Summary
During mitosis, sister chromatids congress to the spindle equator and are subsequently segregated via attachment to dynamic kinetochore microtubule (kMT) plus-ends. A major question is how kMT plus-end assembly is spatially regulated to achieve chromosome congression. Here we find in budding yeast that the widely-conserved kinesin-5 sliding motor proteins, Cin8p and Kip1p, mediate chromosome congression by suppressing kMT plus-end assembly of longer kMTs. Of the two, Cin8p is the major effector and its activity requires a functional motor domain. In contrast, the depolymerizing kinesin-8 motor Kip3p plays a minor role in spatial regulation of yeast kMT assembly. Our analysis identified a model where kinesin-5 motors bind to kMTs, move to kMT plus ends, and upon arrival at a growing plus-end promote net kMT plus-end disassembly. In conclusion, we find that length-dependent control of net kMT assembly by kinesin-5 motors yields a simple and stable self-organizing mechanism for chromosome congression.
Introduction
A central question in biology is how replicated chromosomes are properly segregated during mitosis, such that exactly one copy of each chromosome moves to each of the two daughter cells (Nicklas, 1997). In eukaryotes, chromosome-associated kinetochores attach to dynamic microtubule (MT) plus ends, while MT minus ends in turn generally attach to spindle poles (Inoue and Salmon, 1995). Once properly bioriented, so that one kinetochore is mechanically linked via one or more MTs to one pole and its sister kinetochore linked to the opposite pole, the sister chromosomes move toward the equator of the mitotic spindle, a process known as congression (Fig. 1A). In budding yeast, congression of bioriented sister chromosomes requires that the associated kinetochore microtubule (kMT) plus ends add tubulin subunits efficiently when they are near the poles (i.e. when kMTs are short), but inefficiently when near the equator (i.e. when kMTs are relatively long; (Gardner et al., 2005; Pearson et al., 2006; Sprague et al., 2003) (Fig 1A). The origin of this spatial gradient in net kMT plus end assembly is currently unknown.
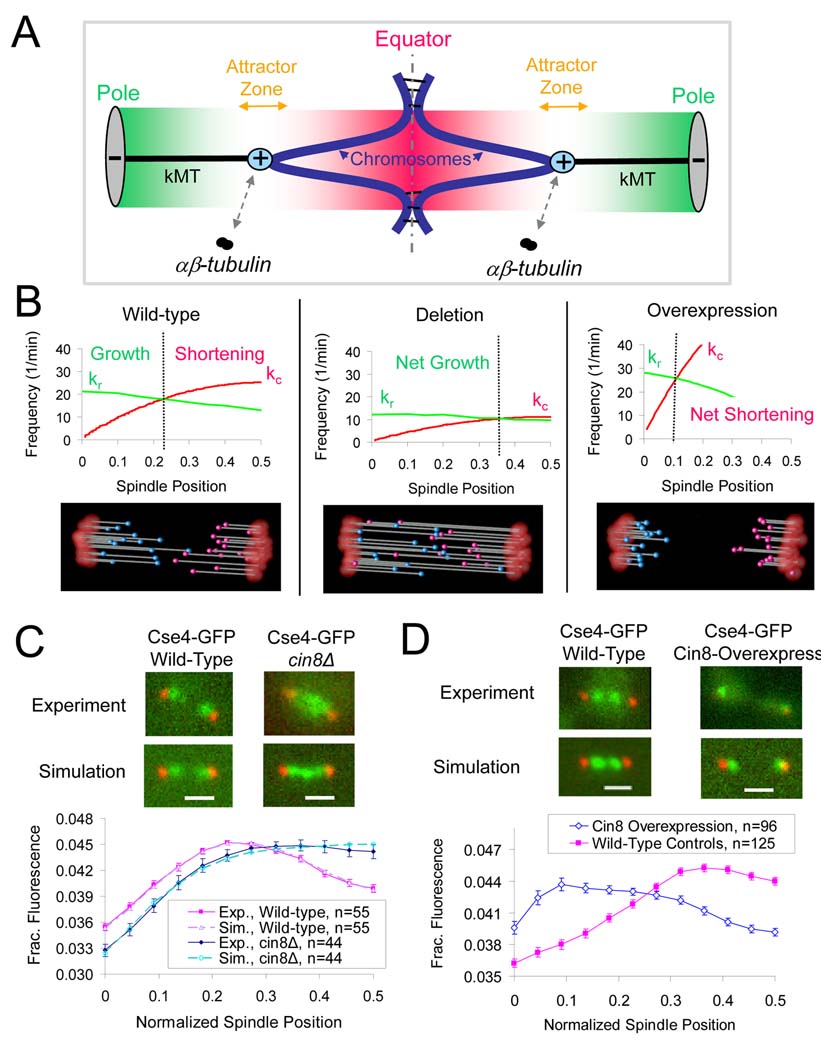
Figure 1. Cin8p organizes metaphase yeast kinetochores in a manner consistent with length-dependent suppression of net kMT plus-end assembly.
(A) A spatial gradient in net kMT plus-end assembly mediates kinetochore congression in yeast. Kinetochores (cyan) congress to attractor zones (yellow arrows) on either side of the spindle equator (dotted line) during yeast metaphase via the plus-end assembly dynamics of kMTs (black). The kMT plus-end assembly dynamics are spatially regulated such that plus-end assembly is favored near the poles (grey) when kMTs are relatively short (favorable assembly zone shown as green gradient), and suppressed near the spindle equator (dotted line) when kMTs are relatively long (assembly suppression zone shown as red gradient). (B) A computational model of yeast metaphase kMT plus end dynamics predicts that deletion of the length-dependent promoter of kMT disassembly will result in longer kMTs and disorganized kinetochores. Conversely, overexpression will result in shorter kMTs and focused kinetochore clusters. All model parameters as in Table S2. (C) CIN8 deletion results in kinetochore disorganization and the net shifting of kinetochores towards the spindle equator, suggesting that kMTs are on average longer than they are in wild-type cells (red, Spc29-CFP pole marker; green, Cse4-GFP kinetochore marker) (scale bar 1000 nm, error bars, s.e.m.) (D) Cin8p overexpression results in clusters of kinetochores near the SPBs, suggesting that kMTs are shorter than in wild-type cells.
One possible explanation for the spatial gradient of kMT net assembly derives from studies with the budding yeast kinesin-8 molecular motor Kip3p, which showed that processive plus-end directed depolymerizing motors could result in length-dependent disassembly of MTs in vitro (Howard and Hyman, 2007; Varga et al., 2006). However, to date there has not been an in vivo observation of length-dependent MT regulation via Kip3p or any other motor. Recent in vivo studies of Kip3p showed that cytoplasmic microtubule dynamics are altered by Kip3p, and that KIP3 deletion results in spindle positioning defects (Gupta et al., 2006). Whether these in vivo effects resulted from the length-dependent control of MT assembly as proposed by Varga et al. (2006) was not clear. As a result, it is not clear at this point whether this interesting phenomenon identified in vitro by Varga et al. plays an important role in a biological process.
We now find that the kinesin-5 molecular motors, mainly Cin8p and, to a lesser extent, Kip1p, exhibit length-dependent control of MT assembly, and so mediate the spatial gradient in kMT plus end net assembly that drives congression during mitosis. Since their discovery, kinesin-5 motors have been viewed as mitosis-specific sliding motors that cross-link antiparallel MTs and exert outward extensional forces on the poles (Hoyt et al., 1992; Kapitein et al., 2005; Roof et al., 1992; Saunders and Hoyt, 1992; Tao et al., 2006). The results were surprising to us because kinesin-5 (Eg5 in vertebrates) is not known to affect microtubule assembly. Surprisingly, the disassembly-promoting activity that we now report is not specific to kMTs, since we found that Cin8p promotes disassembly of cytoplasmic astral MTs (aMTs) as well.
Because the kinesin-5 mediated length-dependent kMT disassembly is similar to that described by Varga et al. for Kip3p on MTs in vitro, we examined the role of Kip3p in controlling kMT assembly in vivo. We found that Kip3p only weakly affects kMT dynamics and is not a major mediator of congression. Our computer simulations show that the difference in kinesin-5 and kinesin-8 behavior can be accounted for simply by the ability of tetrameric kinesin-5 to cross-link microtubules, a property lacking in dimeric kinesin-8. This naturally biases kinesin-5 towards kMT plus ends and kinesin-8 toward interpolar MT plus ends. Together, the results presented here suggest a simple explanation for metaphase chromosome congression in budding yeast by identifying a microtubule length-dependent disassembly-promoting activity in vivo that is associated with kinesin-5 molecular motors.
Results
Simulated phenotypes of a disrupted gradient in net kMT plus end assembly
MTs self-assemble from αβ-tubulin heterodimers via an unusual process called “dynamic instability” where MTs grow at a roughly constant rate, then abruptly and stochastically switch to shortening at a roughly constant rate, and then switch back to growth again and so forth. The switch from growth to shortening is called “catastrophe” and the switch from shortening to growth is called “rescue” (Desai and Mitchison, 1997). Together, the four parameters of dynamic instability, growth rate (Vg [=] µm/min), shortening rate (Vs [=]µm/min), catastrophe frequency (kc [=] 1/min), and rescue frequency (kr [=] 1/min), define the net assembly state of MTs. If the mean length added during a growth phase (Lg=Vg/kc [=] µm) exceeds the mean length lost during shortening (Ls=Vs/kr [=] µm), then, there will be net growth, otherwise there will be net shortening. If the parameters depend upon position in the cell, then there can exist net growth in one part of the cell, and net shortening in another part of the cell. At the transition between these regions there will be no net growth, and this will create an attractor for plus ends, provided net growth is favored for short MTs and net shortening favored for long MTs (Fig. 1A).
Our previous studies showed that net kMT plus-end assembly in budding yeast metaphase spindles is favored for plus-ends located near the spindle pole bodies (SPBs) (i.e. for short kMTs), and inhibited for plus-ends near the equator (i.e. for longer kMTs) (Fig. 1A)(Gardner et al., 2005; Pearson et al., 2006; Pearson et al., 2004; Sprague et al., 2003). This spatial control over net kMT assembly was most readily explained using a catastrophe gradient in kMT plus-end assembly that creates two attractor points where there is no net assembly, one attractor in each half-spindle (Fig. 1B, left, dotted line denotes the location of the attractor point for one half-spindle). These two attractors establish the bilobed distribution of kinetochores into the two distinct clusters that are characteristic of the congressed metaphase spindle (Fig. 1A).
We were interested in identifying the molecules responsible for this net assembly gradient, and so we simulated the expected phenotypes for changes in expression level of a putative spatial regulator of net kMT plus-end assembly. Fig 1B (left) shows a simulation of a wild-type budding yeast metaphase spindle where kMT plus-end assembly is favored near the SPB (where kMTs are short), and suppressed near the spindle equator (where kMTs are longer). Note that in the haploid budding yeast there are 16 kMTs emanating from each SPB, for a total of 32 kMTs, and ~8 longer interpolar MTs (iMTs), for a total of 40 MTs in the metaphase spindle (for the sake of clarity the iMTs are omitted from the animation in Fig.1). If a molecule promoted disassembly of longer kMTs in wild-type cells, and was then deleted, the predicted phenotype would be longer kMTs with kinetochores more broadly distributed along the spindle, as depicted in Fig. 1B, center. Conversely, overexpression of a spatial assembly regulator would produce very short kMTs with highly focused clusters of kinetochores near the SPBs, as depicted in Fig. 1B, right. These model predictions establish specific requirements for experimental identification of a spatial kMT plus-end assembly regulator.
The yeast kinesin-5 motors, Cin8p and Kip1p, control kinetochore positions
While studying various deletion mutants, we observed that cin8Δ mutants lost the clustering of kinetochores within each half spindle (measured by labeling of a kinetochore component, Cse4-GFP, in live cells), as shown in Fig. 1C, consistent with an earlier report (Tytell and Sorger, 2006). Quantitative analysis of experimental Cse4-GFP fluorescence revealed that the peak fluorescence intensity also shifted toward the spindle equator, as predicted by simulations used to model deletion of a molecule that promotes net kMT plus-end disassembly of longer kMTs (Fig. 1C, p= 0.82, where p is the probability that the experimental Cse4-GFP fluorescence distribution curve is consistent with the simulated curve; see Methods for calculation procedure). The simulated Cse4-GFP fluorescence distribution was obtained by convolution of the simulated fluorophore positions with the imaging system point spread function and noise, a computational process we call “model-convolution” (Gardner et al., 2007; Sprague et al., 2003). Deletion of the other yeast kinesin-5 motor, KIP1, had a similar, but weaker, phenotype to cin8Δ, with a moderate shift of kinetochores towards the spindle equator (Fig. S1A). We note that the effects of CIN8 deletion were not due to the well known moderate decrease in steady-state spindle length (Hildebrandt and Hoyt, 2000; Hoyt et al., 1992; Saunders et al., 1997; Saunders and Hoyt, 1992), since we selected spindle lengths that were equal for both wild-type and cin8Δ cells (although results were similar regardless of the spindle length population analyzed (Fig. S2)). To further establish that the effects of CIN8 deletion were not due to changes in metaphase spindle lengths, we performed separate experiments using histone H3 repression mutants, which make centromeric chromatin more compliant and thus increase average spindle length (Bouck and Bloom, 2007). In these longer spindles, wild-type kinetochores were still bilobed while cin8Δ kinetochores were still disorganized (Fig. S1B), showing an insensitivity of the disorganization phenotype to spindle length. In addition, bim1Δ mutant spindles with short spindle lengths (similar to cin8Δ mutant spindle lengths, Fig. S3) did not result in spindle disorganization (Fig. S3). We conclude that kinetochores are declustered in kinesin-5 deletion mutants, independent of the spindle length.
If Cin8p mediates net kMT plus-end disassembly, then Cin8p overexpression will result in short kMTs with focused clusters of kinetochores, one near to each SPB (Fig. 1B, right). As shown in Fig. 1D, kinetochore clusters were indeed tightly focused within each half-spindle and much closer to each SPB. Spindles overexpressing Cin8p also have increased length due to increased motor sliding between oppositely oriented central spindle non-kMTs (also known as interpolar MTs) (Saunders et al., 1997) (Fig. S4). Despite the spindles being longer, kinetochores in Cin8p overexpressing cells were still ~50% closer to SPBs than wild-type controls (Fig. 1D), consistent with Cin8p overexpression resulting in shorter kMTs (Fig. S4). We conclude that Cin8p, and, to a lesser extent, Kip1p, promote net kMT plus-end disassembly as judged by kinetochore position.
GFP-tubulin fluorescence confirms that kMTs are longer in cin8Δ mutants
If Cin8p promotes net kMT plus-end disassembly, then CIN8 deletion will result in longer kMTs, producing a continuous “bar” of fluorescent tubulin along the length of the spindle (Fig. 2A, right), rather than the wild-type fluorescent tubulin “tufts” that emanate from each of the two SPBs (Fig. 2A, left). In experiments with GFP-Tub1, quantitative analysis of tubulin fluorescence in cin8Δ mutants revealed a shift in fluorescence towards the spindle equator, indicating that kMT length was increased (Fig. 2A, bottom). The distribution of GFP-Tub1 was quantitatively predicted in simulations using the same parameter set used to model kinetochore organization in cin8Δ mutants (Fig. 2A, p=0.22, Table S2). In addition, we found that the ratio of spindle tubulin polymer signal to free tubulin signal outside of the spindle area is 2.0:1 in wild-type spindles (n=27), and 3.2:1 in cin8Δ mutant spindles (n=35), which represents an increase in tubulin polymer relative to free tubulin of ~62% in cin8Δ mutants as compared to wild-type cells (p<10−5). This suggests that the increased kMT length in cin8Δ spindles is not the result of an overall increase of tubulin level, but rather reflects a thermodynamic shift toward increased net kMT assembly.
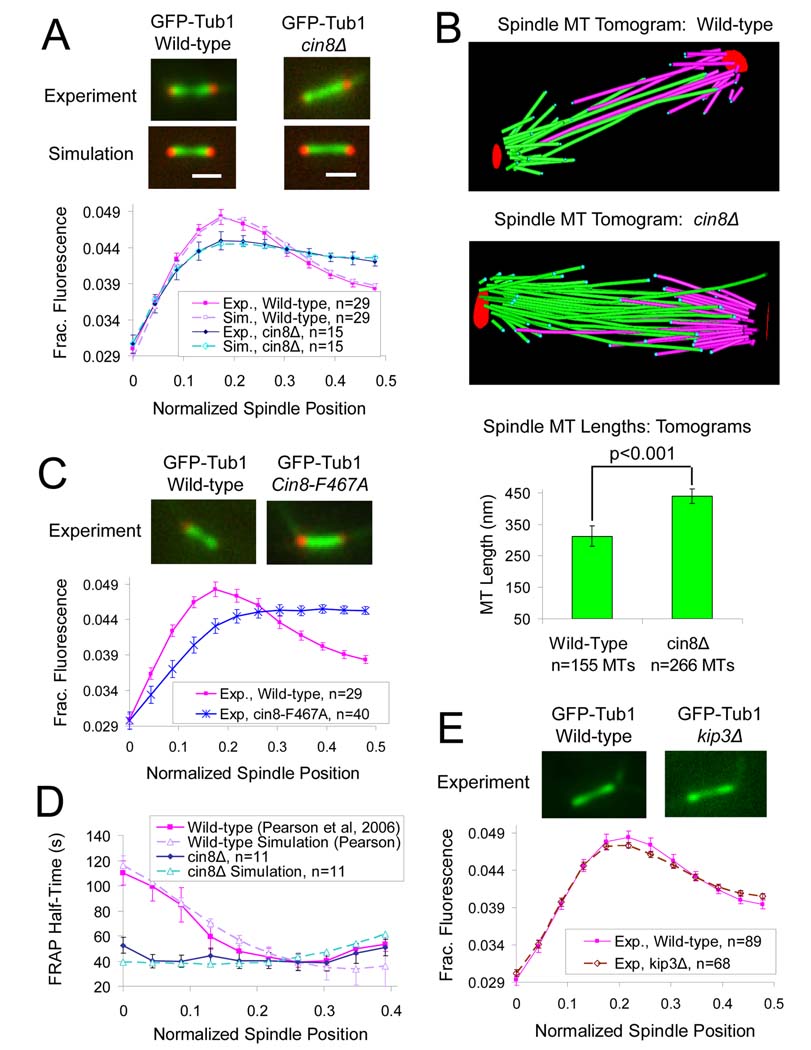
Figure 2. Cin8p promotes kMT disassembly.
(A) CIN8 deletion results in flattening of the GFP-tubulin fluorescence distribution and shifting of fluorescence towards the spindle equator, suggesting that kMT lengths are increased in the mutant. (Red, Spc29 CFP SPB marker; Green, GFP-Tub1 MT marker) (scale bar 1000 nm, error bars, s.e.m.) All model parameters as in Table S2. (B) Cryo-electron tomography reveals increased mean MT length and number in cin8Δ spindles (n=5 spindles, mean spindle length =1387 nm), relative to wild-type spindles (n=4 spindles, mean spindle length=1265 nm). (C) Similar to the CIN8 deletion mutant, the motor domain mutant, cin8-F467A, which has reduced affinity for MTs, results in flattening of the GFP-tubulin fluorescence distribution and shifting of fluorescence towards the spindle equator. (D) CIN8 deletion eliminates the characteristic gradient in GFP-tubulin FRAP recovery half-time, consistent with disruption of the gradient in net kMT assembly. (E) In contrast to the cin8Δ mutants, deletion of the kinesin-8 depolymerase KIP3 does not significantly perturb kinetochore microtubule (kMT) organization.
Cryo-electron tomography confirms that kMTs are longer in cin8Δ mutants
To directly visualize individual spindle MTs, we used cryo-electron tomography to reconstruct complete mitotic spindles from wild-type and cin8Δ mutant spindles (Fig. 2B, supplemental movies 1,2). To control for the moderate spindle length shortening in cin8Δ mutants, we selected spindles of similar length in the wild-type and mutant cell populations. Consistent with model predictions, we found a substantial increase in mean MT length in cin8Δ mutant spindles as compared to wild-type spindles (Fig. 2B, 41% increase in mean overall length, p=0.0007, statistical consistency between cells confirmed by ANOVA in Table S1). Total non-kMT number also increased in cin8Δ as compared to wild-type spindles (42 % increase in total MT number, p=0.002), demonstrating that the total polymer level in the cin8Δ cells is increased relative to wild-type cells. Interestingly, the mean length of the 8 longest MTs in each spindle, presumably interpolar MTs (iMTs), is not statistically different between wild-type and cin8Δ mutant cells (wild-type = 931 +/− 81 nm (mean±s.e.m., n=33 MTs); cin8Δ = 1141 +/− 25 nm (n=41 MTs), p=0.05). This result suggests that deletion of CIN8 most significantly affects the kMT length rather than iMT length.
In summary, the electron microscopy results independently confirm the model predictions and the light microscopy studies by demonstrating that kMTs are indeed longer in cin8Δ cells. We conclude that Cin8p participates in a process that promotes net kMT disassembly.
A Cin8p motor-domain mutant has increased kMT length
To test whether kMT length regulation requires the Cin8p motor domain, the organization of fluorescent tubulin was examined in the cin8-F467A mutant (Gheber et al., 1999). Previous studies found that the cin8-F467A mutation reduced the binding of Cin8p to microtubules in vitro by 10-fold (Gheber et al., 1999). Similar to the cin8Δ mutants, analysis of GFP-Tub1 distribution in the cin8-F467A mutants produced a continuous “bar” of fluorescent tubulin along the length of the spindle (Fig. 2C, right), rather than the wild-type fluorescent tubulin “tufts” (Fig. 2C, left). The similarity between the cin8Δ and the cin8-F467A mutant phenotypes suggests that the kMT disassembly promoting activity of Cin8p requires motor binding to kMTs. Furthermore, since our previous studies showed that net kMT assembly is promoted when kMTs are short and suppressed when kMTs are long (i.e. when kMT plus ends extend into the equatorial region, Fig 1B), we also conclude that Cin8p-mediated suppression of kMT assembly is specific to longer kMTs.
Cin8p mediates the gradient in net kMT assembly as measured by GFP-tubulin FRAP
To further test whether Cin8p specifically suppresses assembly of longer kMTs and thus mediates a gradient in net kMT assembly, we measured the spatial gradient in tubulin turnover within the mitotic spindle. In our previous work, we found that tubulin turnover, as measured by spatially resolved GFP-tubulin Fluorescence Recovery After Photobleaching (FRAP), is most rapid where kMT plus ends are clustered in wild-type cells (Pearson et al., 2006). If Cin8p mediates a gradient in net kMT assembly, then its deletion is predicted to result in loss of the gradient in FRAP half-time. As shown in Fig. 2D, deletion of CIN8 results in loss of the tubulin turnover gradient, as predicted by the model (Fig 1B, center). In general, the kMTs remain dynamic (overall t1/2=46±15 s integrated over the half-spindle in cin8Δ (n=11), compared to t1/2=63±30 s for wild-type (Pearson et al., 2006), (n=22; p=0.03; Fig. S5), and have a high fractional recovery (~90% for cin8Δ, compared to ~70% for wild-type (Maddox et al., 2000; Pearson et al., 2006)).
In simulating the cin8Δ GFP-tubulin FRAP experiment, the best fit between theory and experiment is achieved with a flattened spatial gradient in net kMT plus-end assembly (e.g. a flattened catastrophe gradient), and with values for MT plus-end growth and shortening rates that are slightly higher than in wild-type simulations (Table S2). This result explains why FRAP is relatively rapid along the entire length of the half-spindle in the cin8Δ mutants, and is consistent with recent studies that find increased chromosome oscillation velocities upon deletion of the human kinesin-8 motor Kif18A (Stumpff et al., 2008). (reviewed in (Gardner et al., 2008)). In summary, we find that the tubulin-FRAP studies confirm that Cin8p mediates a spatial gradient in kMT assembly dynamics.
Deletion of the kinesin-8 motor Kip3p does not perturb congression
Recent studies found that the kinesin-8 molecular motor Kip3p acts as a length-dependent depolymerase in vitro (Varga et al., 2006), and further studies have implicated the human kinesin-8 molecular motor Kif18A in the regulation of chromosome oscillation amplitude (Stumpff et al., 2008) and chromosome congression (Mayr et al., 2007). Thus, we asked whether Kip3p deletion would perturb kMT length organization in the yeast mitotic spindle. Interestingly, deletion of KIP3 does not produce the continuous “bar” of fluorescent tubulin along the length of the spindle that is characteristic of cin8Δ spindles, but rather shows wild-type fluorescent tubulin “tufts” (Fig. 2E). Although microtubule organization appears wild-type, spindle lengths are moderately increased in kip3Δ cells, as previously reported (Straight et al., 1998) (mean spindle lengths in Fig. 2E, : WT: 1.60±0.22; kip3Δ: 1.97±0.15 µm). Thus, Kip3p regulates spindle length, possibly through regulation of iMT lengths, while Cin8p regulates kMT length without a measurable effect on iMT lengths. Regardless of effects on iMTs, the effect of Kip3 on kMTs is relatively weak compared to the effect of Cin8p, and is insufficient to establish congression in the absence of Cin8p.
Cin8p promotes shortening of astral MTs in the cytoplasm
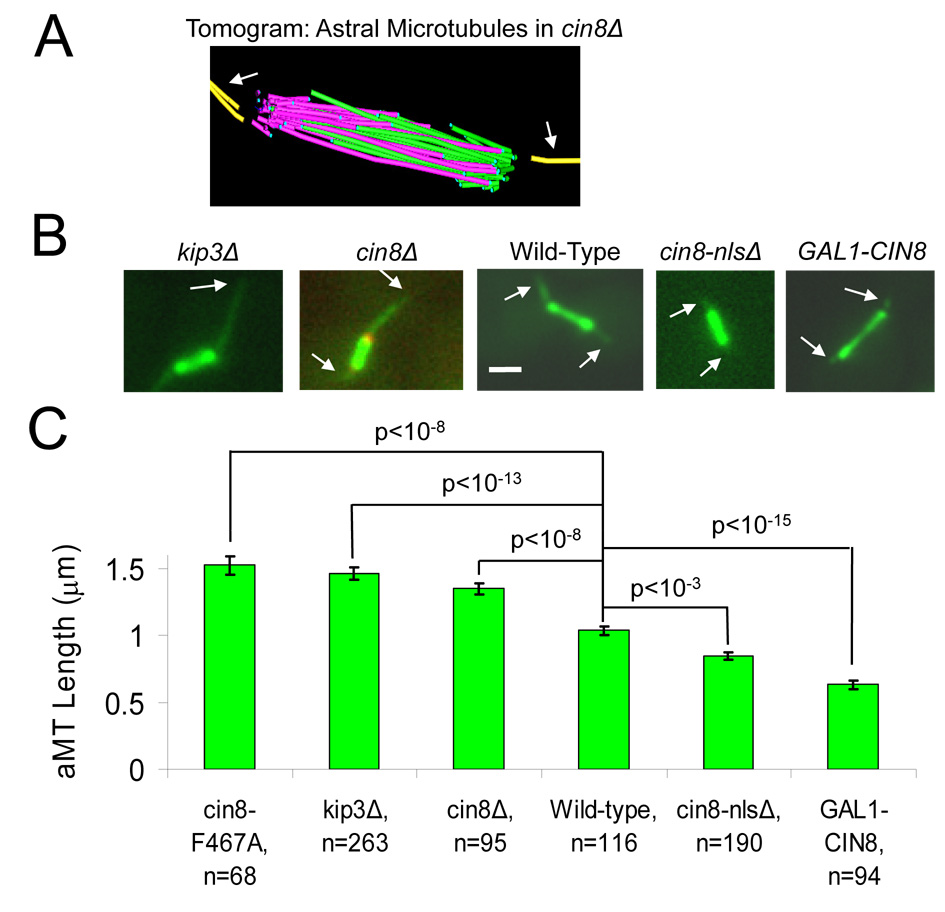
MTs are densely packed in the yeast mitotic spindle (Fig.2B), and so it is difficult to resolve individual spindle MTs via fluorescence microscopy. In contrast, yeast astral MTs (aMTs) are normally much fewer in number (1–3) and splayed apart (Fig. 3A) (Gupta et al., 2002; Shaw et al., 1997). Because Kip3p is known to affect aMT dynamics, (Cottingham and Hoyt, 1997; Gupta et al., 2006; Miller et al., 1998), but the effect of Cin8p is not known, we then examined the effect of motor deletion on aMTs outside of the mitotic spindle. Indeed, we found that aMT lengths were statistically longer in kip3Δ mutants as compared to wild-type cells (Fig. 3B,C), consistent with previous studies (Cottingham and Hoyt, 1997; Gupta et al., 2006).
Figure 3. Cin8p promotes astral MT disassembly.
(A) Astral MTs (aMTs) extend from the SPBs into the cytoplasm, and are fewer in number than spindle microtubules. (B) aMT lengths were measured via GFP-tubulin fluorescence (white arrows point to plus ends; red, Spc29-CFP pole marker; green, GFP-tubulin) (scale bar 500 nm). (C) aMT lengths are increased in cin8Δ cells as compared to wild-type spindles. Cytoplasmic overexpression of Cin8p results in shorter aMTs, whether overexpression is global, as in GAL1-CIN8 overexpression experiments, or if overexpression is local in the cytoplasm, as in experiments with mutant Cin8p lacking the nuclear localization signal (cin8-nlsΔ). Both kip3Δ and cin8-F467A mutants have increased aMT lengths relative to wild-type cells, similar to cin8Δ cells.
A recent report suggests that Cin8p plays a role in spindle positioning through an aMT-dependent mechanism, and so, coupled with our results for kMTs, we hypothesized that Cin8p also suppresses aMT assembly (de Gramont et al., 2007; Geiser et al., 1997). Using GFP-tubulin fluorescence microscopy, we measured the length of individual aMTs in the cytoplasm (Fig. 3B) (Carminati and Stearns, 1997; Gupta et al., 2002; Shaw et al., 1997). Consistent with the behavior of kMTs, aMTs were longer in cin8Δ mutants relative to wild-type cells (Fig. 3 B,C; Fig. S6). In addition, a previous study showed that aMT numbers are not decreased in cin8Δ mutants (de Gramont et al., 2007), consistent with an overall increase in both kMT and aMT polymer in cin8Δ mutants. Interestingly, aMT lengths in the cin8-F467A mutants with reduced binding of Cin8p to microtubules were also statistically longer than wild-type aMTs (Fig 3C). Conversely, overexpression of Cin8p resulted in shorter aMTs (Fig 3 B,C, Fig. S6).
Since the tubulin polymer level in the cin8Δ cells increases in both the nucleus and in the cytoplasm, these results argue against a simple repartitioning of tubulin (or some other Cin8p-dependent assembly-promoting factor) from the cytoplasm into the nucleus in response to CIN8 deletion. Conversely, MT polymer levels decrease in both compartments upon Cin8p overexpression.
One possible reason why both kMTs and aMTs are longer in cin8Δ cells is that CIN8 deletion indirectly promotes MT assembly globally. To test this hypothesis, we shifted Cin8p from the nucleus to the cytoplasm while keeping the overall Cin8p expression level approximately constant. This shift was achieved by deleting the nuclear localization sequence (NLS) of Cin8p (as previously described) (Hildebrandt and Hoyt, 2001). Because budding yeast undergoes a closed mitosis, deleting the NLS decreases the nuclear Cin8p concentration and increases the cytoplasmic Cin8p concentration (Hildebrandt and Hoyt, 2001). We found that cin8-nlsΔ spindle MTs had a flat GFP-Tub1 fluorescence distribution, similar to cin8Δ cells (i.e. no tufts, Fig. S7A). This result is consistent with net kMT assembly in the absence of Cin8p locally in the nucleus (Fig. 3B, Fig S7A). Importantly, and in contrast to cin8Δ cells, aMT lengths in cin8-nlsΔ cells were shorter than in wild-type cells (Fig. 3 B,C; p<0.001, Fig. S7C), consistent with elevation of Cin8p concentration locally in the cytoplasm. These results indicate that Cin8p acts locally in a given cellular compartment, rather than globally throughout the entire cell, to influence the local MT assembly state.
Together, these results suggest that Cin8p and Kip3p act similarly to suppress assembly of aMTs outside of the mitotic spindle. We then asked how Cin8p could act to promote kinetochore congression through suppression of kMT plus-end assembly, while Kip3p appears to regulate spindle length, possibly through suppression of iMT plus-end assembly.
A model for Cin8p motor interaction with kMTs
Because Cin8p requires microtubule binding to promote length-dependent kMT plus-end disassembly, we hypothesized that Cin8p acts directly on kMT plus ends, either by itself or with a binding partner. To test the direct-interaction hypothesis, we first predicted the spatial distribution of Cin8p motors on kMTs via computational modeling. As a starting point, we extended our previous model for individual kMT plus-end dynamics (Gardner et al., 2005) to also include the dynamics of microtubule-associated kinesin-5 molecular motors. This motor model assumes that motors reversibly attach and detach, cross-link MTs, and move toward MT plus ends (supplemental movies 3,4; Fig. S8, S9; Tables S3-S6) (Gheber et al., 1999; Kapitein et al., 2005; Kashina et al., 1996; Valentine et al., 2006).
Cin8-GFP is distributed in a gradient along kMTs
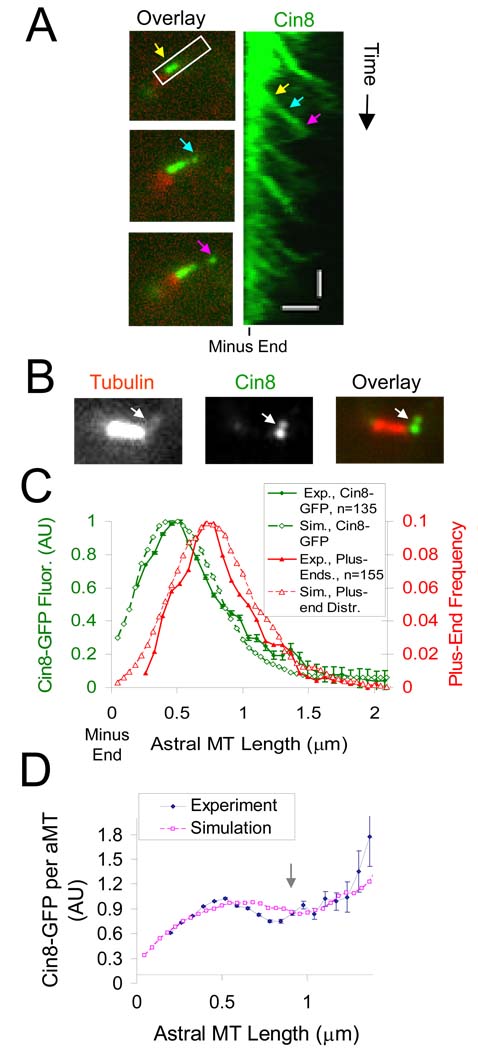
As shown in Fig. 4A, this simple model for motor dynamics predicts that, at steady-state, a substantial fraction of the motors cross-link parallel kMTs emanating from the same SPB (green). In the simulation, these motors walk to a kMT plus end and follow the end as it grows until catastrophe occurs, at which point the motor head detaches (supplemental movie 4). By repetitive photobleaching of Cin8-3XGFP labeled spindles, we were able to experimentally observe spindle-associated Cin8p motor dynamics in the “speckle” regime. Similar to the motor model simulation, we observed motor movement in the spindle with a mean velocity of 58 +/− 24 nm/sec (mean +/− s.d., n=10) (Fig. 4B; see Fig. S8 and supplemental material).
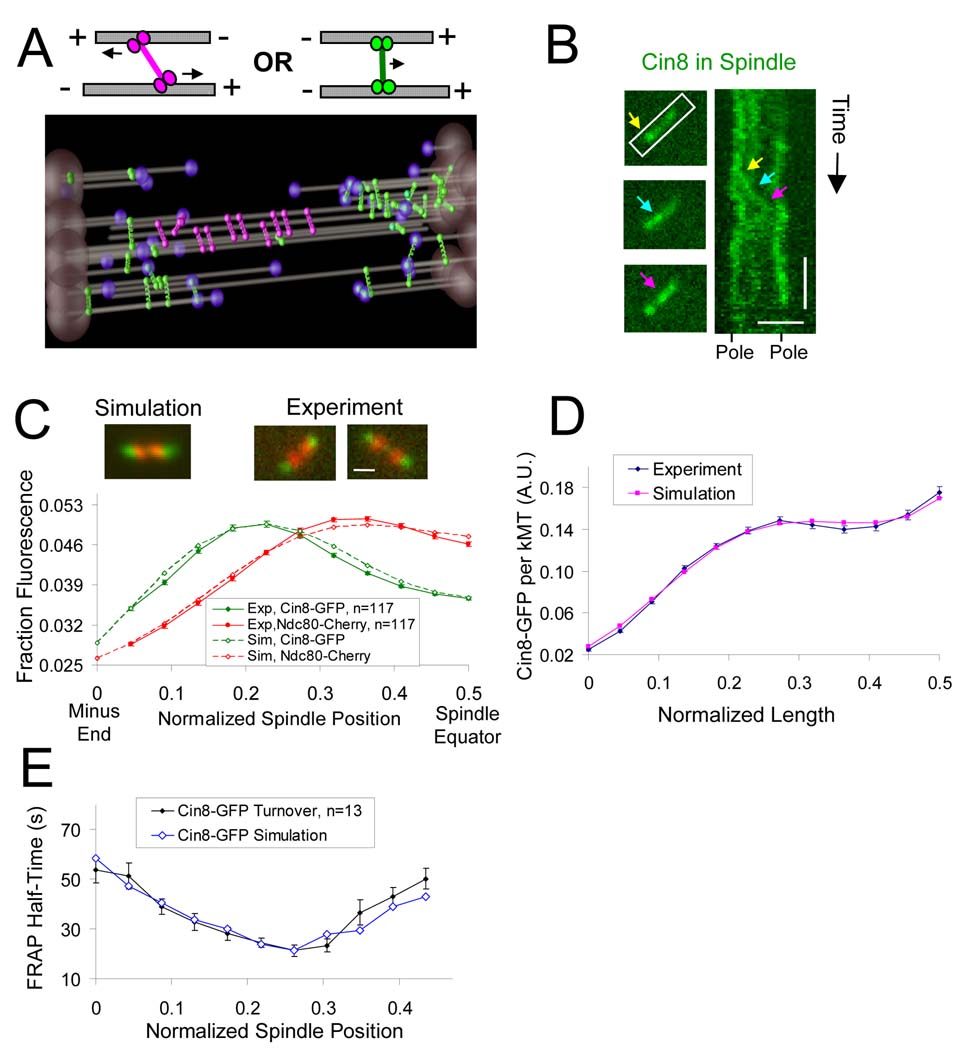
Figure 4. Cin8p accumulates on kMTs in a length-dependent manner and frequently interacts with kMT plus ends.
(A) In simulation, kinesin-5 motors crosslink both anti-parallel-oriented microtubules (left, magenta) and parallel-oriented microtubules (right, green). Simulated motors crosslinking parallel-oriented microtubules move to and frequently interact with kMT plus-ends. (B) Cin8-3XGFP motor movement can be observed in the spindle (horizontal scale bar, 1000 nm; vertical scale bar, 20 sec). The three arrows (yellow, cyan, and magenta) indicate three time points in the movement of a Cin8-3XGFP fluorescent spot that moves in the plus end direction. (C) Cin8p motors concentrate near kinetochores both experimentally and in simulation (Red, Ndc80-Cherry kinetochore marker; green, Cin8-GFP) (scale bar 500 nm, error bars, s.e.m.). (D) Experimentally and in simulation, Cin8-GFP fluorescence normalized to the number of tubulin polymer binding sites increases for longer kMTs. (E) Cin8-GFP FRAP half-time gradient.
To test the motor model, we predicted the distribution of Cin8-GFP fluorescence in live cells as a function of spindle position, and then measured it experimentally. As shown in Fig. 4C, the motor model is able to quantitatively reproduce the experimentally observed Cin8-GFP motor fluorescence distribution (green) relative to kinetochores as measured by Ndc80-cherry fluorescence (red) (p=0.35). We found that in order to correctly reproduce the Cin8-GFP fluorescence distribution, simulated motors are required to track growing but not shortening kMT plus-ends, resulting in a slight, but detectable, shift in the peak of motor-associated fluorescence away from kinetochores and towards SPBs (see supplementary material for analysis of various alternative models, Fig. S9). In the motor model, this shift is the result of the increased motor off-rate imposed by shortening kMT plus-ends within the kinetochore clusters (Fig. 4C, red). In benomyl-treated spindles with stabilized microtubules, this shift is decreased, such that the motor-associated fluorescence is nearly coincident with kinetochores (Fig. S10). Thus, the results with benomyl further support the hypothesis that kMT plus-end assembly dynamics control motor dynamics, in particular motor detachment from kMT plus ends.
The Kip1-GFP fluorescence distribution was qualitatively similar to the Cin8-GFP fluorescence distribution, but was relatively less focused into two clusters, consistent with a weaker affinity of Kip1p for MTs (Fig. S11).
By normalizing the Cin8-GFP fluorescence to the local kMT density, we then calculated the Cin8-GFP density on a per kMT basis (see methods for details). As shown in Fig. 4D, Cin8-GFP concentration per kMT gradually increases with increasing distance from the SPB, as predicted by the motor model. We conclude that αβ-tubulins at the plus ends of longer kMTs are more likely to be associated with Cin8-GFP than αβ-tubulins at the plus ends of shorter kMTs.
Cin8-GFP turnover is most rapid near kMT plus-ends
If motors rapidly detach from shortening kMT plus ends, then the rate of Cin8-GFP turnover on the spindle should be fastest in the vicinity of shortening kMT plus ends. Specifically, the motor model predicts there will be a spatial gradient in Cin8-GFP FRAP half-time that is very similar to the GFP-tubulin FRAP half-time gradient that we reported previously (Pearson et al., 2006) (Fig. 2C). Alternatively, the Cin8-GFP FRAP half-time could be longest where kMT plus-ends are clustered, indicative of high affinity binding to the kinetochore (Tytell and Sorger, 2006) , which would also in principle explain the gradient in Cin8-GFP intensity observed experimentally in Fig. 4C. We performed kinesin-5-GFP FRAP experiments, and found that motor turnover is most rapid in the location of kMT plus-end clustering, (Fig. 4E, Fig S12; p<1×10−10 as compared to half-times near the SPBs), suggesting that Cin8p motors rapidly detach from shortening kMT plus ends. Mean Kip1-GFP FRAP half-times were ~50% faster than Cin8-GFP recovery half-times (Fig. S12), again suggesting that Kip1p has a lower affinity for microtubules than Cin8p. The Cin8-GFP FRAP results are all consistent with a model in which motors frequently interact with kMT plus-ends in a length-dependent manner.
To summarize our studies of spindle-bound kinesin-5 dynamics, we find that kMT-bound kinesin-5 motors are distributed in a spatial gradient along kMTs, with their concentration increasing with increasing distance from the SPB. Consistent with the observed gradient, we also find that kinesin-5 motors frequently interact with, and are controlled in their detachment from the MT lattice by, dynamic kMT plus ends. Because Cin8p suppresses assembly of longer kMTs to mediate kinetochore congression, while Kip3p appears to act primarily to regulate metaphase spindle length, we then asked how the behavior of the kinesin-5 motor Cin8p compares to the behavior of the kinesin-8 motor Kip3p in the mitotic spindle environment.
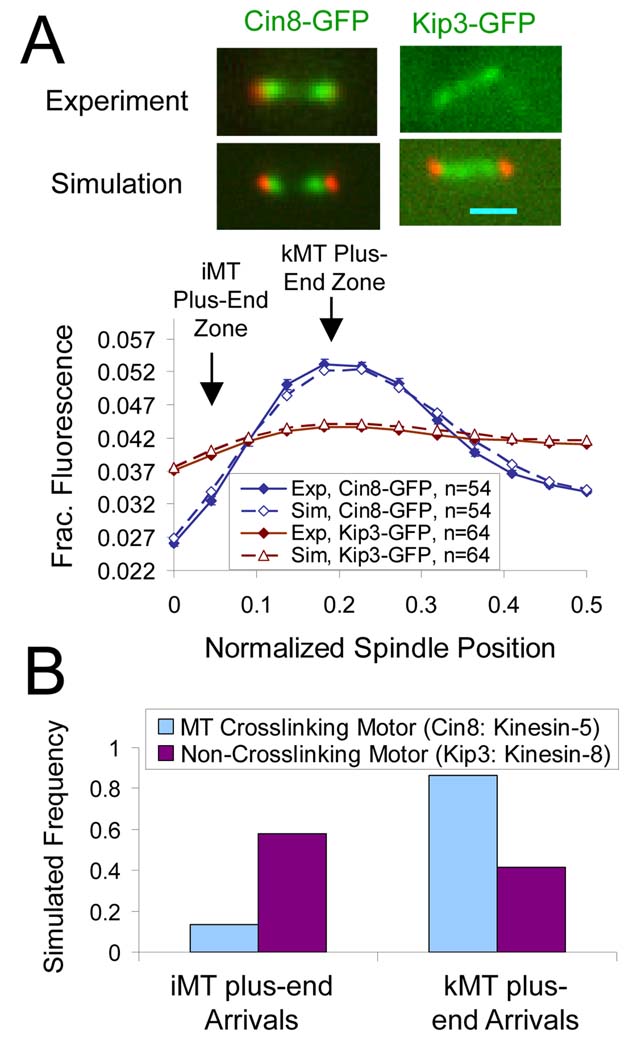
Cin8-GFP concentrates near kMT plus-ends, while Kip3-GFP distribution is diffuse
As shown in Fig. 5A and described above, Cin8-GFP concentrates near kMT plus-ends. This localization is reproduced in simulation because a substantial fraction of the homotetrameric Cin8 motors cross-link parallel kMTs emanating from the same SPB, and then walk efficiently towards the plus-ends of both cross-linked microtubules. In contrast, motors crosslinking anti-parallel microtubules in the simulation (i.e., microtubules attached to opposite SPBs, Fig 4A, magenta) are not able to make progress towards the plus-ends of either microtubule to which they are attached, as the motor heads walk in opposite directions. These anti-parallel attached motors quickly stall, and remain stationary until the detachment of one or both heads. Interestingly, since iMTs run nearly the full length of the spindle, their plus-ends reside in a highly anti-parallel environment, with ~4 iMT plus-ends surrounded by ~20 anti-parallel MTs attached to the opposite SPB. Thus, the cross-linking properties of the tetrameric kinesin-5 motor naturally limit their processivity near to iMT plus-ends.
Figure 5. Relative Distribution of Cin8-GFP and Kip3-GFP.
(A) The experimental and simulated distribution of Cin8-GFP and Kip3-GFP. (B) In simulations, the crosslinking properties of Cin8p frustrates its processivity toward the plus-ends of interpolar (iMT) plus-ends, such that Cin8p visits to iMT plus-ends are rare relative to kMT plus-end visits.
In contrast, dimeric Kip3p motors attach to a single microtubule, and thus their processivity is not hindered by crosslinking to anti-parallel microtubules. In this case, the longer iMTs acts as larger antennas, attracting larger numbers of plus-end directed Kip3p motors to their ends (Varga et al., 2006). As shown in Fig 5A, while Cin8-GFP is specifically concentrated near to kMT plus-ends, Kip3-GFP is more diffusely distributed along the length of the spindle. Importantly, the relative concentration of Kip3-GFP is higher near to where the iMT plus-ends are located (Fig 5A) as compared to Cin8-GFP, whose relative concentration is higher near to where kMT plus-ends are located (Fig 5A).
To test this interpretation of the localization data, simulations were run that reproduced both Cin8-GFP and Kip3-GFP localization in the spindle (Fig 5A). In these simulations, Cin8-GFP is a MT crosslinking motor with simulation parameters as described above, and Kip3p is a slightly faster non-crosslinking motor (Kip3p motor velocity = 75 nm/sec) (Gupta et al., 2006)) with otherwise identical simulation parameters. In these simulations, ~90% of Cin8p motor visits to all MT plus-ends were to kMT plus-ends specifically, while the majority of Kip3p plus-end visits were to iMT plus-ends (~60%, Fig. 5B). Thus, Cin8p molecular motors mediate congression by suppressing the assembly of kMT plus-ends, while Kip3p appears to act primarily to limit assembly of iMT plus-ends. The two different outcomes can be modeled simply by either including cross-linking (Cin8p) or not (Kip3p).
Cin8-GFP motors on aMTs
Because Cin8p promotes aMT disassembly (Fig. 3), we considered whether the motor model applies to cytoplasmic aMTs as well. In contrast to the densely packed kMTs, 1–3 individual aMTs can be readily observed in the cytoplasm so that individual motor interaction with aMTs should also be readily observable. Using time-lapse microscopy we were able to visualize Cin8-NLSΔ-3XGFP interacting with 1–3 individual aMTs. As shown in Fig, 6A (left), Cin8-NLSΔ-3XGFP moves persistently in the plus-end direction along stationary mcherry-Tub1 labeled aMTs (supplemental movie 6). Motors did not move in the minus end direction, consistent with the motor model and the inferred motor behavior on kMTs. The mean motor velocity was 15 +/− 3.3 nm/sec (mean +/− s.d., n=8), which is similar to previously reported aMT plus-end growth rates of 8 – 23 nm/sec (Carminati and Stearns, 1997; Gupta et al., 2002; Gupta et al., 2006; Huang and Huffaker, 2006; Shaw et al., 1997), suggesting that motors track growing aMT plus ends. Since Cin8-GFP was distributed along kMTs in a gradient of increasing motor with increasing distance from the SPB, we were interested to see whether a similar motor gradient occurs on aMTs. As expected from the motor model and similar to the behavior of motors on kMTs, we found that Cin8-NLSΔ-GFP fluorescence is distributed along the length of aMTs with a peak fluorescence shifted slightly away from aMT plus ends (Fig. 6B,C). We then normalized the motor fluorescence to the aMT density, and, as shown in Fig. 6D, found a spatial gradient of Cin8-NLSΔ-GFP that is very similar to the gradient observed on kMTs and to the gradient predicted by the motor model. An interesting feature of both the model and the experiment is a slight dip in the motor concentration at the peak aMT plus ends location (gray arrow in Fig. 5D). This dip was also apparent in the kMT data and simulation, and again suggests that dynamic MT plus ends mediate motor detachment from the MT lattice. We conclude that Cin8p interacts frequently with aMT plus ends in a manner similar to its interaction with kMT plus ends, and consistent with the motor model. We then asked if the spatially regulated interaction of Cin8p with kMT plus-ends, together with the disassembly-promoting activities of Cin8p, could act to mediate chromosome congression in the yeast metaphase spindle.
Figure 6. Cin8p walks processively towards MT plus ends, and its distribution on aMTs mirrors its distribution on kMTs.
(A) Cin8-NLSΔ-3XGFP (left, green) moves in the plus end direction on aMTs, and frequently interacts with aMT plus-ends. The three arrows (yellow, cyan, and magenta) indicate three time points in the movement of a Cin8-GFP fluorescent spot that moves in the plus end direction (red, tubulin-cherry)) (horizontal scale bar, 1500 nm; vertical scale bar, 50 sec) (B) Cin8-NLSΔ-3XGFP (green) on aMTs (tubulin-cherry, red). (C) Cin8-NLSΔ-GFP concentrates near the plus-ends of longer aMTs. (D) Experimentally and in simulation, Cin8-GFP fluorescence normalized to the number of tubulin polymer binding sites increases for longer aMTs.
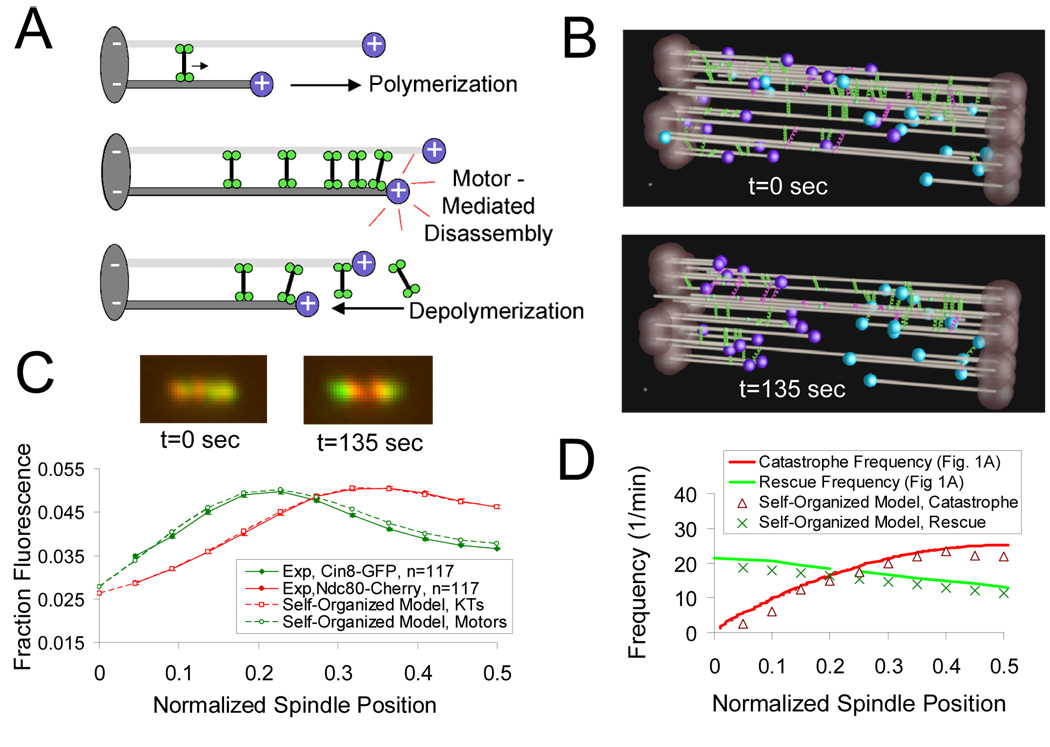
Cin8p-mediated “self-organized” model for metaphase kinetochore organization
The model for kinesin-5 motor dynamics that best agrees with the experimental data described above is one in which motors bind randomly to kMTs (Fig. 7A, top) and then walk toward kMT plus-ends, where they act to promote net kMT disassembly (Fig. 7A, middle). The longer the MT, the more sites there are for motors to attach, which results in more motors at the plus end, and the more that assembly will be disrupted. Once net disassembly occurs (e.g. via a catastrophe), then motors detach from the shortening kMT plus-end (Fig. 7A, bottom). Importantly, because simulated motors bind randomly to microtubules and are plus-end directed, the motor model predicts that the number of motor interactions at kMT plus-ends will increase with increasing kMT length, as shown recently for kinesin-8 molecular motors in vitro (Mayr et al., 2007; Varga et al., 2006) (Fig. 4D, Fig. 6D).
Figure 7. A “self-organized” model for Cin8p motor mediated spindle organization.
(A) A model for interaction of Cin8p motors with kMT plus-ends: Cin8p motors that crosslink parallel microtubules are plus-end directed (top). Cin8p concentrates on longer kMT plus-ends to directly promote kMT disassembly (middle). kMT depolymerization then promotes motor detachment (bottom). (B) Starting with a random distribution of kinetochores and motors in the spindle (top), motor-mediated promotion of kMT plus-end disassembly can organize the spindle into a typical metaphase bi-lobed kinetochore configuration as motors concentrate in a length-dependent fashion onto kMT plus-ends (bottom and Supplemental Movie 5). (C) Simulated images of Cin8-GFP (green) and Ndc80-cherry (red). At simulation start (t=0), motor and kinetochore fluorescence is randomly distributed in the spindle (left). After t=135 sec, simulated fluorescence distributions qualitatively reproduce experimental results (top right, simulated image; bottom, quantitative comparison to experimental data). Model parameters Table S7. (D) The self-organized model results in a gradient of catastrophe frequency (red triangles) that is similar to the theoretical catastrophe gradient depicted in Fig. 1B (red line).
To determine if simulated Cin8p motors could mediate chromosome congression via their disassembly-promoting activity at kMT plus-ends, we allowed the Cin8p motors to promote catastrophe as they concentrate on kMT plus-ends in the motor model simulation as described above. In this “self-organized” model, there is no externally imposed “catastrophe gradient” to regulate kMT lengths (as in Fig. 1B), but rather the catastrophe-promoting motors naturally concentrate on kMT plus-ends in a length-dependent manner. Thus, by starting the simulation with a random organization of motors and kMT lengths (Fig 7B, top), we found that self-organization of kinetochores into a bi-lobed metaphase configuration arises naturally when Cin8p motors promote catastrophe at kMT plus-ends (Fig. 7B, bottom; supplemental movie 5). Here, the catastrophe-promoting Cin8p motors self-organize a simulated bi-lobed spindle in which both Cin8-GFP and Ndc80-cherry fluorescence distributions are qualitatively consistent with experimental results (Fig. 7C). By allowing motor presence at the kinetochore to directly promote catastrophe, the theoretical kMT plus-end “catastrophe gradient” model (Fig 1B) is closely reproduced (Fig. 7D). As in Fig 1B, this gradient serves to establish the bilobed distribution of kinetochores into the two distinct clusters that are characteristic of the congressed metaphase spindle (Fig. 1A). Similar to experimental results in cinΔ mutants, reducing the motor number in the “self-organized” simulation disrupts the gradient in kMT assembly and thus leads to disorganization of the spindle (Fig. S13).
Discussion
Our results demonstrate that kinesin-5 motors, in particular Cin8p, promote net kMT and aMT disassembly in vivo. Since net kMT assembly is specifically suppressed for longer kMTs (i.e. whose plus ends are near the equator) during yeast metaphase, kinesin-5 motors must be mediating their effect, either alone or with binding partners, most strongly on longer kMTs. It is this length-dependent regulation of net kMT plus end assembly that establishes the congressed state of chromosomes that is characteristic of metaphase. These results are surprising to us, since the only established activity of kinesin-5 is its well known antiparallel MT sliding activity, and, to our knowledge, no effect on MT assembly has been reported previously. We also found that Cin8-GFP interacts frequently with MT plus ends in vivo, and exists in a spatial gradient on MTs. The close correspondence of the gradient in net kMT assembly and the gradient in motor distribution strongly suggests that Cin8p, either alone or with a binding partner, directly promotes kMT disassembly via its presence at the kMT plus-end. In addition, this conclusion is strengthened by the close similarity between the cin8Δ mutant and the cin8 motor-domain mutant (F467A) phenotypes.
It is interesting to consider the consequences of the disassembly promotion for spindle length regulation. It is well established that kinesin-5 promotes spindle pole separation by generating an outward extensional force, presumably via cross-linking of antiparallel MTs. The newly identified disassembly-promoting activity shortens kMTs, and thereby should generate an inward pulling force on the spindle poles via stretching of the intervening chromatin between the sister kMT plus ends. Thus, the two activities of Cin8p antagonize each other, and are expected to result in stable spindle pole separation during yeast metaphase.
As described above, the model for kinesin-5 motor dynamics in the metaphase budding yeast mitotic spindle that best agrees with experimental data is one in which motors bind randomly to kMTs (Fig. 7A, top), and then walk toward kMT plus-ends where they act to promote net kMT disassembly (Fig. 7A, middle). Here, because longer kMTs have a larger number of possible motor binding sites, the motor model predicts that the number of motor interactions at kMT plus-ends will increase with increasing kMT length, as shown recently for kinesin-8 molecular motors in vitro (Mayr et al., 2007; Varga et al., 2006). Interestingly, the plus-ends of iMTs, which are the longest MTs in the yeast mitotic spindle, may attract fewer Cin8p motors than the plus-ends of kMTs, simply because iMT plus-ends are surrounded by potential anti-parallel motor attachments, frustrating the plus-end motility of crosslinking Cin8p motors. This explanation is supported by results for the non-crosslinking depolymerizing motor Kip3p, whose localization and deletion phenotypes are distinct from that of Cin8p. We found that Kip3-GFP behavior could be reproduced by taking the Cin8-GFP simulation and turning “off” the cross-linking. In this case, the non-crosslinking motor tends to accumulate more onto iMTs, and less so onto kMTs. The simple physical feature of MT crosslinking, enabled in the tetrameric Cin8p, and disabled in the dimeric Kip3p, is sufficient to explain all the experimental results.
The simplest molecular mechanism for length-dependent MT disassembly is that the kinesin-5 motor itself acts directly to promote MT plus-end disassembly. We speculate that mechanical stress between walking motor head domains would stress tubulin-tubulin bonds to destabilize the lattice and promote MT disassembly. Alternatively, kinesin-5 could carry a disassembly-promoting binding partner to promote net disassembly at MT plus-ends, although to our knowledge there are no known cargoes that are transported by kinesin-5 motors.
Either way, the role of kinesin-5 motors in regulating kMT assembly dynamics is a new property that we have now identified for a motor previously known only as a sliding motor that acts between antiparallel MTs. Because of the potent effect that CIN8 deletion has on kinetochore organization, it seems unlikely that a significant Cin8p-independent pathway will be found to also promote length-dependent disassembly. The effects of kinesin-5 on MT assembly will be important to consider as anticancer drugs directed toward inhibiting kinesin-5 sliding activity are presently in clinical trials (Sudakin and Yen, 2007).
Experimental Procedures
Yeast Strains and Cell Culture
All relevant genotypic information can be found in Table S8. Genes of fusion proteins remained under control of their endogenous promoter. Cell growth techniques and conditions were performed as previously described (Bouck and Bloom, 2007; Gardner et al., 2005; Pearson et al., 2003; Pearson et al., 2004). The CIN8 NLS deletion was performed as previously described (Hildebrandt and Hoyt, 2001). Overexpression of Cin8p from the PGAL1 promoter was performed as previously described (Saunders et al., 1997). The CIN8-3xGFP was made by PCR amplification of 3xGFP from a plasmid, and then the linear PCR product was integrated at the endogenous CIN8 gene. The cin8-F467A strain was made and verified as previously described (Gheber et al., 1999). Benomyl treatment for stabilization of kMT plus-end dynamics was performed as previously described (Pearson et al., 2003).
Fluorescence Imaging and Photobleaching
Fluorescence imaging and photobleaching experiments were performed as previously described (Pearson et al., 2006; Pearson et al., 2001). Astral MT lengths were assessed by measuring the length of GFP-Tub1 or mCherry-Tub1 labeled aMTs in which both the plus-and minus-ends were clearly visible within one focal plane. Counting of Cin8p and Kip1p on the mitotic spindle was completed as previously described (Joglekar et al., 2006).
Image Analysis
Average fluorescence distributions calculated over normalized spindle lengths were obtained as previously described (Gardner et al., 2005).
In FRAP experiments, FRAP half-times were resolved according to spindle position, as previously described (Maddox et al., 2000; Pearson et al., 2006). Reported half-spindle FRAP half-times were calculated by averaging over all spindle positions in the photobleached half-spindle.
Cin8-GFP fluorescence normalized to the number of tubulin binding sites (Fig 4D and Fig 6D) is calculated as follows. For Fig 4D, the tubulin decay function was calculated by assuming a tubulin binding site fraction of 1.3 at the SPBs, which decays inversely with increasing Ndc80-Cherry fluorescence such that there remains a fraction of 0.3 binding sites at the spindle equator (representing interpolar MTs). Then, Cin8-GFP fluorescence was normalized to the number of tubulin binding sites by dividing Cin8-GFP signal minus background by the tubulin decay function at each spindle position. A similar method was used for Fig. 6D, except that the tubulin decay function was calculated directly from the distribution of aMT plus-ends as is shown in Fig. 6C (red).
All p-values for relative comparisons of experimental data sets were calculated using Student’s t-test, unless otherwise noted. Quantitative comparisons of simulations to experimental results were completed as previously described (Sprague et al., 2003).
Simulation Methods
All simulations were run using MATLAB R7.1 (Natick, MA). Detailed simulation methods are provided in the supplemental materials.
Tomography methods
Cells were prepared for electron microscopy using high pressure freezing followed by freeze-substitution as previously described (Winey et al., 1995). Dual axis electron tomography was carried out as described previously (O'Toole et al., 2002).
In total, we recorded 4 wild-type spindles and 5 cin8Δ spindles. Individual microtubules and the position of the SPB central plaque were modeled from the tomographic volumes. A projection of the 3D model was then displayed and rotated to study its 3D geometry. Microtubule lengths were extracted from the model contour data using the program, IMODINFO.
Supplementary Material
Acknowledgments
The authors thank Dominique Seetapun for providing matlab kymograph code, Drs. Tom Hays and Leah Gheber for helpful discussions, Dr. Jeff Molk for comments on the manuscript, and Marybeth Anderson for assistance with bim1Δ images. Plasmids and strains were kindly provided by Drs. S. Reed, B. Errede, M.A. Hoyt, L. Gheber, and D. Pellman. This work was supported by NIH grant GM071522 to D.J.O. K.B. is supported by NIH grant GM32238 and M.K.G. is supported by NIH NRSA grant EB005568. E.T.O. is supported in part by grant RR-00592 from the National Center for Research Resources of the NIH to A. Hoenger. L.V.P. is supported by a SPIRE fellowship (GM00678).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Bouck DC, Bloom K. Pericentric chromatin is an elastic component of the mitotic spindle. Curr Biol. 2007;17:741–748. doi: 10.1016/j.cub.2007.03.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carminati JL, Stearns T. Microtubules orient the mitotic spindle in yeast through dynein-dependent interactions with the cell cortex. J Cell Biol. 1997;138:629–641. doi: 10.1083/jcb.138.3.629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cottingham FR, Hoyt MA. Mitotic spindle positioning in Saccharomyces cerevisiae is accomplished by antagonistically acting microtubule motor proteins. J Cell Biol. 1997;138:1041–1053. doi: 10.1083/jcb.138.5.1041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Gramont A, Barbour L, Ross KE, Cohen-Fix O. The spindle midzone microtubule-associated proteins Ase1p and Cin8p affect the number and orientation of astral microtubules in Saccharomyces cerevisiae. Cell Cycle. 2007;6:1231–1241. doi: 10.4161/cc.6.10.4181. [DOI] [PubMed] [Google Scholar]
- Desai A, Mitchison TJ. Microtubule polymerization dynamics. Annu Rev Cell Dev Biol. 1997;13:83–117. doi: 10.1146/annurev.cellbio.13.1.83. [DOI] [PubMed] [Google Scholar]
- Gardner M, Odde D, Bloom K. Kinesin-8 molecular motors: putting the brakes on chromosome oscillations. Trends Cell Biol. 2008 doi: 10.1016/j.tcb.2008.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gardner MK, Odde DJ, Bloom K. Hypothesis testing via integrated computer modeling and digital fluorescence microscopy. Methods. 2007;41:232–237. doi: 10.1016/j.ymeth.2006.08.002. [DOI] [PubMed] [Google Scholar]
- Gardner MK, Pearson CG, Sprague BL, Zarzar TR, Bloom K, Salmon ED, Odde DJ. Tension-dependent Regulation of Microtubule Dynamics at Kinetochores Can Explain Metaphase Congression in Yeast. Mol Biol Cell. 2005;16:3764–3775. doi: 10.1091/mbc.E05-04-0275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geiser JR, Schott EJ, Kingsbury TJ, Cole NB, Totis LJ, Bhattaacharyya G, He L, Hoyt MA. Saccharomyces cerevisiae genes required in the absence of the CIN8-encoded spindle motor act in functionally diverse mitotic pathways. Molecular Biology of the Cell. 1997;8:1035–1050. doi: 10.1091/mbc.8.6.1035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gheber L, Kuo SC, Hoyt MA. Motile properties of the kinesin-related Cin8p spindle motor extracted from Saccharomyces cerevisiae cells. J Biol Chem. 1999;274:9564–9572. doi: 10.1074/jbc.274.14.9564. [DOI] [PubMed] [Google Scholar]
- Gupta ML, Jr., Bode CJ, Thrower DA, Pearson CG, Suprenant KA, Bloom KS, Himes RH. beta-Tubulin C354 mutations that severely decrease microtubule dynamics do not prevent nuclear migration in yeast. Mol Biol Cell. 2002;13:2919–2932. doi: 10.1091/mbc.E02-01-0003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta ML, Jr., Carvalho P, Roof DM, Pellman D. Plus end-specific depolymerase activity of Kip3, a kinesin-8 protein, explains its role in positioning the yeast mitotic spindle. Nat Cell Biol. 2006;8:913–923. doi: 10.1038/ncb1457. [DOI] [PubMed] [Google Scholar]
- Hildebrandt ER, Hoyt MA. Mitotic motors in Saccharomyces cerevisiae. Biochim Biophys Acta. 2000;1496:99–116. doi: 10.1016/s0167-4889(00)00012-4. [DOI] [PubMed] [Google Scholar]
- Hildebrandt ER, Hoyt MA. Cell cycle-dependent degradation of the Saccharomyces cerevisiae spindle motor Cin8p requires APC(Cdh1) and a bipartite destruction sequence. Mol Biol Cell. 2001;12:3402–3416. doi: 10.1091/mbc.12.11.3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howard J, Hyman AA. Microtubule polymerases and depolymerases. Curr Opin Cell Biol. 2007;19:31–35. doi: 10.1016/j.ceb.2006.12.009. [DOI] [PubMed] [Google Scholar]
- Hoyt MA, He L, Loo KK, Saunders WS. Two Saccharomyces cerevisiae kinesin-related gene products required for mitotic spindle assembly. Journal of Cell Biology. 1992;118:109–120. doi: 10.1083/jcb.118.1.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang B, Huffaker TC. Dynamic microtubules are essential for efficient chromosome capture and biorientation in S. cerevisiae. J Cell Biol. 2006;175:17–23. doi: 10.1083/jcb.200606021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inoue S, Salmon ED. Force generation by microtubule assembly/disassembly in mitosis and related movements. Molecular Biology of the Cell. 1995;6:1619–1640. doi: 10.1091/mbc.6.12.1619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joglekar AP, Bouck DC, Molk JN, Bloom KS, Salmon ED. Molecular architecture of a kinetochore-microtubule attachment site. Nat Cell Biol. 2006;8:581–585. doi: 10.1038/ncb1414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapitein LC, Peterman EJ, Kwok BH, Kim JH, Kapoor TM, Schmidt CF. The bipolar mitotic kinesin Eg5 moves on both microtubules that it crosslinks. Nature. 2005;435:114–118. doi: 10.1038/nature03503. [DOI] [PubMed] [Google Scholar]
- Kashina AS, Baskin RJ, Cole DG, Wedaman KP, Saxton WM, Scholey JM. A bipolar kinesin. Nature. 1996;379:270–272. doi: 10.1038/379270a0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maddox P, Bloom K, Salmon ED. Polarity and Dynamics of Microtubule Assembly in the Budding Yeast Saccharomyces cerevisiae. Nature Cell Biology. 2000;2:36–41. doi: 10.1038/71357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayr MI, Hummer S, Bormann J, Gruner T, Adio S, Woehlke G, Mayer TU. The human kinesin Kif18A is a motile microtubule depolymerase essential for chromosome congression. Curr Biol. 2007;17:488–498. doi: 10.1016/j.cub.2007.02.036. [DOI] [PubMed] [Google Scholar]
- Miller RK, Heller KK, Rose MD. The Kinesin-related Proteins, Kip2p and Kip3p, Function Differently in Nuclear Migration in Yeast. Molecular Biology of the Cell. 1998;9:2051. doi: 10.1091/mbc.9.8.2051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicklas RB. How cells get the right chromosomes. Science. 1997;275:632–637. doi: 10.1126/science.275.5300.632. [DOI] [PubMed] [Google Scholar]
- O'Toole ET, Winey M, McIntosh JR, Mastronarde DN. Electron tomography of yeast cells. Methods Enzymol. 2002;351:81–95. doi: 10.1016/s0076-6879(02)51842-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearson CG, Gardner MK, Paliulis LV, Salmon ED, Odde DJ, Bloom K. Measuring Nanometer Scale Gradients in Spindle Microtubule Dynamics Using Model Convolution Microscopy. Mol Biol Cell. 2006;17:4069–4079. doi: 10.1091/mbc.E06-04-0312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearson CG, Maddox PS, Salmon ED, Bloom K. Budding Yeast Chromosome Structure and Dynamics during Mitosis. J Cell Biol. 2001;152:1255–1266. doi: 10.1083/jcb.152.6.1255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearson CG, Maddox PS, Zarzar TR, Salmon ED, Bloom K. Yeast kinetochores do not stabilize Stu2p-dependent spindle microtubule dynamics. Mol Biol Cell. 2003;14:4181–4195. doi: 10.1091/mbc.E03-03-0180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearson CG, Yeh E, Gardner M, Odde D, Salmon ED, Bloom K. Stable kinetochore-microtubule attachment constrains centromere positioning in metaphase. Curr Biol. 2004;14:1962–1967. doi: 10.1016/j.cub.2004.09.086. [DOI] [PubMed] [Google Scholar]
- Roof DM, Meluh PB, Rose MD. Kinesin-related proteins required for assembly of the mitotic spindle. Journal of Cell Biology. 1992;118:95–108. doi: 10.1083/jcb.118.1.95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saunders W, Lengyel V, Hoyt MA. Mitotic Spindle Function in Saccharomyces cerevisiae Requires a Balance between Different Types of Kinesin-related Motors. Molecular biology of the cell. 1997;8:1025–1033. doi: 10.1091/mbc.8.6.1025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saunders WS, Hoyt MA. Kinesin-related proteins required for structural integrity of the mitotic spindle. Cell. 1992;70:451–458. doi: 10.1016/0092-8674(92)90169-d. [DOI] [PubMed] [Google Scholar]
- Shaw SL, Yeh E, Maddox P, Salmon ED, Bloom K. Astral microtubule dynamics in yeast: A microtubule-based searching mechanism for spindle orientation and nuclear migration into the bud. Journal of Cell Biology. 1997;139:985–994. doi: 10.1083/jcb.139.4.985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sprague BL, Pearson CG, Maddox PS, Bloom KS, Salmon ED, Odde DJ. Mechanisms of Microtubule-Based Kinetochore Positioning in the Yeast Metaphase Spindle. Biophys J. 2003;84:1–18. doi: 10.1016/S0006-3495(03)75087-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Straight AF, Sedat JW, Murray AW. Time-lapse microscopy reveals unique roles for kinesins during anaphase in budding yeast. The Journal of Cell Biology. 1998;143:687. doi: 10.1083/jcb.143.3.687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stumpff J, von Dassow G, Wagenbach M, Asbury C, Wordeman L. The kinesin-8 motor Kif18A suppresses kinetochore movements to control mitotic chromosome alignment. Dev Cell. 2008;14:252–262. doi: 10.1016/j.devcel.2007.11.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sudakin V, Yen TJ. Targeting mitosis for anti-cancer therapy. BioDrugs. 2007;21:225–233. doi: 10.2165/00063030-200721040-00003. [DOI] [PubMed] [Google Scholar]
- Tao L, Mogilner A, Civelekoglu-Scholey G, Wollman R, Evans J, Stahlberg H, Scholey JM. A homotetrameric kinesin-5, KLP61F, bundles microtubules and antagonizes Ncd in motility assays. Curr Biol. 2006;16:2293–2302. doi: 10.1016/j.cub.2006.09.064. [DOI] [PubMed] [Google Scholar]
- Tytell JD, Sorger PK. Analysis of kinesin motor function at budding yeast kinetochores. J Cell Biol. 2006;172:861–874. doi: 10.1083/jcb.200509101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valentine MT, Fordyce PM, Krzysiak TC, Gilbert SP, Block SM. Individual dimers of the mitotic kinesin motor Eg5 step processively and support substantial loads in vitro. Nat Cell Biol. 2006;8:470–476. doi: 10.1038/ncb1394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varga V, Helenius J, Tanaka K, Hyman AA, Tanaka TU, Howard J. Yeast kinesin-8 depolymerizes microtubules in a length-dependent manner. Nat Cell Biol. 2006;8:957–962. doi: 10.1038/ncb1462. [DOI] [PubMed] [Google Scholar]
- Winey M, Mamay CL, O'Toole ET, Mastronarde DN, Giddings TH, Jr., McDonald KL, McIntosh JR. Three-dimensional ultrastructural analysis of the Saccharomyces cerevisiae mitotic spindle. Journal of Cell Biology. 1995;129:1601–1615. doi: 10.1083/jcb.129.6.1601. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.