Abstract
DNA microarray technology is a powerful tool for monitoring gene expression or for finding the location of DNA-bound proteins. DNA microarrays can suffer from gene-specific dye bias (GSDB), causing some probes to be affected more by the dye than by the sample. This results in large measurement errors, which vary considerably for different probes and also across different hybridizations. GSDB is not corrected by conventional normalization and has been difficult to address systematically because of its variance. We show that GSDB is influenced by label incorporation efficiency, explaining the variation of GSDB across different hybridizations. A correction method (Gene- And Slide-Specific Correction, GASSCO) is presented, whereby sequence-specific corrections are modulated by the overall bias of individual hybridizations. GASSCO outperforms earlier methods and works well on a variety of publically available datasets covering a range of platforms, organisms and applications, including ChIP on chip. A sequence-based model is also presented, which predicts which probes will suffer most from GSDB, useful for microarray probe design and correction of individual hybridizations. Software implementing the method is publicly available.
Keywords: DNA microarrays, fluorescent dye labelling, mRNA expression profiling, normalization, two channel
Introduction
DNA microarrays are applied widely throughout the life sciences for a variety of purposes including mRNA analysis and genome-wide localization studies (Young, 2000). The accuracy of microarray experiments depends on appropriate normalization of signals derived from different samples or arrays (Quackenbush, 2002). For dual-channel microarrays, two samples are labelled with different fluorescent dyes and hybridized to a single microarray. The differences in the properties and in the detection of the fluorescent dyes result in measurement bias. Global and/or intensity-dependent bias can be normalized by locally weighted linear regression (Yang et al, 2002). A different type of bias is gene specific and is not corrected by such methods. Various such gene- or probe-specific biases have been reported for the majority of platforms (Dombkowski et al, 2004; Rosenzweig et al, 2004; Dobbin et al, 2005; Martin-Magniette et al, 2005; Kelley et al, 2008), including single-channel platforms (Hekstra et al, 2003; Naef and Magnasco, 2003; Zhang et al, 2003; Schuster et al, 2007). Such biases interfere with accurate determination of differential expression.
For two-channel microarray experiments, gene-specific dye bias (GSDB) is easily observed in dye-swap replicates. It results in probes being affected mainly by the dyes rather than the samples, when dyes are swapped between two samples. Because of its variable nature, GSDB is difficult to address systematically and can result in deviations of more than two-fold from the correct ratio (Martin-Magniette et al, 2005; Kelley et al, 2008). GSDB is likely caused by differential fluorescence quenching within labelled material (Cox et al, 2004). A recent study showed that GSDB is dependent on the labelled nucleoside and presented a maximum-likelihood error model for correcting GSDB. The VERA method (Kelley et al, 2008) works well if the degree of GSDB is uniform across different hybridizations. Here, we show that the degree of GSDB is strongly linked to sample labelling efficiency. This agrees with the variable nature of GSDB across different hybridizations, which has complicated earlier attempts to model and correct GSDB effectively. We present a method that consists of a gene-specific correction that is modulated by the degree of overall bias observed in individual hybridizations. This significantly alleviates GSDB and results in greater accuracy of DNA microarray experiments. A sequence-based model is also presented.
Results and discussion
GSDB varies across hybridizations
As part of a project to determine differential expression between various mutant yeast strains, a number of control experiments were carried out. These controls consisted of labelling and hybridizing a single reference wild-type (wt) RNA sample against other wt RNA samples, each processed on different days. These hybridizations show diverse degrees of variation (Figure 1A and B).
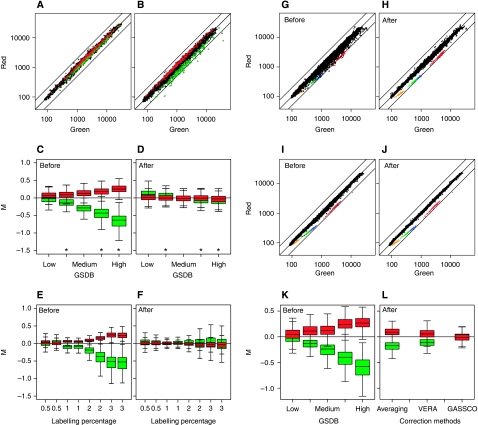
Figure 1.
Gene-specific dye bias and its correction. The degree of GSDB varies from one hybridization to another. Examples of a reference wt (green) versus other wt (red) scatterplot showing very little GSDB (A) or a large degree of GSDB (B). Each dot represents a single probe from the microarray. Green and red dots belong to the 5th and 95th percentiles of the iGSDB, respectively. The numbers along the axis represent normalized fluorescent intensities. The solid black lines mark two-fold up, no change and two-fold down. Boxplot of M-values (log2-ratio Cy5/Cy3) of the probes that suffer from the highest degree of GSDB, before (C) and after applying the correction method (D). The results of five different wt versus reference wt hybridizations are shown that suffer from increasing degrees of GSDB (low–high). These boxplots are derived from hybridizations with different dye orientations, showing that the outliers depend on the dye, rather than on the sample. From left to right the common reference wt sample was labelled with Cy5, Cy3, Cy5, Cy3 and Cy3, respectively, and is indicated with an asterisk (Cy3). The genes represented in these boxplots are identical to those coloured red and green in (A) and (B). Boxplots before (E) and after (F) GSDB correction derived from self versus self hybridizations, whereby only the degree of fluorescent label incorporation was varied for both dyes in each hybridization. A labelling percentage of 1 indicates that both Cy5 and Cy3 were incorporated at a determined efficiency of 1 fluorescent dye per 100 bases of amplified RNA. The correction applied to these arrays is derived from the independent set of 12 hybridizations also used to correct the data shown in (C, D). Scatterplot of self versus self hybridization labelled: at 3% efficiency before (G) and after (H) GSDB correction; at 2% before (I) and after (J) GSDB correction. These scatterplots are from two of the hybridizations depicted in (E) and (F). The coloured dots represent probes from four different external controls, whose RNAs were spiked in to achieve a two-fold molar difference between channels. Each external control is represented by multiple probes on the arrays. Boxplot of M-values before (K) and after (L) applying three different correction methods. Performance of the methods is measured as the change in variance of M-values compared with averaging. Averaging: simple averaging of dye swaps; VERA: (Kelley et al, 2008). This actually results in an overall 3% increase variance compared with averaging. However, the variance of the most extremely affected probes does decrease. GASSCO: the method described here, which results in 25% variance decrease.
When samples are labelled in reverse dye orientations, the outliers do not also reverse (Figure 1C), which is a hallmark of GSDB. The variable nature of GSDB (Figure 1A–C) complicates correction by methods that assume a uniform degree of bias. However, the bias seems monotonous for individual genes. That is, for all the genes that exhibit GSDB, it increases or decreases to similar degrees in different hybridizations (Figure 1C). We, therefore, expressed GSDB as the product of an intrinsic, GSDB factor, iGSDB, and a slide-dependent factor, F (see Materials and methods). The iGSDB for each individual probe was first estimated from 12 control hybridizations, reference wt versus other wt. Corrections were then applied to other hybridizations, using the iGSDB estimated from the controls multiplied by a slide-dependent factor derived from each of the new hybridizations separately. Applying this Gene- And Slide-Specific Correction (GASSCO) significantly reduces the variation in hybridizations that show different degrees of bias, even though these hybridizations were not used for estimating the iGSDB (Figure 1C and D).
GSDB is linked to the degree of label incorporation
To validate GASSCO further and to investigate the cause and variable nature of GSDB, a dataset was generated that consisted of self versus self hybridizations, whereby the degree of label incorporation was varied from 0.5 to 3%. Strikingly, the degree of GSDB shows strong association with the degree of label incorporation (Figure 1E). As the degree of label incorporation tends to vary between different samples, this is, therefore, a likely cause of the variable nature of GSDB between different hybridizations and further rationalizes the inclusion of a slide-dependent factor in GSDB correction methods.
GSDB correction based on the previously described control hybridizations greatly reduces self–self variation in this dataset with variable label incorporations (Figure 1F). To ensure that the correction does not simply flatten all differential ratios regardless of whether they are true or false, a series of external RNA controls was spiked in as true positive differentials. The correction applied to the external controls was identical. For hybridizations with a high degree of GSDB (Figure 1G) or for hybridizations with moderate GSDB (Figure 1I), the correction works well, reducing the variation in self–selfs but without affecting spiked in true positive ratios (Figure 1H–J). This conclusion also holds for higher degrees of GSDB, as well as for experiments with low dye incorporation that initially do not seem to suffer from GSDB (Supplementary information). Note that alleviating GSDB by reducing the amount of incorporated dye has the disadvantage of lowering sensitivity (Supplementary information).
Comparison with existing methods
Several earlier studies have explicitly addressed the problem of GSDB in two-colour arrays (Dombkowski et al, 2004; Rosenzweig et al, 2004; Dobbin et al, 2005; Martin-Magniette et al, 2005; Kelley et al, 2008). In most cases, these methods do not take any slide-specific GSDB into account, or make the assumption that this is constant within a batch (Rosenzweig et al, 2004). The method reported by Dobbin et al (2005) does model slide-specific effects of GSDB. However in their linear model, the slide effect is allowed to vary for each probe individually and, therefore, requires inordinate numbers of hybridizations to be estimated properly. In GASSCO, the slide-specific effect is constant for all probes. Another way of countering GSDB is to average the results of dye-swap replicate hybridizations, in an attempt to cancel out GSDB.
We compared the performance of GASSCO with the latest available one, VERA (Kelley et al, 2008), as well as with simply averaging dye swaps (Figure 1K and L), using the data presented in Figure 1C. As both VERA and averaging result in a single merged dataset, we also averaged our individually GSDB-corrected hybridizations for the sake of comparison. The results show clearly that GASSCO outperforms both methods. Importantly, as GASSCO corrects hybridizations individually (Figure 1A–J), it has the additional advantage of conserving statistical power, which is lost by methods that merge. The new method is also computationally less challenging than VERA (500-fold faster in our tests). As the degree of GSDB is associated with labelling efficiency (Figure 1E), both simple merging of dye swaps and methods that do not take into account the slide dependency of dye bias will only work well if label incorporation is identical between dye-swap replicates. In this study, ‘control' of sample labelling efficiency (Figure 1E) was only achieved by performing a number of reactions under varying conditions, matching labelled samples that happened to have the same incorporation. In practice, it is too challenging to control the degree of label incorporation precisely enough, especially for prolonged projects, which is one reason why the degree of GSDB varies (see Figure 3 for other examples of this variation from independent studies).
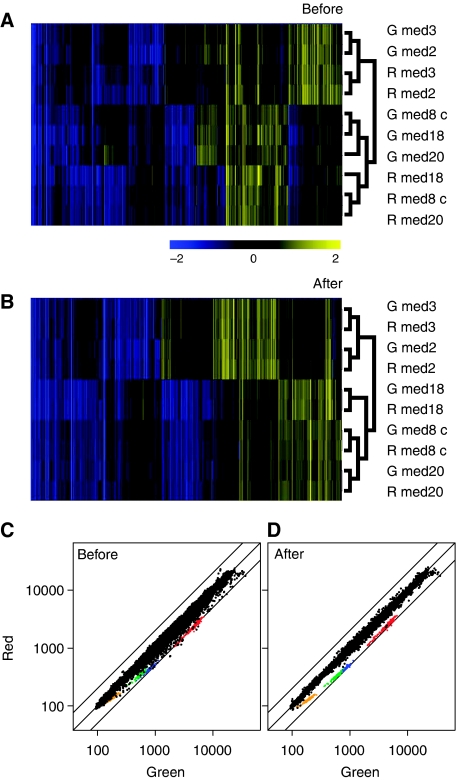
Figure 2.
GSDB correction for small-scale experiments without additional controls. (A) Cluster diagram of uncorrected expression profiles derived from five yeast strains carrying whole gene deletions of MED2, MED3, MED18, MED20 and a carboxy-terminal truncation mutation of MED8 (Lariviere et al, 2008), all of which encode subunits of the transcription complex Mediator. Med2 protein (Med2p) and Med3p form part of the Tail submodule within this complex. Med18p, Med20p and Med8p form part of the Head submodule. Loss of MED2 results in loss of Med3p from the complex and vice versa, resulting in similar expression profiles. Likewise, loss of MED18 results in loss of Med20p from the complex and vice versa. Loss of part of Med8p protein (med8c) also results in loss of both Med18p and Med20p. Together, these physical relationships underlie the similarity of the ensuing expression profiles (Lariviere et al, 2008). Two biological replicates of each of the five strains were hybridized to a common reference wt RNA sample. G and R refer to the dye orientation, with G indicating that the mutant sample was labelled with Cy3. In this cluster diagram, dye-swap replicates of individual strains do not cluster together. (B) As (A) but after applying GSDB correction based only on these 10 hybridizations. Now the replicates of the individual strains do cluster together. (C) Scatterplot of the self versus self hybridization shown in Figure 1g before (C) and after (D) applying the GSDB correction that is derived from the 10 hybridizations shown in the cluster diagrams.
Figure 3.
Correcting GSDB in previously published studies. Examples of GSDB determined from earlier studies that included dye swaps or self versus self hybridizations. Graphs as in Figure 1; left: before correction; right: after correction. In the left panel, the relevant publication, the organism and the platform. In the right panel, the number of hybridizations corrected and the achieved average and maximum performance.
GSDB correction for small-scale projects
The GSDB correction presented above is based on a set of 12 control hybridizations, reference wt versus other wt, undertaken as part of a large-scale expression-profiling project. GASSCO can also be independently applied to projects that do not include any same versus same controls (Figure 2). Here, the GSDB correction is first derived from, and then applied to, a series of 10 hybridizations from five mutant yeast strains, each hybridized as a biological replicate in dye swap against a common reference wt RNA (Lariviere et al, 2008). Before applying GASSCO, clustering of the replicates is strongly influenced by the dye orientation (Figure 2A). After applying GASSCO, the dye-swap replicates properly cluster together as pairs and the correct relationship between the different mutant strains is revealed (Figure 2B). The iGSDB estimates derived from this experiment were also used in the correction of the earlier self versus self hybridizations, showing similar results (compare Figures 2C, D with 1G, H).
Identical clustering results were also obtained with iGSDB estimates based on leave-one-out cross-validation (data not shown). To approximate the minimum number of dye-swapped slides needed to get trustworthy iGSDB estimates, we compared estimates derived from all combinations to the original that was based on all five pairs. Using just two pairs of hybridizations resulted in a correlation between the different iGSDB estimates always >0.94, suggesting that three pairs of dye swaps may generally suffice. Although such iGSDB estimates are slightly less accurate than those obtained from self versus self hybridizations, Figure 2 shows that the GSDB correction method can successfully be applied to smaller projects without self versus self control hybridizations.
Applying GASSCO to previously published data
To assess the generality of our method, we applied GASSCO to five publically available datasets from different laboratories. These include cDNA and Agilent oligo arrays, for both expression profiling and ChIP on chip studies, for yeast, mouse and human, using different labelling protocols (Dobbin et al, 2005; Chua et al, 2006; Chen et al, 2007; Tan et al, 2008; Tuteja et al, 2008). The criterion for selection of these datasets was that they included dye-swap replicates, which were used to estimate the iGSDB and subsequently correct the data. The results are shown in Figure 3 demonstrating that GSDB is widespread, in some cases to very high degrees and with a large degree of variation within single studies. GASSCO is able to correct all these data efficiently, as it takes into account that the degree of GSDB varies from one hybridization to another. It is therefore evident that the method described here is appropriate for a wide variety of experimental designs, platforms and applications.
Sequence-only based model
Differences between Cy3 and Cy5 with regard to dye–dye quenching contribute to GSDB (Cox et al, 2004). Using linear regression, several probe characteristics were scrutinized for correlation with GSDB. In agreement with an earlier study (Kelley et al, 2008), the adenine content of the probe, which corresponds to the aminoallyl-UTP used here to label the target RNA, has the strongest influence (Supplementary information). This fits with the finding that a higher degree of dye incorporation results in a higher degree of GSDB (Figure 1C). A linear regression model that also includes melting temperature is able to predict the iGSDB with a 0.76 correlation (Supplementary information). Although this is less accurate than empirically determining GSDB from a series of experiments (Figures 1 and 2), it offers the possibility of predicting which probes will suffer most from GSDB. This is useful for probe design, as well as for correction of individual hybridizations. This is illustrated by using the sequence-based linear regression model to correct a genome-wide chromatin immunoprecipitation experiment performed on an Agilent tiling array (Supplementary information).
In summary, the correction method described here works robustly, alleviates the GSDB artefact to a great extent, in a wide variety of experimental designs, without loss of statistical power and is therefore likely to be beneficial to most two-colour microarray studies.
Materials and methods
Full details about samples, microarrays, datasets, hybridization, scanning, normalization and clustering are described in Supplementary information. All microarray data and full protocols have been deposited in the public microarray database ArrayExpress (http://www.ebi.ac.uk/microarray) under accession E-MTAB-462.
GSDB correction
The total GSDB is expressed as the product of two factors, the iGSDB and a slide-dependent factor (F). That is,
GSDBij is the GSDB of gene i on slide j. iGSDBi is the intrinsic gene-specific dye bias of gene i and Fj is the slide-dependent factor of slide j. The method consists of the following steps: estimation of the iGSDBs once for an entire project; estimation of the slide-dependent factor for each hybridization; and lastly application of the individual corrections. The full R source code, including detailed documentation and worked examples, is available in the dyebias package from www.holstegelab.nl/publications/margaritis_lijnzaad/ or through BioConductor (www.bioconductor.org).
Estimation of iGSDB
For the first step, iGSDBi is arbitrarily defined as the average dye effect of probe i in the set of hybridizations used to estimate iGSDB. The set of hybridizations used to estimate iGSDB may consist of same versus same hybridizations (Figure 1) but may also consist of slides with real experimental factors, as long as in this case it includes technical or biological dye-swap replicates (Figures 2 and 3). After normalization of the raw data, the dye effect (log2-ratio red over green) is computed by simple averaging in the case of balanced designs. Fitting a linear model, for instance using the LIMMA package (Smyth, 2005), can also estimate iGSDB of unbalanced designs.
Estimation of the slide-dependent factor
The slide-dependent factor (F) is most accurately determined from probes that show the strongest GSDB. Firstly, probes mapping to highly variable transcripts are discarded. For instance, Ty-elements and mitochondrial genes were ignored in Figures 1 and 2, because of their high biological variability in yeast cultures. Next, the slide-dependent factor (F) is defined as the average of two ratios: the median of the M-values (i.e. the log2-ratio of the red over the green signal) of the strongly green-biased probes, divided by the median of their iGSDB, and likewise the median of the M-values of the strongly red-biased probes divided by the median of their iGSDB. The strongly green- or red-biased probes are defined as those having an iGSDB in the 0–5th (green) or 95–100th (red) percentile of all iGSDBs.
Application of GASSCO
The dye bias correction for each probe in an individual hybridization is the product of the probe's iGSDB and the slide-dependent factor (Formula 1). The correction is subtracted from M but only for probes unlikely to be affected by border effects due to the dynamic range of the technology. Probes with a log2(intensity) in either channel, >15 or intensity <1.5-fold above the local area background are not corrected. If the local area background is not available, 1.5-fold the minimum intensity of all probes can be used as a threshold instead.
Performance evaluation
To measure the performance of the correction method, we compared the variances of the apparent M (i.e. the log2-ratio (Cy5 over Cy3)) of the hybridization before and after the correction. In Figure 3, the average and maximum variance reduction are given.
Conflict of interest
The authors declare that they have no conflict of interest.
Supplementary Material
GSDB correction of very high or very low label incorporation data Prediction of GSDB from sequence Correction of an Agilent array ChIP experiment using sequence-predicted iGSDB Materials and methods not described in the main text
Details regarding dye bias correction of previously published data sets
References
- Chen S, de Vries MA, Bell SP (2007) Orc6 is required for dynamic recruitment of Cdt1 during repeated Mcm2–7 loading. Genes Dev 21: 2897–2907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chua G, Morris QD, Sopko R, Robinson MD, Ryan O, Chan ET, Frey BJ, Andrews BJ, Boone C, Hughes TR (2006) Identifying transcription factor functions and targets by phenotypic activation. Proc Natl Acad Sci USA 103: 12045–12050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox WG, Beaudet MP, Agnew JY, Ruth JL (2004) Possible sources of dye-related signal correlation bias in two-color DNA microarray assays. Anal Biochem 331: 243–254 [DOI] [PubMed] [Google Scholar]
- Dobbin KK, Kawasaki ES, Petersen DW, Simon RM (2005) Characterizing dye bias in microarray experiments. Bioinformatics 21: 2430–2437 [DOI] [PubMed] [Google Scholar]
- Dombkowski AA, Thibodeau BJ, Starcevic SL, Novak RF (2004) Gene-specific dye bias in microarray reference designs. FEBS Lett 560: 120–124 [DOI] [PubMed] [Google Scholar]
- Hekstra D, Taussig AR, Magnasco M, Naef F (2003) Absolute mRNA concentrations from sequence-specific calibration of oligonucleotide arrays. Nucleic Acids Res 31: 1962–1968 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelley R, Feizi H, Ideker T (2008) Correcting for gene-specific dye bias in DNA microarrays using the method of maximum likelihood. Bioinformatics 24: 71–77 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lariviere L, Seizl M, van Wageningen S, Röther S, van de Pasch L, Feldmann H, Strässer K, Hahn S, Holstege FC, Cramer P (2008) Structure-system correlation identifies a gene regulatory Mediator submodule. Genes Dev 22: 872–877 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin-Magniette ML, Aubert J, Cabannes E, Daudin JJ (2005) Evaluation of the gene-specific dye bias in cDNA microarray experiments. Bioinformatics 21: 1995–2000 [DOI] [PubMed] [Google Scholar]
- Naef F, Magnasco MO (2003) Solving the riddle of the bright mismatches: labeling and effective binding in oligonucleotide arrays. Phys Rev E Stat Nonlin Soft Matter Phys 68: 011906. [DOI] [PubMed] [Google Scholar]
- Quackenbush J (2002) Microarray data normalization and transformation. Nat Genet 32(Suppl): 496–501 [DOI] [PubMed] [Google Scholar]
- Rosenzweig BA, Pine PS, Domon OE, Morris SM, Chen JJ, Sistare FD (2004) Dye bias correction in dual-labeled cDNA microarray gene expression measurements. Environ Health Perspect 112: 480–487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuster EF, Blanc E, Partridge L, Thornton JM (2007) Correcting for sequence biases in present/absent calls. Genome Biol 8: R125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smyth GK (2005) Limma: Linear Models for Microarray Data. New York: Springer [Google Scholar]
- Tan K, Feizi H, Luo C, Fan SH, Ravasi T, Ideker TG (2008) A systems approach to delineate functions of paralogous transcription factors: role of the Yap family in the DNA damage response. Proc Natl Acad Sci USA 105: 2934–2939 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuteja G, Jensen ST, White P, Kaestner KH (2008) Cis-regulatory modules in the mammalian liver: composition depends on strength of Foxa2 consensus site. Nucleic Acids Res 36: 4149–4157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang YH, Dudoit S, Luu P, Lin DM, Peng V, Ngai J, Speed TP (2002) Normalization for cDNA microarray data: a robust composite method addressing single and multiple slide systematic variation. Nucleic Acids Res 30: e15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young RA (2000) Biomedical discovery with DNA arrays. Cell 102: 9–15 [DOI] [PubMed] [Google Scholar]
- Zhang L, Miles MF, Aldape KD (2003) A model of molecular interactions on short oligonucleotide microarrays. Nat Biotechnol 21: 818–821 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
GSDB correction of very high or very low label incorporation data Prediction of GSDB from sequence Correction of an Agilent array ChIP experiment using sequence-predicted iGSDB Materials and methods not described in the main text
Details regarding dye bias correction of previously published data sets