Abstract
Crosstalk mechanisms have not been studied as thoroughly as individual signaling pathways. We exploit experimental and computational approaches to reveal how a concordant interplay between the insulin and epidermal growth factor (EGF) signaling networks can potentiate mitogenic signaling. In HEK293 cells, insulin is a poor activator of the Ras/ERK (extracellular signal-regulated kinase) cascade, yet it enhances ERK activation by low EGF doses. We find that major crosstalk mechanisms that amplify ERK signaling are localized upstream of Ras and at the Ras/Raf level. Computational modeling unveils how critical network nodes, the adaptor proteins GAB1 and insulin receptor substrate (IRS), Src kinase, and phosphatase SHP2, convert insulin-induced increase in the phosphatidylinositol-3,4,5-triphosphate (PIP3) concentration into enhanced Ras/ERK activity. The model predicts and experiments confirm that insulin-induced amplification of mitogenic signaling is abolished by disrupting PIP3-mediated positive feedback via GAB1 and IRS. We demonstrate that GAB1 behaves as a non-linear amplifier of mitogenic responses and insulin endows EGF signaling with robustness to GAB1 suppression. Our results show the feasibility of using computational models to identify key target combinations and predict complex cellular responses to a mixture of external cues.
Keywords: computational modeling, crosstalk, epidermal growth factor receptor, insulin receptor, networks
Introduction
Cells respond to a myriad of external cues using a limited number of signaling pathways that convert multiple inputs into diverse cellular decisions. Although individual receptors and pathways have been extensively studied, it is not understood how signaling networks integrate multiple cues. The epidermal growth factor (EGF) receptor (EGFR) and the insulin receptor (IR) belong to the family of receptors with intrinsic tyrosine kinase activity (referred to as receptor tyrosine kinases, RTKs), which regulate pivotal cellular processes, including proliferation, differentiation, cell metabolism, survival, and apoptosis (Schlessinger, 2000; Taniguchi et al, 2006). The main physiological function of insulin signaling is metabolic, involving the control of glucose metabolism and stimulation of protein and lipid syntheses, whereas EGF induces proliferative responses.
The EGFR and IR networks share many downstream components. Under some conditions, EGF can evoke metabolic responses, e.g., GLUT4 translocation (Ishii et al, 1994; Gogg and Smith, 2002), whereas insulin can be mitogenic, especially in cancer cells (Ish-Shalom et al, 1997; Papa et al, 1997). There is evidence that insulin can enhance EGF-stimulated extracellular signal-regulated kinase (ERK) activation, DNA synthesis, and cell proliferation responses (Crouch et al, 2000; Ediger and Toews, 2000; Chong et al, 2004), whereas growth factors, cytokines, and other hormones can negatively regulate insulin signaling (Gual et al, 2003). Yet, despite experimental evidence of crosstalk between insulin and growth factor pathways, it is unknown how combined EGF and insulin inputs are processed into integrative cellular response. This is at least in part due to the combinatorial complexity of molecular interactions and a variety of feedback and feed-forward loops, whose concerted operation is difficult to comprehend intuitively.
Ligand binding and subsequent autophosphorylation of tyrosine residues on the EGFR and IR triggers mobilization of multiple adaptors, such as Src (src avian sarcoma viral oncogene homolog) homology and collagen domain protein (Shc), growth factor receptor binding protein 2 (Grb2), insulin receptor substrate family members (IRS1–6, GAB1–3), and enzymes that contain characteristic domains recognizing receptor phosphotyrosines, such as phosphatidylinositol 3-kinase (PI3K), soluble tyrosine kinase c-Src, protein tyrosine phosphatases (e.g., SHP2 and PTP1B) and others (White, 1998; Sebastian et al, 2006). Subsequently, EGF and insulin-induced signals propagate through multiple interacting branches, including the mitogen-activated protein kinase (MAPK) cascade downstream of the small membrane-anchored GTPase Ras and the PI3K/AKT cell survival pathway.
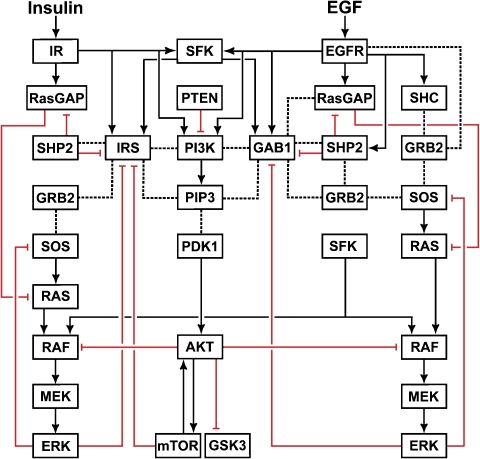
The flow chart in Figure 1 shows that the same key signal transducers are activated by both EGFR and IR, either directly or indirectly. Importantly, although either receptor can stimulate both the Ras/ERK and PI3K/AKT pathways, the major routes of activation are different. IR phosphorylates IRS proteins, which are linked to the activation of Ras/ERK pathway through binding to the Grb2–SOS complex (SOS is a guanine nucleotide exchange factor for Ras), whereas EGFR activates the same pathway either by direct binding of Grb2-SOS or by binding and phosphorylation of Shc, which then recruits the Grb2–SOS complex (White, 1998; Sebastian et al, 2006). Likewise, the PI3K/AKT pathway is activated by IR via either direct or IRS-mediated recruitment of PI3K, whereas EGFR-mediated PI3K activation occurs mainly via a more intricate route that involves EGFR- and Src-induced phosphorylation of the Grb2-associated binder 1 (GAB1) (Rodrigues et al, 2000; Gu and Neel, 2003; Kiyatkin et al, 2006).
Figure 1.
Flow chart of signal propagation through the EGFR and IR signaling networks. Solid lines with arrows show the activation or tyrosine phosphorylation of proteins and lipids. Dotted lines represent direct protein–protein and protein–lipid interactions. Red lines with blunt ends show inhibition.
There are several points of crosstalk between the EGFR and IR signaling networks (Figure 1). Following the initial activation of PI3K, the production of phosphatidylinositol-3,4,5-triphosphate (PIP3) in the plasma membrane leads to the membrane recruitment of GAB1 and IRS proteins through their pleckstrin homology (PH) domains. The membrane-recruited and subsequently tyrosine-phosphorylated GAB1 and IRS influence the Ras/ERK and PI3K/AKT pathways in multiple ways. They bind p85, the regulatory subunit of PI3K, and alleviate the intrinsic inhibition of PI3K, which further increases PIP3 production, thereby generating a positive feedback and a crosstalk point between EGFR and IR (Ogawa et al, 1998; Gu and Neel, 2003). Both GAB1 and IRS can recruit Grb2–SOS, leading to Ras activation (Myers et al, 1994; Lewitzky et al, 2001; Weng et al, 2001), or can bind the GTPase-activating protein RasGAP, which catalyzes Ras deactivation (Montagner et al, 2005). An important crosstalk point emerges because of the binding of SH2 domain-containing tyrosine protein phosphatase 2 (SHP2) to the phosphorylated GAB1 and IRS proteins, which results in both positive and negative regulations of downstream signaling (Noguchi et al, 1994; Yamauchi et al, 1995; Myers et al, 1998; Yart et al, 2001; Asante-Appiah and Kennedy, 2003). Intriguingly, although tyrosine phosphatase SHP2 negatively regulates IRS, GAB1 and PI3K/AKT signaling (Noguchi et al, 1994; Asante-Appiah and Kennedy, 2003), it positively influences ERK activity, which is partly explained by dephosphorylation of the specific sites involved in RasGAP binding (Cunnick et al, 2002; Agazie and Hayman, 2003; Montagner et al, 2005; Stoker, 2005). Downstream targets of EGFR and IR, such as ERK, GSK3 (glycogen synthase kinase 3) or mTOR (the mammalian target of rapamycin), feedback and phosphorylate GAB1 and IRS on serine/threonine residues, which disable tyrosine phosphorylation at sites engaging PI3K, Grb2, RasGAP or SHP2 (Paz et al, 1997; Gu and Neel, 2003; Johnston et al, 2003). These negative feedback loops generate additional crosstalk points between EGFR and IR.
Although many mechanisms of EGFR–IR crosstalk are well characterized at the molecular level, this knowledge is insufficient to understand cellular responses to EGF plus insulin at the systems level, owing to the multitude of interpathway interactions and feedback loops. This paper brings together experimental studies of combined EGF and insulin signaling with computational modeling of the interactive EGFR and IR networks. We show that, although in HEK293 cells insulin by itself is a poor activator of ERK, it greatly enhances MAPK pathway activation by physiological EGF concentrations. The computational model elucidates the function of feedback loops and crosstalk nodes in combined EGF and insulin signaling. We demonstrate that synergistic activation of the mitogenic pathway by EGF plus insulin primarily occurs upstream of and at the Ras/Raf level. This potentiation of Ras/ERK response is initiated by insulin-induced PIP3 increase, which leads to subsequent increases in membrane recruitment of Grb2–SOS and SHP2 by GAB1 and IRS. The computational model unveils that insulin makes the mitogenic EGFR signal more robust toward GAB1 knockdown. Our results may have important ramifications for the identification of therapeutic targets aimed at eliminating insulin-induced amplification of mitogenic and survival signaling stimulated by low growth factor levels in tumor cells.
Results
Building a computational model of the EGF and insulin signaling networks
We have developed a computational model to describe in quantitative terms how cell stimulation by EGF and insulin is linked to the activation of Ras/ERK and PI3K/AKT pathways. The current model stems from our previously developed EGFR network models that were based on in vitro and in vivo measurements of signaling kinetics. A number of EGFR signaling model predictions were validated in our own studies (Kholodenko et al, 1999; Moehren et al, 2002; Kiyatkin et al, 2006; Birtwistle et al, 2007) and, in addition, tested by other groups (Schoeberl et al, 2002; Hatakeyama et al, 2003; Resat et al, 2003; Blinov et al, 2006). This paper extends our previous models to incorporate IR signaling and regulatory processes involved in EGFR–IR crosstalk. However, we do not create a combinatorially complex in silico replica of all distinct biochemical species and interactions, which would be impractical (Borisov et al, 2005; Hlavacek et al, 2006; Birtwistle et al, 2007). Instead, we construct a basic, minimal model of the combined EGFR and IR networks. The goal of this model is to provide an insight into the mechanisms of cellular responses to combined EGF and insulin treatment that can account for our data.
The model involves 78 variables for different molecular species, 111 chemical reactions (processes) and more than 200 parameters. A list of reactions, rate equations and parameter values is given in the Supplementary Table S1, and the model SBML file is provided. For many reactions, the parameter values are quantitatively consistent with the previously published values, whose derivation is fully documented in the papers by Kholodenko et al (1999), Moehren et al (2002), and Markevich et al (2004a, 2004b). For additional processes and parameters that describe multi-step processes as single reactions, Supplementary Table S1 cites the corresponding references or indicates that the parameter value was optimized using a training set of data (see Materials and methods). Below, we describe the major signaling processes that are considered and analyzed by this model.
In the model, signal transduction is initiated by ligand (EGF or/and insulin) binding to their cognate receptors. This causes dimerization and autophosphorylation of EGFR, or an allosteric transition and autophosphorylation of the kinase activation loop of the predimerized IR, which leads to activation of the IR kinase and autophosphorylation of its cytoplasmic domain (De Meyts and Whittaker, 2002; Sebastian et al, 2006). The model considers that phosphorylated EGFR can directly bind Shc, Grb2–SOS, PI3K, protein phosphatase(s) and RasGAP (Jones et al, 2006; Sebastian et al, 2006). The membrane recruitment of cytoplasmic SOS is critical for the initiation of the Ras/ERK pathway by both EGFR and IR (Medema et al, 1993; Aronheim et al, 1994; Kholodenko, 2000). Interestingly, the direct recruitment of the Grb2–SOS complex by EGFR was shown to be a less effective route of Ras activation than the EGFR–Shc–Grb2–SOS mediated pathway owing to the corresponding binding affinities (Ravichandran et al, 1995; Kholodenko et al, 1999). At the membrane, SOS catalyzes the transformation of Ras-GDP into active Ras-GTP, whereas RasGAP catalyzes the reverse process of Ras deactivation.
In the model, phosphorylated IR can directly associate with IRS, PI3K, phosphatase(s), and RasGAP (Staubs et al, 1994). The IRS family of major IR docking proteins consists of at least six members, IRS1-6; however, the model considers only a single ‘representative' IRS protein. Importantly in the model, Src is strongly activated by EGFR and more weakly by IR (Schmelzle et al, 2006). Following initial PI3K stimulation and production of PIP3, IRS and GAB1 bind PIP3 and get phosphorylated by IR or EGFR/Src. Phosphorylated IRS and GAB1 recruit cytoplasmic proteins PI3K, Grb2–SOS, SHP2, and RasGAP to the plasma membrane, which results in additional PIP3 production and both activatory and inhibitory regulations of Ras activity (Myers et al, 1994; Ogawa et al, 1998; Gu and Neel, 2003). PIP3 is converted back to phosphatidylinositol-4,5-diphosphate (PIP2) by PTEN (phosphatase and tensin homologue) (Weng et al, 2001). Although both IRS and GAB1 have multiple tyrosine phosphorylation sites, the minimal model represents them by a single, virtual phosphorylation site (Birtwistle et al, 2007). There are some experimental data supporting this assumption and showing that binding of multiple SH2 domain-containing proteins correlates with the overall phosphorylation levels of GAB1 (Figure 4D and E below) and IRS (Goldstein et al, 2000).
Where appropriate, we describe complex yet sequential multi-step processes as a single, semi-mechanistic step. As these condensed processes are sequential, our simplifications allow the reduced model to retain the original network topology. For instance, the activation of Raf by Ras includes a conformational change in Raf caused by binding to Ras-GTP, followed by the dissociation of 14-3-3 protein, dephosphorylation of inhibitory S259 and phosphorylation of activatory S338 sites (Dhillon et al, 2002). In the model, all these processes are considered as a single partial step of Raf activation. The complete Raf activation requires tyrosine phosphorylation by kinases, which are thought to belong to the Src family kinases (SFK) (Wellbrock et al, 2004). In the model, these kinases are linked to Src activity, which is differently stimulated by EGFR or IR. Likewise, in the absence of evidence for a distributed mechanism of ERK kinase (MEK) activation by Raf (Kolch, 2005), we use a simplified, one-step description, whereas a distributed mechanism of ERK activation by MEK and deactivation by MKP3 is described as a two-step process (Markevich et al, 2004a).
The model also incorporates and analyzes complex feedback circuitry of the EGFR and IR networks. For instance, PIP3-dependent positive feedback circuits in the model involve GAB1–PI3K and IRS–PI3K interactions (Rodrigues et al, 2000; Johnston et al, 2003; Mattoon et al, 2004). Activated ERK inhibits SOS (Dong et al, 1996; Fucini et al, 1999), GAB1 (Lehr et al, 2004) and IRS (De Fea and Roth, 1997) by direct phosphorylation. Activated mTOR mediates multiple modes of feedback, including positive feedback to AKT and negative feedback loops to IRS (Gual et al, 2003; Sarbassov et al, 2005). Although AKT-induced inhibitory phosphorylation of Raf (Zimmermann and Moelling, 1999; Wellbrock et al, 2004) is included in the model, we assume this inhibition to be weak in HEK293 cells, as no noticeable MEK or ERK activation was detected experimentally, following inhibition of AKT activity (see Supplementary Figure S4). The current model involves many parameters that have no analogs in our previously published models. We used the experimental data that are shown in Figures 2 and 3 (excluding experiments with PI3K inhibitor) as a training data set to obtain reasonable fit between the model simulations and data by manually varying the parameter values (see Supplementary Table S1). However, when parameters were fitted, their upper and lower bounds were in agreement with experimental observations for similar reaction types. In addition, reaction rates were always constrained not to be faster than the diffusion limit.
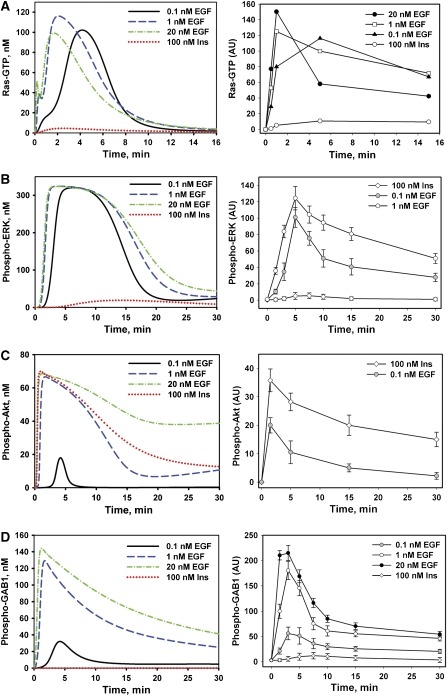
Figure 2.
Dynamics of EGF or insulin-induced Ras-GTP, ERK and AKT activation, and GAB1 phosphorylation. The left panels show the time courses calculated in silico and right panels show the corresponding time courses measured in HEK293 cells stimulated with insulin (Ins, 100 nM) or EGF (0.1, 1 or 20 nM) for the indicated time intervals (min). Active GTP-bound Ras was immunoprecipitated (IP) from total cell lysates (TCL) by the agarose-conjugated Ras-binding domain (RBD) of Raf as described in Materials and methods. Proteins from Ras-IP or TCL were separated on LDS-PAGE (4–12%), transferred to nitrocellulose membrane, and immunoblotted (IB) with anti-Ras (A) or anti-phospho-ERK1/2 (T202/Y204), anti-phospho-AKT (S473) or anti-phospho-GAB1 (Y627) antibodies (B–D), respectively. The signal intensities of phosphorylated ERK1/2, AKT, or GAB1 normalized against the appropriate signal of α-tubulin protein level are expressed in arbitrary units (AU). Data shown are the mean of normalized signal intensities±s.d. from five independent experiments each performed in triplicates.
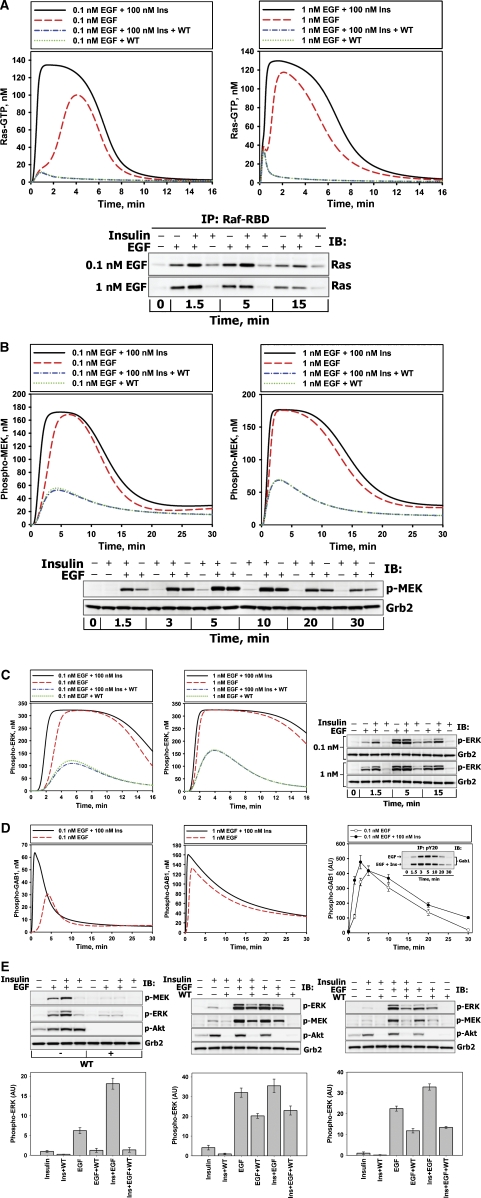
Figure 3.
Insulin amplifies EGF-induced Ras/MAPK pathway activation at low EGF doses. Comparison of the calculated in silico dynamics of Ras-GTP (A), phospho-MEK (B), phospho-ERK1/2 (C), and phospho-GAB1 (D) stimulated with EGF (0.1 or 1 nM) or EGF plus insulin (EGF+Ins) in the absence or presence of PI3K inhibitor wortmannin (WT) with the corresponding kinetic measurements (shown in bottom (A, B) or right (C, D) panels) carried out in HEK293 cells stimulated with EGF (0.1, 1 or 20 nM) or co-stimulated with insulin (100 nM) plus EGF (+ or − indicate the presence or absence of the ligand). Grb2 levels serve as a loading control to show that equal amounts of protein were loaded per lane. Representative blots are shown (n=3). (E) HEK293 cells were pretreated with 100 nM WT (+) or equivalent amounts of solvent DMSO (−) for 30 min and stimulated with 0.1 nM EGF or 100 nM insulin or both ligands simultaneously for 1.5 min (left panel), 5 min (middle panel) or 15 min (right panel). Immunoblots were analyzed for phosphorylated MEK (S217/221), ERK1/2 (T202/Y204), or AKT (S473) (representative blots on the upper part of each panel). The ligand-induced ERK responses are expressed in arbitrary units (AU) (mean±s.d., n=7).
Dose dependence of responses to EGF and insulin
Steady-state plasma concentrations of EGF are reported to be in the range of 0.05–0.2 nM, which is much lower than the concentration range commonly used in the studies on isolated cells (Jansson et al, 1993; Lemos-Gonzalez et al, 2007). For HEK293 cells, which express endogenous EGFR and IR, we measured both EGF and insulin dose dependencies. We found that half-maximal ERK activation is attained at about 0.1–0.2 nM EGF (Supplementary Figure S1A). Half-maximal phosphorylation of IR was reached at about 50–100 nM insulin, which was sufficient to saturate ERK activation (Supplementary Figure S1B). To exclude crosstalk at the receptor level, we measured EGFR activation by insulin (and vice versa), and found that neither EGF transactivated IR, nor insulin transactivated EGFR (Supplementary Figure S2A). Although EGF was reported to induce phosphorylation of IRS proteins in A431 cells that express high levels of EGFR (Fujioka et al, 2001), EGF could not induce significant IRS phosphorylation in HEK293 cells (Supplementary Figure S2B), which is consistent with other studies (Kadowaki et al, 1987). By contrast, insulin treatment resulted in potent phosphorylation of IRS1 at Y612 (Supplementary Figure S2B), which is one of the nine putative PI3K binding sites (White, 1998). Robust phosphorylation of these sites by IR correlates with a stronger activation of the PI3K target AKT by insulin than by EGF, although the basal level of AKT phosphorylation in HEK293 cells is significant (Supplementary Figure S2B). However, even at saturating concentrations, insulin is a much weaker activator of ERK than physiological, low EGF (Supplementary Figure S2B), which is also supported by independent observations (Weng et al, 2001).
Signaling dynamics of EGF and insulin-induced responses
To analyze the signaling dynamics, we measured experimentally and simulated in silico the time course of activation of Ras/ERK and PI3K/AKT pathways stimulated by step changes in the EGF and insulin concentrations. First, cells were stimulated with 100 nM insulin or with several EGF concentrations that ranged from low concentrations of 0.1 nM to saturating concentrations of 20 nM (Figure 2). Both the experimental data and simulations showed that the activation of the Ras/ERK and PI3K/AKT pathways by persistent EGF or insulin stimulation was transient (Figure 2 and Supplementary Figure S3). The model explains this transient behavior by multiple negative feedback regulations mediated by ERK, AKT and mTOR. In fact, disruption of negative feedback loops in silico transforms transient Ras/ERK and PI3K/AKT pathway responses into sustained responses (Supplementary Figure S5).
Our data demonstrate weak Ras and ERK (Figure 2A and B) activation by insulin compared with EGF. The model provides several arguments to explain these observations, which involve signaling processes both upstream and downstream of Ras. First, IR binds and phosphorylates Shc with very low efficacy compared with EGFR ((Paz et al, 1996, 1997; Weng et al, 2001) and Supplementary Figure S2B), whereas in EGF signaling, the Shc-dependent pathways contribute substantially to the Grb2–SOS membrane recruitment and subsequent Ras activation (Kholodenko et al, 1999). Second, IRS1-6 docking proteins, when phosphorylated by IR, bind preferably PI3K, whereas their binding to Grb2–SOS is weak. Therefore, IRS phosphorylation does not lead to significant Ras activation in the model (Sasaoka et al, 1996; Fucini et al, 1999). Third, IR is a much weaker stimulator of Src activity than EGFR (Schmelzle et al, 2006), which has the following ramifications in the model: (a) although insulin triggers the PIP3-mediated membrane recruitment of GAB1, the phospho-GAB1 level remains much lower than this level for EGF stimulation, when GAB1 is phosphorylated by both EGFR and Src (see Figure 2D, left and right panels compare the model simulations and data); (b) the lower level of GAB1 phosphorylation by insulin results in less effective Grb2–SOS recruitment and Ras activation (Figure 2A); and (c) finally, downstream of Ras in the model, full Raf activation requires Src activity, which is weaker for insulin than for EGF. Although a higher AKT activation by insulin than by EGF may contribute to additional Raf inhibition, our data suggest that this inhibition is insignificant in HEK293 cells (Supplementary Figure S4). Overall, the simulations qualitatively agree with the data (Figure 2) for a wide range of parameter values for the proposed regulatory structure of the model.
Insulin potentiates MAPK responses to physiological EGF concentrations
Our experimental and simulation results show that co-stimulation by low, physiological EGF (0.1 nM) and insulin synergistically activates the Ras/ERK pathway (Figure 3A–C, quantitation of the blots shown in Figure 3A–C is presented in Supplementary Figure S6A–C). Figure 3E demonstrates that at 1.5 min (left panel) and 15 min (right panel) following co-stimulation with insulin and EGF, the concentration of active ERK is 2.57±0.44 and 1.4±0.1 times greater, respectively, than the sum of the active ERK levels observed with each ligand (this synergistic increase is statistically significant, P<0.01, n=5). Importantly, the synergy effect is not a consequence of additional activation of EGFR and/or IR by co-stimulation with two ligands (Supplementary Figure S2A). Neither can it be explained by activation of insulin-like growth factor receptor-1 (IGF-1R) by insulin. Although 100 nM insulin weakly activates IGF-1 receptor at 1.5 min (Supplementary Figure S2C), 10 nM insulin, which does not transactivate IGF-1R (Supplementary Figure S2C), induces the comparable synergistic effect when combined with 0.1 or 1 nM EGF (Supplementary Figure S2D). When ERK activation is maximal, the synergy becomes insignificant (P>0.05) for any studied EGF dose (Figure 3C and E). Furthermore, the synergistic activation of ERK is less pronounced at higher EGF (1 nM, Figure 3C), and disappears at saturating EGF (20 nM, Supplementary Figure S7).
One group of crosstalk interactions that facilitate the ERK response is located upstream of Ras. These interactions affect multiple Ras activation and deactivation routes involving the adaptor proteins GAB1 and IRS and the protein phosphatase SHP2. In the model, co-stimulation of cells by EGF and insulin increases the amount of PIP3 produced by PI3K (relative to EGF stimulation alone) and further facilitates the GAB1 membrane recruitment and its subsequent tyrosine phosphorylation (Figure 3D, left and middle panels). An increase in membrane-bound phospho-GAB1 is corroborated by our data and it is more appreciable at physiological EGF levels (Supplementary Figure S8B, Figure 3D, right panel, and Supplementary Figure S8A).
An increase in the phosphorylation level of membrane-bound GAB1 promotes Grb2–SOS binding, increasing the SOS concentration in close proximity to Ras (Kholodenko et al, 2000). At the same time, this gain in phospho-GAB1 also increases the amount of RasGAP (Ras deactivator) and the phosphatase SHP2 bound to GAB1. In the model, this phosphatase negatively regulates the level of phosphorylated receptors, IRS, and GAB1; yet it has the pronounced positive effect on Ras activation, as we showed using a specific SHP2 inhbitor, NSC-87877 (Supplementary Figure S9A). This positive effect is related to the formation of the GAB1–SHP2 and IRS–SHP2 complexes in close proximity to the plasma membrane and subsequent dephosphorylation of the docking sites on EGFR, IR, GAB1, and IRS, which are involved in RasGAP binding. Thus, SHP2 has an essential role in inhibiting RasGAP-catalyzed Ras deactivation, only if there is a strong activatory signal to Ras arising from EGF co-stimulation, whereas the IRS–SHP2 complexes cannot effectively activate Ras on their own. Simulations predict that the net result of these insulin plus EGF-induced interactions is an increase in positive signaling and decrease in negative signaling to Ras. This amplifies the Ras-GTP level, which is corroborated by the experimental data (Figure 3A).
The second group of crosstalk interactions occurs downstream of Ras and involves processes with positive and negative effects on ERK responses. In the model, at any given Ras-GTP load, the simultaneous exposure to insulin and EGF increases Raf activity relative to insulin alone, owing to EGF-induced stimulation of SFK, which further activates Raf. At the same time, an increase in AKT activity brought about by simultaneous stimulation with insulin and EGF should negatively influence Raf. Yet, the experimental data (Supplementary Figure S4) and simulations suggest that in HEK293 cells the negative influence of AKT on Raf is negligible in comparison with increased Raf activation, owing to increases in Src and Ras activities. Thus, the net effect of insulin-EGF co-stimulation on MEK and ERK activation is positive (Figure 3B and C).
PI3K inhibition suppresses insulin–EGF synergy in Ras/ERK responses
To test the model predictions on the PIP3/GAB1-mediated signaling routes that result in synergistic Ras/MAPK activation by insulin and EGF, the cells were pretreated with the PI3K inhibitor wortmannin (WT). As expected, 100 nM WT prevented AKT phosphorylation in response to insulin and EGF (Figure 3E). By contrast, tyrosine phosphorylation of EGFR, IR and Shc was not affected by PI3K inhibition (Supplementary Figure S10 and Kiyatkin et al, 2006).
Although the model is trained to describe the experimental time-series data for unperturbed cells, simulations of pharmacological interventions, such as inhibition of network nodes and small interfering RNA (siRNA) experiments (see below), were not fitted to the data. Rather, the model predictions are merely compared with the experimental data. The simulations and data suggest that EGF-induced MEK/ERK activation is inhibited by WT due to the disruption of GAB1–PI3K positive feedback. The model predicts that owing to inhibition of the GAB1 and IRS–SHP2 membrane recruitment, WT suppresses synergistic amplification of Ras-GTP/MEK/ERK responses, which is supported by our experimental data (Figure 3A–C and E). The model simulations suggest that although WT disrupts the EGF–insulin synergy, the maximal activation of MEK and ERK remains significant, following 5-min stimulation with EGF or EGF plus insulin. The data presented in Figure 3E (middle panel) validate the model prediction and also show that insulin is unable to enhance MEK and ERK when PI3K is inhibited (Figure 3E, all panels). The agreement between the model predictions and experimental observations substantiates the computational model.
GAB1 complexes with Grb2, SHP2 and PI3K measured by immunoprecipitation analysis
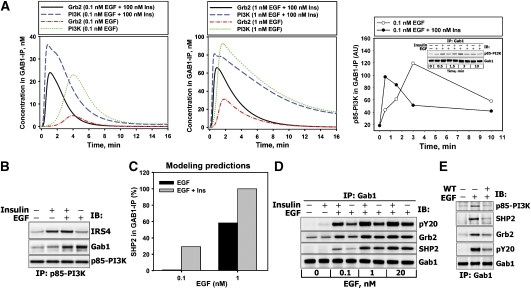
Quantitative data on the dynamics of protein–protein complexes can further validate the model and provide insight into mechanisms of integrative cellular responses to insulin–EGF co-stimulation. As the data (Figure 3D and Supplementary Figure S8) suggest that GAB1 is one of the key mediators of insulin–EGF crosstalk, we determined GAB1 interactions with other network nodes. Although it is not immediately obvious, the simulations predict that following co-stimulation with insulin and EGF, the amount of GAB1-bound PI3K will increase (compared with EGF stimulation) only on a short time scale (Figure 4A, blue dashed lines versus green dotted lines), whereas the post-peak concentration of GAB1-bound PI3K will decrease owing to dephosphorylation of GAB1 phosphotyrosines by GAB1-bound SHP2 and the competitive recruitment of PI3K by phosphorylated IRS. To test this prediction, the binding of PI3K and SHP2 to GAB1 and of PI3K to IRS4 (the major IRS protein in these cells (Fantin et al, 1998)) was measured by immunoprecipitation analysis following stimulation of HEK293 cells with EGF, insulin and their combination. The data confirm that insulin–EGF co-stimulation increased the PI3K–GAB1 complex concentration at 30 s and 1.5 min, whereas the post-peak levels were decreased compared with EGF only stimulation, in agreement with in silico predictions (Figure 4A, right panel). At 3 min of stimulation with insulin and EGF, the data show robust PI3K–IRS binding response and decreased PI3K–GAB1 binding, compared with EGF alone (Figure 4B). The predicted increase in GAB1-bound Grb2 (Figure 4A, solid versus dash–dotted lines) and SHP2 (Figure 4C) correlates with an increase in the total level of GAB1 tyrosine phosphorylation (Figures 3D and 4E) and is supported by the data shown in Figure 4D for 1.5-min stimulation (quantitation of the blots shown in Figure 4B, D and E is presented in Supplementary Figure S11A–C).
Figure 4.
Association of Grb2, PI3K, or SHP2 with GAB1 in response to EGF, insulin, or their combination. (A) The left and middle panels show simulated dynamics of GAB1-containing complexes (scales on the Y axis are different). The right panel shows quantitation of p85–PI3K in GAB1-IP (see Materials and methods). (B) HEK293 cells were stimulated with 100 nM insulin and/or 1 nM EGF for 1.5 min. p85–PI3K–IP was analyzed for IRS4, GAB1, or p85–PI3K proteins. (C) Modeling predictions of relative SHP2 levels in GAB1-IP at 1.5 min after stimulation with 100 nM insulin and/or 0.1 and 1 nM EGF. (D) HEK293 cells were stimulated with 100 nM insulin and/or 0.1, 1, and 20 nM EGF for 1.5 min. GAB1-IP was analyzed for total phosphotyrosines (pY20), Grb2, SHP2, or GAB1 proteins. (E) HEK293 cells were pretreated with 100 nM WT (+) or equivalent amounts of solvent DMSO (−) for 30 min and stimulated with 1 nM EGF for 3 min. GAB1-IP was analyzed for total phosphotyrosines (pY20), Grb2, SHP2, p85–PI3K, or GAB1 proteins. Representative blots are shown in each panel (n=3).
As WT disrupts GAB1–PI3K positive feedback, the total GAB1 phosphorylation level and the concentrations of GAB1-bound Grb2, SHP2, and PI3K decrease dramatically in WT-treated cells (Figure 4E). We conclude that the loss of insulin–EGF synergy caused by WT arises from the loss of the GAB1-mediated membrane recruitment of signaling molecules. The following section presents direct in silico and experimental analyses of GAB1 depletion.
Computational predictions and experimental validation of the GAB1, SHP2, and Src control of ERK responses to EGF and insulin
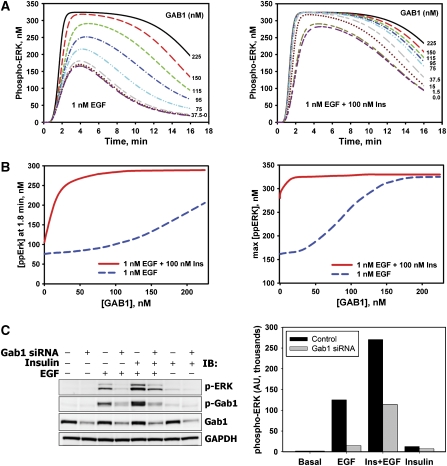
We have recently shown that RNA interference (RNAi)-mediated GAB1 depletion can reduce the peak amplitude and decrease the duration of ERK activation (Kiyatkin et al, 2006). These findings are also supported by independent data (Meng et al, 2005). To get further insight into the mechanisms of crosstalk, we simulated the dynamics of ERK responses to EGF versus EGF plus insulin in cells where the GAB1 protein level was held at different levels with respect to control (Figure 5A). As expected, GAB1 suppression reduces the phospho-ERK level, and its decline is more pronounced for EGF (Figure 5A, left panel) than for EGF plus insulin (Figure 5A, right panel). To test the model, HEK293 cells were transfected with siRNA against human GAB1, resulting in ∼75% reduced GAB1 protein level relative to control (non-targeting siRNA-transfected cells) (Figure 5C, quantitation of blots is shown in Supplementary Figure S11D). The phospho-ERK levels measured at 1.5 min following stimulation (Figure 5C) are consistent with the simulations (Figure 5B, left). These results confirm the in silico prediction of the larger influence of GAB1 depletion on EGF- rather than on EGF plus insulin-induced ERK phosphorylation.
Figure 5.
Effects of GAB1 depletion on ERK activation induced by EGF, insulin, or their combination. (A) Computational analysis of ERK activation kinetics in response to 0.1 nM EGF in the absence (left panel) or presence (right panel) of 100 nM insulin at the indicated levels of GAB1 protein. Time courses are shown for 225 nM (control; black—solid line), 150 nM (red—long dash line), 115 nM (dark yellow—short–long–short dash line), 95 nM (dark pink—long–short–short dash line), 75 nM (green—short dash line), 37.5 nM (blue—dash–dot line), 15.0 nM (cyan—dash–dot–dot line), 1.5 nM (gray—long–short dash line) and 0 nM (dark red—dotted line) GAB1 concentrations. (B) Simulated dependences of phospho-ERK level at 1.8 min (left panel) and maximal phospho-ERK level (right panel) on the GAB1 abundance for cells stimulated with 0.1 nM EGF in the presence (red solid line) or absence (blue dashed line) of 100 nM insulin. (C) HEK293 cells transfected with specific siRNA against GAB1 (+) or non-targeting siRNA (−) were stimulated with 100 nM insulin and/or 1 nM EGF for 1.5 min. Immunoblots were analyzed for phosphorylated ERK1/2 (T202/Y204) or GAB1 (Y627). GAB1 protein levels demonstrate the efficacy of GAB1 suppression. GAPDH was used as a loading control. Representative blot (left panel) and the bar graph of respective numerical values (right panel) are shown.
The computational results unveil that insulin endows the mitogenic EGFR pathway with increased robustness towards GAB1 downregulation. The simulations show that for EGF plus insulin stimulation, the peak level of phospho-ERK decreases only slightly with GAB1 depletion, whereas for EGF-induced ERK activation, the peak level decreases significantly (Figure 5B, right panel). The difference between EGF- and insulin plus EGF-induced maximal ERK activity is larger in cells with low GAB1 expression than in cells with high GAB1 expression (Figure 5B, right panel), although the absolute value of the peak ERK response is greater for larger GAB1 abundance (Figure 5A). This dependence of the gain in ERK signaling on the GAB1 abundance resembles the behavior of a non-linear amplifier, in which a change in the input does not produce a proportional change in the output. Moreover, the GAB1-mediated amplification of ERK signaling depends on the particular cellular context.
The model predicts that expression of GAB1 PH domain deletion mutant, which is unable to bind PIP3, affects ERK activation in a similar manner as GAB1 knockdown (Supplementary Figure S12A and Figure 5A, left panel). Likewise, co-stimulation with EGF and insulin makes ERK activation less sensitive to such GAB1 mutation (Supplementary Figure S12A and Figure 5A, right panel). The effects of mutant GAB1 expression or GAB1 suppression differ from the inhibition of PIP3 production by wortmannin (Figure 3C and E). When PIP3 is present in the membrane, but GAB1 is depleted or mutated, PIP3 facilitates the formation of PIP3–IRS–SHP2 complexes, which partially compensate the lack of PIP3–GAB1–Grb2–SOS and PIP3–GAB1–SHP2 complexes by IRS–SHP2-dependent suppression of RasGAP signaling. The simulations show that when IRS–SHP2 interactions are disrupted, synergistic increase in ERK activity by EGF plus insulin co-stimulation can be observed before, but not after the time when ERK reaches its peak activity (Supplementary Figure S12B). Disruption of GAB1–SHP2 interactions substantially decreases the duration of ERK signaling induced by EGF, while the insulin plus EGF synergy is observed (Supplementary Figure S12C). The effects of other perturbations to the system are presented in Supplementary Figures S12D and S12E. The lack of GAB1 phosphorylation by EGFR leads to phospho-ERK changes that are qualitatively similar to the changes induced by the lack of GAB1–PIP3 binding (Supplementary Figure S12D), whereas the effects of IRS depletion are qualitatively similar to the effects of disruption of IRS–SHP2 interactions (Supplementary Figure S12E).
The phospho-ERK levels measured in Src-depleted cells (Supplementary Figure S13A) agree reasonably well with the corresponding model predictions (Supplementary Figure S13C). Neither EGFR nor IR autophosphorylation levels were affected by Src downregulation (data not shown). Moreover, we found that RNAi-mediated suppression of Src in HEK293 cells led to a decrease in GAB1 phosphorylation that was more pronounced for EGF than for insulin. Considering a substantial Src contribution to GAB1 phosphorylation under various conditions (Nishida et al, 1999; Bisotto and Fixman, 2001; Chan et al, 2003; Jin et al, 2005), the model explains this observation by the fact that Src is preferably activated by EGFR and not by IR.
At the same time, the model predicts that the influence of SHP2 activity inhibition on ERK phosphorylation will be pronounced after 5 or more minutes following the onset of stimulation (Supplementary Figure S13D), whereas the experimental data show that this effect is already substantial at 1.5 min after stimulation (Supplementary Figure S13B). This shows the limitations of the minimal model, which should be improved in the future studies.
Discussion
Physiological stimuli never act in isolation, and cells in the body are often exposed to insulin and EGF at the same time. Receptors, adaptors, and enzymes involved in insulin and EGF signaling are relatively well-described and form complex networks with intricate interaction circuitry. Downstream of IR and EGFR, these networks significantly overlap. This paper employs a combined experimental and computational approach to address the question of how multiple crosstalk mechanisms in the IR and EGFR networks operate together in a cell-dependent context.
The dynamics of ERK activation depends on a complex interplay of a multitude of regulatory signals and interactions induced by combined insulin and EGF stimulation. Using only qualitative arguments, it is almost impossible to predict the ERK dynamics that results from multiple non-linear interactions and feedback loops, and a testable computational model helps us provide insights into key causative relationships (Kholodenko, 2006). Previous experimental studies indicated that insulin enhances growth-factor-dependent mitogenesis, but the molecular mechanisms remained obscure, as insulin treatment alone fails to induce mitogenesis in most cell types. Using ERK phosphorylation as a marker for mitogenic signaling, we show that insulin leads to the co-localization of mitogenic signaling intermediates at the plasma membrane, whereas EGF-dependent processes contribute to tyrosine phosphorylation of key intermediates. Insulin thereby indirectly boosts signaling events induced by physiological, low EGF doses, despite insulin itself being unable to directly trigger mitogenic signaling. Our results suggest that the regulation of PI3K signaling by non-mitogenic stimuli might be an important means to control the key quantitative aspects of growth-factor-induced MAPK signaling (e.g., signal amplitude and duration).
Our results suggest that co-stimulation with low, physiological EGF doses and insulin endows the mitogenic EGFR pathway with more sustained ERK signaling. Early observations showed that the duration of ERK activation is crucial for cell fate decisions (Marshall, 1995). In these classical experiments, PC12 cells proliferated after transient ERK activation by EGF, but differentiated after more sustained ERK activation by the nerve growth factor. Subsequent work revealed that the duration of ERK activation is sensed by a network of immediate early genes, including the transcription factor c-Fos (Murphy et al, 2002), while the differential timing of ERK signaling is explained by subtle alterations in signaling circuits, induced by distinct ligands (Kholodenko, 2007).
Crosstalk interactions in receptor-induced Ras-MAPK signaling typically occur upstream or at the level of Raf (Katz et al, 2007). Our computational and experimental results also confirm that the major interactions involved in insulin–EGF crosstalk and responsible for amplification of ERK signaling in HEK293 cells operate at the level of protein adaptors, such as GAB1 and IRS, and at the Ras/Raf level. However, in other cellular systems PI3K-induced crosstalk interactions can occur downstream of Ras/Raf. For instance, PIP3 generated by PI3K enables Rac activation. Subsequently, Rac activates PAK, which in turn stimulates Raf by S338 phosphorylation and MEK by S298 phosphorylation (Chaudhary et al, 2000; Xiang et al, 2002). An alternative pathway of PIP3-dependent ERK activation involves phosphoinositide-dependent kinase-1 (PDK1), a target of PI3K, which can activate MEK directly (Sato et al, 2004) and ERK indirectly through the activation of certain PKC isoforms (Myers et al, 1996; Schonwasser et al, 1998; Newton, 2003), although it has been reported that PKCs are not involved in EGF-induced ERK activation (Xu et al, 2002; Robin et al, 2004). However, in HEK293 cells, we found that the inhibition of c-Raf by GW-5074 eliminated MEK and ERK activities (Supplementary Figure S14), which supports the conclusion that major insulin–EGF crosstalk mechanisms to amplify ERK signaling are localized upstream of Ras and at the Ras/Raf level.
The model analyzed interactions that involve five key network nodes, the adaptor proteins GAB1 and IRS, PI3K/PIP3, soluble tyrosine kinase Src and protein tyrosine phosphatase SHP2, which are all localized upstream of MEK/ERK. Together, these nodes contribute to the amplification of ERK signaling in the context of multiple interactions, which involve insulin and EGFRs, Grb2–SOS, RasGAP, as well as positive and negative feedback controls from downstream targets of insulin and EGF. The model explains how the network structures, which involve (i) GAB1-mediated insulin and EGF coincidence detection, (ii) coherent feed-forward loop from EGFR to Raf via Src, and (iii) multiple positive (PI3K–PIP3–GAB1–PI3K) and negative (ERK–GAB1, ERK–SOS, mTOR–IRS) feedback loops contribute to the control of insulin plus EGF signaling (a network motif where an initial input signal (A) induces an intermediate input signal (B), and both the initial and intermediate inputs are needed to generate the final output (C), is referred as a coherent feed-forward loop (Mangan et al, 2003)). The simplified scheme shown in Figure 6 illustrates how insulin enhances EGF-induced mitogenesis through two partially redundant and compensating signaling branches via IRS and GAB1. The goal of our computational model is to predict and explain a large number of diverse patterns of signaling dynamics obtained from our own experiments and literature data. We applied in silico perturbations to our model and, wherever possible, verified the predicted temporal dynamics in vivo. Our strategy is similar to the so-called pattern-oriented modeling, in which a large number of quantitative and also qualitative patterns help us exclude models that are too simple in structure or too uncertain experimentally (Grimm et al, 2005).
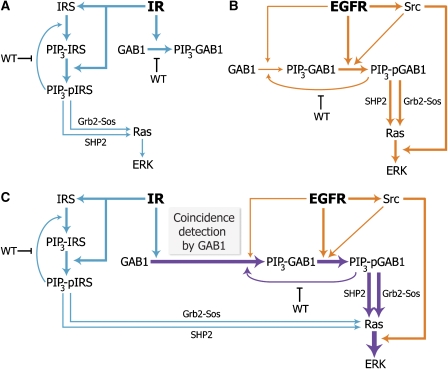
Figure 6.
Mechanisms of insulin–EGF signal integration. (A) Weak ERK activation in response to insulin. IR is a strong activator of PI3K signaling, and insulin stimulation induces pronounced adaptor recruitment to the membrane (GAB1 → PIP3-GAB1; IRS → PIP3-IRS). The membrane-associated IRS proteins (but not GAB1) are the preferred substrates of the IR kinase (PIP3-IRS → PIP3-pIRS). However, phosphorylated IRS proteins do not effectively recruit Ras activators, such as Grb2–SOS, implying that insulin is a weak activator of ERK signaling. (B) ERK activation in response to physiological EGF doses. Active EGFR phosphorylates the membrane-associated GAB1 (PIP3-GAB1 → PIP3-pGAB1), which in turn efficiently recruits Grb2–SOS and SHP2, and thereby activates Ras-MAPK signaling (for simplicity, the Shc-dependent pathway of EGFR-induced MAPK activation is not shown). However, ERK phosphorylation induced by low doses of EGF may remain suboptimal, as the GAB1 recruitment to the membrane is submaximal and highly transient (GAB1 → PIP3-GAB1). GAB1 phosphorylation and Raf activation are enhanced by EGFR-induced Src kinase activity. (C) ERK activation can increase upon insulin and EGF co-stimulation. Synergistic ERK activation predominantly arises from coincidence detection of insulin and EGF stimuli at the level of GAB1; the GAB1 adaptor is massively recruited to the membrane by IR signaling (GAB1 → PIP3-GAB1) and subsequently phosphorylated by EGFR and active Src (PIP3-GAB1 → PIP3-pGAB1). The relative activities of the processes are illustrated by the boldness of the arrows.
We tested the model by using perturbations, including different ligand concentrations, specific inhibitors, and RNAi-mediated suppression of key protein levels. These perturbations corresponded to single-parameter changes, which represent the greatest challenge for any model, because a multitude of temporal patterns have to be described without changes in the rate constants. Our model generated qualitative predictions of the temporal dynamics for diverse cellular responses. In particular, downregulation of GAB1 and Src expression levels using siRNA, or inhibition of PI3K resulted in a significant decrease in synergistic ERK activation by insulin plus EGF. The simulated ERK dynamics in cells with different GAB1 expression (Figure 5A and B) suggest that insulin significantly increases robustness of ERK activation to GAB1 downregulation. The experimental data for GAB1-depleted HEK293 cells demonstrate a significantly larger decrease in the ERK active fraction for EGF than for EGF plus insulin, supporting the modeling results (Figure 5C).
Yet, the systems biology approach, while revealing a critical role of GAB1, also points to the fact that this adaptor is just one of several critical players in a complex set of interactions that generate crosstalk between the EGFR and IR networks. Despite the fact that RNAi-mediated knockdown of GAB1 expression significantly decreases ERK signaling (Figure 5C), the downregulation of additional critical nodes is required to uncouple and completely suppress insulin and EGF-induced Ras/ERK activation. Overall, the analysis presented here demonstrates the feasibility of using computational models to identify critical combinations of therapeutic targets and predict their effects on complex cellular responses to concurrent external cues.
Materials and methods
Antibodies
Rabbit anti-phospho-MEK (S217/S221, anti-phospho-Shc (Y317), anti-IGF-1 receptor β (all from Cell Signaling)), anti-phospho-IR β (Y1162/1163), anti-GRB2 (C-23), anti-GAB1 (H-198), anti-phospho-GAB1 (Y627), anti-SH-PTP2 (C-18) (all from Santa Cruz Biotechnology), anti-phospho-EGFR (Y1173), anti-phospho-IRS1 (Y612) (BD Biosciences), anti-PI3K–p85 α, anti-GAB1 (CT) (all from Millipore); mouse anti-phospho-ERK1/2 (T202/185 and Y204/187), anti-phospho-AKT1 (S473) (587F11) (both from Cell Signaling), anti-α-tubulin (DM1A), anti-Src (GD11) (both from Millipore), anti-Ras (BD Biosciences), anti-glyceraldehyde-3-phosphate dehydrogenase (GAPDH) (6C5) (Chemicon) and anti-phosphotyrosine (pY20) (BioLegend). Secondary horse anti-mouse HRP-linked IgG antibodies were from Cell Signaling, HRP-conjugated ImmunoPure goat anti-rabbit IgG antibodies were from Pierce. HRP-conjugated Protein A was from Millipore.
Other reagents
Human recombinant EGF and IGF-1 were from PeproTech, and insulin was from SIGMA. Wortmannin (WT, PI3K inhibitor) (Cell Signaling), NSC-87877 (SHP1/2 inhibitor), AKT-VIII (AKT 1/2/3 inhibitor) (both from Calbiochem) and GW-5074 (Tocris Biosciences) were dissolved in DMSO and used at indicated concentrations. Annealed, desalted GAB1 and c-Src siGENOME SMARTpool siRNAs and siCONTROL Non-Targeting siRNA Pool were from Dharmacon. siRNA stock solution was prepared according to the manufacturer's instructions and stored at −80°C. Other chemical reagents were from Fisher Scientific.
Cell culture and transfection
HEK293 (ATCC No. CRL-1573) were cultured in DMEM/F-12 (GIBCO) supplemented with 10% FBS and penicillin–streptomycin solution (Mediatech). The protocol of RNAi-mediated transient gene silencing using synthetic siRNA was described previously (Kiyatkin et al, 2006).
Cell stimulation and protein extraction
For ligand-response studies, the cells were starved from 12 to 16 h in FBS-free medium and stimulated with the indicated ligand concentrations. Preparation of total cell lysates and subcellular fractionation for signaling analysis was performed as described previously (Kiyatkin et al, 2006).
Protein immunoprecipitation
50 μl of recombinant protein A–Sepharose 4B (Sigma-Aldrich) 50% beads slurry was preincubated with the indicated primary antibody (8 μg) while gently mixing for 2 h at room temperature. The beads were incubated with 500 μg of total lysate protein for 4 h at 4°C. After the incubation, the beads were washed twice with ice-cold IP buffer (20 mM HEPES (pH 7.4), 150 mM NaCl, 0.1% Triton-X and 10% glycerol) and once with PBS. The proteins were eluted in NuPAGE LDS Sample Buffer (25% 4 × NuPAGE LDS+10% 10 × NuPAGE Sample Reducing Agent + 65% PBS), heated for 5 min at 75°C and spun down.
Ras activation assay
Active (GTP-bound) Ras from 500 μg total cell lysates was captured with 30 μl of Raf-1 Ras binding domains (RBD) bound to glutathione-agarose beads (Millipore) for 3 h at 4°C. Protein complexes were collected by brief centrifugation and washed three times with ice-cold IP buffer supplemented with 10 mM MgCl2. Ras-GTP was released from agarose beads with NuPAGE LDS sample buffer.
Electrophoresis and immunoblotting procedures were conducted according to the methods described in Aksamitiene et al (2007).
Data evaluation
Chemiluminescence signals from immunoreactive bands were detected on a KODAK Image Station 440CF after treatment with a working solution of SuperSignal West Dura Extended Duration Substrate (Pierce Biotechnology) and quantified using KODAK Digital Science software (Kodak Scientific Imaging Systems). To enable comparison, the capture times and number of frames were set equal for each separately exposed membrane, or the whole membranes were exposed simultaneously. Results were plotted and statistically analyzed using SigmaPlot and Microsoft Excel software.
Computational modeling analysis
Numerical integration was performed using Dbsolve and PottersWeel. Dbsolve (Goryanin et al, 1999) is based on numerical techniques, developed by Khibnik et al (1993). The program files are freely available together with the model SBML file. Ranges of kinetic parameters were constrained based on the literature data and in vivo measurements of the signaling kinetic. The list of the quantitative parameters is presented in Supplementary Table S1, which documents how these parameters were determined. The parameter values marked ‘fitted' were varied manually within the upper and lower bounds taken from the experiment to describe a training data set, which includes the experimental time courses in Figures 2 and 3 obtained for an unperturbed HEK293 system. The simulations of the perturbation results, such as inhibition of PI3K, SHP2 and EGFR, and GAB1, IRS or Src depletion, were not fitted to the data. Rather, the model predictions were merely compared with the experimental data.
Supplementary Material
Supplementary Figures 1 - 14, Supplementary Table 1
Supplementary Figure 6
Supplementary Figure 6
Supplementary Figure 6
Supplementary Figure 8
Acknowledgments
This work was supported by the NIH Grants GM059570 and R33HL088283 (a part of the NHLBI Exploratory Program in Systems Biology).
Footnotes
The authors declare that they have no conflict of interest.
References
- Agazie YM, Hayman MJ (2003) Molecular mechanism for a role of SHP2 in epidermal growth factor receptor signaling. Mol Cell Biol 23: 7875–7886 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aksamitiene E, Hoek JB, Kholodenko B, Kiyatkin A (2007) Multistrip Western blotting to increase quantitative data output. Electrophoresis 28: 3163–3173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aronheim A, Engelberg D, Li N, al-Alawi N, Schlessinger J, Karin M (1994) Membrane targeting of the nucleotide exchange factor Sos is sufficient for activating the Ras signaling pathway. Cell 78: 949–961 [DOI] [PubMed] [Google Scholar]
- Asante-Appiah E, Kennedy BP (2003) Protein tyrosine phosphatases: the quest for negative regulators of insulin action. Am J Physiol Endocrinol Metab 284: E663–E670 [DOI] [PubMed] [Google Scholar]
- Birtwistle MR, Hatakeyama M, Yumoto N, Ogunnaike BA, Hoek JB, Kholodenko BN (2007) Ligand-dependent responses of the ErbB signaling network: experimental and modeling analyses. Mol Syst Biol 3:144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bisotto S, Fixman ED (2001) Src-family tyrosine kinases, phosphoinositide 3-kinase and Gab1 regulate extracellular signal-regulated kinase 1 activation induced by the type A endothelin-1 G-protein-coupled receptor. Biochem J 360: 77–85 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blinov ML, Faeder JR, Goldstein B, Hlavacek WS (2006) A network model of early events in epidermal growth factor receptor signaling that accounts for combinatorial complexity. Biosystems 83: 136–151 [DOI] [PubMed] [Google Scholar]
- Borisov NM, Markevich NI, Hoek JB, Kholodenko BN (2005) Signaling through receptors and scaffolds: independent interactions reduce combinatorial complexity. Biophys J 89: 951–966 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan PC, Chen YL, Cheng CH, Yu KC, Cary LA, Shu KH, Ho WL, Chen HC (2003) Src phosphorylates Grb2-associated binder 1 upon hepatocyte growth factor stimulation. J Biol Chem 278: 44075–44082 [DOI] [PubMed] [Google Scholar]
- Chaudhary A, King WG, Mattaliano MD, Frost JA, Diaz B, Morrison DK, Cobb MH, Marshall MS, Brugge JS (2000) Phosphatidylinositol 3-kinase regulates Raf1 through Pak phosphorylation of serine 338. Curr Biol 10: 551–554 [DOI] [PubMed] [Google Scholar]
- Chen L, Sung SS, Yip ML, Lawrence HR, Ren Y, Guida WC, Sebti SM, Lawrence NJ, Wu J (2006) Discovery of a novel shp2 protein tyrosine phosphatase inhibitor. Mol Pharmacol 70: 562–570 [DOI] [PubMed] [Google Scholar]
- Chong MP, Barritt GJ, Crouch MF (2004) Insulin potentiates EGFR activation and signaling in fibroblasts. Biochem Biophys Res Commun 322: 535–541 [DOI] [PubMed] [Google Scholar]
- Crouch MF, Davy DA, Willard FS, Berven LA (2000) Insulin induces epidermal growth factor (EGF) receptor clustering and potentiates EGF-stimulated DNA synthesis in Swiss 3T3 cells: a mechanism for costimulation in mitogenic synergy. Immunol Cell Biol 78: 408–414 [DOI] [PubMed] [Google Scholar]
- Cunnick JM, Meng S, Ren Y, Desponts C, Wang HG, Djeu JY, Wu J (2002) Regulation of the mitogen-activated protein kinase signaling pathway by SHP2. J Biol Chem 277: 9498–9504 [DOI] [PubMed] [Google Scholar]
- De Fea K, Roth RA (1997) Modulation of insulin receptor substrate-1 tyrosine phosphorylation and function by mitogen-activated protein kinase. J Biol Chem 272: 31400–31406 [DOI] [PubMed] [Google Scholar]
- De Meyts P, Whittaker J (2002) Structural biology of insulin and IGF1 receptors: implications for drug design. Nat Rev Drug Discov 1: 769–783 [DOI] [PubMed] [Google Scholar]
- Dhillon AS, Meikle S, Yazici Z, Eulitz M, Kolch W (2002) Regulation of Raf-1 activation and signalling by dephosphorylation. EMBO J 21: 64–71 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong C, Waters SB, Holt KH, Pessin JE (1996) SOS phosphorylation and disassociation of the Grb2-SOS complex by the ERK and JNK signaling pathways. J Biol Chem 271: 6328–6332 [DOI] [PubMed] [Google Scholar]
- Ediger TL, Toews ML (2000) Synergistic stimulation of airway smooth muscle cell mitogenesis. J Pharmacol Exp Ther 294: 1076–1082 [PubMed] [Google Scholar]
- Fantin VR, Sparling JD, Slot JW, Keller SR, Lienhard GE, Lavan BE (1998) Characterization of insulin receptor substrate 4 in human embryonic kidney 293 cells. J Biol Chem 273: 10726–10732 [DOI] [PubMed] [Google Scholar]
- Fucini RV, Okada S, Pessin JE (1999) Insulin-induced desensitization of extracellular signal-regulated kinase activation results from an inhibition of Raf activity independent of Ras activation and dissociation of the Grb2-SOS complex. J Biol Chem 274: 18651–18658 [DOI] [PubMed] [Google Scholar]
- Fujioka T, Kim JH, Adachi H, Saito K, Tsujimoto M, Yokoyama S, Ui M (2001) Further evidence for the involvement of insulin receptor substrates in epidermal growth factor-induced activation of phosphatidylinositol 3-kinase. Eur J Biochem 268: 4158–4168 [DOI] [PubMed] [Google Scholar]
- Gogg S, Smith U (2002) Epidermal growth factor and transforming growth factor alpha mimic the effects of insulin in human fat cells and augment downstream signaling in insulin resistance. J Biol Chem 277: 36045–36051 [DOI] [PubMed] [Google Scholar]
- Goldstein BJ, Bittner-Kowalczyk A, White MF, Harbeck M (2000) Tyrosine dephosphorylation and deactivation of insulin receptor substrate-1 by protein-tyrosine phosphatase 1B. Possible facilitation by the formation of a ternary complex with the Grb2 adaptor protein. J Biol Chem 275: 4283–4289 [DOI] [PubMed] [Google Scholar]
- Goryanin I, Hodgman TC, Selkov E (1999) Mathematical simulation and analysis of cellular metabolism and regulation. Bioinformatics 15: 749–758 [DOI] [PubMed] [Google Scholar]
- Grimm V, Revilla E, Berger U, Jeltsch F, Mooij WM, Railsback SF, Thulke HH, Weiner J, Wiegand T, DeAngelis DL (2005) Pattern-oriented modeling of agent-based complex systems: lessons from ecology. Science 310: 987–991 [DOI] [PubMed] [Google Scholar]
- Gu H, Neel BG (2003) The ‘Gab' in signal transduction. Trends Cell Biol 13: 122–130 [DOI] [PubMed] [Google Scholar]
- Gual P, Gremeaux T, Gonzalez T, Le Marchand-Brustel Y, Tanti JF (2003) MAP kinases and mTOR mediate insulin-induced phosphorylation of insulin receptor substrate-1 on serine residues 307, 612 and 632. Diabetologia 46: 1532–1542 [DOI] [PubMed] [Google Scholar]
- Hatakeyama M, Kimura S, Naka T, Kawasaki T, Yumoto N, Ichikawa M, Kim JH, Saito K, Saeki M, Shirouzu M, Yokoyama S, Konagaya A (2003) A computational model on the modulation of mitogen-activated protein kinase (MAPK) and Akt pathways in heregulin-induced ErbB signalling. Biochem J 373: 451–463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hlavacek WS, Faeder JR, Blinov ML, Posner RG, Hucka M, Fontana W (2006) Rules for modeling signal-transduction systems. Sci STKE 2006: re6. [DOI] [PubMed] [Google Scholar]
- Ish-Shalom D, Christoffersen CT, Vorwerk P, Sacerdoti-Sierra N, Shymko RM, Naor D, De Meyts P (1997) Mitogenic properties of insulin and insulin analogues mediated by the insulin receptor. Diabetologia 40(Suppl 2): S25–S31 [DOI] [PubMed] [Google Scholar]
- Ishii K, Kamohara S, Hayashi H, Todaka M, Kanai F, Imanaka T, Ebina Y (1994) Epidermal growth factor triggers the translocation of insulin-responsive glucose transporter (GLUT4). Biochem Biophys Res Commun 205: 857–863 [DOI] [PubMed] [Google Scholar]
- Jansson PA, Fowelin JP, von Schenck HP, Smith UP, Lonnroth PN (1993) Measurement by microdialysis of the insulin concentration in subcutaneous interstitial fluid. Importance of the endothelial barrier for insulin. Diabetes 42: 1469–1473 [DOI] [PubMed] [Google Scholar]
- Jin ZG, Wong C, Wu J, Berk BC (2005) Flow shear stress stimulates Gab1 tyrosine phosphorylation to mediate protein kinase B and endothelial nitric-oxide synthase activation in endothelial cells. J Biol Chem 280: 12305–12309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnston AM, Pirola L, Van Obberghen E (2003) Molecular mechanisms of insulin receptor substrate protein-mediated modulation of insulin signalling. FEBS Lett 546: 32–36 [DOI] [PubMed] [Google Scholar]
- Jones RB, Gordus A, Krall JA, MacBeath G (2006) A quantitative protein interaction network for the ErbB receptors using protein microarrays. Nature 439: 168–174 [DOI] [PubMed] [Google Scholar]
- Kadowaki T, Koyasu S, Nishida E, Tobe K, Izumi T, Takaku F, Sakai H, Yahara I, Kasuga M (1987) Tyrosine phosphorylation of common and specific sets of cellular proteins rapidly induced by insulin, insulin-like growth factor I, and epidermal growth factor in an intact cell. J Biol Chem 262: 7342–7350 [PubMed] [Google Scholar]
- Katz M, Amit I, Yarden Y (2007) Regulation of MAPKs by growth factors and receptor tyrosine kinases. Biochim Biophys Acta 1773: 1161–1176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khibnik AI, Kuznetsov YA, Levitin VV, Nikolaev EV (1993) Continuation techniques and interactive software for bifurcation analysis of ODEs and iterated maps. Physica D 62: 360–371 [Google Scholar]
- Kholodenko BN (2000) Negative feedback and ultrasensitivity can bring about oscillations in the mitogen-activated protein kinase cascades. Eur J Biochem 267: 1583–1588 [DOI] [PubMed] [Google Scholar]
- Kholodenko BN (2006) Cell-signalling dynamics in time and space. Nat Rev Mol Cell Biol 7: 165–176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kholodenko BN (2007) Untangling the signalling wires. Nat Cell Biol 9: 247–249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kholodenko BN, Demin OV, Moehren G, Hoek JB (1999) Quantification of short term signaling by the epidermal growth factor receptor. J Biol Chem 274: 30169–30181 [DOI] [PubMed] [Google Scholar]
- Kholodenko BN, Hoek JB, Westerhoff HV (2000) Why cytoplasmic signalling proteins should be recruited to cell membranes. Trends Cell Biol 10: 173–178 [DOI] [PubMed] [Google Scholar]
- Kiyatkin A, Aksamitiene E, Markevich NI, Borisov NM, Hoek JB, Kholodenko BN (2006) Scaffolding protein Grb2-associated binder 1 sustains epidermal growth factor-induced mitogenic and survival signaling by multiple positive feedback loops. J Biol Chem 281: 19925–19938 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolch W (2005) Coordinating ERK/MAPK signalling through scaffolds and inhibitors. Nat Rev Mol Cell Biol 6: 827–837 [DOI] [PubMed] [Google Scholar]
- Lehr S, Kotzka J, Avci H, Sickmann A, Meyer HE, Herkner A, Muller-Wieland D (2004) Identification of major ERK-related phosphorylation sites in Gab1. Biochemistry 43: 12133–12140 [DOI] [PubMed] [Google Scholar]
- Lemos-Gonzalez Y, Rodriguez-Berrocal FJ, Cordero OJ, Gomez C, Paez de la Cadena M (2007) Alteration of the serum levels of the epidermal growth factor receptor and its ligands in patients with non-small cell lung cancer and head and neck carcinoma. Br J Cancer 96: 1569–1578 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewitzky M, Kardinal C, Gehring NH, Schmidt EK, Konkol B, Eulitz M, Birchmeier W, Schaeper U, Feller SM (2001) The C-terminal SH3 domain of the adapter protein Grb2 binds with high affinity to sequences in Gab1 and SLP-76 which lack the SH3-typical P-x-x-P core motif. Oncogene 20: 1052–1062 [DOI] [PubMed] [Google Scholar]
- Mangan S, Zaslaver A, Alon U (2003) The coherent feedforward loop serves as a sign-sensitive delay element in transcription networks. J Mol Biol 334: 197–204 [DOI] [PubMed] [Google Scholar]
- Markevich NI, Hoek JB, Kholodenko BN (2004a) Signaling switches and bistability arising from multisite phosphorylation in protein kinase cascades. J Cell Biol 164: 353–359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Markevich NI, Moehren G, Demin O, Kiyatkin A, Hoek JB, Kholodenko BN (2004b) Signal processing at the Ras circuit: What shapes Ras activation patterns? IEE Syst Biol 1: 104–113 [DOI] [PubMed] [Google Scholar]
- Marshall CJ (1995) Specificity of receptor tyrosine kinase signaling: transient versus sustained extracellular signal-regulated kinase activation. Cell 80: 179–185 [DOI] [PubMed] [Google Scholar]
- Mattoon DR, Lamothe B, Lax I, Schlessinger J (2004) The docking protein Gab1 is the primary mediator of EGF-stimulated activation of the PI-3K/Akt cell survival pathway. BMC Biol 2: 24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medema RH, de Vries-Smits AM, van der Zon GC, Maassen JA, Bos JL (1993) Ras activation by insulin and epidermal growth factor through enhanced exchange of guanine nucleotides on p21ras. Mol Cell Biol 13: 155–162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meng S, Chen Z, Munoz-Antonia T, Wu J (2005) Participation of both Gab1 and Gab2 in the activation of the ERK/MAPK pathway by epidermal growth factor. Biochem J 391: 143–151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moehren G, Markevich N, Demin O, Kiyatkin A, Goryanin I, Hoek JB, Kholodenko BN (2002) Temperature dependence of the epidermal growth factor receptor signaling network can be accounted for by a kinetic model. Biochemistry 41: 306–320 [DOI] [PubMed] [Google Scholar]
- Montagner A, Yart A, Dance M, Perret B, Salles JP, Raynal P (2005) A novel role for Gab1 and SHP2 in epidermal growth factor-induced Ras activation. J Biol Chem 280: 5350–5360 [DOI] [PubMed] [Google Scholar]
- Murphy LO, Smith S, Chen RH, Fingar DC, Blenis J (2002) Molecular interpretation of ERK signal duration by immediate early gene products. Nat Cell Biol 4: 556–564 [DOI] [PubMed] [Google Scholar]
- Myers MG Jr, Mendez R, Shi P, Pierce JH, Rhoads R, White MF (1998) The COOH-terminal tyrosine phosphorylation sites on IRS-1 bind SHP-2 and negatively regulate insulin signaling. J Biol Chem 273: 26908–26914 [DOI] [PubMed] [Google Scholar]
- Myers MG Jr, Wang LM, Sun XJ, Zhang Y, Yenush L, Schlessinger J, Pierce JH, White MF (1994) Role of IRS-1-GRB-2 complexes in insulin signaling. Mol Cell Biol 14: 3577–3587 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myers MG Jr, Zhang Y, Aldaz GA, Grammer T, Glasheen EM, Yenush L, Wang LM, Sun XJ, Blenis J, Pierce JH, White MF (1996) YMXM motifs and signaling by an insulin receptor substrate 1 molecule without tyrosine phosphorylation sites. Mol Cell Biol 16: 4147–4155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newton AC (2003) Regulation of the ABC kinases by phosphorylation: protein kinase C as a paradigm. Biochem J 370: 361–371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishida K, Yoshida Y, Itoh M, Fukada T, Ohtani T, Shirogane T, Atsumi T, Takahashi-Tezuka M, Ishihara K, Hibi M, Hirano T (1999) Gab-family adapter proteins act downstream of cytokine and growth factor receptors and T- and B-cell antigen receptors. Blood 93: 1809–1816 [PubMed] [Google Scholar]
- Noguchi T, Matozaki T, Horita K, Fujioka Y, Kasuga M (1994) Role of SH-PTP2, a protein-tyrosine phosphatase with Src homology 2 domains, in insulin-stimulated Ras activation. Mol Cell Biol 14: 6674–6682 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogawa W, Matozaki T, Kasuga M (1998) Role of binding proteins to IRS-1 in insulin signalling. Mol Cell Biochem 182: 13–22 [PubMed] [Google Scholar]
- Papa V, Costantino A, Belfiore A (1997) Insulin receptor what role in breast cancer? Trends Endocrinol Metab 8: 306–312 [DOI] [PubMed] [Google Scholar]
- Paz K, Hemi R, LeRoith D, Karasik A, Elhanany E, Kanety H, Zick Y (1997) A molecular basis for insulin resistance. Elevated serine/threonine phosphorylation of IRS-1 and IRS-2 inhibits their binding to the juxtamembrane region of the insulin receptor and impairs their ability to undergo insulin-induced tyrosine phosphorylation. J Biol Chem 272: 29911–29918 [DOI] [PubMed] [Google Scholar]
- Paz K, Voliovitch H, Hadari YR, Roberts CT Jr, LeRoith D, Zick Y (1996) Interaction between the insulin receptor and its downstream effectors. Use of individually expressed receptor domains for structure/function analysis. J Biol Chem 271: 6998–7003 [DOI] [PubMed] [Google Scholar]
- Ravichandran KS, Lorenz U, Shoelson SE, Burakoff SJ (1995) Interaction of Shc with Grb2 regulates association of Grb2 with mSOS. Mol Cell Biol 15: 593–600 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Resat H, Ewald JA, Dixon DA, Wiley HS (2003) An integrated model of epidermal growth factor receptor trafficking and signal transduction. Biophys J 85: 730–743 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robin P, Boulven I, Bole-Feysot C, Tanfin Z, Leiber D (2004) Contribution of PKC-dependent and -independent processes in temporal ERK regulation by ET-1, PDGF, and EGF in rat myometrial cells. Am J Physiol Cell Physiol 286: C798–C806 [DOI] [PubMed] [Google Scholar]
- Rodrigues GA, Falasca M, Zhang Z, Ong SH, Schlessinger J (2000) A novel positive feedback loop mediated by the docking protein Gab1 and phosphatidylinositol 3-kinase in epidermal growth factor receptor signaling. Mol Cell Biol 20: 1448–1459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarbassov DD, Guertin DA, Ali SM, Sabatini DM (2005) Phosphorylation and regulation of Akt/PKB by the rictor-mTOR complex. Science 307: 1098–1101 [DOI] [PubMed] [Google Scholar]
- Sasaoka T, Ishihara H, Sawa T, Ishiki M, Morioka H, Imamura T, Usui I, Takata Y, Kobayashi M (1996) Functional importance of amino-terminal domain of Shc for interaction with insulin and epidermal growth factor receptors in phosphorylation-independent manner. J Biol Chem 271: 20082–20087 [DOI] [PubMed] [Google Scholar]
- Sato S, Fujita N, Tsuruo T (2004) Involvement of 3-phosphoinositide-dependent protein kinase-1 in the MEK/MAPK signal transduction pathway. J Biol Chem 279: 33759–33767 [DOI] [PubMed] [Google Scholar]
- Schlessinger J (2000) Cell signaling by receptor tyrosine kinases. Cell 103: 211–225 [DOI] [PubMed] [Google Scholar]
- Schmelzle K, Kane S, Gridley S, Lienhard GE, White FM (2006) Temporal dynamics of tyrosine phosphorylation in insulin signaling. Diabetes 55: 2171–2179 [DOI] [PubMed] [Google Scholar]
- Schoeberl B, Eichler-Jonsson C, Gilles ED, Muller G (2002) Computational modeling of the dynamics of the MAP kinase cascade activated by surface and internalized EGF receptors. Nat Biotechnol 20: 370–375 [DOI] [PubMed] [Google Scholar]
- Schonwasser DC, Marais RM, Marshall CJ, Parker PJ (1998) Activation of the mitogen-activated protein kinase/extracellular signal-regulated kinase pathway by conventional, novel, and atypical protein kinase C isotypes. Mol Cell Biol 18: 790–798 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sebastian S, Settleman J, Reshkin SJ, Azzariti A, Bellizzi A, Paradiso A (2006) The complexity of targeting EGFR signalling in cancer: from expression to turnover. Biochim Biophys Acta 1766: 120–139 [DOI] [PubMed] [Google Scholar]
- Staubs PA, Reichart DR, Saltiel AR, Milarski KL, Maegawa H, Berhanu P, Olefsky JM, Seely BL (1994) Localization of the insulin receptor binding sites for the SH2 domain proteins p85, Syp, and GAP. J Biol Chem 269: 27186–27192 [PubMed] [Google Scholar]
- Stoker AW (2005) Protein tyrosine phosphatases and signalling. J Endocrinol 185: 19–33 [DOI] [PubMed] [Google Scholar]
- Taniguchi CM, Emanuelli B, Kahn CR (2006) Critical nodes in signalling pathways: insights into insulin action. Nat Rev Mol Cell Biol 7: 85–96 [DOI] [PubMed] [Google Scholar]
- Wellbrock C, Karasarides M, Marais R (2004) The RAF proteins take centre stage. Nat Rev Mol Cell Biol 5: 875–885 [DOI] [PubMed] [Google Scholar]
- Weng LP, Smith WM, Brown JL, Eng C (2001) PTEN inhibits insulin-stimulated MEK/MAPK activation and cell growth by blocking IRS-1 phosphorylation and IRS-1/Grb-2/Sos complex formation in a breast cancer model. Hum Mol Genet 10: 605–616 [DOI] [PubMed] [Google Scholar]
- White MF (1998) The IRS-signalling system: a network of docking proteins that mediate insulin action. Mol Cell Biochem 182: 3–11 [PubMed] [Google Scholar]
- Xiang X, Zang M, Waelde CA, Wen R, Luo Z (2002) Phosphorylation of 338SSYY341 regulates specific interaction between Raf-1 and MEK1. J Biol Chem 277: 44996–45003 [DOI] [PubMed] [Google Scholar]
- Xu KP, Dartt DA, Yu FS (2002) EGF-induced ERK phosphorylation independent of PKC isozymes in human corneal epithelial cells. Invest Ophthalmol Vis Sci 43: 3673–3679 [PubMed] [Google Scholar]
- Yamauchi K, Milarski KL, Saltiel AR, Pessin JE (1995) Protein-tyrosine-phosphatase SHPTP2 is a required positive effector for insulin downstream signaling. Proc Natl Acad Sci USA 92: 664–668 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yart A, Laffargue M, Mayeux P, Chretien S, Peres C, Tonks N, Roche S, Payrastre B, Chap H, Raynal P (2001) A critical role for phosphoinositide 3-kinase upstream of Gab1 and SHP2 in the activation of ras and mitogen-activated protein kinases by epidermal growth factor. J Biol Chem 276: 8856–8864 [DOI] [PubMed] [Google Scholar]
- Zimmermann S, Moelling K (1999) Phosphorylation and regulation of Raf by Akt (protein kinase B). Science 286: 1741–1744 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figures 1 - 14, Supplementary Table 1
Supplementary Figure 6
Supplementary Figure 6
Supplementary Figure 6
Supplementary Figure 8