a weak predictor of coronary artery disease. Nat Med 12: 705–710 [DOI] [PubMed] [Google Scholar]
- Koves TR, Li P, An J, Akimoto T, Slentz D, Ilkayeva O, Dohm GL, Yan Z, Newgard CB, Muoio DM (2005) Peroxisome proliferator-activated receptor-gamma co-activator 1alpha-mediated metabolic remodeling of skeletal myocytes mimics exercise training and reverses lipid-induced mitochondrial inefficiency. J Biol Chem 280: 33588–33598 [DOI] [PubMed] [Google Scholar]
- Koves TR, Ussher JR, Noland RC, Slentz D, Mosedale M, Ilkayeva O, Bain J, Stevens R, Dyck JR, Newgard CB, Lopaschuk GD, Muoio DM (2008) Mitochondrial overload and incomplete fatty acid oxidation contribute to skeletal muscle insulin resistance. Cell Metab 7: 45–56 [DOI] [PubMed] [Google Scholar]
- Lawlor DA, Ebrahim S, May M, Davey SG (2004) (Mis)use of factor analysis in the study of insulin resistance syndrome. Am J Epidemiol 159: 1013–1018 [DOI] [PubMed] [Google Scholar]
- Millington DS, Kodo N, Norwood DL, Roe CR (1990) Tandem mass spectrometry: a new method for acylcarnitine profiling with potential for neonatal screening for inborn errors of metabolism. J Inherit Metab Dis 13: 321–324 [DOI] [PubMed] [Google Scholar]
- Muoio DM, Newgard CB (2008) Mechanisms of disease: molecular and metabolic mechanisms of insulin resistance and beta-cell failure in type 2 diabetes. Nat Rev Mol Cell Biol 9: 193–205 [DOI] [PubMed] [Google Scholar]
- Patterson BW, Zhao G, Elias N, Hachey DL, Klein S (1999) Validation of a new procedure to determine plasma fatty acid concentration and isotopic enrichment. J Lipid Res 40: 2118–2124 [PubMed] [Google Scholar]
- Petersen KF, Befroy D, Dufour S, Dziura J, Ariyan C, Rothman DL, DiPietro L, Cline GW, Shulman GI (2003) Mitochondrial dysfunction in the elderly: possible role in insulin resistance. Science 300: 1140–1142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rissanen AM (1979) Familial occurrence of coronary heart disease: effect of age at diagnosis. Am J Cardiol 44: 60–66 [DOI] [PubMed] [Google Scholar]
- Sabatine MS, Liu E, Morrow DA, Heller E, McCarroll R, Wiegand R, Berriz GF, Roth FP, Gerszten RE (2005) Metabolomic identification of novel biomarkers of myocardial ischemia. Circulation 112: 3868–3875 [DOI] [PubMed] [Google Scholar]
- Shah S, Kraus W, Crossman D, Granger C, Haines J, Jones C, Mooser V, Huang L, Haynes C, Dowdy E, Vega G, Grundy S, Vance J, Hauser E (2006) Serum lipids in the GENECARD study of coronary artery disease identify quantitative trait loci and phenotypic subsets on chromosomes 3q and 5q. Ann Hum Genet 70: 738–748 [DOI] [PubMed] [Google Scholar]
- Shea S, Ottman R, Gabrieli C, Stein Z, Nichols A (1984) Family history as an independent risk factor for coronary artery disease. J Am Coll Cardiol 4: 793–801 [DOI] [PubMed] [Google Scholar]
- World Health Organization. The World Health Report 2002—Reducing Risks, Promoting Healthy Life. Geneva: World Health Organization, 2002 [Google Scholar]
- Wu JY, Kao HJ, Li SC, Stevens R, Hillman S, Millington D, Chen YT (2004) ENU mutagenesis identifies mice with mitochondrial branched-chain aminotransferase deficiency resembling human maple syrup urine disease. J Clin Invest 113: 434–440 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Materials This file contains: Supplement 1. Pedigrees of the eight multiplex GENECARD families. Supplement 2. Clinical characteristics of GENECARD families. Supplement 3. Heritabilities, clinical covariates, and household effects for individual metabolites. Supplement 4. Nomenclature and Intra-Individual Variability of Metabolites. Supplement 5. Internal spiked standards for acylcarnitine and amino acid measurements. Supplement 6. Correlation matrix for metabolites. Supplementary Information
Dataset Supplementary Data