Abstract
The large degree of phenotypic fluctuation among isogenic cells highlighted by recent studies on stochastic gene expression confers fitness on some individuals through a ‘bet-hedging' strategy, when faced with different selective environments. Under a single selective environment, the fluctuation may be suppressed through evolution, as it prevents maintenance of individuals around the fittest state and/or function. However, as fluctuation can increase phenotypic diversity, similar to mutation, it may contribute to the survival of individuals even under a single selective environment. To discuss whether the fluctuation increases over the course of evolution, cycles of mutation and selection for higher GFP fluorescence were carried out in Escherichia coli. Mutant genotypes possessing broad GFP fluorescence distributions with low average values emerged under strong selection pressure. These ‘broad mutants' appeared independently on the phylogenetic tree and increased fluctuations in GFP fluorescence were attributable to the variance in mRNA abundance. In addition to the average phenotypic change by genetic mutation, the observed increase in phenotypic fluctuation acts as an evolutionary strategy to produce an extreme phenotype under severe selective environments.
Keywords: evolutionary experiment, individual cell selection, inherent heterogeneity, phenotypic fluctuation
Introduction
Recently, a great deal of attention has been focused on fluctuations in biochemical processes within the cell. The magnitude of fluctuations in protein abundance among the isogenic bacterial cells have been shown to be quite large, and these fluctuations are not only because of the external noise from the environment but also because of the stochasticity in intracellular reaction processes (Oosawa, 1975; Hasty et al, 2000; Ueda et al, 2001; Elowitz et al, 2002; Ozbudak et al, 2002; Kaern et al, 2004; Pedraza and Oudenaarden, 2005; Rosenfeld et al, 2005). The relevance of phenotypic fluctuations has recently been examined in relation to biological responses (Arkin et al, 1998; Korobkova et al, 2004; Paulsson, 2004), adaptation (Balaban et al, 2004; Kashiwagi et al, 2006; Acar et al, 2008), differentiation (Kaneko and Yomo, 1999; Maamar et al, 2007; Suel et al, 2007) and evolution (Sato et al, 2003), along with mutational robustness (Wagner, 2000; Kaneko, 2007).
By contrast, the fluctuation could be an obstacle to the maintenance of a state with higher function. Under a fixed set of environmental conditions, fluctuation around an optimal state could reduce fitness. Hence, how the magnitude of phenotypic fluctuation changes through evolution is an important issue that must be examined experimentally. In fact, it was recently shown in our laboratory that the magnitude of phenotypic fluctuation is positively correlated with the rate of evolution under a fixed set of environmental conditions (Sato et al, 2003; Kaneko and Furusawa, 2006). In directed evolution experiments, in order to enhance a given function, both the fluctuation and the rate of evolution decrease over the course of evolution. Our earlier study indicated that phenotypic fluctuation is reduced when the fitness is increased with generations (Sato et al, 2003). This decrease in fluctuation through evolution is natural, as it would be advantageous to reduce fluctuations around the fittest state to maintain optimal function (Landry et al, 2007; Lehner, 2008). As the decrease in fluctuation reduces the potential for the changing parameters of the system to be tuned for further optimisation of the function, it may be natural that the potential or the rate of evolution would also decrease. However, as mentioned above, extant cells still show relatively large fluctuations derived from external noise and/or stochasticity in intracellular reaction processes, and evolution does not necessarily cease.
These observations raise the question of how selection affects phenotypic fluctuations. In nature, the environment is not fixed but fluctuates over time. To cope with such fluctuating external environmental conditions, a certain level of phenotypic fluctuation is advantageous as it can produce various phenotypes, each of which is fit for the stochastically appearing environments (Thattai and Oudenaarden, 2004; Kussell and Leibler, 2005; Acar et al, 2008). This ‘bet-hedging' strategy can explain why extant cells maintain a large degree of phenotypic fluctuation. However, this may not be the only mechanism for maintaining fluctuations. Even under a fixed set of environmental conditions, amplification of isogenic phenotypic fluctuations may be possible through evolution. To address these points, an evolutionary experiment was carried out to enhance the fluorescence of GFP protein expressed in Escherichia coli following essentially the same procedure as described earlier (Sato et al, 2003). The main difference between the present and earlier experiments is that individual cells with phenotypic diversity because of both mutation and fluctuation were selected, rather than selection based on the average phenotype of a range of cells with fluctuating phenotypes for each individual mutant genotype. In contrast to the decrease in fluctuation observed earlier (Sato et al, 2003), some mutants with a large degree of phenotypic fluctuation emerged during the process of evolution in this study. Indeed such amplification of fluctuation was shown to be possible under tight selection of individual cells.
Results and discussion
In our earlier study (Ito et al, 2004), the gene for green fluorescent protein (GFP) fused with an artificial random 149-amino-acid polypeptide (RP) RP3-34H was used as the starting material. The variant with the highest fluorescence intensity (FI) in each generation was selected from a mutant library, where part of the artificial random-polypeptide gene was mutated randomly. This iterative process of mutation and selection was repeated over 10 generations. In the evolutionary experiment, the variant was selected on the basis of the average fluorescence intensity of 109 cells.
Here, with mutagenesis of the selected clone at the 10th generation, HLG10-6, as the starting material, selection was done on the basis of individual cells, where the fluctuation could influence the clones that evolve. GFP, FI and forward scattering (FS) of the cells were measured by flow cytometry (FCM) for selection at the individual cell level (Figure 1A). The intensity of FS is generally regarded as proportional to the size of the cell (Bouvier et al, 2001). Thus, the GFP fluorescence density was estimated by FI/FS, which was used as an index of selection. Specifically, the selected genes were amplified from 20 000 cells (0.2% of total cells measured) in region R1 in Figure 1A, reinserted into fresh vector, and reintroduced into fresh E. coli cells to exclude unexpected mutations in other regions of the vector. After repeating the selection and amplification (FCM selection cycle) several times, the selected genes were mutated in the next generation. This evolutionary process was carried out for three generations. In this selection procedure, cells that happened to possess a higher value of FI/FS because of phenotypic fluctuations could be selected. Thus, the genes or clones causing larger fluctuations in FI/FS, as well as those with a higher average value, may evolve.
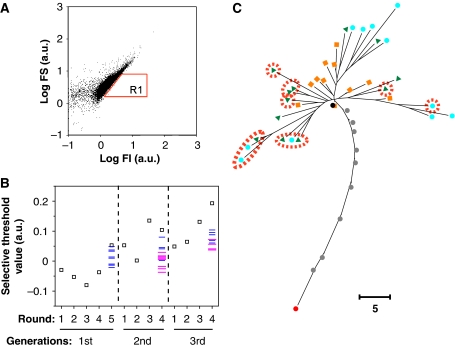
Figure 1.
Evolutionary experiment. (A) GFP fluorescence intensity (FI) and forward scattering (FS) of individual cells measured by FCM (those of HLG10-6 are also shown). Cells in the R1 region (top 0.2% of the total in fluorescence density (FI/FS)) were sorted for the next selection cycle. (B) The selection threshold value, log(FI/FS) at the 99.8th percentile in the cell population, was measured. Open squares indicate values at each round of selection. Values of 12 clones picked at random from the last cycle of each generation are plotted (some clones had the same value). Blue and magenta bars indicate narrow and broad mutants, respectively. The selection threshold values in clone analysis (blue and magenta bars) were slightly lower than those in population analysis (open squares). This decrease was probably because of the differences in culture conditions. (C) Phylogenetic tree of the selected clones, 12 clones sampled from each generation (as in B) and 11 clones in our earlier experiment (Ito et al, 2004). Nodes of the 12 clones sampled from each of the first, second and third generations are indicated in orange, green and light blue, respectively. Broad mutants are encircled in red. Nodes of the clone selected at each generation, RP3-34H, first to ninth, and HLG10-6, of our earlier experiment, are indicated in red, grey and black, respectively. The evolutionary experiment was started from the clone of the 10th generation (black). Scale bar: five nucleotide substitutions.
Figure 1B and C show an outline of the evolutionary process used in this study. The selection threshold value, defined as log(FI/FS) at 99.8 percentile among the cells in a population, increased during the evolutionary experiment. Twelve clones were picked at random from the last selection cycle of each generation. The cell distribution of FI/FS was measured (Figure 2 and Supplementary Figure S1) and the RP gene for each clone was sequenced (Supplementary Figure S2). The selection threshold values for these randomly picked clones also increased with the generations (Figure 1B). Sequence analysis of these clones revealed a phylogenetic tree with diverged branches. These observations indicated that the evolutionary experiment succeeded in increasing the selection threshold value as designed but retained some diversity in the evolved population.
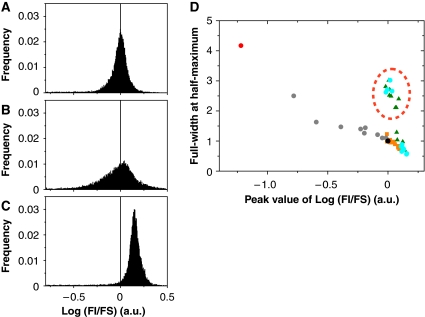
Figure 2.
Phenotypic distributions of broad and narrow mutants. (A–C) Histograms of log(fluorescence intensity (FI)/forward scattering (FS)) measured by FCM for each of the population of HLG10-6 (A), examples of broad mutants HLG2nd-5 (B) and narrow mutants HLG2nd-2 (C). The histograms (A–C) show the results of the re-cloning experiments. The peak position of HLG10-6 is indicated with the vertical line as a reference. (D) Peak value versus full-width at half-maximum. These values were obtained from the histogram of log(FI/FS) for each clone represented in Figure 1C. Colours and symbols are as in Figure 1C. Broad mutants are encircled by a red broken line.
Surprisingly, two distinct groups were found among the selected clones constituting the phylogeny: one with a large fluctuation and another with a small fluctuation (Figure 2D and Supplementary Figure S1), which are referred to as ‘broad mutants' and ‘narrow mutants', respectively.
To examine whether the distinct phenotypes with respect to fluctuation are reproducible, the RP gene was recloned from each of the variants in each round into a fresh vector carrying the GFP gene and its promoter sequence. The cell distributions of FI/FS before and after re-cloning were almost identical (Figure 2A–C and Supplementary Figure S1). These re-cloning experiments showed that the two distinct levels of fluctuation were reproducible phenotypes controlled by the genotype.
These observations raise the question about how the broad-mutant genes with lower average FI survived through the evolution experiment. In this evolution experiment, the fitness of selection consisted mainly of two parameters: FI/FS in FCM and the cellular growth rate in cell culture followed by FCM selection. Under the conditions used in this evolution experiment, however, the growth rate differences did not significantly affect the total fitness and relative survival rate (as shown in Supplementary information). Therefore, the evolution was considered to have proceeded in a single selective environment where phenotypic selection was carried out with respect to FI/FS. Even the selection threshold values for the broad mutants, selected in each generation, were almost the same as those of narrow mutants (Figure 1B), indicating that the former had the same number of cells as the latter above the threshold. Although the broad mutants had lower average fluorescence than the narrow mutants, larger distribution of the broad mutants could result in a similar survival probability. Moreover, the broad mutants appeared on different branches in the phylogenetic tree (Figure 1C), indicating that the emergence of a gene showing large phenotypic fluctuation would occur several times in the course of evolution.
These findings raise the question about how such a large fluctuation was produced in the broad mutants. To determine the possible origin of the fluctuation, the plasmid copy number and global noise level (Pedraza and Oudenaarden, 2005) in each cell were estimated by introducing the red fluorescent protein (RFP) gene into the plasmids with an independent promoter to that for GFP expression (Supplementary Figure S4A). HLG10-6 and one of the broad mutants showed narrow and broad distributions, respectively, in FI/(RFP fluorescence intensity (RFP FI)) (Figure 3A and B, Supplementary Figure S4B and C), and the same trend in FI/FS (Supplementary Figure S4D and E). These observations indicated that the broad mutant had a wide distribution of GFP fusion-protein expression level.
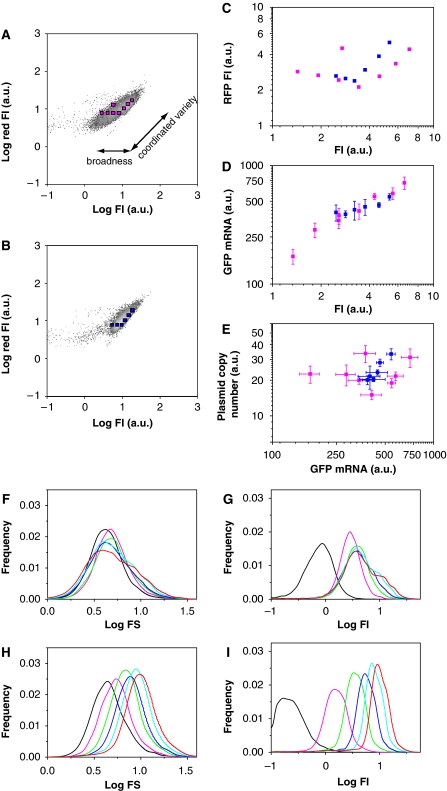
Figure 3.
Differences between broad and narrow mutants. (A–E) Fluctuations in GFP gene copy number, transcript and protein levels in broad and narrow mutants. Fluorescence intensity (FI) and red fluorescence intensity of individual cells of the broad mutant HLG3rd-1 (A) and HLG10-6 (B), in which the RFP gene was introduced in the plasmid, indicated by density plots. The colour gradient from grey to light grey represents an increase in cell density. The two-dimensional fluorescence distribution of diagonal direction on which GFP and RFP are equal, corresponds to ‘coordinated variety', whereas the horizontal direction corresponds to ‘broadness'. The cells in each region indicated by magenta (in A) and blue squares (in B) were sorted for the quantification of plasmid copy number and amounts of GFP mRNA by FCM. (C) Fluorescence comparison of the broad mutant (magenta) and HLG10-6 (blue), with each sorting region described in (A) and (B). The RFP fluorescence was determined by normalization for leakage of GFP fluorescence. (D, E) Quantification of GFP mRNA and plasmid copy number are shown as GFP fluorescence intensity versus GFP mRNA (D) and GFP mRNA versus plasmid copy number (E). The colour code is the same as in (C). Bars indicate s.d. from three or four experiments. (F–I) Time-course analysis of FI and FS for the broad mutant HLG3rd-1 (F, G) and HLG10-6 (H, I). Histograms of log FS (F, H) and log FI (G, I) at 0.5 h (black), 1 h (magenta), 1.5 h (green), 2 h (blue), 2.5 h (light blue) and 3 h (red) after IPTG induction, are shown.
Next, whether the broad phenotype of the broad mutants was mediated at the level of transcription or translation was investigated. As shown in Figure 3A and B, the two-dimensional fluorescence distribution showed two types of variety, that is, coordinated variety (diagonal correlation) and broadness (horizontal width). To examine the coordinated variety and the broadness in HLG10-6 and the broad mutant, 5000 cells in each of the small regions were sorted (Figure 3A and B) and the amounts of GFP mRNA and the plasmid were determined (Figure 3D and E). The average fluorescence over the cells for each region is shown in Figure 3C. As shown in Figure 3D, there was a positive correlation between GFP fluorescence and the GFP mRNA level in both HLG10-6 and the broad mutant, indicating that the variation in GFP fluorescence between the cells of the two genotypes was attributable to the variation in mRNA abundance. The mRNA abundance was then found to be partly correlated with the plasmid copy number (Figure 3E). HLG10-6 showed a clear correlation between GFP mRNA level and plasmid copy number, whereas the data from the broad mutant constituted two separated positive correlations: one for the small regions on the right edge in the two-dimensional distribution and the other for those on the left edge. The plots of FI versus RFP FI (Figure 3C) and GFP mRNA versus plasmid copy number (Figure 3E) resembled each other. This indicated that the coordinated variety, which affected the fluctuations in both GFP and RFP fluorescence equally, could be attributed to the variation in plasmid copy number, whereas the broadness cannot be explained by the plasmid copy number. The broad-mutant cells from the right and left edges but with the same plasmid copy number showed higher and lower mRNA abundance, respectively (Figure 3E). These observations indicated that the large broadness of GFP fluorescence in the broad mutant reflects a larger fluctuation in transcription and/or degradation of the GFP mRNA.
The broadness described above was also analysed with regard to temporal changes in FS and FI (Figure 3F–I). Although the HLG10-6 clone constituted a homogeneous population with regard to the increases in FS and FI, the cells of the broad mutant were divided into two populations from 1.5 h after induction: a major population with persistence and a minor population with an increase similar to that seen in HLG10-6. The heterogeneity among the cells in FS and FI corresponds to the broadness, and can arise from the physiological heterogeneity of the cells, which may be amplified by the toxicity of fused GFP protein of the broad mutant. A detailed discussion of the temporal changes and a possible model for the increase in heterogeneity of the broad mutants at the transcriptional level is presented in the Supplementary information.
There are two strategies to leave offspring with an increased chance of survival even under a fixed set of conditions: one is to maintain a high average value in the property under selection by tightly suppressing fluctuation, and the other is to leave a larger degree of fluctuation while having a lower average. For example, in the process of selecting cells with a threshold value of log(FI/FS) larger than 0.25 in Figure 2B and C, several cells of the narrow mutant were selected, but those of the broad mutant could also be selected as the distribution was extended to a much higher value than the peak. If there are two mutants of comparable population size with the same average properties but with different levels of fluctuation, the mutant with larger fluctuation will become dominant when the selection threshold value is higher than the average value, that is, less than half of the cells survive through the selection process. The effects of phenotypic selection stringency on the fitness of the broad mutants were calculated and are discussed quantitatively in the Supplementary information. In general, the strategy of large fluctuation is preferable as the survival fraction is smaller. Note that this strategy works even under a fixed environment, in contrast to the strategy to maintain stochasticity by environmental fluctuation (Thattai and Oudenaarden, 2004; Kussell and Leibler, 2005; Acar et al, 2008). Indeed, the broad mutant was considered to have evolved in a single environment under which phenotypic selection was carried out, as the growth rate selection in our evolution experiment contributed little to the total fitness (Supplementary information).
In summary, an increase in phenotypic fluctuation was observed during an evolutionary experiment based on the individual phenotypes under a fixed set of environmental conditions, whereas the fluctuation decreased gradually in an earlier evolutionary experiment based on selection with the average phenotype of each mutant genotype. The present evolutionary experiment simulated the situation wherein organisms are faced with a severe environment allowing only a small fraction of individuals to survive. When severe selection works at the individual level as in this study or in nature, the increase in phenotypic fluctuation is a relevant evolutionary strategy (Clayton and Robertson, 1957; Hill and Zhang, 2004). As the phenotypic fluctuation is correlated with the potential of evolution (Sato et al, 2003; Kaneko, 2007), this increase results in recovery of evolutionary potential.
Organisms require a large degree of phenotypic diversity in which only a small number of phenotypes survive, especially when they encounter a severe environment. This phenotypic diversity is derived from both phenotypic fluctuation and genetic diversity. Hence, the phenotypic fluctuation may be recruited and maintained in the evolutionary history of encountering severe environments.
Materials and methods
This study was performed using the same E. coli strain DH5α(DE3), plasmid pETHLGT1 and enzymes as in our earlier study (Ito et al, 2004).
Evolutionary experiments
The top clone at the 10th generation in our earlier study, HLG10-6 (Ito et al, 2004), was used as the initial gene sequence in the present experiment. Mutagenesis on HLG10-6 was performed with ΔTth DNA polymerase and the same error-prone PCR conditions as in our earlier paper, except for the number of PCR cycles, which was 15 here. The number of rounds of FCM selection and PCR amplification (FCM selection cycle, see below) was 5 times in the first generation and 4 times in the second and third generations. After each FCM selection cycle, the selected and amplified genes were ligated into fresh vector and electroporated into E. coli. Twelve transformants chosen at random were examined by FCM for each clone (see below). The remaining clones (about 106 colonies) were pooled, and plasmids were extracted from these collected cells to introduce mutagenesis for the next generation.
FCM selection cycle
The artificial random polypeptide genes were amplified by PCR, separated by electrophoresis through agarose gels and the corresponding DNA fragments isolated. The fragments were digested with NheI and SfiI, ligated into fresh vector and then electroporated into E. coli cells. The cells were cured in SOB medium at 37°C for 30 min and then cultured at 37°C for 12 h with addition of ampicillin (100 μg/ml). The cells were washed with LB medium to remove β-lactamase in the SOB medium, and grown in LB medium supplemented with ampicillin (75 μg/ml) at 37°C for 1 h. After addition of 1 mM isopropylthiogalactoside (IPTG), culture was continued to overexpress GFP-fused artificial polypeptides for 3 h. An aliquot of the culture was diluted 200-fold with phosphate-buffered saline, and the cells were examined by FCM (ALTRA; Beckman Coulter) with an argon-ion laser tuned to 488 nm and a band-pass filter of 525±12.5 nm for GFP fluorescence. A total of 20 000 cells were sorted from the region ‘R1' in Figure 1A, defined as the region of 0.2% of the total cells with higher GFP fluorescence per forward scatter. The sorted cells were boiled for 10 min, and selected genes were amplified by PCR for the next FCM selection cycle.
FCM measurement for fluorescence intensity and the distribution of E. coli clones
The selected clones were cultured individually in LB medium with 75 μg/ml ampicillin at 37°C. After addition of 1 mM IPTG at OD660=0.4, culture was continued for a further 3 h. Aliquots of 2 × 104 cells were examined by FCM with band-pass filters of 525±12.5 nm for green fluorescence and 610±10 nm for red fluorescence if necessary. All flow event data were converted to text format using WinMDI ver.2.8 to analyse the fluorescence distributions of the selected clones. Fluorescence was normalized relative to the leakage of green fluorescence into the red filter. GFP fluorescence of the selected clones in each generation was normalized relative to that of HLG10-6, which was examined after the selected clones in every generation.
Phylogenetic tree
The artificial random polypeptide (RP) genes fused with GFP of all the selected clones were sequenced. The phylogenetic tree was constructed by the neighbour-joining method (Saitou and Nei, 1987) implemented in MEGA version 4 (Tamura et al, 2007) using the distance between the genes (nucleotide sequence between NheI and SfiI sites in the RP gene) of the clones.
Protein purification and characterisation of its fluorescence
A His6-tag was introduced into the C-termini of GFP-fused polypeptides and GFP, and the tagged proteins were purified from E. coli lysate with a Qiagen Ni-NTA Spin kit in accordance with the manufacturer's protocol (Qiagen). Each purified protein was confirmed to run as a single band on SDS–PAGE. The concentrations of these purified proteins were quantified from the absorbance at 280 nm measured with a spectrometer (U-3000; Hitachi). The fluorescence spectrum with excitation at 488 nm was measured with a spectrofluorometer (FluoroMax-3; Horiba). These data were normalized relative to the protein concentration determined as described above.
Protein solubility and expression level
Protein solubility and expression level were measured in E. coli cells cultured under the same conditions with FCM measurement as described above by quantification of band density on SDS–PAGE at equal cell density, as described earlier (Ito et al, 2004). These measurements were performed in triplicate.
Plasmid construction for co-expression with RFP
The red fluorescent protein and constitutive promoter used here were DsRedT4 (Bevis and Glick, 2002) and PT5N25 (Gentz and Bujard, 1985), respectively. The sequence of the RFP gene with the promoter was constructed by PCR amplification of the RFP gene, inserted into pET21a (Novagen), with the following primers (the promoter sequence is underlined):
5′-TCATAAAAAATTTATTTGCTTTCAGGAAAATTTTTCTGTATAATAGATTCA TATCTAGAAATAATTTTGTTTAACTTTAAGAAGGAGATATACATA-3′
5′-TGCAGTGAATTCTTACAGGAACAGGTGGTGGCGGCCCT-3′.
The DNA fragment was purified and inserted into the SphI site with blunt ending of the original plasmid pETHLGT1 (Ito et al, 2004) in the opposite orientation to the GFP fusion polypeptide (see Supplementary Figure S3A).
Estimation of plasmid copy number and GFP mRNA quantity from sorted E. coli cells
Cells were grown and induced as described above. The cells were examined by FCM (FACSAria; BD Biosciences) with an argon-ion laser tuned to 488 nm and band-pass filters of 530±15 nm for GFP fluorescence and 610±10 nm for RFP fluorescence. Five thousand cells in small regions of the FCM measurement (see Figure 3A and B) were sorted into 10 mM Tris buffer, pH 8.5, and into a mixture of phenol and distilled water for estimation of plasmid copy number and GFP mRNA quantity.
For quantification of plasmid copy number, the sorted cells were sonicated for 6 s at room temperature, followed by RNaseA treatment to decrease contamination by any transcripts. Quantitative PCR was performed using a real-time PCR machine (Mx3005P; Stratagene) with SYBR green premix (SYBR PremixExTaq; TAKARA) and the following primers: 5′-CTACGATACGGGAGGGCTTA-3′ and 5′-ATAAATCTGGAGCCGGTGAG-3′.
For quantification of GFP mRNA, the sorted cells were disrupted by phenol extraction. Total RNA was isolated using an RNeasy Micro kit in accordance with the manufacturer's instructions (Qiagen), including on-column DNaseI treatment. Quantitative RT–PCR was performed using the same real-time PCR machine described above with a SuperScript III Platinum SYBR Green One-Step qRT–PCR Kit (Invitrogen) and the following primers: 5′-AGAGTGCCATGCCCGAAGG-3′ and 5′-GAGTTTGTGTCCGAGAATGTTTCC-3′.
These measurements were performed in triplicate or in quadruplicate.
Supplementary Material
Nucleotide sequences of the selected RP genes Average properties of the selected GFP fusion mutants A possible scenario for emergence of the broad mutants Fitness of selection in this evolution experiment Supplementary figures
Acknowledgments
We are grateful to T Kiguchi for technical assistance and Drs DS Tawfik, S Sawai, N Tokuriki, C Furusawa, K Sato and the members of the Yomo lab for fruitful discussions and comments. This study was partially conducted in the Open Laboratories for Advanced Bioscience and Biotechnology (OLABB), Osaka University. This research was supported in part by the ‘Special Coordination Funds for Promoting Science and Technology: Yuragi Project' and ‘Global (Centers of Excellence) Program' of the Ministry of Education, Culture, Sports, Science and Technology, Japan.
Footnotes
The authors declare that they have no conflict of interest.
References
- Acar M, Mettetal JT, Oudenaarden AV (2008) Stochastic switching as a survival strategy in fluctuating environments. Nat Genet 40: 471–475 [DOI] [PubMed] [Google Scholar]
- Arkin A, Ross J, McAdams HH (1998) Stochastic kinetic analysis of developmental pathway bifurcation in phage lambda-infected Escherichia coli cells. Genetics 149: 1633–1648 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balaban NQ, Merrin J, Chait R, Kowalik L, Leibler S (2004) Bacterial persistence as a phenotypic switch. Science 305: 1622–1625 [DOI] [PubMed] [Google Scholar]
- Bevis BJ, Glick BS (2002) Rapidly maturing variants of the Discosoma red fluorescent protein (DsRed). Nat Biotechnol 20: 83–87 [DOI] [PubMed] [Google Scholar]
- Bouvier T, Troussellier M, Anzil A, Coueties C, Servais P (2001) Using light scatter signal to estimate bacterial biovolume by flow cytometry. Cytometry 44: 188–194 [DOI] [PubMed] [Google Scholar]
- Clayton GA, Robertson A (1957) An experimental check on quantitative genetical theory. J Genet 55: 152–170 [Google Scholar]
- Elowitz MB, Levine AJ, Siggia ED, Swain PS (2002) Stochastic gene expression in a single cell. Science 297: 1183–1186 [DOI] [PubMed] [Google Scholar]
- Gentz R, Bujard H (1985) Promoters recognized by Escherichia coli RNA polymerase selected by function: highly efficient promoters from bacteriophage T5. J Bacteriol 164: 70–77 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hasty J, Pradines J, Dolnik M, Collins JJ (2000) Noise-based switches and amplifiers for gene expression. Proc Natl Acad Sci USA 97: 2075–2080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill W, Zhang X (2004) Effect on phenotypic variability of directional selection arising through genetic differences in residual variability. Genet Res Camb 83: 121–132 [DOI] [PubMed] [Google Scholar]
- Ito Y, Kawama T, Urabe I, Yomo T (2004) Evolution of an arbitrary sequence in solubility. J Mol Evol 58: 196–202 [DOI] [PubMed] [Google Scholar]
- Kaern M, Elston TC, Blake WJ, Collins JJ (2004) Stochasticity in gene expression: from theories to phenotypes. Nat Rev Genet 6: 451–464 [DOI] [PubMed] [Google Scholar]
- Kaneko K (2007) Evolution of robustness to noise and mutation in gene expression dynamics. PLoS ONE 2: e434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaneko K, Furusawa C (2006) An evolutionary relationship between genetic variation and phenotypic fluctuation. J Theor Biol 240: 78–86 [DOI] [PubMed] [Google Scholar]
- Kaneko K, Yomo T (1999) Isologous diversification for robust development of cell society. J Theor Biol 199: 243–256 [DOI] [PubMed] [Google Scholar]
- Kashiwagi A, Urabe I, Kaneko K, Yomo T (2006) Adaptive response of a gene network to environmental changes by attractor selection. PLoS ONE 1: e49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korobkova E, Emonet T, Vilar JM, Shimidzu TS, Cluzel P (2004) From molecular noise to behavioural variability in a single bacterium. Nature 428: 574–578 [DOI] [PubMed] [Google Scholar]
- Kussell E, Leibler S (2005) Phenotypic diversity, population growth, and information in fluctuating environments. Science 309: 2075–2078 [DOI] [PubMed] [Google Scholar]
- Landry CR, Lemos B, Dickinson WJ, Hartle DL (2007) Genetic properties influencing the evolvability of the gene expression. Science 317: 118. [DOI] [PubMed] [Google Scholar]
- Lehner B (2008) Selection to minimize noise in living systems and its implications for the evolution of gene expression. Mol Syst Biol 4: 170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maamar H, Raj A, Dubnau D (2007) Noise in gene expression determines cell fate in Bacillus subtilitis. Science 317: 526–529 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oosawa F (1975) The effect of field fluctuation on a macromolecular system. J Theor Biol 52: 175–186 [DOI] [PubMed] [Google Scholar]
- Ozbudak EM, Thattai M, Kurtser I, Grossman AD, Oudenaarden AV (2002) Regulation of noise in the expression of a single gene. Nat Genet 31: 69–73 [DOI] [PubMed] [Google Scholar]
- Paulsson J (2004) Summing up the noise in gene networks. Nature 427: 415–418 [DOI] [PubMed] [Google Scholar]
- Pedraza JM, Oudenaarden AV (2005) Noise propagation in gene network. Science 307: 1965–1969 [DOI] [PubMed] [Google Scholar]
- Rosenfeld N, Young JW, Alon U, Swain PS, Elowitz MB (2005) Gene regulation at the single-cell level. Science 307: 1962–1965 [DOI] [PubMed] [Google Scholar]
- Saitou N, Nei M (1987) The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol 4: 406–425 [DOI] [PubMed] [Google Scholar]
- Sato K, Ito Y, Yomo T, Kaneko K (2003) On the relation between fluctuation and response in biological systems. Proc Natl Acad Sci USA 100: 14086–14090 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suel GM, Kulkarni RP, Dworkin J, Garcia-Ojalvo J, Elowitz MB (2007) Tunability and noise dependence in differentiation dynamics. Science 315: 1716–1719 [DOI] [PubMed] [Google Scholar]
- Tamura N, Dudley J, Nei M, Kumar S (2007) MEGA4: molecular evolutionary genetics analysis (MEGA) software version 4.0. Mol Biol Evol 24: 1596–1599 [DOI] [PubMed] [Google Scholar]
- Thattai M, Oudenaarden AV (2004) Stochastic gene expression in fluctuating environments. Genetics 167: 523–530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ueda M, Sako Y, Tanaka T, Devreotes P, Yanagida T (2001) Single-molecule analysis of chemotactic signaling in Dictyostelium cells. Science 294: 864–867 [DOI] [PubMed] [Google Scholar]
- Wagner A (2000) Robustness against mutations in genetic networks of yeast. Nature 24: 355–361 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Nucleotide sequences of the selected RP genes Average properties of the selected GFP fusion mutants A possible scenario for emergence of the broad mutants Fitness of selection in this evolution experiment Supplementary figures