Abstract
The ubiquitously expressed amyloid precursor-like protein 2 (APLP2) has been previously found to regulate cell surface expression of the MHC class I molecule Kd and bind strongly to Kd. In the study reported here, we demonstrated that APLP2 binds, in varied degrees, to several other mouse MHC class I allotypes, and that the ability of APLP2 to affect cell surface expression of an MHC class I molecule is not limited to Kd. Ld, like Kd, was found associated with APLP2 in the Golgi, but Kd was also associated with APLP2 within intracellular vesicular structures. We also investigated the effect of β2m on APLP2/MHC interaction, and found that human β2m transfection increased the association of APLP2 with mouse MHC class I molecules, likely by affecting H2 class I heavy chain conformation. APLP2 was demonstrated to bind specifically to the conformation of Ld having folded outer domains, consistent with our previous results with Kd and indicating APLP2 interacts with the α1α2 region on each of these H2 class I molecules. Furthermore, we observed that binding to APLP2 involved the MHC α3/transmembrane/cytoplasmic region, suggesting that conserved as well as polymorphic regions of the H2 class I molecule may participate in interaction with APLP2. In summary, we demonstrated that APLP2′s binding, co-localization pattern, and functional impact vary among H2 class I molecules, and that APLP2/MHC association is influenced by multiple domains of the MHC class I heavy chain and by β2m’s effects on the conformation of the heavy chain.
Keywords: amyloid precursor-like protein 2, antigen presentation, beta 2-microglobulin, major histocompatibility complex class I, mouse
Introduction
Major histocompatibility complex (MHC) class I molecules trigger the lysis of cells bearing viral or tumor-specific antigens by presenting peptide fragments of the antigens at the cell surface to T lymphocytes. The peptides first associate with beta 2-microglobulin (β2m)-associated MHC class I heavy chains in the endoplasmic reticulum (ER), with the assistance of several proteins in an intricately regulated process (Cresswell et al. 1999; Serwold et al. 2002; Saric et al. 2002; Paquet et al. 2004; Park et al. 2006). Beyond the ER, there is additional regulation of MHC class I molecule cell surface expression (Paquet et al. 2004; Joyce 1997; Marguet et al. 1999; Spiliotis et al. 2000; Sohn et al. 2001; Pencheva et al. 2001; Paulsson et al. 2002). A protein that has been shown to bind to MHC class I molecules beyond the ER is amyloid precursor-like protein 2 (APLP2) (Feuerbach and Burgert 1993; Sester et al. 2000; Morris et al. 2003). In addition to its effects on MHC class I molecules, involvement in several other cellular processes (e.g., mitosis, nerve growth, and cell migration) has been reported for APLP2 (Thinakaran et al. 1995; Rassoulzadegan et al. 1998; Guo et al. 1998; Cappai et al. 1999; Li et al. 1999). Structurally, APLP2 is a ubiquitously expressed transmembrane protein bearing an ectodomain that can be proteolytically clipped off and then secreted (Slunt et al. 1994).
APLP2 has been shown to down-regulate the surface expression of the MHC class I molecule Kd (Morris et al. 2003). For APLP2 to associate with Kd, β2m expression is required (Sester et al. 2000). Evidence indicates that APLP2 interacts with the region of the MHC class I molecule that includes the peptide-binding groove. For example, supplementing cell lysates with Kd peptide ligands results in the release of APLP2 from Kd (Feuerbach and Burgert 1993). In addition, APLP2 has been shown to associate preferentially with the folded conformation of the MHC class I molecule Kd (Morris et al. 2003), which suggests that APLP2 binds to the Kd N-terminal domains. Here, we report our findings that APLP2 binds to H2 class I molecules in addition to Kd, with the strength of interaction dependent on the particular MHC allotype. Our results show that APLP2 affects the cell surface expression of Ld, but to a lesser extent than Kd. In addition, we demonstrated that APLP2 interacts with Kd in vesicular compartments and in the Golgi, but Ld only associates with APLP2 in the Golgi. We also discovered that multiple domains of the MHC class I heavy chain influence MHC binding to APLP2, and that the transfection of human β2m into mouse cells alters the avidity of MHC/APLP2 interaction. Thus, both polymorphic and conserved portions of MHC class I molecules influence APLP2′s binding and regulation of MHC class I cell surface expression.
Materials and Methods
Antibodies
The 34-1-2 monoclonal antibody binds to the α1 and/or α2 domains of Kd, Dd, and Dq (Ozato et al. 1983). The 34-1-2 antibody also weakly recognizes Db and Ld, but can bind more strongly to Ld heavy chains that are associated with human β2m, as well as to certain Ld mutants with substitutions located in the peptide-binding groove (Ozato et al. 1983; Nieto et al 1989; Solheim et al. 1995). The SF1.1.1 monoclonal antibody binds to the α3 domain of Kd (Cox et al. 1991). The 64-3-7 monoclonal antibody recognizes Ld that has an open, peptide-free groove (Cox et al. 1991; Smith et al. 1993; Smith et al. 1995), and 64-3-7 can also bind to open forms of other MHC class I heavy chains into which the 64-3-7 epitope has been inserted (Yu et al. 1999; Myers et al. 2000; Harris et al. 2001; Lybarger et al. 2001). The addition of the 64-3-7 epitope into Kd and other MHC class I molecules has been shown not to interfere with peptide ligand binding or expression at the cell surface (Yu et al. 1999; Myers et al. 2000; Harris et al. 2001; Lybarger et al. 2001). The 30-5-7 antibody binds to the α2 domain of folded Ld (Smith et al. 1993; Smith et al. 1995; Ozato et al. 1980; Evans et al. 1982; Solheim et al. 1993) and the 28-14-8 antibody binds to the α3 domain of Ld, Db, Dq, and Lq (Evans et al. 1982; Solheim et al. 1993; Ozato and Sachs 1981). BBM1.1 recognizes human β2m (Brodsky et al. 1979). All the anti-H2 antibodies used in this study, as well as the BBM1.1 antibody, were gifts from Dr. T. Hansen (Washington University, St. Louis, MO). The antibody recognizing APLP2 that was used in these studies was prepared by rabbit immunization with full-length mouse APLP2; this antibody was purchased from Calbiochem. The PanAb5 antibody against β-actin was purchased from Novus Biologicals.
Cell lines
For these studies, the cell lines were cultured in RPMI 1640 medium (Invitrogen) that was supplemented with 15% fetal bovine serum, pyruvate, glutamine, and penicillin/streptomycin. The L-Ld, L-Lq, L-Db, and L-Dq cell lines are H-2k L cell fibroblast (Ltk DAP-3k) cell lines that have been transfected with the L-Ld, L-Lq, L-Db, or L-Dq gene, respectively (Cox et al. 1991; Lee et al. 1988). The HeLa cell line was a gift from Dr. W. Maury (University of Iowa, Iowa City, IA). HeLa cells were transfected with a cDNA for Kd or for a Kd heavy chain bearing the 64-3-7 epitope (Yu et al. 1999) in the RSV.5neo vector (a gift from Dr. E. Long, NIH, Bethesda, MD) and were selected in G418 to create the HeLa-Kd or HeLa-etKd cell line, respectively. To generate the HeLa-Ld cell line, HeLa cells were transfected with an Ld cDNA in the pRSV.5-neo vector (Solheim et al. 1993). The 721.221 cell line is a B lymphoblastoid cell line that does not express HLA-A, -B, or —C (Shimizu et al. 1988; Shimizu and DeMars 1989). The 721.221 cell line, as well as the L, L-Ld, L-Lq, L-Db, and L-Dq cell lines, was a kind gift from Dr. T. Hansen (Washington University, St. Louis, MO). The L-Kd and L-etKd cell lines were made by transfecting the cDNA for Kd or for a Kd heavy chain with the 64-3-7 epitope (Yu et al. 1999), respectively, in the pRSV.5neo vector into L cells with Effectene, and selecting with G418. The L-etKd+hβ2m cell line was created by co-transfecting the 64-3-7-epitope-tagged Kd cDNA along with a cDNA encoding human β2m (both in pRSV.5neo) into L cells by the use of Effectene and selecting with G418.
Transient transfection of APLP2 siRNA into HeLa-etKd and HeLa-Ld cells was performed in the following manner. Control siRNA (ON-TARGETplus siCONTROL non-targeting pool) and APLP2-specific siRNA (Dharmacon ON-TARGETplus SMARTpool siRNA for human APLP2) were purchased from Thermo Fisher Scientific. Mock transfections (with transfection reagent but no siRNA), transfection with control siRNA, or APLP2-specific siRNA were done using DharmaFECT 1 Transfection Reagent (Thermo Fisher Scientific) according to the manufacturer’s suggested protocol. Down-regulation of APLP2 expression was verified by Western blotting.
The Kdα1α2/Ldα3 chimera (referred to as Kd/Ld) was constructed by joining the sequence encoding the leader peptide, α1 domain, and α2 domain of Kd to the sequence encoding the α3 domain, transmembrane domain, and cytoplasmic tail (α3/TM/CYT) of Ld. The construct was cloned into the pIRES-puro vector and transfected into HeLa cells with Effectene, and the positive stable transfectant was selected with puromycin.
Immunoprecipitations and Western blots
In these studies, immunoprecipitations and Western blotting were performed by protocols similar to published methods (Turnquist and Solheim, 2001). Before immunoprecipitations, cells were washed in 20 mM iodoacetamide in PBS (Sigma-Aldrich, St. Louis, MO) three times and then lysed in 3-[(3-cholamidopropyl)dimethylammonio]-1-propanesulfonate (CHAPS) lysis buffer. The lysis buffer was composed of 1% CHAPS (Roche Applied Science, Indianapolis, IN) in Tris-buffered saline (pH 7.4) with freshly added 0.2 mM PMSF, 20 mM iodoacetamide, and excess antibody. After a 1 h incubation on ice, the lysates were centrifuged to pellet the cell nuclei and the lysate supernatants were added to Protein A-Sepharose beads (Amersham Biosciences, Piscataway, NJ) and mixed intermittently for 45 min. The beads were then washed four times in 0.1% CHAPS/20 mM iodoacetamide in TBS (pH 7.4) and boiled for 5 min in 0.125 M Tris (pH 6.8)/2% SDS/12% glycerol/0.02% bromphenol blue to elute the proteins from the beads.
Prior to some of the immunoprecipitations of mouse MHC class I molecules from transfected L cells, the cells were grown in methionine- and cysteine-free medium for 30 min and then radiolabeled with [35S]-methionine/cysteine for 30 min. The additional steps in the immunoprecipitation procedure were performed as described above. Following electrophoresis, the radiolabeled proteins were transferred to Immobilon-P membranes (Millipore), which were subsequently dried and autoradiographed to visualize the immunoprecipitated MHC class I heavy chain bands.
For Western blotting, after elution the immunoprecipitates were electrophoresed on precast SDS-PAGE gels (Invitrogen) and then the proteins were transferred to Immobilon-P membranes (Millipore). Following overnight blocking in 10% (w/v) dry milk dissolved in 0.05% Tween 20 in PBS, the blotting membranes were incubated in primary antibody diluted in milk for 2 h, washed with 0.05% Tween 20/PBS three times, and incubated for 1 h in biotin-conjugated goat anti-mouse or anti-rabbit IgG (Caltag Laboratories) diluted in 0.05% Tween 20/PBS. After three 0.05% Tween 20/PBS washes, the membranes were incubated with streptavidin-conjugated horseradish peroxidase (Zymed) diluted in 0.05% Tween 20/PBS for 1 h. The membranes were then washed three times with 0.3% Tween 20/PBS, and treated with enhanced chemiluminescence Western blot developing reagents (Amersham Biosciences). The blots were exposed to Kodak BioMax film (Eastman Kodak Co.) to allow visualization of the bands. For some experiments, protein bands were quantified by multiplying the mean of the luminosity by the pixel number as determined by the histogram function of Adobe Photoshop, as previously described (Ng et al. 2003; Eck et al. 2006).
Immunofluorescence analysis
To assess the co-localization of Kd and APLP2 at steady state, HeLa-etKd or HeLa-Ld cells were grown on glass cover slips, transiently transfected for 24 h with APLP2-FLAG using Effectene reagent, and fixed with 4% (vol/vol) paraformaldehyde in PBS for 10 minutes. Fixed cells were incubated with primary antibodies (34-1-2 for Kd or 30-5-7 for Ld) and rabbit anti-FLAG antibody prepared in staining solution [0.2% saponin (wt/vol) and 0.5% (wt/vol) bovine serum albumin in PBS] for 1 h at room temperature. After 3 washes in PBS (5 min/wash), the cells were incubated with the appropriate fluorochrome-conjugated secondary antibody mixture (Alexa Fluor goat anti-mouse 568-nm and Alexa Fluor donkey anti-rabbit 488-nm antibodies) in staining solution for 30 min at room temperature. After 3 more washes in PBS (5 min/wash), the cells were mounted and analyzed with a Zeiss LSM 5 Pascal confocal microscope, using a 63X lens with 1.4 numerical aperture. The degree of co-localization of APLP2 with either Kd or Ld was quantified with the Image J software, utilizing the JACoP program (http://rsb.info.nih.gov/ij/plugins/track/jacp.html) for the calculation of Pearson’s coefficients. Ten sets of images (acquired using the same optical settings) were used for each pairwise comparison, and the Pearson coefficients and standard errors of the mean were calculated.
To determine whether a large, compact intracellular compartment in which APLP2 and Kd were co-localized was the Golgi, the approach was to transfect transiently etKd, APLP2-FLAG, and YFP-Golgi (Clontech) into HeLa cells using Effectene(Qiagen), fix the HeLa cells with 4% (vol/vol) paraformaldehyde in PBS for 10 min, and then incubate the fixed cells with anti-Kd antibody 34-1-2 and rabbit anti-FLAG in staining solution for 1 h at room temperature. After 3 PBS washes of 5 min each, the cells were incubated with secondary antibodies (Alexa Fluor goat anti-mouse 430 nm and Alexa Fluor goat anti-rabbit 568-nm antibodies) in staining solution for 30 min at room temperature, washed 3 times in PBS, and mounted for image analysis with a Zeiss LSM 5 Pascal confocal microscope, using a 63X lens with 1.4 numerical aperture.
Flow cytometry assays
In flow cytometry assays, cells were suspended at 5 × 106/ml in PBS with 0.2% BSA and 0.1% sodium azide. Cell suspension aliquots in volumes of 0.1 ml were distributed to wells in a 96-well plate. The cells were incubated with excess monoclonal antibody or with BSA/azide/PBS alone (as a control) at 4°C for 30 min, washed twice, and incubated with a PE-conjugated, Fc-specific F(ab’)2 portion of goat anti-mouse IgG (Jackson ImmunoResearch) at 4°C for 30 min. The cells were washed 3 times, resuspended in BSA/azide/PBS, and analyzed with the use of a FACSCalibur flow cytometer and Cell Quest software (BD Biosciences).
Results
The polymorphism of H2 class I molecules influences their association with APLP2
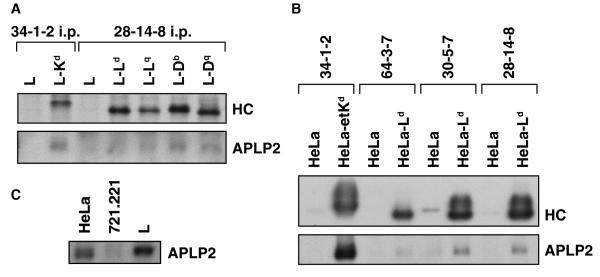
Our laboratory and others have shown that APLP2 binds to Kd (Sester et al. 2000; Morris et al. 2003). In this study, we investigated the association of APLP2 with a diverse set of mouse MHC class I molecules individually expressed in the mouse L fibroblast cell line. As shown in Figure 1A, in lysates of mouse L cells there was detectable association of APLP2 with immunoprecipitated Kd and with certain other H2 class I molecules. Consistent with these findings with mouse cells, immunoprecipitation and Western blotting revealed that in HeLa cells Kd molecules bound strongly to APLP2, whereas Ld expressed in HeLa also bound APLP2 but more weakly (Fig. 1B). The conformation of Ld associating with APLP2 was demonstrated to be the folded form (immunoprecipitated by antibody 30-5-7) and not the open form (immunoprecipitated by antibody 64-3-7) (Fig. 1B). APLP2 was also found associated with Ld molecules immunoprecipitated by the 28-14-8 antibody; this antibody recognizes both folded and open forms of Ld (Fig. 1B). These observations that APLP2 bound preferentially to folded Ld are consistent with our previous finding that APLP2 preferentially interacts with the folded form of Kd (Morris et al. 2003).
Figure 1.
(A) Kd, Db, and Dq expressed in L cells were weakly associated with APLP2. The indicated cell types were radiolabeled with 35 S-methionine/cysteine and immunoprecipitations were performed on lysates of the cells with 34-1-2 (for Kd) or 28-14-8 (for Ld, Lq, Db, or Dq). After electrophoresis on a 4→20% acrylamide gel, the immunoprecipitated proteins were transferred to a blotting membrane and autoradiographed to reveal the immunoprecipitated heavy chain (HC) or probed with antiserum recognizing APLP2 to reveal the co-immunoprecipitated APLP2. (B) Much more APLP2 was found associated with Kd than Ld in transfected HeLa cells. EtKd or Ld molecules expressed in HeLa cells were immunoprecipitated from cell lysates with 34-1-2 (for folded Kd), 64-3-7 (for open, peptide-free Ld), 30-5-7 (for folded Ld), or 28-14-8 (for both folded and open Ld). The immunoprecipitates were electrophoresed on 4-20% acrylamide gels and the proteins were transferred to a membrane and probed with 64-3-7 for the immunoprecipitated, denatured etKd or Ld MHC class I heavy chain (HC, top panel) or with antibody against APLP2 (bottom panel). Protein bands were quantified by multiplying the luminosity mean by the pixel number (as determined by Adobe Photoshop), and the ratio of the co-precipitated APLP2 band intensity to the Ld or Kd heavy chain band intensity was calculated. The results for the APLP2/30-5-7+ Ld ratio were 20% of the APLP2/34-1-2+ Kd ratio. (C) Expression levels of APLP2 in HeLa and L cells were demonstrated to be very similar. Samples of HeLa, 721.221, and L cell lysates were electrophoresed on a 4→20% gel, transferred to a blotting membrane, and probed with anti-APLP2 antiserum. Bands were quantified by multiplying the luminosity mean by the pixel number (as determined by Adobe Photoshop), and the ratio of the L cell APLP2 band to the HeLa APLP2 band was calculated to be 1.3.
Notably, Kd expressed in mouse L cells was weakly associated with APLP2 compared to Kd expressed in human HeLa cells, as can be observed by comparison of Figures 1A and B. A plausible explanation for greater APLP2 association with mouse MHC class I molecules in HeLa cells compared to L cells could be that HeLa cells might express more APLP2 than do L cells. However, when the expression levels of APLP2 in L cells and HeLa cells were compared (by Western blotting for APLP2), there was no indication that HeLa expresses more APLP2 (Fig. 1C). (Interestingly, the intracellular expression of APLP2 in the human HLA-A,B,C class I-deficient cell line 721.221 was much lower than in L cells or HeLa cells (Fig. 1C). Figure 2A provides results from a direct, side-by-side comparison of the APLP2 associated with Kd in HeLa cells versus the APLP2 associated with Kd in L cells. Densitometry of the APLP2 and MHC heavy chain bands on the Western blot shown in Figure 2A revealed that the APLP2/Kd ratio in L-etKd is only 28% of the APLP2/Kd ratio in HeLa-etKd, providing further confirmation that APLP2 associates more strongly with Kd in HeLa cells than in L cells.
Figure 2.
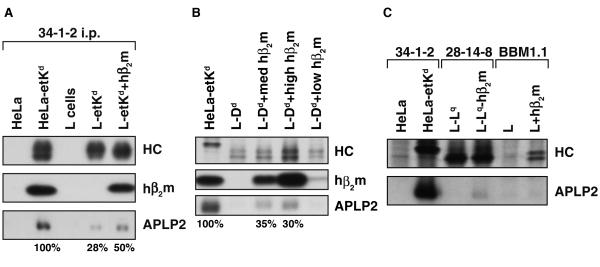
(A)The presence of human β2m increased association of Kd with APLP2 in mouse L cells. Kd was immunoprecipitated from cell lysates with 34-1-2. The Kd immunoprecipitates were electrophoresed on 4→20% gels, and the proteins were transferred to blotting membranes. The membranes were probed with 64-3-7, which recognizes the Kd heavy chain (HC) denatured on the blot (top panel), with BBM1.1 that recognizes human β2m (middle panel), or with antiserum against APLP2 (bottom panel). Protein bands were quantified by multiplying the luminosity mean by the pixel number (as determined by Adobe Photoshop). The APLP2/etKd ratios for L-etKd and L-etKd+hβ2m are presented as percentages of the APLP2/Kd ratio for HeLa-etKd. (B) The presence of human β2m increased association of Dd with APLP2 in mouse L cells. Dd was immunoprecipitated from radiolabeled cell lysates with 34-1-2, electrophoresed on a 4→20% gel, and transferred to blotting membranes that were subsequently dried and autoradiographed to visualize the immunoprecipitated HC (top panel). The Dd immunoprecipitates were also electrophoresed on 4→20% gels for protein transfer to blotting membranes that were probed with BBM1.1 for human β2m (middle panel) or with antiserum against APLP2 (bottom panel). The APLP2/Dd ratios for L-Dd+med hβ2m and L-Dd+high hβ2m are presented as percentages of the APLP2/Kd ratio for HeLa-etKd. (C) APLP2 association was detectable for Lq after human β2m transfection into L-Lq, and β2m itself does not strongly associate with APLP2. The indicated immunoprecipitations from radiolabeled cell lysates were performed, and samples of the immunoprecipitates were electrophoresed on a 4→20% gel. The separated proteins were transferred to blotting membranes that were subsequently dried and autoradiographed to visualize the immunoprecipitated HC (top panel). The immunoprecipitates were also electrophoresed on 4→20% gels for protein transfer to blotting membranes that were probed with antiserum against APLP2 (bottom panel).
β2m has been shown to be a requirement for APLP2 interaction with Kd (Sester et al. 2000) and we postulated that the presence of human versus murine β2m in HeLa and L cells could affect the degree of APLP2 association with mouse MHC class I molecules. We transfected human β2m into L-etKd cells, and Kd immunoprecipitated from L-etKd or L-etKd+hβ2m was compared for levels of associated APLP2 (and for associated human β2m, as a control) by Western blotting. The transfection of human β2m into L cells increased APLP2 association with Kd by about 2 fold, though not fully to the level of APLP2/Kd association in HeLa cells (Fig. 2A). Transfecting human β2m into L-Dd cells also increased mouse APLP2 association with Dd (Fig. 2B). At the highest level of transfected human β2m, Dd folding was also facilitated, as shown by the increased amount of immunoprecipitated folded Dd heavy chain (Fig. 2B). At a medium level of human β2m transfection, however, there was little or no change in the level of folded Dd, but there was still evidence of increased APLP2 association (Fig. 2B). Human β2m transfection also slightly increased APLP2 association with Lq and Db, but had virtually no effect on APLP2 association with Dq in L cell transfectants (Fig. 2C and data not shown). Thus, transfection of human β2m was able to increase APLP2 interaction with several mouse MHC class I allotypes in mouse cells.
Potentially, direct association between APLP2 and β2m could be occurring and this interaction could be stronger for human β2m than mouse β2m. To investigate whether there was evidence of direct binding of APLP2 to human β2m, we performed human β2m immunoprecipitations on L+hβ2m cells and L cells, and probed the immunoprecipitates for associated APLP2. Immunoprecipitation of human β2m from L+hβ2m lysates did co-immunoprecipitate some proteins of the appropriate molecular weight to be the endogenous Kk and Dk molecules (Fig. 2C, top panel), but little or no APLP2 was co-immunoprecipitated (Fig. 2C, bottom panel). Thus, the effect of human β2m on APLP2 association with mouse MHC class I molecules was likely not due to direct interaction between β2m and APLP2, but rather to indirect effects of human β2m on the MHC class I heavy chain conformation. In addition, these data suggest that Kk and Dk (the endogenous H2 class I molecules in L cells) are not strongly associated with APLP2 in L cells, again indicating that there is allotypic specificity in MHC class I binding to APLP2.
MHC polymorphism influenced APLP2 co-localization with H2 class I molecules
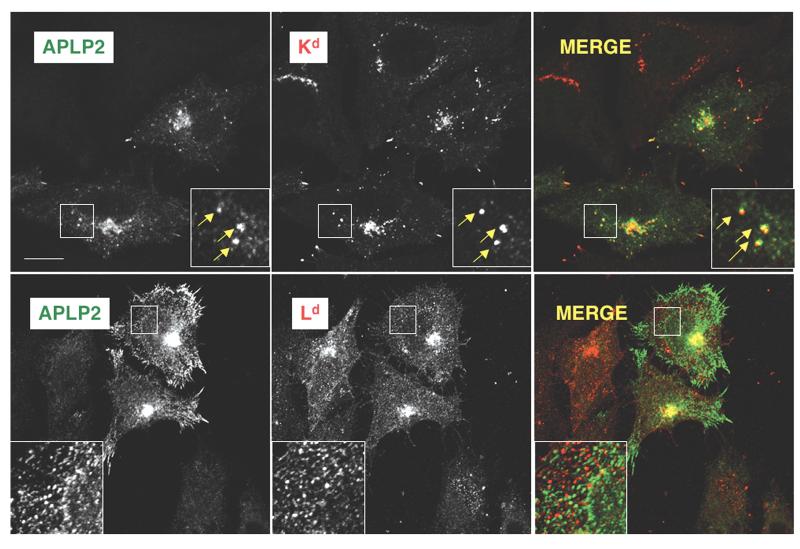
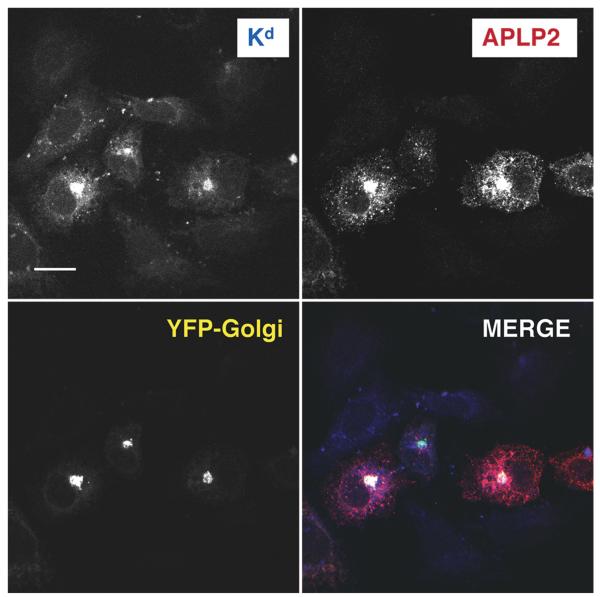
In addition to comparing the co-immunoprecipitation of APLP2 with Kd and Ld expressed in HeLa cells (Fig. 1B), we also compared the co-localization of Kd and Ld in HeLa cells. Ld, like Kd, was associated with APLP2 in a large, compact cellular compartment (Fig. 3). We have identified this large compartment containing associated APLP2 and Kd as the Golgi (Fig. 4). Kd and APLP2 were also associated in intracellular vesicles, but the interaction between Ld and APLP2 was limited to the large compartment and was not observed within the vesicles (Fig. 3).
Figure 3.
Both Kd and Ld were found to co-localize with APLP2 in a large intracellular compartment, but only Kd was found to co-localize with APLP2 in vesicles. (Top panel) At steady state, folded (34-1-2+) Kd was co-localized with APLP2. The co-localization was noted in a compact cellular compartment (identified as the Golgi in our earlier studies on Kd and APLP2) and also seen in peripheral vesicular structures. (Bottom panel) Folded (30-5-7+) Ld was co-localized with APLP2, but the co-localization could only be seen in the large compact compartment. For these experiments, HeLa-etKd or HeLa-Ld cells were transiently transfected with APLP2-FLAG, fixed, and incubated with 34-1-2 or 30-5-7 and rabbit anti-FLAG antibody and fluorescently labeled secondary antibodies in staining solution. Images were analyzed on a Zeiss LSM 5 Pascal confocal microscope. Green = APLP2; red = folded Kd or Ld; yellow = co-localized APLP2 and Kd or Ld molecules (single staining of APLP2 or Kd or Ld is shown in black and white in the figure). Bar, 10 μm.
Figure 4.
The Golgi is the large compartment in which Kd and APLP2 co-localized. APLP2-FLAG, etKd, and YFP-Golgi were transiently transfected into HeLa cells. The cells were fixed and then incubated with anti-Kd antibody 34-1-2 and rabbit anti-FLAG and fluorescently labeled secondary antibodies in staining solution. Images were analyzed with a Zeiss LSM 5 Pascal confocal microscope. Blue = folded Kd; red=APLP2; yellow = YFP-Golgi; white = co-localized APLP2 and Kd (fluorescence from Kd, APLP2, and YFP-Golgi, individually, is shown in black and white in the figure). Bar, 10 μm.
We also quantified co-localization of APLP2/Kd and APLP2/Ld with Image J software, utilizing JACoP to calculate Pearson’s coefficients. (The higher the Pearson’s coefficient, the higher the degree of co-localization.) Each pairwise comparison was done on ten sets of images acquired with the same optical settings. The Pearson coefficient for co-localization of APLP2 with Kd was 0.701±0.095 (standard error of the mean), and for co-localization of APLP2 with Ld it was 0.333±0.017. Thus, this quantitative analysis confirmed that there was a higher degree of intracellular co-localization between APLP2 and Kd than between APLP2 and Ld.
MHC polymorphism influenced the impact of APLP2 on MHC class I cell surface expression
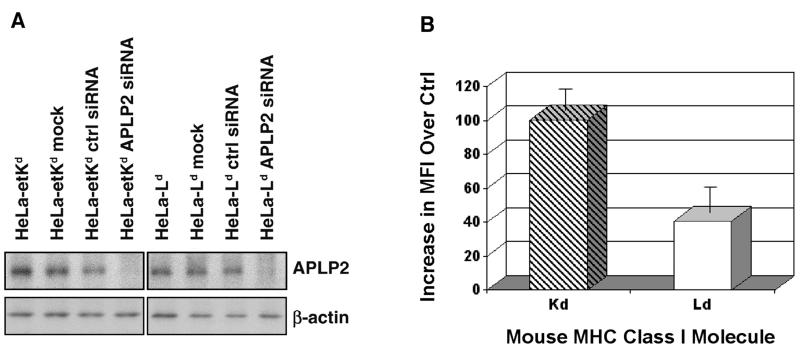
Previously, we demonstrated that transient transfection with APLP2-specific siRNA causes an increase in the expression of Kd at the plasma membrane of HeLa-etKd cells (Morris et al. 2003). In this earlier work, we showed that by day 3 of transient transfection with APLP2 siRNA or control siRNA, the cells that had been transfected with APLP2 siRNA and stained with the 34-1-2 antibody had a mean fluorescence intensity 83.2 channels higher than control cells that had been transfected with inverse siRNA and stained with 34-1-2 (Morris et al. 2003). In this current study, we have performed siRNA transfection studies with Ld, and included Kd as a control. Transient transfections of HeLa-etKd and HeLa-Ld cells were performed with APLP2 siRNA or control siRNA for 48 or 72 hours, and Western blotting confirmed down-regulation of APLP2 but not β-actin by the APLP2 siRNA (Fig. 5A). Our flow cytometric results with Kd were similar to those we previously obtained with transient APLP2 siRNA expression (Fig. 5B; Morris et al. 2003). In addition, we found that expression of Ld, like Kd, was increased at the cell surface by APLP2 siRNA transient transfection, although Ld expression was increased only 40% as much as Kd (Fig. 5B).
Figure 5.
Transient down-regulation of APLP2 slightly increased cell surface expression of Ld. (A) Samples from lysed HeLa-etKd or HeLa-Ld cells that had been untreated, mock treated, transfected with a control siRNA pool for 48 or 72 hours, or transfected with APLP2-specific siRNA for 48 or 72 hours were electrophoresed on a 4→20% gel, transferred to a blotting membrane, and probed with anti-APLP2 antiserum. Results from a 48 hour transfection are shown here, and results from 72 hours of transfection also showed down regulation (data not shown). (B) HeLa-etKd or HeLa-Ld cells that had been transfected with a control siRNA pool or with APLP2-specific siRNA for 48 or 72 hours were stained with 34-1-2 (for Kd) or 30-5-7 (for Ld) and a phycoerythrin-labeled secondary antibody, and then the fluorescence was assessed on a BD FACS Calibur. For each cell type, the mean fluorescence intensity (MFI) results in each of three experiments for cells expressing control siRNA were subtracted from the MFI results in the same experiment for cells transfected with APLP2-specific siRNA. The mean increases in MFI (i.e., for specific siRNA minus control siRNA) are shown in the graph with error bars indicating the standard error of the mean.
Strong binding to APLP2 required the α3/TM/CYT region of the Kd heavy chain
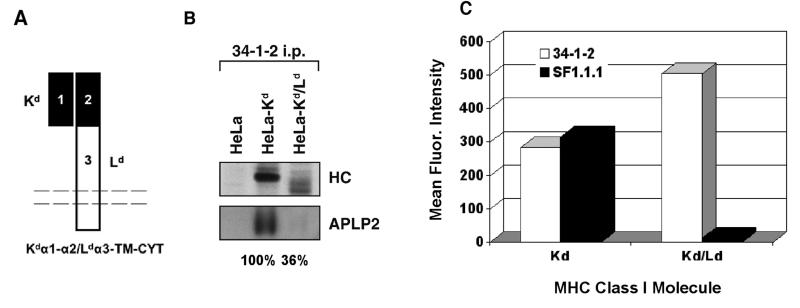
To determine the importance of regions outside the folded outer domains of the Kd molecule to the strong interaction of Kd with APLP2 and to the functional effects of APLP2 on Kd, a chimeric Kd/Ld molecule was constructed and expressed in HeLa cells. This chimeric molecule included the first 214 amino acids of Kd (the N-terminus of the Kd leader peptide through the α1 and α2 domains) joined to the last 145 amino acids of Ld (including the α3 domain, TM, and CYT domains) (Fig. 6A). The Kd/Ld chimera, along with wild type Kd for comparison, was immunoprecipitated from transfected HeLa cell lysates, and the immunoprecipitates were probed for associated APLP2. Although APLP2 interacted strongly with wild type Kd, relatively weak association was noted between APLP2 and Kd/Ld(Fig. 6B).
Figure 6.
Substitution of the Ld α3/TM/CYT domain for the Kd α3/TM/CYT domain caused loss of APLP2 co-immunoprecipitation and increased cell surface expression. (A) A diagram of the Kd/Ld chimera. (B) Kd or Kd/Ld was immunoprecipitated from the indicated radiolabeled cell lysates with 34-1-2. The immunoprecipitates were electrophoresed on 4→20% gels, and the proteins were transferred to blotting membranes that were autoradiographed (HC, top panel) or probed with antiserum against APLP2 (bottom panel). (C) Shown on the bar graph are the mean fluorescence intensity values for Kd and Kd/Ld, as detected with antibody 34-1-2 (recognizing the Kd α1α2 domains) or SF1.1.1 (recognizing the Kd α3 domain) on Kd- or Kd/Ld-transfected HeLa cells. The MFI for Kd and Kd/Ld with fluorescently labeled secondary antibody only was less than 10 channels.
We also examined the cell surface expression of the Kd/Ld chimeric molecule, comparing it to the surface expression of Kd. By flow cytometry with the 34-1-2 antibody (recognizing the Kd α1α2 region), we found that the surface expression of Kd/Ld was almost twice as high as the expression of Kd (Fig. 6C). The higher expression of Kd/Ld at the plasma membrane was not attributable to higher intracellular expression of Kd/Ld, as Kd/Ld exhibited somewhat lower expression than Kd in immunoprecipitation experiments (Fig. 6B). Furthermore, in testing of the Kd/Ld-HeLa and Kd-HeLa cells by flow cytometry with the SF1.1.1 antibody (specific for the Kd α3 region), the SF1.1.1 antibody bound to the Kd-HeLa but not the Kd/Ld-HeLa cells, thus verifying the substitution of the region that includes the α3 domain (Fig. 6C).
Discussion
In this study, we demonstrated that APLP2 interacts with certain other murine MHC class I molecules in addition to Kd (Fig. 1). In our comparisons of APLP2 interaction with Ld and Kd, we found that APLP2 bound to Kd more strongly than to Ld(Fig. 1A and B). APLP2 could be seen co-localized with both Kd and Ld in a large, compact cellular compartment (identified as the Golgi), but APLP2 co-localization with Ld in vesicular cellular compartments was not observed (Figs. 3 and 4). These data imply that the strong binding between APLP2 and Kd might be necessary to maintain the interaction during transport into these vesicles.
The fact that the binding of APLP2 to MHC class I molecules was found to be influenced by MHC class I heavy chain polymorphism (Fig. 1A and B) suggests that APLP2 may interact with the α1α2 region of the MHC class I molecule. In addition, the specificity of APLP2 for folded, and not open, Ld and Kd indicates that the α1α2 region of both H2 molecules is likely involved in APLP2 binding (Fig. 1B; Morris et al. 2003). Further support for the notion that APLP2 binds to the α1α2 region is that the addition of Kd peptide ligands to cell lysates causes release of APLP2 from Kd (Feuerbach and Burgert 1993). However, our experiments with the Kd/Ld chimera demonstrate that the α3/TM/CYT region of Kd is also important to Kd’s ability to associate tightly with APLP2 (Fig. 6). Thus, APLP2 presumably binds to more than one site on the MHC class I heavy chain. Evidence that binding is influenced by MHC class I polymorphism (directly or indirectly) has also been shown for tapasin (Peh et al., 2000; Turnquist et al., 2000; Turnquist et al., 2002a; Hildebrand et al., 2002; Park et al., 2003; Zernich et al., 2004; Thammavongsa et al., 2006). Furthermore, like APLP2, tapasin interacts with more than one domain on the MHC class I heavy chain; there are data supporting tapasin interaction with both the α2 and α3 domains of the MHC class I molecule (Turnquist et al. 2002b).
The expression of human β2m in L cells increased the association of H2 class I molecules with APLP2 (Fig. 2). A previous study found that the co-precipitation of a 105 kDa protein with Db was facilitated by the presence of excess human β2m (Townsend et al. 1990). As first theorized by others (Feuerbach and Burgert 1993; Sester et al. 2000), this large protein co-precipitating with Db could be APLP2; our findings are supportive of this notion. The results shown in Figure 2C suggest that human β2m does not interact directly and strongly with APLP2. Instead, human β2m may increase APLP2 binding to mouse MHC class I heavy chains indirectly by influencing the conformation of the MHC class I heavy chain α1/α2 region. The conformation of mouse MHC class I molecules (including Kd, Dd, and Ld) has been shown to be stabilized better by human β2m than mouse β2m (Shields et al. 1998; Shields et al. 1999). For Kd, the stabilizing effect of human β2m has been tracked to aspartate 53; mutation of this position decreases the stabilization approximately to the level of mouse β2m (Shields et al. 1998). The aspartate at position 53 also influences Dd stabilization by human β2m, and other positions in human β2m play a role in Dd stabilization as well (Shields et al. 1998; Shields et al. 1999). Several studies have shown that xenogeneic β2m can alter the conformation of the outer domains of the MHC class I molecule and affect T cell recognition (Nieto et al. 1989; Shields et al. 1998; Rocca et al. 1992; Kahn-Perles et al. 1987; Achour et al. 2006; Benoit and Tan 2007). Thus, it is most likely that human β2m alters mouse MHC class I heavy chain conformation in some way that strengthens APLP2 binding to the MHC heavy chain. As a distant possibility, it should also be kept in mind that our results do not exclude that some mouse MHC class I heavy chains (e.g., Kd) could, by binding to human β2m, cause human β2m to adopt a conformation that allows it to bind well to APLP2 directly.
APLP2 was expressed at similar levels in the mouse L cells and human HeLa cells, but the APLP2 level in 721.221 cells was much lower than in either L or HeLa cells (Fig. 1C). Since 721.221 cells lack expression of HLA-A, B, and C, this observation raises the question of whether MHC class I heavy chains normally increase the expression of APLP2. To address this point, the level of APLP2 in MHC class I-transfected 721.221 cells could be compared to the level in untransfected 721.221 cells.
Following transfection with APLP2 siRNA, the expression of Ld, like Kd, was increased at the cell surface, with Ld expression increased to a lesser extent than Kd (Fig. 5). These surface expression results parallel the weaker binding of Ld to APLP2, relative to Kd binding to APLP2 (Fig. 1A and B). When we assessed expression of Kd and Kd/Ld molecules at the plasma membrane, we found that Kd/Ld had higher cell surface expression than Kd (Fig. 6C), even though the intracellular expression of Kd/Ld was lower than the intracellular expression of Kd (Fig. 6B). Furthermore, Kd/Ld, unlike Kd, was weakly associated with APLP2 (Fig. 6B). The results shown in Figures 5 and 6 extend our previous finding that APLP2 has a down-regulatory effect on Kd surface expression (Morris et al. 2003) by showing that partial reduction of the strength of MHC/APLP2 binding (as seen for Ld and Kd/Ld, relative to Kd) is paralleled by a lessening of the effect of APLP2 on the surface expression of the MHC molecule. These findings further support a model in which the binding of APLP2 to the MHC class I molecule is necessary for APLP2’s effect on MHC class I expression at the plasma membrane, as opposed to a model in which APLP2 exercises its effect by a more indirect means (e.g., via binding to a third protein). New findings from our laboratory indicate that APLP2′s effect on MHC class I expression is due to an APLP2-induced increase in MHC class I endocytosis (data not shown). Thus, the role of the APLP2-MHC class I interaction appears to be to control MHC class I cell surface expression via regulation of the process of internalization of MHC class I molecules.
These new data show that APLP2 interacts differentially with several mouse MHC class I allotypes, displaying disparate affinity, intracellular co-localization, and functional impact. We have also demonstrated that APLP2 association with H2 class I molecules is influenced by more than one domain of the MHC class I heavy chain and by the sequence of the β2m light chain with which the heavy chain is associated. Our new studies support a role for APLP2 in influencing MHC class I molecules to varied extents, by binding to multiple molecular sites on the MHC heavy chain.
Acknowledgments
This work was supported by National Institutes of Health Grant GM57428 (to J.C.S.) and GM74876 (to S.C.), UNMC Graduate Studies Fellowships (to A.T. and M.S.), and a Nebraska Center for Cellular Signaling Fellowship (to M.S.). We thank Dr. Ted Hansen, Dr. Wendy Maury, and Dr. Eric Long for gifts of antibodies, cell lines, and cloning vectors. We also gratefully acknowledge the assistance of Haley Capek, Vivek Gautam, Mary McIlhaney, and the personnel of the University of Nebraska Medical Center Cell Analysis Facility and Monoclonal Antibody Facility.
References
- Achour A, Michaelsson J, Harris RA, Ljunggren H-G, Karre K, Schnieder G, Scandalova T. Structural basis of the differential stability and receptor-specificity of H-2Db in complex with murine versus human β2-microglobulin. J Mol Biol. 2006;356:382–396. doi: 10.1016/j.jmb.2005.11.068. [DOI] [PubMed] [Google Scholar]
- Benoit LA, Tan R. Xenogeneic β2-microglobulin substitution affects functional binding of MHC class I molecules by CD8+ T cells. J Immunol. 2007;179:3588–3595. doi: 10.4049/jimmunol.179.6.3588. [DOI] [PubMed] [Google Scholar]
- Brodsky FM, Bodmer WF, Parham P. Characterization of a monoclonal anti-beta 2-microglobulin antibody and its use in the genetic and biochemical analysis of major histocompatibility antigens. Eur J Immunol. 1979;9:536–545. doi: 10.1002/eji.1830090709. [DOI] [PubMed] [Google Scholar]
- Cappai R, Mok SS, Galatis D, Tucker DF, Henry A, Beyreuther K, Small DH, Masters CL. Recombinant human amyloid precursor-like protein 2 (APLP2) expressed in the yeast Pichia pastoris can stimulate neurite outgrowth. FEBS Lett. 1999;442:95–98. doi: 10.1016/s0014-5793(98)01635-4. [DOI] [PubMed] [Google Scholar]
- Cox JH, Bennick J, Yewdell JW. Retention of adenovirus E19 glycoprotein in the endoplasmic reticulum is essential to its ability to block antigen presentation. J Exp Med. 1991;174:1629–1637. doi: 10.1084/jem.174.6.1629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cresswell P, Bangia N, Dick T, Diedrich G. The nature of the MHC class I peptide loading complex. Immunol Rev. 1999;172:21–28. doi: 10.1111/j.1600-065x.1999.tb01353.x. [DOI] [PubMed] [Google Scholar]
- Eck SC, Zhu P, Petter M, Bensinger SJ, Freedman BD, Laufer TM. Developmental alterations in thymocyte sensitivity are actively regulated by MHC class II expression in the thymic medulla. J Immunol. 2006;176:2229–2237. doi: 10.4049/jimmunol.176.4.2229. [DOI] [PubMed] [Google Scholar]
- Evans GA, Margulies DH, Shykind B, Seidman JG, Ozato K. Exon shuffling: mapping polymorphic determinants on hybrid mouse transplantation antigens. Nature. 1982;300:755–757. doi: 10.1038/300755a0. [DOI] [PubMed] [Google Scholar]
- Feuerbach D, Burgert H-G. Novel proteins associated with MHC class I antigens in cells expressing adenovirus protein E3/19K. EMBO J. 1993;12:3153–3161. doi: 10.1002/j.1460-2075.1993.tb05984.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo J, Thinakaran G, Guo Y, Sisodia SS, Yu FX. A role for amyloid precursor-like protein 2 in corneal epithelial wound healing. Invest Ophthalmol Vis Sci. 1998;39:292–300. [PubMed] [Google Scholar]
- Harris MR, Lybarger L, Myers NB, Hilbert C, Solheim JC, Hansen TH, Yu YY. Interactions of HLA-B27 with the peptide loading complex as revealed by heavy chain mutations. Int Immunol. 2001;13:1275–1282. doi: 10.1093/intimm/13.10.1275. [DOI] [PubMed] [Google Scholar]
- Hildebrand WH, Turnquist HR, Prilliman KR, Hickman HD, Schenk EL, McIlhaney MM, Solheim JC. HLA class I polymorphism has a dual impact on ligand binding and chaperone interaction. Hum Immunol. 2002;63:248–255. doi: 10.1016/s0198-8859(02)00364-6. [DOI] [PubMed] [Google Scholar]
- Joyce S. Traffic control of completely assembled MHC class I molecules beyond the endoplasmic reticulum. J Mol Biol. 1997;266:993–1001. doi: 10.1006/jmbi.1996.0822. [DOI] [PubMed] [Google Scholar]
- Kahn-Perles B, Boyer C, Arnold B, Sanderson AR, Ferrier P, Lemonnier FA. Acquisition of HLA class I W6/32 defined antigenic determinant by heavy chains from different species following association with bovine β2-microglobulin. J Immunol. 1987;138:2190–2196. [PubMed] [Google Scholar]
- Lee DR, Rubocki RJ, Lie W-R, Hansen TH. The murine MHC class I genes, H-2Dq and H-2Lq, are strikingly homologous to each other, H-2Ld, and two genes reported to encode tumor-specific antigens. J Exp Med. 1988;268:1719–1724. doi: 10.1084/jem.168.5.1719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li XF, Thinakaran G, Sisodia SS, Yu FS. Amyloid precursor-like protein 2 promotes cell migration toward fibronectin and collagen IV. J Biol Chem. 1999;274:27249–27256. doi: 10.1074/jbc.274.38.27249. [DOI] [PubMed] [Google Scholar]
- Lybarger L, Yu YYL, Chun T, Wang C-R, Grandea AG, III, Van Kaer L, Hansen TH. Tapasin enhances peptide-induced expression of H2-M3 molecules, but is not required for the retention of open conformers. J Immunol. 2001;167:2097–2105. doi: 10.4049/jimmunol.167.4.2097. [DOI] [PubMed] [Google Scholar]
- Marguet D, Spiliotis ET, Pentcheva T, Lebowitz M, Schneck J, Edidin M. Lateral diffusion of GFP-tagged H2Ld molecules and of GFP-TAP1 reports on the assembly and retention of these molecules in the endoplasmic reticulum. Immunity. 1999;11:231–240. doi: 10.1016/s1074-7613(00)80098-9. [DOI] [PubMed] [Google Scholar]
- Morris CR, Petersen JL, Vargas SE, Turnquist HR, McIlhaney MM, Sanderson SD, Bruder JT, Yu YYL, Burgert H-G, Solheim JC. The amyloid precursor-like protein 2 and the adenoviral E3/19K protein both bind to a conformational site on H-2Kd and regulate H-2Kd expression. J Biol Chem. 2003;278:12618–12623. doi: 10.1074/jbc.M208203200. [DOI] [PubMed] [Google Scholar]
- Myers NB, Harris MR, Connolly JM, Lybarger L, Yu YY, Hansen TH. Kb, Kd, and Ld molecules share common tapasin dependencies as determined using a novel epitope tag. J Immunol. 2000;165:5656–5663. doi: 10.4049/jimmunol.165.10.5656. [DOI] [PubMed] [Google Scholar]
- Ng YP, He W, Ip NY. Leukemia inhibitory factor receptor signaling negatively modulates nerve growth factor-induced neurite outgrowth in PC12 cells and sympathetic neurons. J Biol Chem. 2003;278:38731–38739. doi: 10.1074/jbc.M304623200. [DOI] [PubMed] [Google Scholar]
- Nieto M, Song ES, McKinney D, McMillan M, Goodenow RS. The association of H-2Ld with human beta-2 microglobulin induces localized conformational changes in the alpha-1 and -2 superdomain. Immunogenetics. 1989;30:361–369. doi: 10.1007/BF02425276. [DOI] [PubMed] [Google Scholar]
- Ozato K, Hansen TH, Sachs DH. Monoclonal antibodies to mouse MHC antigens. II. Antibodies to the H-2Ld antigen, the product of a third polymorphic locus of the mouse major histocompatibility complex. J Immunol. 1980;125:2473–2477. [PubMed] [Google Scholar]
- Ozato K, Sachs DH. Monoclonal antibodies to mouse MHC antigens. III. Hybridoma antibodies reacting to antigens of the H-2b haplotype reveal genetic control of isotype expression. J Immunol. 1981;126:317–321. [PubMed] [Google Scholar]
- Ozato K, Evans GA, Shykind B, Margulies D, Seidman JG. Hybrid H-2 histocompatibility gene products assign domains recognized by alloreactive T cells. Proc Natl Acad Sci USA. 1983;80:2040–2043. doi: 10.1073/pnas.80.7.2040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paquet ME, Cohen-Doyle M, Shore GC, Williams DB. Bap29/31 influences the intracellular traffic of MHC class I molecules. J Immunol. 2004;172:7548–7555. doi: 10.4049/jimmunol.172.12.7548. [DOI] [PubMed] [Google Scholar]
- Park B, Lee S, Kim E, Cho K, Riddell SR, Cho S, Ahn K. Redox regulation facilitates optimal peptide selection by MHC class I during antigen processing. Cell. 2006;127:369–382. doi: 10.1016/j.cell.2006.08.041. [DOI] [PubMed] [Google Scholar]
- Paulsson KM, Kleijmeer MJ, Griffith J, Jevon M, Chen S, Anderson PO, Sjogren HO, Li S, Wang P. Association of tapasin and COPI provides a mechanism for the retrograde transport of major histocompatibility complex (MHC) class I molecules from the Golgi complex to the endoplasmic reticulum. J Biol Chem. 2002;277:18266–18271. doi: 10.1074/jbc.M201388200. [DOI] [PubMed] [Google Scholar]
- Park B, Lee S, Kim E, Ahn K. A single polymorphic residue within the peptide-binding cleft of MHC class I molecules determines spectrum of tapasin dependence. J Immunol. 2003;170:961–968. doi: 10.4049/jimmunol.170.2.961. [DOI] [PubMed] [Google Scholar]
- Peh CA, Laham N, Burrows SR, Zhu Y, McCluskey J. Distinct functions of tapasin revealed by polymorphism in MHC class I peptide loading. J Immunol. 2000;164:292–299. doi: 10.4049/jimmunol.164.1.292. [DOI] [PubMed] [Google Scholar]
- Pencheva T, Edidin M. Clustering of peptide-loaded MHC class I molecules for endoplasmic reticulum export imaged by fluorescence resonance energy transfer. J Immunol. 2001;166:6625–6632. doi: 10.4049/jimmunol.166.11.6625. [DOI] [PubMed] [Google Scholar]
- Rassoulzadegan M, Yang Y, Cuzin F. APLP2, a member of the Alzheimer precursor protein family, is required for correct genomic segregation in dividing mouse cells. EMBO J. 1998;17:4647–4656. doi: 10.1093/emboj/17.16.4647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rocca A, Opolski A, Samaan A, Frangoulis B, Degos L, Pla M. Localization of the conformational alteration of MHC molecules induced by the association of mouse class I heavy chain with a xenogeneic β2-microglobulin. Mol Immunol. 1992;29:481–487. doi: 10.1016/0161-5890(92)90005-i. [DOI] [PubMed] [Google Scholar]
- Saric T, Chang SC, Hattori A, York IA, Markant S, Rock KL, Tsujimoto M, Goldberg AL. An IFN-γ-induced aminopeptidase in the ER, ERAP1, trims precursors to MHC class I-presented peptides. Nat Immunol. 2002;3:1169–1176. doi: 10.1038/ni859. [DOI] [PubMed] [Google Scholar]
- Serwold T, Gonzalez F, Kim J, Jacon R, Shastri N. ERAAP customizes peptides for MHC class I molecules in the endoplasmic reticulum. Nature. 2002;419:480–483. doi: 10.1038/nature01074. [DOI] [PubMed] [Google Scholar]
- Sester M, Feuerbach D, Frank R, Preckel T, Gutermann A, Burgert H-G. The amyloid precursor-like protein 2 associates with the major histocompatibility complex class I molecule Kd. J Biol Chem. 2000;275:3645–3654. doi: 10.1074/jbc.275.5.3645. [DOI] [PubMed] [Google Scholar]
- Shields MJ, Hodgson W, Ribaudo RK. Differential association of β2-microglobulin mutants with MHC class I heavy chains and structural analysis demonstrate allele-specific interactions. Mol Immunol. 1999;36:561–573. doi: 10.1016/s0161-5890(99)00077-2. [DOI] [PubMed] [Google Scholar]
- Shields MJ, Assefi N, Hodgson W, Kim EJ, Ribaudo RK. Characterization of the interactions between MHC class I subunits: a systematic approach for the endgineering of higher affinity variants of β2-microglobulin. J Immunol. 1998;160:2297–2307. [PubMed] [Google Scholar]
- Shimizu Y, Geraghty DE, Koller BH, Orr HT, DeMars R. Transfer and expression of three cloned human non-HLA-A,B,C class I major histocompatibility complex genes in mutant lymphoblastoid cells. Proc Natl Acad Sci USA. 1988;85:227–231. doi: 10.1073/pnas.85.1.227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimizu Y, DeMars R. Production of human cells expressing individual transferred HLAA,-B,-C genes using an HLA-A,-B,-C null human cell line. J Immunol. 1989;142:3320–3328. [PubMed] [Google Scholar]
- Slunt HH, Thinakaran G, Von Koch C, Lo ACY, Tanzi RE, Sisodia SS. Expression of a ubiquitous, cross-reactive homologue of the mouse β-amyloid precursor protein. J Biol Chem. 1994;269:2637–2644. [PubMed] [Google Scholar]
- Smith JD, Myers NB, Gorka J, Hansen TH. Model for the in vivo assembly of nascent Ld class I molecules and for the expression of unfolded Ld molecules at the cell surface. J Exp Med. 1993;178:2035–2046. doi: 10.1084/jem.178.6.2035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith JD, Solheim JC, Carreno BM, Hansen TH. Characterization of class I MHC folding intermediates and their disparate interactions with peptide and β2-microglobulin. Mol Immunol. 1995;32:531–540. doi: 10.1016/0161-5890(95)00013-5. [DOI] [PubMed] [Google Scholar]
- Sohn HW, Shin YK, Lee IS, Bae YM, Suh YH, Kim MK, Kim TJ, Jung KC, Park WS, Park C-S, Chung DH, Ahn K, Kim IS, Ko YH, Bang YJ, Kim CW, Park SH. CD99 regulates the transport of MHC class I molecules from the Golgi complex to the cell surface. J Immunol. 2001;166:787–794. doi: 10.4049/jimmunol.166.2.787. [DOI] [PubMed] [Google Scholar]
- Solheim JC, Carreno BM, Smith JD, Gorka J, Myers NB, Wen Z, Martinko JM, Lee DR, Hansen TH. Binding of peptides lacking consensus anchor residue alters H-2Ld serologic recognition. J Immunol. 1993;151:5387–5397. [PubMed] [Google Scholar]
- Solheim JC, Carreno BM, Myers NB, Lee DR, Hansen TH. Peptide-induced rescue of serologic epitopes on class I MHC molecules. J Immunol. 1995;154:1188–1197. [PubMed] [Google Scholar]
- Spiliotis ET, Manley H, Osorio M, Zuniga MC, Edidin M. Selective export of MHC class I molecules from the ER after their dissociation from TAP. Immunity. 2000;13:841–851. doi: 10.1016/s1074-7613(00)00081-9. [DOI] [PubMed] [Google Scholar]
- Thammavongsa V, Raghuraman G, Filzen TM, Collins KL, Raghavan M. HLA-B44 polymorphisms at position 116 of the heavy chain influence TAP complex binding via an effect on peptide occupancy. J Immunol. 2006;177:3150–3161. doi: 10.4049/jimmunol.177.5.3150. [DOI] [PubMed] [Google Scholar]
- Thinakaran G, Kitt CA, Roskams AJ, Slunt HH, Masliah E, von Koch C, Ginsberg SD, Ronnett GV, Reed RR, Price DL. Distribution of an APP homolog, APLP2, in the mouse olfactory system: a potential role for APLP2 in axogenesis. J Neurosci. 1995;15:6314–6326. doi: 10.1523/JNEUROSCI.15-10-06314.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Townsend A, Elliott T, Cerundolo V, Foster L, Barber B, Tse A. Assembly of MHC class I molecules analyzed in vitro. Cell. 1990;62:285–295. doi: 10.1016/0092-8674(90)90366-m. [DOI] [PubMed] [Google Scholar]
- Turnquist HR, Thomas HJ, Prilliman KR, Lutz CT, Hildebrand WH, Solheim JC. HLAB polymorphism affects interactions with multiple endoplasmic reticulum proteins. Eur J Immunol. 2000;30:3021–3028. doi: 10.1002/1521-4141(200010)30:10<3021::AID-IMMU3021>3.0.CO;2-U. [DOI] [PubMed] [Google Scholar]
- Turnquist HR, Solheim JC. Analysis of MHC class I interactions with endoplasmic reticulum proteins. Methods Mol Biol. 2001;156:165–173. doi: 10.1385/1-59259-062-4:165. [DOI] [PubMed] [Google Scholar]
- Turnquist HR, Schenk EL, McIlhaney MM, Hickman HD, Hildebrand WH, Solheim JC. Disparate binding of chaperone proteins by HLA-A subtypes. Immunogenetics. 2002a;53:830–834. doi: 10.1007/s00251-001-0404-x. [DOI] [PubMed] [Google Scholar]
- Turnquist HR, Vargas SE, Schenk EL, McIlhaney MM, Reber AJ, Solheim JC. The interface between tapasin and MHC class I: identification of amino acid residues in both proteins that influence their interaction. Immunol Res. 2002b;25:261–269. doi: 10.1385/ir:25:3:261. [DOI] [PubMed] [Google Scholar]
- Yu YYL, Myers NB, Hilbert CH, Harris MR, Balendiran GK, Hansen TH. Definition and transfer of a serological epitope specific for peptide-empty forms of MHC class I. Int Immunol. 1999;11:1897–1906. doi: 10.1093/intimm/11.12.1897. [DOI] [PubMed] [Google Scholar]
- Zernich D, Purcell AW, Macdonald WA, Kjer-Nielsen L, Ely LK, Laham N, Crockford T, Mifsud NA, Bharadwaj M, Chang L, Tait BD, Holdsworth R, Brooks AG, Bottomley SP, Beddoe T, Peh CA, Rossjohn J, McCluskey J. Natural HLA class I polymorphism controls the pathway of antigen presentation and susceptibility to viral evasion. J Exp Med. 2004;200:13–24. doi: 10.1084/jem.20031680. [DOI] [PMC free article] [PubMed] [Google Scholar]