Abstract
High quality datasets are needed to understand how global and local properties of protein-protein interaction, or “interactome”, networks relate to biological mechanisms, and to guide research on individual proteins. Evaluations of existing curation of protein interaction experiments reported in the literature find that curation can be error prone and possibly of lower quality than commonly assumed.
An essential component of systems biology is discovery of the network of all possible physical protein-protein interactions (PPIs), the “interactome” network1-3. There are two complementary ways to obtain comprehensive PPI information. One is to systematically test all pairwise combinations of protein pairs for physical interactions at proteome-scale with a high-throughput assay3. The alternative is to curate all publications in the literature, each describing one (or a few) PPI(s) assayed at low-throughput4, then making the curation accessible in interaction databases. As neither strategy is yet able to come close to discovering the full interactome5-7, the matter arises as to which strategy can best fill in the missing pieces.
High-throughput Protein Interaction Assays
Two approaches are in frequent use for high-throughput mapping of protein interactions at proteome-scale. Yeast two-hybrid assays attempt to identify binary interactions8,9, whereas co-affinity purification followed by mass spectrometry identifies membership in a protein complex10, but may not accurately determine the binary interactions between proteins within a complex7. Other technologies exist for mapping both binary interactions and co-complex memberships11, but none can yet be routinely scaled up for high-throughput assays, although recently a protein complementation assay allowed a large-scale mapping of the yeast interactome12.
Curating Protein Interactions
The onset of manual curation of protein interactions from literature began with pioneering curation for the yeast Saccharomyces cerevisiae by the Yeast Proteome Database (YPD)13. Those early efforts demonstrated that effective curation was possible, and also broadly aimed to capture all types of functional and genomic information, not only PPIs. Genomic databases specific to a single model organism arose in parallel with genome sequencing projects, e.g. the Saccharomyces Genome Database (SGD)14 and The Arabidopsis Information Resource (TAIR)15. Although initially devoted to sequence information, many of these databases eventually added many types of literature-curated information, including PPI data. In time the great many publications reporting PPIs exceeded the capacity of specialized genome databases and led to the creation of databases dedicated to PPIs, e.g., the Munich Information Center for Protein Sequence (MIPS) protein interaction database16, the Biomolecular Interaction Network Database (BIND)17, the Database of Interacting Proteins (DIP)18, the Molecular INTeraction database (MINT)19, and the protein InterAction database (IntAct)20. More recent PPI curation efforts, the Biological General Repository for Interaction Datasets (BioGRID)21 and the Human Protein Reference Database (HPRD)22, have attempted larger scale curation of more papers and more interactions, reporting higher productivity.
High-throughput Efforts versus Literature Curation
High-throughput approaches contrast in several attributes with literature-curated strategies (Table 1). Literature-curated collections represent the accumulation of thousands of small-scale, “hypothesis-driven” investigations, while high-throughput experiments are “discovery-based”, designed to discover new biology without a priori expectations of what could be learned. Because literature-curated datasets are hypothesis-driven, biological functions of interacting proteins can be often, though not always, inferred from the actual study design. Discovery-based high-throughput datasets do not present this advantage, though function can sometimes be inferred through further analyses23. Hypothesis-driven studies set up an inevitable study bias7, in that what has been successfully investigated before tends to get investigated again, whereas high-throughput screens avoid study bias24. The completeness, or the portion of the proteome that has been tested for interactions5, can be precisely estimated in a carefully designed high-throughput study5,7,25, but this is not so even for the largest literature-curated datasets, since negative results, the pairs tested but not found to interact, are almost never reported.
Table 1.
Comparison of s trategies towards completing an interactome map
| Attribute | High-throughput | Literature Curated |
|---|---|---|
| Investigation | discovery based | hypothesis driven |
| Functional inference | determinable from network? | determinable from study design? |
| Study bias | unbiased | biased |
| Completeness | estimable | inestimable |
| Reliability | determinable | indeterminable |
Estimating reliability, the portion of reported interactions that are valid (and hence reproducible), is daunting. For high-throughput datasets the introduction of an empirical framework for interactome mapping now allows experimental estimation of reliability parameters5. Previously, reliability of high-throughput datasets was routinely estimated by measuring the overlaps with a reference set of gold standard positives (GSP). Several caveats must be taken into account when constructing GSPs. The assays used to generate a GSP have to match as closely as possible the assays used to generate the experimental dataset, especially indirect co-complex vs. binary representation7. A GSP should be as unbiased as possible, sampling all, or at least most, parts and processes of the cell26, and a GSP must be of the highest reliability and reproducibility27.
Literature-curated datasets are used for appraisal of the reliability of experimental PPI datasets, for predicting PPIs, for predicting other features such as protein function, and for benchmarking data mining methodologies28-30. In these efforts the superior reliability of literature-curated PPI datasets, versus high-throughput datasets, is generally presumed. High-quality reference datasets of PPIs are integral for empirical estimation of the reliability and size of interactome maps5,7,25,27. Confidence in literature curation is accordingly prerequisite for generation of useful reference datasets. Whether literature-curated PPI datasets really have exceptional reliability has not been thoroughly investigated.
Completeness and Reproducibility of Literature-Curated Datasets
Since PPIs supported by multiple publications should be more reliable than those supported by only a single publication we assessed the proportion of multiply supported PPIs for yeast. We ranked the 11,858 literature curated yeast PPIs in BioGRID21 (LC-all). Only 25% of LC-all PPIs have been described in multiple publications (Fig. 1), with just 5% and 2% of these pairs described in ≥3 or ≥5 publications, respectively. More than 75% of LC-all PPIs are thus supported by only a single publication. Consistent with this low portion of multiply supported PPIs, experimental retests have demonstrated a significantly lower quality for singly-supported versus multiply-supported literature curated PPIs for yeast7.
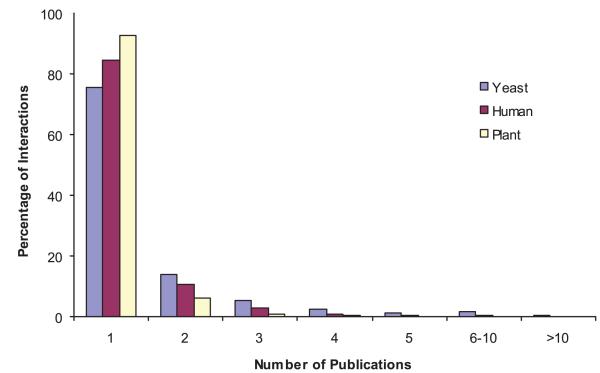
Figure 1.
Distribution of the number of published papers supporting each interaction: in the dataset of yeast protein interactions downloaded from the BioGRID21 database; in the literature-curated dataset of human protein interactions; and in the literature-curated dataset of Arabidopsis protein interactions.
Similar investigations for human and for Arabidopsis showed comparably low proportions of multiply supported PPIs. In the initial search space of ∼7,000 × 7,000 genes for a first-draft human interactome mapping project there are 4,067 binary literature-curated interactions31. Only 15% of these PPIs have been described in multiple publications (Fig. 1), with just 5% and 1% described in ≥3 or ≥5 publications, respectively. More than 85% of human PPIs in the literature curated set are supported by a single publication, higher than the 75% for yeast. The set of Arabidopsis PPIs was taken from the only two interaction databases that curate Arabidopsis interactions, TAIR15 and IntAct20. The Arabidopsis PPI dataset has many fewer interactions supported by multiple papers than yeast or human (Fig. 1), with just 1% and 0.1% described in ≥3 or ≥5 publications, respectively, with 93% of available Arabidopsis literature-curated PPIs supported by only a single publication. All told, the number of PPIs supported by multiple publications is small.
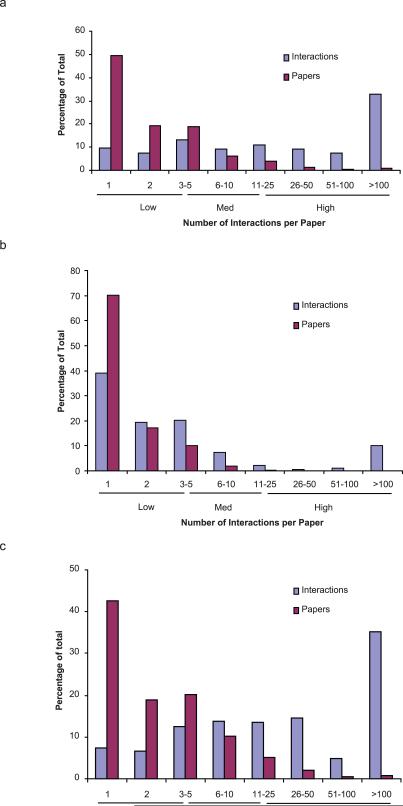
Literature-curated datasets are reported to be composed primarily of small-scale experiments21,32. To assess the presumption that PPI databases are small-scale we therefore measured the proportion of total PPIs coming from high-throughput experiments. For yeast we ranked the 8,933 interactions supported by a single publication by the number of distinct PPIs reported in each corresponding publication (Fig. 2a). More than 60% of protein pairs were curated from papers that described more than 10 interactions, altogether extracted from only 6% of all the papers curated. One-third of the total interactions come from less than 1% of all papers that each describe 100 or more interactions (Fig. 2a), which would reasonably be considered high-throughput. Thus, the yeast literature curated dataset of PPIs supported by a single publication record is not composed solely of validated interactions from small-scale studies, but has a marked portion of PPIs derived from high-throughput experiments. A similar analysis done with a dataset of human curated PPIs31 showed that this human PPI dataset is predominantly low-throughput (Fig. 2b), possibly because at the time these PPIs were downloaded from the databases few medium- to high-throughput experiments were published. For Arabidopsis the proportion of the total curated interactions derived from medium to high-throughput papers is about the same as yeast (Fig. 2c). In sum, many available literature-curated PPI datasets are populated widely by PPIs from high-throughput experiments.
Figure 2.
Distribution of the publications in literature-curated datasets by the number of interactions reported in the publication. Distribution in the (a) yeast (b) human and (c) Arabidopsis literature-curated PPI datasets supported by a single publication.
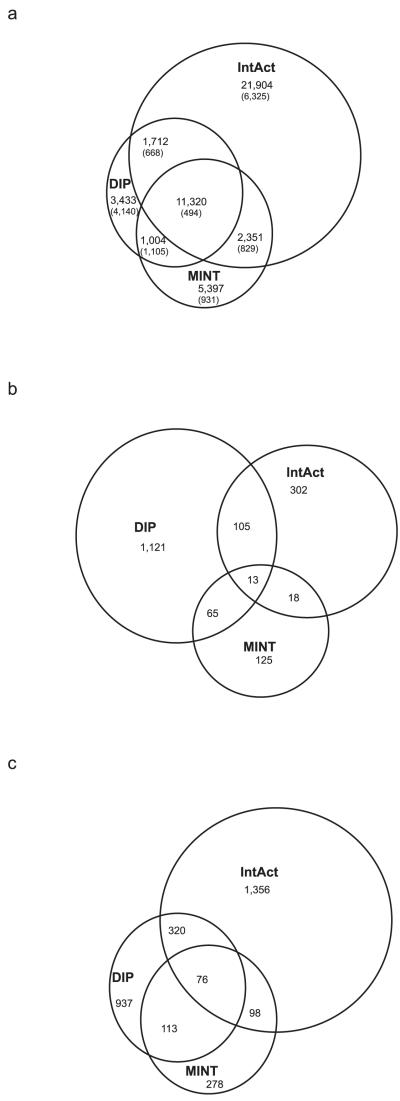
Since assessment of completeness for literature curated datasets is not possible (Table 1), we evaluated database overlaps as a surrogate for completeness, on the argument that different PPI databases should curate from much the same set of PubMed reports. BioGRID reports the greatest completeness for yeast, but is not yet a participating member of the IMEx consortium33,34, so we could not use it for this analysis. The three IMEx members that do substantial curation of yeast PPIs (MINT, IntAct, and DIP) show surprisingly low overlaps of curated PPIs (Fig. 3a). That the overlap is so small following years of intense curation of protein interactions is reason for concern. The small overlap is not due to differential curation of high-throughput data, since removal of the six largest PPI reports35-40 still leaves small overlaps, especially of IntAct with the other two databases (Fig. 3a).
Figure 3.
Overlaps of reported curation for yeast PPIs (a) Overlaps of the total number of reported binary PPIs, or after removing the largest high-throughput yeast PPI reports (numbers in parentheses). (b) Overlaps of the Pubmed reports curated. (c) Overlaps after removing multiply supported interactions.
Are the small overlaps due to curating different papers or to differential curation of data from overlapping sets of papers? The answer seems to be that vastly different sets of papers are curated, since the coverage of PubMed reports also shows small overlap (Fig. 3b). For multiply supported interactions (those reported in two or more papers) the low overlap remains (Fig. 3c), though the number of interactions drops greatly. Hence even the most heavily investigated interactions, those most likely to be multiply curated, do not seem to be comprehensively covered. To sum, surrogate estimates of completeness of literature-curated datasets, at least for yeast, suggest that coverage of curated literature is far from comprehensive.
These investigations suggest, but in no way demonstrate, that literature curated PPIs may not have the highest reliability often attributed to them. There has not yet been any intensive investigation of the actual reliability of literature-curated PPI datasets. To do so we recurated representative samples of existing literature-curated PPI datasets for three model organisms — yeast Saccharomyces cerevisiae, human, and plant Arabidopsis thaliana — finding that the literature curation of PPI publications can be less than impeccable.
Estimating Curation Reliability by Recuration
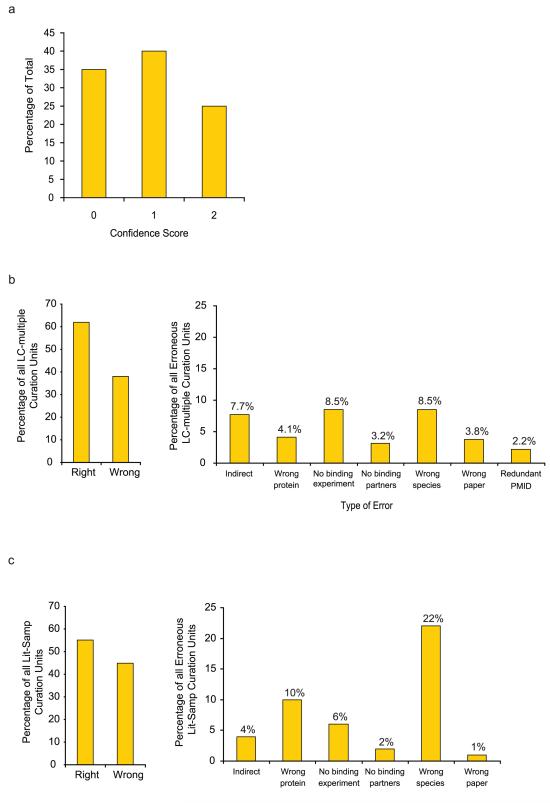
For yeast we undertook a detailed recuration of a sample of 100 pairs selected randomly from the yeast dataset of singly supported interactions (Fig. 1). After evaluation of several relevant criteria, each interaction was assigned a score of 0 (no confidence), 1 (low confidence or unsubstantiated), or 2 (substantiated or of high confidence) (see detailed protocol below).
The results of this recuration (Fig. 4a and Supplementary Table 1 online) showed that only 25% of the sampled interactions could be substantiated, while three-quarters were not. Of the interacting pairs in the sample 35% were incorrectly curated. These observations explain the poor reliability, relative to high-throughput datasets, of the singly supported literature curated dataset in both computational and experimental comparative analyses7.
Figure 4.
Summary of recuration results. (a) 100 interacting pairs randomly drawn from the yeast literature curated dataset supported by only a single publication. Score 0: erroneous, not reported in the associated publication; score 1: reported in the associated publication but not verified; score 2: reported and verified. (b) Recuration results of the literature curated sample for human PPIs reported in multiple publications. Proportion of correct and erroneous curation units (left panel) and a distribution of different types of curation errors (right panel). (c) Summary of curation results of randomly sampled sets from human literature curated interacting pairs reported in a single publication. Correct and erroneous curation units (left side); distribution of different types of curation errors (right side).
For human PPI recuration two curation datasets were prepared. One is a presumed high-confidence literature-curated dataset of interactions (LC-multiple). within the initial search space of ∼7,000 × 7,000 genes for a first-draft human interactome mapping project31) corresponding to pairs reported two or more times (i.e. two different PubMed IDs) and curated in two or more databases (the five databases used were HPRD22, BIND41, MINT19, MIPS mammalian database16, and DIP18). From within this small (only 275 multiply supported interactions) ‘hypercore’ set of protein interactions31 188 interactions were left for recuration, after excluding homodimers.
The other dataset is a lower confidence Literature Sampled dataset of 188 interactions, generated by randomly selecting interactions from initial search space5. Most of these interactions have one publication linked to the interaction, but since sampling was random several interactions were reported in more than one publication.
In the LC-multiple recuration set 38% of the initial curation units (defined in Table 2) were wrong (Fig. 4b and Supplementary Table 2 online). The most common errors were wrong species, i.e., a species other than human, and absence of a binding experiment supporting the interactions. Although 40% of the human LC-multiple interactions were no longer supported by multiple publications after recuration, most of these now have only one supporting paper instead of two or more, perhaps constituting a “secondary” dataset of reduced confidence (Supplementary Table 2 online).
Table 2.
Summary of curation results for human and Arabidopsis
| Sampled Dataset | Interaction Units | Curation Units* |
|---|---|---|
| Human LC-multiple | Correct: 172 (91.5%) Incorrect: 16 (8.5%) |
Correct: 362 (62%) Incorrect: 223 (38%) |
| Human Literature Sampled |
Correct: 88 (55%) Incorrect:72 (45%) |
Correct: 88 (55%) Incorrect:72 (45%) |
| Arabidopsis | Correct: 94 (94%) Incorrect: 6 (6%) |
Correct: 201 (89.3%) Incorrect: 24 (10.7%) |
For human a Curation Unit is an interaction reported in one publication regardless of the number of databases curating the interaction. An interaction reported in three distinct papers and curated in two databases represents three Curation Units. For Arabidopsis a Curation Unit is an interaction reported in one publication or one database. An interaction reported in three distinct papers and with all three curated in the two Arabidopsis PPI databases represents six Curation Units.
For the presumably lower confidence Literature Sampled dataset of 160 interacting pairs (after removing interactions that had more than one supporting publication), 45% of interactions were not validated (Fig. 4c and Supplementary Table 3 online) and 55% were validated. Almost half of the randomly sampled interactions are not supported by recuration. The most common errors here were wrong species and wrong protein name (Fig. 4c).
Yeast and human have the largest amount of curated literature in interaction databases21,42. A model organism with significantly fewer curated interactions might present different results. We curated 100 higher-confidence protein interactions of Arabidopsis thaliana from the two interaction databases that curate Arabidopsis, TAIR15 and IntAct20. The results (Supplementary Table 4 online) were significantly better than the yeast or human results, as only 6 interactions and 24 curation units were scored incorrect (Table 2). The 24 errors were scored as: 9 “No binding experiment”; 6 “No binding partner”; 6 “Indirect”; and 3 “Wrong protein”. The better results for Arabidopsis likely reflect a smaller research community which can maintain uniformity in gene/protein names15.
Why is Reliability of Literature Curation so Low?
The findings here of large error rates in curated protein interaction databases, at least for yeast and human, are consistent with recent hints that the quality of literature-curated datasets may not be as high as widely perceived23,29,43-45. Perhaps occasionally curator error is responsible. However, we suggest that the errors are due not so much to curators, but to the simple reality that extracting accurate information out of a long free-text document can be extremely difficult. Gene name confusion is particularly thorny30,46. An example from our curated yeast sample illustrates the difficulties. A TAP-tag affinity purification with Vps71/Swc6 as bait47 does pull down a protein named Swc3, but double-checking finds that this ORF is actually SWC3/YAL011w, and not the ALR1/SWC3/YOL130w ORF curated in the database. A shared synonym thoroughly muddled the curation.
Common curation practice has been to score equally every interaction reported in a publication21,48, even though common experimental practice consists of first screening for new interacting proteins, then focusing on and substantiating one or a few of the most interesting interactions while leaving the others aside. Perhaps more curator judgment is needed, applying higher ranking to verified interactions and accordingly lower ranking to unverified “along for the ride” interactions. Users can then choose the confidence level suitable to their needs. Given the demands of systems biology, perhaps biological databases should no longer serve as mere repositories of data but ought to upgrade to appraisals of data49. Recent small incremental steps at developing a confidence score for curated PPIs have been taken50,51.
The difficulty of literature curation is often underappreciated4,21,30. The lack of formal representation of PPIs in published papers makes it difficult, if not impossible, to extract the PPI data in usable form. Designation of the species of origin of the protein interactors, an absolutely critical piece of information, is often buried or lacking altogether; protein/gene name synonyms used in a particular paper are hard to trace back to the canonical protein/gene names, especially in older papers; and standardized descriptions, sometimes any description, of the methods used are absent. Faced with these difficulties, the curator is forced either to omit the information altogether (curated false negative), or make a diligent effort at an educated guess, even though guesses, albeit educated ones, are often erroneous (curated false positive). The small overlaps noted between curated yeast interactions in different databases (Fig. 3) might be due to differential treatments of potential curated false negatives.
The observations here that literature curated datasets have inherent reliability difficulties should influence thinking about proper generation of positive reference sets29. Already the human positive reference sets generated in our sampled recuration efforts have proven useful in multiple investigations5,27,52.
It is still rarely doubted that literature-curated interactions are better than datasets generated with any high-throughput technology6,21,53,54. Our findings lead us to argue otherwise. If rigorously carried out, high-throughput experimental PPIs can be of higher quality than literature-curated interactions5,25,27.
Improving Reliability of Literature-Curated PPI Datasets
The difficulty of curation arises partly because PPI data are not submitted to databases in standardized format upon publication55,56, unlike DNA sequence or protein structure data. The difficulty that curators have in extracting PPI information from papers has led to the promulgation of the MIMIx (Minimal Information about a Moleular Interaction experiment) initiative55. MIMIx standardizes the presentation of PPI information in published papers regarding species, protein names, methodological descriptions, and protein identifiers, making it easier for curators to extract the pertinent information33. Once widely promulgated, which should come about sooner if the Structured Digital Abstract57,58 project gains traction, MIMIx will greatly improve curation such that the erroneous curation uncovered here will be lessened. Other Minimal Information initiatives for large-scale biology data are under development59, and their further development is wholeheartedly endorsed by the biocuration community so as to reduce curation error30.
Our findings, while possibly critical of the quality of existing PPI curation, must not be taken for quality evaluation of the underlying scientific literature. Actually, some PPI publications do warn of possible cross contamination60, or even occasionally provide heuristic confidence scores61, warnings that should be taken into account in the curation.
Curation Protocols
Yeast PPI recuration
For each randomly selected protein pair the reporting paper was read in detail (text, figures, and supporting information), searching for all supporting information about the presumed interaction. Five questions were answered for each protein pair. (1) Is there any information in the paper that supports the interaction? (2) Has the experiment supporting the interaction been done at low throughput? Since the perception persists that low-throughput experiments have greater reliability21 knowing this is important. (3) Are the interacting proteins mentioned together in the text? Lack of co-citation indicates that the authors did not actually focus on that particular interaction. (4) Is the interaction supported by multiple methods? (5) Is the interaction likely direct? That is, did the method(s) used gauge binary interaction or co-complex membership. Lastly, an overall score of 0 (no confidence: no mention of the interacting pair, negative answer to the other 4 questions), 1 (low confidence: interacting pair is mentioned but the interaction is not substantiated by alternative methods), or 2 (high confidence: multiple validations by alternative methods) was given to each interacting pair.
All interactions were curated and scored independently by two different curators. The few scoring conflicts were resolved by a third independent curator.
Human PPI recuration
The human PPI dataset was compiled as previously described31. First, the method codes used by each database were classified as binary (e.g. two hybrid methods) or indirect (e.g. co-affinity purification)25. Then only protein interactions with binary support were selected for further analysis31. The multiply supported literature-curated dataset comprised 585 ‘curation units’ (one interaction scored from one publication) representing 188 protein-protein interactions, each reported in two or more publications and curated in two or more PPI databases. The dataset randomly selected from the full human literature-curated dataset31 comprised 240 curation units representing 188 protein-protein interactions.
Among the types of information collected during recuration were the gene symbols and GeneID of each interactor, the associated Pubmed ID, the name and the ID# of the interaction assay following the standard “interaction detection method” vocabulary implemented in PSI-MI34; the region of each protein used for the interaction assay (marked full-length if the entire protein sequence was used); the species for each interacting protein; and clarifying free-text comments used by the curator when needed. Several interpretative fields included: an assessment whether the interaction is bona fide, i.e. not erroneous; an assessment whether the interaction is indeed binary and an error field, using a simple controlled vocabulary to classify erroneous curation units such as ‘wrong protein’, ‘wrong species’, ‘no binding experiment’, ‘no binding partner’ (interaction between the proteins is not shown), ‘indirect’ (no direct interaction is shown), ‘Redundant PMID’ (some papers (usually crystallographic structure determination papers) have two distinct PMID numbers in PPI databases, and thus do not constitute two distinct papers supporting an interaction).
If there was no information about the region of the protein responsible for the interaction then the default was to full-length. If the species of the interacting proteins was not stated in a paper, a distressingly common occurrence, the default was to record the species as human. Thus, many interactions that did not involve human proteins might have been curated as human, so we may have underestimated the actual error rate. If the interaction was legitimate yet one or the other protein partner was a species other than human, then this interaction was not called bona fide. An interaction supported by multiple methods had to have just one bona fide and binary method to be recorded as legitimate; other methods apart from this one could be not binary or erroneous and not affect the final scoring.
Generally yeast two-hybrid and other protein complementation assays as well as structural determinations were labeled binary. Immunoprecipitation and co-affinity purification methodologies done in vivo that assess co-complex membership were considered not binary, while those done in vitro with e.g. recombinant proteins were considered binary. If a tagged protein was heterologously expressed in a cell to pull down endogenous proteins then such an interaction was called not binary. However, if both proteins of an interacting pair were heterologously expressed in a cell and shown to interact by e.g. pull down, then such an interaction was considered binary, since it is unlikely that an endogenous host protein mediates the interaction between the two heterologous proteins. If a co-immunoprecipitation done in vivo occurred in both orientations (protein A immunoprecipitation pulls down protein B and protein B immunoprecipitation pulls down protein A) then this interaction was judged binary. Since experimental procedures are often not described in sufficient detail to enable judgment of binary interaction, consistent policies in this regard were difficult to achieve.
Structural determinations were usually binary, except for protein complexes of more than two proteins where the interacting protein pairs did not actually contact each other in the solved structure. Solved structures that required a small third entity for crystal formation (e.g. GTP, phosphatidylinositol) were scored as binary, even though the interaction does not occur unless the small molecule is present.
A particular curation unit could have more than one error, though only the most prominent error was counted.
Arabidopsis PPI recuration
For Arabidopsis high-confidence interactions were defined as those supported by two papers or by two databases. In the initial search space of an ongoing Arabidopsis interactome mapping project 100 such interactions were collected. The union (OR) instead of the intersection (AND) was chosen for Arabidopsis, in contrast to human, so that a sufficiently sized sample of interactions was available. Otherwise, curation policies were as for human, including the error codes, but adding the name and the ID# of the “participant identification method” vocabulary implemented in PSI-MI34.
Box 1 Interologs
Interologs are in silico predictions of protein interactions in one species between a pair of proteins whose orthologs are known to interact in another species62-64. The assumption that interologs are more likely true than not is widely held65,66. Recent evaluations have now revisited this assumption in several ways24,67.
The most important question is where to draw the line for inter-species transfer. For instance, is mouse-human transfer close enough, but more evolutionarily distant mammals not? Actually, it is not the species relatedness but the sequence relatedness that really matters. Interolog transfers are only accurate for especially high sequence similarity24,64. Hence, interolog predictions with low sequence conservation should not be accepted, even between closely related species24.
Investigations of intrinsic disorder in proteins have also unsettled the certainty that protein interactions are highly conserved. There are two types of interacting surfaces in proteins. Domain-domain interactions are more prevalent in stable protein complexes, whereas domain-disorder interactions are more transient2,68,69. Domain-disorder interactions evolve much faster than domain-domain interactions70. The proportion of protein interactions that are of the domain-disorder type versus the domain-domain type is not known, even approximately, for any species. Still, the likely significant proportion of poorly conserved domain-disorder interactions means that the proportion of non-conserved interactions is substantial24.
In the one experimental test of interologs so far only one-third of the sample set of yeast interactions found by yeast two-hybrid were reproduced by yeast two-hybrid between the C. elegans orthologs63. A significant proportion perhaps, but not high enough to confirm the interolog hypothesis. Perhaps the large evolutionary distance between yeast and worm precluded a higher success rate, and mouse-human interologs might have a better success rate, but that supposition has not been experimentally tested.
In light of all these reappraisals curation policies are changing. For instance, one interaction database has stopped transferring non-human interactions to human19, a change from earlier practice48. Other interaction databases may follow suit. Alternatively, those interactions predicted by interolog extrapolation could be explicitly delineated in databases from those experimentally demonstrated, so the user could chose the appropriate data to examine. Either policy becomes complicated because species of the interactors are not often provided in publications30,33. Overall, it would seem best practice to only curate the species for which there is direct experimental evidence; in reality doing so is difficult.
Supplementary Material
ACKNOWLEDGEMENTS
This work was supported by National Human Genome Research Institute grants R01 HG001715 awarded to M.V. and F.P.R., P50 HG004233 awarded to M.V., and R01 HG003224 awarded to F.P.R., by funds from the W.M. Keck Foundation awarded to M.V., by an award (DBI-0703905) from the National Science Foundation to M.V. and J.R.E. and D.E.H., and by Institute Sponsored Research funds from the Dana-Farber Cancer Institute Strategic Initiative awarded to M.V. and CCSB. M.V. is a “Chercheur Qualifié Honoraire” from the Fonds de la Recherche Scientifique (FRS-FNRS, French Community of Belgium). We thank all members of CCSB for constructive discussions.
Footnotes
COMPETING INTERESTS STATEMENT The authors declare that they have no competing financial interests.
REFERENCES
- 1.Cusick ME, Klitgord N, Vidal M, Hill DE. Interactome: Gateway into systems biology. Hum. Mol. Genet. 2005;14:R171–81. doi: 10.1093/hmg/ddi335. [DOI] [PubMed] [Google Scholar]
- 2.Bader S, Kuhner S, Gavin AC. Interaction networks for systems biology. FEBS Lett. 2008;582:1220–4. doi: 10.1016/j.febslet.2008.02.015. [DOI] [PubMed] [Google Scholar]
- 3.Vidal M. Interactome modeling. FEBS Lett. 2005;579:1834–8. doi: 10.1016/j.febslet.2005.02.030. [DOI] [PubMed] [Google Scholar]
- 4.Roberts PM. Mining literature for systems biology. Brief. Bioinform. 2006;7:399–406. doi: 10.1093/bib/bbl037. [DOI] [PubMed] [Google Scholar]
- 5.Venkatesan K, et al. An empirical framework for binary interactome mapping. Nat. Methods. 2008 doi: 10.1038/nmeth.1280. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Stumpf MP, et al. Estimating the size of the human interactome. Proc. Natl. Acad. Sci. USA. 2008;105:6959–64. doi: 10.1073/pnas.0708078105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yu H, et al. High-quality binary protein interaction map of the yeast interactome network. Science. 2008;322:104–10. doi: 10.1126/science.1158684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Parrish JR, Gulyas KD, Finley RL., Jr. Yeast two-hybrid contributions to interactome mapping. Curr. Opin. Biotechnol. 2006;17:387–93. doi: 10.1016/j.copbio.2006.06.006. [DOI] [PubMed] [Google Scholar]
- 9.Ito T, et al. Roles for the two-hybrid system in exploration of the yeast protein interactome. Mol. Cell. Proteomics. 2002;1:561–6. doi: 10.1074/mcp.r200005-mcp200. [DOI] [PubMed] [Google Scholar]
- 10.Köcher T, Superti-Furga G. Mass spectrometry-based functional proteomics: from molecular machines to protein networks. Nat. Methods. 2007;4:807–15. doi: 10.1038/nmeth1093. [DOI] [PubMed] [Google Scholar]
- 11.Suter B, Kittanakom S, Stagljar I. Interactive proteomics: what lies ahead? Biotechniques. 2008;44:681–91. doi: 10.2144/000112799. [DOI] [PubMed] [Google Scholar]
- 12.Tarassov K, et al. An in vivo map of the yeast protein interactome. Science. 2008;320:1465–70. doi: 10.1126/science.1153878. [DOI] [PubMed] [Google Scholar]
- 13.Garrels JI. YPD—A database for the proteins of Saccharomyces cerevisiae. Nucleic Acids Res. 1996;24:46–9. doi: 10.1093/nar/24.1.46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hong EL, et al. Gene Ontology annotations at SGD: new data sources and annotation methods. Nucleic Acids Res. 2008;36:D577–81. doi: 10.1093/nar/gkm909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Swarbreck D, et al. The Arabidopsis Information Resource (TAIR): gene structure and function annotation. Nucleic Acids Res. 2007 doi: 10.1093/nar/gkm965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pagel P, et al. The MIPS mammalian protein-protein interaction database. Bioinformatics. 2005;21:832–4. doi: 10.1093/bioinformatics/bti115. [DOI] [PubMed] [Google Scholar]
- 17.Bader GD, Betel D, Hogue CW. BIND: the Biomolecular Interaction Network Database. Nucleic Acids Res. 2003;31:248–50. doi: 10.1093/nar/gkg056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Salwinski L, et al. The Database of Interacting Proteins: 2004 update. Nucleic Acids Res. 2004;32:D449–51. doi: 10.1093/nar/gkh086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chatr-aryamontri A, et al. MINT: the Molecular INTeraction database. Nucleic Acids Res. 2007;35:D572–4. doi: 10.1093/nar/gkl950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kerrien S, et al. IntAct—open source resource for molecular interaction data. Nucleic Acids Res. 2007;35:D561–5. doi: 10.1093/nar/gkl958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Reguly T, et al. Comprehensive curation and analysis of global interaction networks in Saccharomyces cerevisiae. J. Biol. 2006;5:11. doi: 10.1186/jbiol36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mishra GR, et al. Human protein reference database—2006 update. Nucleic Acids Res. 2006;34:D411–4. doi: 10.1093/nar/gkj141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Myers CL, Barrett DR, Hibbs MA, Huttenhower C, Troyanskaya OG. Finding function: evaluation methods for functional genomic data. BMC Genomics. 2006;7:187. doi: 10.1186/1471-2164-7-187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mika S, Rost B. Protein-protein interactions more conserved within species than across species. PLoS Comput. Biol. 2006;2:e79. doi: 10.1371/journal.pcbi.0020079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Simonis N, et al. Empirically-controlled mapping of the Caenorhabditis elegans protein-protein interaction network. Nat. Methods. 2008 doi: 10.1038/nmeth.1279. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jansen R, Gerstein M. Analyzing protein function on a genomic scale: the importance of gold-standard positives and negatives for network prediction. Curr. Opin. Microbiol. 2004;7:535–45. doi: 10.1016/j.mib.2004.08.012. [DOI] [PubMed] [Google Scholar]
- 27.Braun P, et al. An experimentally derived confidence score for binary protein-protein interactions. Nat. Methods. 2008 doi: 10.1038/nmeth.1281. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bader GD, Hogue CW. Analyzing yeast protein-protein interaction data obtained from different sources. Nat. Biotechnol. 2002;20:991–7. doi: 10.1038/nbt1002-991. [DOI] [PubMed] [Google Scholar]
- 29.Ramírez F, Schlicker A, Assenov Y, Lengauer T, Albrecht M. Computational analysis of human protein interaction networks. Proteomics. 2007;7:2541–52. doi: 10.1002/pmic.200600924. [DOI] [PubMed] [Google Scholar]
- 30.Howe D, et al. The future of biocuration. Nature. 2008;455:47–50. doi: 10.1038/455047a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rual JF, et al. Towards a proteome-scale map of the human protein-protein interaction network. Nature. 2005;437:1173–8. doi: 10.1038/nature04209. [DOI] [PubMed] [Google Scholar]
- 32.Peri S, et al. Development of Human Protein Reference Database as an initial platform for approaching systems biology in humans. Genome Res. 2003;13:2363–71. doi: 10.1101/gr.1680803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Orchard S, et al. Submit your interaction data the IMEx way. A step by step guide to trouble-free deposition. Proteomics. 2007;7:28–34. doi: 10.1002/pmic.200700286. [DOI] [PubMed] [Google Scholar]
- 34.Kerrien S, et al. Broadening the horizon - Level 2.5 of the HUPO-PSI format for molecular interactions. BMC Biol. 2007;5:44. doi: 10.1186/1741-7007-5-44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gavin AC, et al. Proteome survey reveals modularity of the yeast cell machinery. Nature. 2006;440:631–6. doi: 10.1038/nature04532. [DOI] [PubMed] [Google Scholar]
- 36.Gavin AC, et al. Functional organization of the yeast proteome by systematic analysis of protein complexes. Nature. 2002;415:141–7. doi: 10.1038/415141a. [DOI] [PubMed] [Google Scholar]
- 37.Ho Y, et al. Systematic identification of protein complexes in Saccharomyces cerevisiae by mass spectrometry. Nature. 2002;415:180–3. doi: 10.1038/415180a. [DOI] [PubMed] [Google Scholar]
- 38.Krogan NJ, et al. Global landscape of protein complexes in the yeast Saccharomyces cerevisiae. Nature. 2006;440:637–43. doi: 10.1038/nature04670. [DOI] [PubMed] [Google Scholar]
- 39.Ito T, et al. A comprehensive two-hybrid analysis to explore the yeast protein interactome. Proc. Natl. Acad. Sci. USA. 2001;98:4569–74. doi: 10.1073/pnas.061034498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Uetz P, et al. A comprehensive analysis of protein-protein interactions in Saccharomyces cerevisiae. Nature. 2000;403:623–7. doi: 10.1038/35001009. [DOI] [PubMed] [Google Scholar]
- 41.Alfarano C, et al. The Biomolecular Interaction Network Database (BIND) and related tools 2005 update. Nucleic Acids Res. 2005;33:D418–24. doi: 10.1093/nar/gki051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mathivanan S, et al. An evaluation of human protein-protein interaction data in the public domain. BMC Bioinformatics. 2006;7:S19. doi: 10.1186/1471-2105-7-S5-S19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gentleman R, Huber W. Making the most of high-throughput protein-interaction data. Genome Biol. 2007;8:112. doi: 10.1186/gb-2007-8-10-112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mackay JP, Sunde M, Lowry JA, Crossley M, Matthews JM. Protein interactions: is seeing believing? Trends Biochem. Sci. 2007;32:530–1. doi: 10.1016/j.tibs.2007.09.006. [DOI] [PubMed] [Google Scholar]
- 45.Mackay JP, Sunde M, Lowry JA, Crossley M, Matthews JM. Response to Chatr-aryamontri et al.: Protein interactions: to believe or not to believe? Trends Biochem. Sci. 2008;33:242–3. [Google Scholar]
- 46.Nelson DR. Gene nomenclature by default, or BLASTing to Babel. Hum. Genomics. 2005;2:196–201. doi: 10.1186/1479-7364-2-3-196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Krogan NJ, et al. A Snf2 family ATPase complex required for recruitment of the histone H2A variant Htz1. Mol. Cell. 2003;12:1565–76. doi: 10.1016/s1097-2765(03)00497-0. [DOI] [PubMed] [Google Scholar]
- 48.Zanzoni A, et al. MINT: a Molecular INTeraction database. FEBS Lett. 2002;513:135–40. doi: 10.1016/s0014-5793(01)03293-8. [DOI] [PubMed] [Google Scholar]
- 49.Philippi S, Kohler J. Addressing the problems with life-science databases for traditional uses and systems biology. Nat. Rev. Genet. 2006;7:482–8. doi: 10.1038/nrg1872. [DOI] [PubMed] [Google Scholar]
- 50.Kiemer L, Costa S, Ueffing M, Cesareni G. WI-PHI: a weighted yeast interactome enriched for direct physical interactions. Proteomics. 2007;7:932–43. doi: 10.1002/pmic.200600448. [DOI] [PubMed] [Google Scholar]
- 51.Chatr-Aryamontri A, Ceol A, Licata L, Cesareni G. Protein interactions: integration leads to belief. Trends Biochem. Sci. 2008;33:241–2. doi: 10.1016/j.tibs.2008.04.002. [DOI] [PubMed] [Google Scholar]
- 52.Boxem M, et al. A protein domain-based interactome network for C. elegans early embryogenesis. Cell. 2008;134:534–45. doi: 10.1016/j.cell.2008.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.von Mering C, et al. Comparative assessment of large-scale data sets of protein-protein interactions. Nature. 2002;417:399–403. doi: 10.1038/nature750. [DOI] [PubMed] [Google Scholar]
- 54.Batada NN, Hurst LD, Tyers M. Evolutionary and physiological importance of hub proteins. PLoS Comput. Biol. 2006;2:e88. doi: 10.1371/journal.pcbi.0020088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Orchard S, et al. The minimum information required for reporting a molecular interaction experiment (MIMIx) Nat. Biotechnol. 2007;25:894–8. doi: 10.1038/nbt1324. [DOI] [PubMed] [Google Scholar]
- 56.Hermjakob H, et al. The HUPO PSI’s molecular interaction format—a community standard for the representation of protein interaction data. Nat. Biotechnol. 2004;22:177–83. doi: 10.1038/nbt926. [DOI] [PubMed] [Google Scholar]
- 57.Ceol A, Chatr-Aryamontri A, Licata L, Cesareni G. Linking entries in protein interaction database to structured text: The FEBS Letters experiment. FEBS Lett. 2008;582:1171–7. doi: 10.1016/j.febslet.2008.02.071. [DOI] [PubMed] [Google Scholar]
- 58.Gerstein M, Seringhaus M, Fields S. Structured digital abstract makes text mining easy. Nature. 2007;447:142. doi: 10.1038/447142a. [DOI] [PubMed] [Google Scholar]
- 59.Taylor CF, et al. Promoting coherent minimum reporting guidelines for biological and biomedical investigations: the MIBBI project. Nat. Biotechnol. 2008;26:889–96. doi: 10.1038/nbt.1411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Stevens SW, et al. Composition and functional characterization of the yeast spliceosomal penta-snRNP. Mol. Cell. 2002;9:31–44. doi: 10.1016/s1097-2765(02)00436-7. [DOI] [PubMed] [Google Scholar]
- 61.Fromont-Racine M, Rain JC, Legrain P. Toward a functional analysis of the yeast genome through exhaustive two-hybrid screens. Nat. Genet. 1997;16:277–82. doi: 10.1038/ng0797-277. [DOI] [PubMed] [Google Scholar]
- 62.Walhout AJ, et al. Protein interaction mapping in C. elegans using proteins involved in vulval development. Science. 2000;287:116–22. doi: 10.1126/science.287.5450.116. [DOI] [PubMed] [Google Scholar]
- 63.Matthews LR, et al. Identification of potential interaction networks using sequence-based searches for conserved protein-protein interactions or “interologs”. Genome Res. 2001;11:2120–6. doi: 10.1101/gr.205301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Yu H, et al. Annotation transfer between genomes: protein-protein interologs and protein-DNA regulogs. Genome Res. 2004;14:1107–18. doi: 10.1101/gr.1774904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Ramani AK, Bunescu RC, Mooney RJ, Marcotte EM. Consolidating the set of known human protein-protein interactions in preparation for large-scale mapping of the human interactome. Genome Biol. 2005;6:R40. doi: 10.1186/gb-2005-6-5-r40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Sharan R, et al. Conserved patterns of protein interaction in multiple species. Proc. Natl. Acad. Sci. USA. 2005;102:1974–9. doi: 10.1073/pnas.0409522102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Levy ED, Pereira-Leal JB. Evolution and dynamics of protein interactions and networks. Curr. Opin. Struct. Biol. 2008;18:349–57. doi: 10.1016/j.sbi.2008.03.003. [DOI] [PubMed] [Google Scholar]
- 68.Tompa P, Fuxreiter M. Fuzzy complexes: polymorphism and structural disorder in protein-protein interactions. Trends Biochem. Sci. 2008;33:2–8. doi: 10.1016/j.tibs.2007.10.003. [DOI] [PubMed] [Google Scholar]
- 69.Fuxreiter M, Tompa P, Simon I. Local structural disorder imparts plasticity on linear motifs. Bioinformatics. 2007;23:950–6. doi: 10.1093/bioinformatics/btm035. [DOI] [PubMed] [Google Scholar]
- 70.Beltrao P, Serrano L. Specificity and evolvability in eukaryotic protein interaction networks. PLoS Comput. Biol. 2007;3:e25. doi: 10.1371/journal.pcbi.0030025. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.