Abstract
Methods that allow for the manipulation of genes or their products have been highly fruitful for biomedical research. Here, we describe a method that allows the control of protein abundance by a genetically encoded regulatory system. We developed a dormant N-degron that can be attached to the N-terminus of a protein of interest. Upon expression of a site-specific protease, the dormant N-degron becomes deprotected. The N-degron then targets itself and the attached protein for rapid proteasomal degradation through the N-end rule pathway. We use an optimized tobacco etch virus (TEV) protease variant combined with selective target binding to achieve complete and rapid deprotection of the N-degron-tagged proteins. This method, termed TEV protease induced protein inactivation (TIPI) of TIPI-degron (TDeg) modified target proteins is fast, reversible, and applicable to a broad range of proteins. TIPI of yeast proteins essential for vegetative growth causes phenotypes that are close to deletion mutants. The features of the TIPI system make it a versatile tool to study protein function in eukaryotes and to create new modules for synthetic or systems biology.
Keywords: development, protein inactivation, regulated protein inactivation, tissue-specific
Introduction
Regulation of protein activity by gene disruption/deletion, RNAi, promoter shut-off, temperature sensitive alleles, chemical inhibition/inactivation, constitutive protein destabilization, and heat or small molecule-regulated N-degrons for induced protein degradation (Dohmen et al, 1994; Stack et al, 2000; Mnaimneh et al, 2004; Dohmen and Varshavsky, 2005; Banaszynski et al, 2006; Suter et al, 2006; Boutros and Ahringer, 2008) are highly useful tools to study protein function. A restriction immanent to these methods is the difficulty to induce protein degradation/inactivation in a highly selective manner, for example, in a specific tissue or during a particular cell-cycle or developmental stage. Methods to overcome some of these limitations exist, for example, the Cre/lox system for the genetically encoded and regulated excision of a gene from the chromosome (Sauer, 2002). In this case, the time required to establish the phenotype is decided mainly by the stability of the protein. Another method relies on regulated cleavage of target proteins by the tobacco etch virus (TEV) protease inside living cells (Henrichs et al, 2005; Pauli et al, 2008; Satoh and Warren, 2008). This system requires a priori knowledge about the structure of a protein to be able to introduce the TEV protease cleavage site into a target protein and to render its function sensitive to site-directed proteolysis. Alternatively, proteolysis sensitive sites could be identified by means of functional assays. However, ectopically driven proteolytic fragmentation may only affect a specific subset of the functions of a protein.
Complete degradation of proteins has been achieved using N-degrons; this degradation mechanism is conserved from bacteria to higher eukaryotes (Varshavsky, 1997). N-degrons constitute natural or artificial amino-terminal tags that are proteolytically processed, thereby leading to the exposure of an amino acid other than methionine at the amino terminus of a protein. The exposed amino acid serves as a recognition signal for poly-ubiquitylation and subsequent proteasomal degradation through the N-end rule pathway in eukaryotes (Bachmair et al, 1986). It determines the degradation rate of the protein with half-life times ranging between a few minutes (e.g. 2–3 min for arginine, phenylalanine, and aspartic acid) up to >20 h (e.g. serine or methionine) (Bachmair et al, 1986; Mogk et al, 2007).
Results and discussion
To create an N-degron that is activated only upon the conditional expression of a specific activator, we developed a degron that is protected at its N-terminus by an attached peptide that can be removed by proteolysis using the site-specific TEV protease (Parks et al, 1994). The TEV protease has been used in vivo in many different organisms (bacteria, yeast cells, drosophila, and mammalian cell culture) without negative side effects (Uhlmann et al, 2000; Kapust et al, 2002; Wehr et al, 2006; Pauli et al, 2008). Initially, we generated a fusion of a seven amino acid long TEV protease recognition site to the N terminus of an earlier developed N-degron (Suzuki and Varshavsky, 1999). The TEV protease cleaves between positions 6 and 7 of the recognition site. The enzymatic activity of TEV is somewhat flexible towards changes in the sequence, especially at position 7 (Kapust et al, 2002), which becomes the new N-terminal amino acid (in the following termed residue X) after proteolytic cleavage. Destabilizing amino-acid residues at the amino terminus (position X) target a protein for rapid destruction if the N-degron contains a sequence that allows the attachment of ubiquitin (Varshavsky, 1997). To monitor the cleavage, we fused a fluorescent protein to the N terminus of the TEV protease recognition site. To improve the processivity of the TEV protease, we enhanced the binding of the TEV protease to its substrate. We fused the N-degron construct with the TEV protease recognition site to the SF3b155381−424 protein domain. This domain is specifically recognized by the human spliceosome subunit p14 (Spadaccini et al, 2006), which we in turn fused to the TEV protease (named p14–TEV). Furthermore, we identified, by chance, a mutated allele of p14 (called p14*), which enhanced cleavage significantly. In summary, we have constructed a dormant N-degron that is constituted of a reporter, followed by a TEV protease recognition site (including residue X), an N-degron and SF3b155381−424 (in the following termed Reporter–TDegX-tag, e.g. GFP–TDegF-tag). This dormant N-degron can be deprotected by the expression of the p14*–TEV fusion protein (pTEV). An overview of the TEV protease induced protein inactivation (TIPI) system is shown in Figure 1. The mechanism underlying the enhanced activity of the p14*–TEV fusion versus p14–TEV is not clear, as the responsible mutation lies within a stretch of amino acids in p14 that is not involved in binding of p14 to SF3b155381−424 (data not shown; Schellenberg et al, 2006; Spadaccini et al, 2006). For details on the development of the TIPI system, see Supplementary information.
Figure 1.
TEV protease induced protein instability (TIPI). The principle of TIPI, a method to genetically control abundance of proteins with an N terminus exposed to the cytoplasm or nucleus. (A) The GFP–TDegX-tag is fused to the 5′-end of the target open reading frame (target ORF), directly in front of the ATG. The gene for pTEV expression is regulated by a controllable promoter; in this study, we used the galactose responsive GAL1-promoter in yeast. (B) Upon expression of pTEV, the pTEV protease binds to the GFP–TDegX-target protein. Binding is mediated by interaction of p14 with SF3B155381−424. This interaction directs efficient cleavage of the GFP–TDegX-tag by the TEV protease at its consensus site (ENLYFQ-X). (C) Cutting of the GFP–TDeg-tag leads to deprotection of the dormant N-degron that is part of the GFP–TDegX-tag. The N-degron is constituted by the new N-terminal amino acid X and a sequence that promotes efficient poly-ubiquitylation by Ubr1p (Suzuki and Varshavsky, 1999). The exposed amino acid X determines the fate of the protein. In yeast, X=A, C, G, M, P, S, T, and V lead to stable proteins, whereas X = D, E, F, H, I, K, L, N, Q, R, W, and Y render proteins instable (half lives=2–30 min) (Bachmair et al, 1986). (D) The target protein is poly-ubiquitylated by Ubr1p and degraded by the proteasome.
We used Saccharomyces cerevisiae as a model organism to develop and test the TIPI system. As a target protein, we used the non-essential, soluble, and freely diffusible protein Don1p (Maeder et al, 2007). Don1p is a protein with a role only in yeast sporulation, and it is absent in vegetatively growing cells (Knop and Strasser, 2000). We monitored the processing and degradation of the GFP–TDegX–Don1p fusion proteins as a function of pTEV expression (driven by the inducible GAL1-promoter) using western blotting and antibodies specific for GFP or Don1p. The amino acid at position X of the GFP–TDegX-tag is predicted to influences both, the cleavage efficiency of pTEV and the half-life of the target protein. We found that X=Phe (F; GFP–TDegF) and X=Asp (D; GFP–TDegD) provide optimal combinations of both, excellent cleavage followed by rapid protein degradation resulting in very low Don1p protein amounts upon pTEV expression (Figure 2A–C). Degradation is dependent on the E3 protein, which is encoded by the ubiquitin-protein ligase gene UBR1 (Figure 2A), indicating proteasomal degradation by the N-end rule pathway (Bartel et al, 1990). Furthermore, repression of pTEV expression rapidly restores protein levels of the target protein (Figure 2B). The TEV protease cleaved target protein is not degraded in strains lacking Ubr1p or if the TDegM-tag is fused to the target protein (Figure 2A and B). This excludes that addition of the TDegF-tag or expression of the TEV protease caused side effects that act on target protein production. The use of different residues at position X enables specific modulation of the cleavage efficiency (e.g. TDegK) and the degradation rate (e.g. TDegH) (Figure 2C). In summary, TIPI is a new method suitable for the precise post-translational regulation of protein abundance.
Figure 2.
TIPI mediates rapid degradation of proteins in yeast. (A) TIPI leads to rapid degradation of GFP–TDegD-tagged proteins. GFP–TDegD–DON1 was expressed chromosomally using the constitutive ADH1 promoter. Expression of pTEV or GFP–pTEV was induced by the addition of galactose (2% final concentration) to the culture. Samples of logarithmically growing yeast cells were removed from the culture at the indicated time points and subjected to western blotting. For detection of reporter constructs, anti-GFP and anti-Don1p antibodies were used. Detection of tubulin was used as a loading control. Positions of cleaved and uncleaved species are indicated in the figure. Strains used were either wild type or deleted for the gene UBR1 (ubr1Δ) as indicated. The strains we used in this experiment are described in Supplementary Table I, and their construction is indicated in Supplementary Table III. (B) Depletion of proteins by TIPI is reversible. GFP–TDegF–DON1 and GFP–TDegM–DON1 were expressed chromosomally using the constitutive ADH1 promoter. To induce pTEV expression galactose was added (at time point 0 h), repression of pTEV expression was done by adding glucose (at time point 3 h). Western blotting was performed as described in panel A. A # indicates the position of a non-specific band. (C) Modulation of protein abundance using different versions of GFP–TDegX. Protein levels of cleaved and uncleaved GFP–TDegF-, GFP–TDegM-, GFP–TDegK-, or GFP–TDegH–Don1p were assessed in crude extracts of yeast cells before and after 3 h of pTEV expression. GFP–TDegX constructs were expressed chromosomally from the ADH1-promoter. Western blotting was performed as described in Figure 2A. (D) C-terminal truncation of pTEV protease enhances proteolytic activity. Protein levels of cleaved and uncleaved GFP–TDeg–Don1p were assessed before and after 3 h of pTEV or C-terminally truncated pTEV+ expression. Strong overexpression of GFP–TDegD–DON1 constructs was achieved using the strong GPD-promoter. Western blotting was performed as described in panel A. (E) Protein depletion by TIPI can be followed by live cell imaging. Plasmid encoded CFP–TDegF–mKATE and CFP–TDegM–mKATE were expressed constitutively under control of the ADH1 promoter in wild-type cells and cells lacking UBR1 (ubr1Δ). Expression of pTEV+ (plasmid encoded) was induced by the addition of galactose (2% final concentration) to the cells. Images of the cells were taken at the indicated time points. (F) Quantification of the experiment shown in (E). Images from the cells used in (E) were recorded after induction of YFP–pTEV+. Automated quantitative image analysis was used to measure the cellular fluorescence of the different fluorescent protein reporters in 1000 to 3000 cells per strain (error bars represent the standard error of the mean). The yeast strains that were used to perform the experiments (A–F) are listed in Supplementary information. The genotypes are given in Supplementary Table I, the plasmids are described in Supplementary Table II.
The published crystal structure of the TEV protease (Phan et al, 2002; Nunn et al, 2005) allowed us to predict a TEV protease mutation, which improved its proteolytic activity by redefining the protein border at the C terminus. Using this modified pTEV, called pTEV+, efficient cleavage of the protease was enhanced, as observed by improved processing of the GFP–TDegX–Don1p reporter in cells where its expression was driven by the very strong GPD-promoter (Figure 2D).
We used live cell imaging to obtain the kinetics of protein depletion by TIPI. We used the red fluorescent protein mKate as a target, N-terminally tagged with the CFP–TDegX-tag, yielding the construct CFP–TDegX–mKate. The pTEV+ protease (under control of the inducible GAL1 promoter) was N-terminally tagged with the yellow fluorescent protein (YFP) citrine (YFP–pTEV+). Using this setup, we observed rapid depletion of mKate fluorescence upon YFP–pTEV+ expression in wild type, but not in ubr1Δ cells or if a CFP–TDegM–mKate construct was used (Figure 2E). Quantification of the cellular fluorescence intensities in the yellow and red channel revealed rapid depletion of TDegF–mKate within the first hour of YFP–pTEV+ expression (Figure 2F). After 3–4 h of YFP–pTEV+ expression, we noticed some residual red fluorescence. This is because of the loss of YFP–pTEV+ encoding plasmids in a subpopulation of cells (as confirmed by microscopy) and, therefore, incomplete processing of CFP–TDegF–mKate. Such residual levels do not occur in the experiments where chromosomally integrated constructs were used (Figure 2A–C).
Together, these results show that TIPI is a valuable method to induce the depletion of a protein. We used a controllable promoter to induce pTEV protease expression, but the method is easily adapted to a developmental process using distinct drivers for pTEV expression, which are only active at specific stages or in specific cell types. Protein depletion is easily followed in live cells; this allows correlating protein abundance with phenotype establishment.
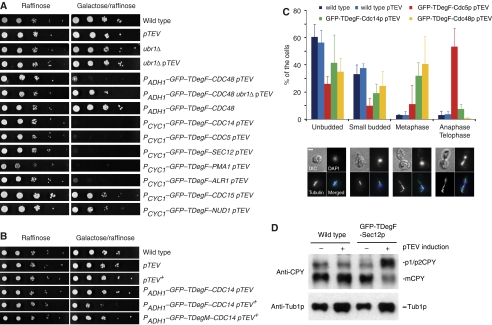
To test whether TIPI is able to deplete S. cerevisiae proteins sufficiently to cause a phenotype similar to the corresponding gene-deletion, we fused the GFP–TDegF-tag to several soluble (nuclear and cytoplasmic) and membrane proteins, which are all essential for vegetative growth of S. cerevisiae. The amino terminus of all chosen proteins is either exposed to the cytoplasm or the nucleoplasm. The GFP–TDegX–tagged fusion proteins revealed localizations that were comparable to the corresponding C-terminally GFP-tagged proteins (Huh et al, 2003) (Supplementary Figure 2A). Expression of pTEV led to the cleavage of the fusion proteins (Supplementary Figure 2B) and to the inhibition of cell growth, which was either completely abolished (in 6/8 tested proteins) or reduced (Cdc15p and Nud1p) (Figure 3A). Growth was rescued by a UBR1 deletion or by using GFP–TDegM that contains a stabilizing amino acid (Figure 3A and data not shown). Importantly, expression of pTEV or pTEV+ protease alone did not affect growth of yeast cells (Figure 3A and B). Strong production of the target protein GFP–TDeg–Cdc14p using the ADH1 promoter (Janke et al, 2004) required the presence of the more active pTEV+ protease to result in a growth phenotype (Figure 3B). The use of the pTEV protease was sufficient to abrogate Cdc14 function, if the target protein was expressed from the weaker CYC1 promoter (Janke et al, 2004) (Figure 3A). Surprisingly, TIPI of the integral membrane proteins GFP–TDegF–Sec12p, GFP–TDegF–Pma1p, and GFP–TDegF–Alr1p resulted in non-viable cells. This indicates that these proteins are accessible to the degradation machinery, which may be the case prior or during their insertion into the membrane, or at their final localization. We analyzed whether pTEV is able to cut efficiently near the plasma membrane and found complete cutting within 3–4 h after induction of pTEV expression (Supplementary Figure 3). Up to now, there is no report of membrane proteins being degraded through the N-end rule pathway. It may be that degradation of these proteins is assisted by other ubiquitylation triggered degradation pathways, for example, through endocytosis and vacuolar degradation (Hicke, 1997; Hicke and Dunn, 2003).
Figure 3.
TIPI of essential yeast proteins causes lethal phenotypes. (A) TIPI of essential proteins leads to impaired growth phenotypes. Serial dilutions (1:10) of yeast cultures (genotypes of yeast strains are indicated) were spotted on synthetic complete media containing either raffinose or galactose/raffinose and incubated at 30°C for 3 days. GFP–TDegF fusions were expressed either from the ADH1 (PADH1) or the CYC1 (PCYC1) promoter (as indicated). (B) pTEV+ protease exhibits increased activity as compared with pTEV. Experimental conditions were the same as described in (A) using strains that express the indicated constructs. (C) TIPI of Cdc5p, Cdc14p or Cdc48p leads to cell-cycle defects. Cell-cycle phenotypes were assessed after 3 h of pTEV expression in GFP–TDegF–CDC5, GFP–TDegF–CDC14 and GFP–TDegF–CDC48 expressing strains. Wild-type cells with and without expression of pTEV were used as controls. Samples were fixed and cell-cycle stages assessed based on bud size, spindle morphology, and DNA segregation. (D) TIPI of Sec12p leads to impaired secretion. Samples of control cells and TDegF–Sec12p expressing cells were taken before (−) and after 3 h (+) of pTEV protease induction and subjected to western blotting. The secretory marker protein carboxypeptidase Y (CPY) was detected. mCPY, mature, vacuolar form of CPY; p1+p2CPY, ER and Golgi glycosylated forms of CPY. The yeast strains that were used to perform the experiments (A–D) are listed in Supplementary information. The genotypes of these strains are given in Supplementary Table I.
TIPI of Cdc5p, Cdc14p, and Cdc48p, three proteins involved in cell-cycle regulation, led to cell-cycle defects: predominantly anaphase arrest in the case of GFP–TDegF–Cdc5p (55±3% versus 3±2% in the wild type) and increased frequency of metaphase arrested cells in the case of GFP–TDegF–Cdc14p (32±10% versus 3±3% in the wild type) and GFP–TDegF–Cdc48p (41±21% versus 3±3% in the wild type) (Figure 3C). The cell-cycle defects of GFP–TDegF–Cdc5p and GFP–TDegF–Cdc48p match the ones reported in the literature for temperature sensitive mutants (Hartwell et al, 1973; Frohlich et al, 1991). For GFP–TDegF–Cdc14p, we noticed a predominant arrest in metaphase (Figure 3C), whereas cdc14 temperature sensitive mutants arrest in anaphase and exhibit defects in exit of mitosis (Charles et al, 1998). This phenotype may indicate that deprotection of the degron in GFP–TDegF–Cdc14p leads to a Cdc14p species that interferes with an early mitotic function (Bloom and Cross, 2007) or that the commonly used cdc14 temperature sensitive mutants do not block this function completely. In the absence of TEV protease, no difference between GFP–TDegF-tagged and control cells was found (data not shown). TIPI of Sec12p, a membrane protein involved in ER to Golgi transport caused a prominent enrichment of the high molecular weight precursor forms of carboxypeptidase Y (CPY) (Figure 3D), indicative for impaired transport of proCPY to the vacuole where it is proteolytically processed (Kaiser and Schekman, 1990).
Our experiments demonstrate that TIPI enables the construction of conditional mutants: the regulated induction of pTEV expression enables the depletion of proteins from cells by targeted degradation. In conclusion, our yeast work demonstrates that protein depletion by TIPI is very quick; another advantage is that live cell imaging can be used to follow protein inactivation. This allows the comparison of protein abundance to phenotype establishment.
The high conservation of the N-end rule pathway in eukaryotic organisms (Mogk et al, 2007) suggests that TIPI could be useful in many different cell types and model organisms. Alternative implementations of TIPI could be developed, for example, using small molecule-regulated binding (Bayle et al, 2006) of the TEV protease to the N-degron. This, or the use of another site-specific protease for which a specific inhibitor is available, may provide additional ways to control the deprotection of the N-degron. Instead of using the TEV protease to deprotect the N-degron, one could also use the split-ubiquitin-system. This system was originally developed to detect protein–protein interactions (Wittke et al, 2000). Here, conditional expression of one half of ubiquitin and its specific targeting to a substrate carrying the other half, for example, using the p14–SF3b155 interaction, would trigger the removal of the reconstituted ubiquitin by the ubiquitin proteases (Johnsson and Varshavsky, 1994) and would lead to target protein destabilization.
One attractive application of TIPI is to use it for developmental studies in higher eukaryotes. Distinct drivers or promoters specific to developmental stages, tissues, and cell types are available for model organisms like worm, Drosophila, or mice. They should be applicable for selective induction of pTEV or pTEV+ protease expression. The specificity of the expression system used will decide how precisely the effects of TIPI-mediated protein inactivation can be linked to the process under investigation. In addition, it is essential to conduct the experiments in animals that lack the functional wild-type protein (e.g. by using a mutant, gene knock-out animal, or gene downregulation by RNAi-based methods). Control experiments without expressed TEV protease will report whether the TIPI-tag constricts the function of the target protein. Overproduction of the target protein should be avoided, as it may interfere with the function of the protein and with its downregulation by the TIPI system. The use of stable variants of the TIPI tag, for example, TDegM, provide further controls for the effect of the N-degron.
In conclusion, the TIPI system provides a method for efficient regulation of protein abundance in functional studies and for the creation of regulatory modules in synthetic biology.
Materials and methods
Yeast strains, plasmids, and growth conditions
All yeast strains used in this study were derived from the S288C strain ESM356-1 (Pereira et al, 2001). Genotypes are listed in Supplementary Table I. Manipulation of yeast strains using PCR targeting was performed as described (Janke et al, 2004). Standard methods for yeast strain construction were used otherwise (Sherman, 2002). Supplementary Table II lists the plasmids used to construct the yeast strains (Supplementary Table I), as indicated in Supplementary Table III. The gene encoding the TEV protease was isolated from TEV-infected tobacco leaves by PCR. The mutation S219V that inhibits autoproteolysis (Kapust et al, 2001) was introduced along with mutations that increase the solubility of the TEV protease (van den Berg et al, 2006). The codons for leucine and arginine were exchanged to optimize expression in yeast cells. The amino-acid sequences of the GFP–TDegX-tag and p14–TEV protease are provided in Supplementary Figure 4. Cloning details and nucleotide sequences are available upon request.
Standard preparations of growth media were used as described (Sherman, 2002). Growth tests were performed on synthetic complete media plates supplemented either with 2% raffinose or with 2% raffinose and 2% galactose. Cells used for immunodetection of tagged proteins by western blotting were grown in liquid synthetic complete media supplemented with 2% raffinose. TEV protease expression was induced by adding 2% galactose. TEV protease expression was repressed by the addition of 2% glucose. Cells used for fluorescence microscopy were grown in low-fluorescence media (Sheff and Thorn, 2004) supplemented with 2% raffinose.
Western blotting, antibodies, and immunofluorescence
Aliquots of cells from growing cultures were taken for crude protein extract preparation and western blotting using the protocol described in Janke et al (2004). Polyclonal rabbit anti-CPY, rabbit anti-tubulin, and rabbit anti-GFP antibodies were used to detect CPY, tubulin, and GFP (Finger et al, 1993; Maier et al, 2008). Immunofluorescence microscopy was performed as described in Maier et al, 2008.
Light microscopy and quantification
Live cell imaging was performed as described earlier (Taxis et al, 2006). Briefly, the cells were grown to logarithmic growth phase, adhered to concanavalin A coated glass-bottom-dishes (MaTek Corp.) and imaged in bright field and green fluorescence (Supplementary Figures 2A and 3) or bright field, cyan, yellow, and red fluorescence (Figure 2E). For quantification, cells were segmented from image backgrounds using yellow fluorescence images. Subsequently, cell outlines were transformed into masks used to measure pixel intensities of all fluorescent channels. Images of yeast cells expressing no fluorescent proteins were used to measure background fluorescence. Quantification data shown in Figure 2E were obtained by imaging between 1000 and 3000 cells for each time point and each yeast strain. Image processing, segmentation, and quantification were performed using the software imageJ. Background subtraction, normalization (time point 0 h for RFP, time point 4 h for YFP), calculation of the mean fluorescence and the standard error of the mean were done using the software Excel (Microsoft Corp.).
Supplementary Material
Supplementary Results Supplementary Discussion Supplementary Figures and Legends Yeast strains used for the figures Supplementary Tables Supplementary References
Acknowledgments
C Taxis was supported in parts by the ‘Forschung Optische Technologien 2003/2004' research program of the ‘Landesstiftung Baden-Württemberg GmbH'. E Schiebel, S Trautmann as well as M Jungbluth are acknowledged for helpful comments on the manuscript and D Störmer for technical assistance. We kindly acknowledge Michael Sattler for supporting G Stier and R Spadaccini. The authors declare that they have no conflict of interest.
Footnotes
The authors declare that they have no conflict of interest.
References
- Bachmair A, Finley D, Varshavsky A (1986) In vivo half-life of a protein is a function of its amino-terminal residue. Science 234: 179–186 [DOI] [PubMed] [Google Scholar]
- Banaszynski LA, Chen LC, Maynard-Smith LA, Ooi AG, Wandless TJ (2006) A rapid, reversible, and tunable method to regulate protein function in living cells using synthetic small molecules. Cell 126: 995–1004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartel B, Wunning I, Varshavsky A (1990) The recognition component of the N-end rule pathway. EMBO J 9: 3179–3189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bayle JH, Grimley JS, Stankunas K, Gestwicki JE, Wandless TJ, Crabtree GR (2006) Rapamycin analogs with differential binding specificity permit orthogonal control of protein activity. Chem Biol 13: 99–107 [DOI] [PubMed] [Google Scholar]
- Bloom J, Cross FR (2007) Novel role for Cdc14 sequestration: Cdc14 dephosphorylates factors that promote DNA replication. Mol Cell Biol 27: 842–853 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boutros M, Ahringer J (2008) The art and design of genetic screens: RNA interference. Nat Rev Genet 9: 554–566 [DOI] [PubMed] [Google Scholar]
- Charles JF, Jaspersen SL, Tinker-Kulberg RL, Hwang L, Szidon A, Morgan DO (1998) The Polo-related kinase Cdc5 activates and is destroyed by the mitotic cyclin destruction machinery in S. cerevisiae. Curr Biol 8: 497–507 [DOI] [PubMed] [Google Scholar]
- Dohmen RJ, Varshavsky A (2005) Heat-inducible degron and the making of conditional mutants. Methods Enzymol 399: 799–822 [DOI] [PubMed] [Google Scholar]
- Dohmen RJ, Wu P, Varshavsky A (1994) Heat-inducible degron: a method for constructing temperature-sensitive mutants. Science 263: 1273–1276 [DOI] [PubMed] [Google Scholar]
- Finger A, Knop M, Wolf DH (1993) Analysis of two mutated vacuolar proteins reveals a degradation pathway in the endoplasmic reticulum or a related compartment of yeast. Eur J Biochem 218: 565–574 [DOI] [PubMed] [Google Scholar]
- Frohlich KU, Fries HW, Rudiger M, Erdmann R, Botstein D, Mecke D (1991) Yeast cell cycle protein CDC48p shows full-length homology to the mammalian protein VCP and is a member of a protein family involved in secretion, peroxisome formation, and gene expression. J Cell Biol 114: 443–453 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartwell LH, Mortimer RK, Culotti J, Culotti M (1973) Genetic control of the cell division cycle in yeast: V. Genetic analysis of cdc mutants. Genetics 74: 267–286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henrichs T, Mikhaleva N, Conz C, Deuerling E, Boyd D, Zelazny A, Bibi E, Ban N, Ehrmann M (2005) Target-directed proteolysis at the ribosome. Proc Natl Acad Sci USA 102: 4246–4251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hicke L (1997) Ubiquitin-dependent internalization and down-regulation of plasma membrane proteins. FASEB J 11: 1215–1226 [DOI] [PubMed] [Google Scholar]
- Hicke L, Dunn R (2003) Regulation of membrane protein transport by ubiquitin and ubiquitin-binding proteins. Annu Rev Cell Dev Biol 19: 141–172 [DOI] [PubMed] [Google Scholar]
- Huh WK, Falvo JV, Gerke LC, Carroll AS, Howson RW, Weissman JS, O'Shea EK (2003) Global analysis of protein localization in budding yeast. Nature 425: 686–691 [DOI] [PubMed] [Google Scholar]
- Janke C, Magiera MM, Rathfelder N, Taxis C, Reber S, Maekawa H, Moreno-Borchart A, Doenges G, Schwob E, Schiebel E, Knop M (2004) A versatile toolbox for PCR-based tagging of yeast genes: new fluorescent proteins, more markers and promoter substitution cassettes. Yeast 21: 947–962 [DOI] [PubMed] [Google Scholar]
- Johnsson N, Varshavsky A (1994) Split ubiquitin as a sensor of protein interactions in-vivo. Proceedings of the National Academy of Sciences of the United States of America 91: 10340–10344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaiser CA, Schekman R (1990) Distinct sets of SEC genes govern transport vesicle formation and fusion early in the secretory pathway. Cell 61: 723–733 [DOI] [PubMed] [Google Scholar]
- Kapust RB, Tozser J, Copeland TD, Waugh DS (2002) The P1' specificity of tobacco etch virus protease. Biochem Biophys Res Commun 294: 949–955 [DOI] [PubMed] [Google Scholar]
- Kapust RB, Tozser J, Fox JD, Anderson DE, Cherry S, Copeland TD, Waugh DS (2001) Tobacco etch virus protease: mechanism of autolysis and rational design of stable mutants with wild-type catalytic proficiency. Protein Eng 14: 993–1000 [DOI] [PubMed] [Google Scholar]
- Knop M, Strasser K (2000) Role of the spindle pole body of yeast in mediating assembly of the prospore membrane during meiosis. EMBO J 19: 3657–3667 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maeder CI, Hink MA, Kinkhabwala A, Mayr R, Bastiaens PI, Knop M (2007) Spatial regulation of Fus3 MAP kinase activity through a reaction-diffusion mechanism in yeast pheromone signalling. Nat Cell Biol 9: 1319–1326 [DOI] [PubMed] [Google Scholar]
- Maier P, Rathfelder N, Maeder CI, Colombelli J, Stelzer EHK, Knop M (2008) The SpoMBe pathway drives membrane bending necessary for cytokinesis and spore formation in yeast meiosis. EMBO J 27: 2363–2374 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mnaimneh S, Davierwala AP, Haynes J, Moffat J, Peng WT, Zhang W, Yang X, Pootoolal J, Chua G, Lopez A, Trochesset M, Morse D, Krogan NJ, Hiley SL, Li Z, Morris Q, Grigull J, Mitsakakis N, Roberts CJ, Greenblatt JF et al. (2004) Exploration of essential gene functions via titratable promoter alleles. Cell 118: 31–44 [DOI] [PubMed] [Google Scholar]
- Mogk A, Schmidt R, Bukau B (2007) The N-end rule pathway for regulated proteolysis: prokaryotic and eukaryotic strategies. Trends Cell Biol 17: 165–172 [DOI] [PubMed] [Google Scholar]
- Nunn CM, Jeeves M, Cliff MJ, Urquhart GT, George RR, Chao LH, Tscuchia Y, Djordjevic S (2005) Crystal structure of tobacco etch virus protease shows the protein C terminus bound within the active site. J Mol Biol 350: 145–155 [DOI] [PubMed] [Google Scholar]
- Parks TD, Leuther KK, Howard ED, Johnston SA, Dougherty WG (1994) Release of proteins and peptides from fusion proteins using a recombinant plant virus proteinase. Anal Biochem 216: 413–417 [DOI] [PubMed] [Google Scholar]
- Pauli A, Althoff F, Oliveira RA, Heidmann S, Schuldiner O, Lehner CF, Dickson BJ, Nasmyth K (2008) Cell-type-specific TEV protease cleavage reveals cohesin functions in Drosophila neurons. Dev Cell 14: 239–251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pereira G, Tanaka TU, Nasmyth K, Schiebel E (2001) Modes of spindle pole body inheritance and segregation of the Bfa1p-Bub2p checkpoint protein complex. EMBO J 20: 6359–6370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phan J, Zdanov A, Evdokimov AG, Tropea JE, Peters HK III, Kapust RB, Li M, Wlodawer A, Waugh DS (2002) Structural basis for the substrate specificity of tobacco etch virus protease. J Biol Chem 277: 50564–50572 [DOI] [PubMed] [Google Scholar]
- Satoh A, Warren G (2008) In situ cleavage of the acidic domain from the p115 tether inhibits exocytic transport. Traffic 9: 1522–1529 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sauer B (2002) Cre/lox: one more step in the taming of the genome. Endocrine 19: 221–228 [DOI] [PubMed] [Google Scholar]
- Schellenberg MJ, Edwards RA, Ritchie DB, Kent OA, Golas MM, Stark H, Luhrmann R, Glover JN, MacMillan AM (2006) Crystal structure of a core spliceosomal protein interface. Proc Natl Acad Sci USA 103: 1266–1271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheff MA, Thorn KS (2004) Optimized cassettes for fluorescent protein tagging in Saccharomyces cerevisiae. Yeast 21: 661–670 [DOI] [PubMed] [Google Scholar]
- Sherman F (2002) Getting started with yeast. Methods Enzymol 350: 3–41 [DOI] [PubMed] [Google Scholar]
- Spadaccini R, Reidt U, Dybkov O, Will C, Frank R, Stier G, Corsini L, Wahl MC, Luhrmann R, Sattler M (2006) Biochemical and NMR analyses of an SF3b155-p14-U2AF-RNA interaction network involved in branch point definition during pre-mRNA splicing. RNA 12: 410–425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stack JH, Whitney M, Rodems SM, Pollok BA (2000) A ubiquitin-based tagging system for controlled modulation of protein stability. Nat Biotechnol 18: 1298–1302 [DOI] [PubMed] [Google Scholar]
- Suter B, Auerbach D, Stagljar I (2006) Yeast-based functional genomics and proteomics technologies: the first 15 years and beyond. Biotechniques 40: 625–644 [DOI] [PubMed] [Google Scholar]
- Suzuki T, Varshavsky A (1999) Degradation signals in the lysine-asparagine sequence space. EMBO J 18: 6017–6026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taxis C, Maeder C, Reber S, Rathfelder N, Miura K, Greger K, Stelzer EH, Knop M (2006) Dynamic organization of the actin cytoskeleton during meiosis and spore formation in budding yeast. Traffic 7: 1628–1642 [DOI] [PubMed] [Google Scholar]
- Uhlmann F, Wernic D, Poupart MA, Koonin EV, Nasmyth K (2000) Cleavage of cohesin by the CD clan protease separin triggers anaphase in yeast. Cell 103: 375–386 [DOI] [PubMed] [Google Scholar]
- van den Berg S, Lofdahl PA, Hard T, Berglund H (2006) Improved solubility of TEV protease by directed evolution. J Biotechnol 121: 291–298 [DOI] [PubMed] [Google Scholar]
- Varshavsky A (1997) The N-end rule pathway of protein degradation. Genes Cells 2: 13–28 [DOI] [PubMed] [Google Scholar]
- Wehr MC, Laage R, Bolz U, Fischer TM, Grunewald S, Scheek S, Bach A, Nave KA, Rossner MJ (2006) Monitoring regulated protein-protein interactions using split TEV. Nat Methods 3: 985–993 [DOI] [PubMed] [Google Scholar]
- Wittke S, Dunnwald M, Johnsson N (2000) Sec62p, a component of the endoplasmic reticulum protein translocation machinery, contains multiple binding sites for the Sec-complex. Mol Biol Cell 11: 3859–3871 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Results Supplementary Discussion Supplementary Figures and Legends Yeast strains used for the figures Supplementary Tables Supplementary References