Abstract
Coenzyme Q10 (which is also designated as CoQ10, ubiquinone-10, UQ10, CoQ, UQ or simply as Q) plays an important role in energy metabolism. For NADH-Q oxidoreductase (complex I), Ohnishi and Salerno proposed a hypothesis that the proton pump is operated by the redox-driven conformational change of a Q-binding protein, and that the bound form of semiquinone (SQ) serves as its gate [FEBS Letters 579 (2005) 45–55]. This was based on the following experimental results: (i) EPR signals of the fast-relaxing SQ anion (designated as ) are observable only in the presence of the proton electrochemical potential ( ); (ii) iron-sulfur cluster N2 and are directly spin-coupled; and (iii) their center-to-center distance was calculated as 12Å, but is only 5Å deeper than N2 perpendicularly to the membrane. After the priming reduction of Q to , the proton pump operates only in the steps between the semiquinone anion ( ) and fully reduced quinone (QH2). Thus, by cycling twice for one NADH molecule, the pump transports 4H+ per 2e−.
This hypothesis predicts the following phenomena: (a) Coupled with the piericidin A sensitive NADH-DBQ or Q1 reductase reaction, would be established; (b) would enhance the SQ EPR signals; and (c) the dissipation of with the addition of an uncoupler would increase the rate of NADH oxidation and decrease the SQ signals.
We reconstituted bovine heart complex I, which was prepared at Yoshikawa’s laboratory, into proteoliposomes. Using this system, we succeeded in demonstrating that all of these phenomena actually took place. We believe that these results strongly support our hypothesis.
Keywords: NADH-CoQ oxidoreductase (complex I), proteoliposome, reconstitution, membrane potential, iron-sulfur cluster N2, semiquinone, EPR
1. Introduction
NADH-Q oxidoreductase (complex I) is one of energy transducing enzyme complexes in the mitochondrial respiratory chain. It is located at the entry point of electrons from NADH, and catalyses the following reaction:
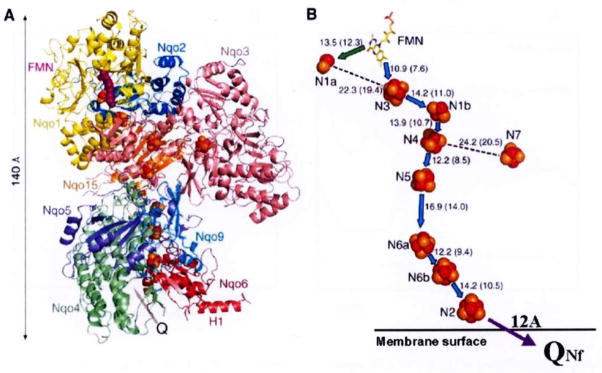
where Q stands for coenzyme Q10 (CoQ10 or ubiquinone-10) [1,33]. In this exergonic redox reaction, the transfer of 2 electrons from one molecule of NADH to Q is coupled to the translocation of 4 protons from the negative side (abbreviated as N-side) to the positive side (P-side) of the mitochondrial inner membrane [6,32]. Complex I is the largest energy transducing complex (Molecular weight is ~1 megadalton [7], composed of 45 subunits [3] and contains 1 non-covalently bound FMN, and 8 or 9 iron-sulfur (Fe/S) clusters [19]. Recently the first atomic structure at 3.3Å resolution of the peripheral arm (hydrophilic domain) from Thermus thermophilus HB-8 complex I was determined by Sazanov and Hinchliffe [27] (See Fig. 1). These authors revealed the presence of a long electron transfer pathway of NADH → FMN – N3 – N1b – N4 – N5 – N6a – N6b – N2 → Q which is composed of one FMN and 7 Fe/S clusters. It stretches to a total of ~95Å with individual clusters having an edge-to-edge distance of < 14Å [12]. It is likely to be the main route for the electron transfer within complex I [28]. These authors also proposed a possible role of cluster N1a as an anti-oxidant. N1a has, in general, a very low redox midpoint potential (Em7.0 = −370 mV) [19], and its edge-to-edge distance to FMN is 12.3Å and that to the closest iron-sulfur cluster N3 is 19.4Å. Therefore, N1a seems to play a role as a temporary electron reservoir. Cluster N7 was found only in a limited number of bacteria (which include Thermus thermophilus and Escherichia coli) [13,14], and too far from all other clusters (> 20.5Å). Thus, N7 is located away from the main electron transfer chain [27]. This cluster was reported not being reducible with NADH, and perhaps, it plays a role in stabilizing the complex I molecule [24]. The highest potential cluster N2 is only 8Å from the surface of the isolated hydrophilic arm, and is partly exposed to an elongated cavity which is probably the binding site for ubiquinone (which corresponds to QNf as will be described below). These results are in good agreement with our predictions of N2 ↔ QNf distance of 12Å based on our spin-spin interaction analysis [35].
Fig. 1.
Architecture of the hydrophilic domain of Thermus thermophilus HB-8 NADH-Q oxidoreductase (complex I). A, side view, possible quinone binding site (Q) is indicated by an arrow. B, arrangement of redox centers, (by Sazanov and Hinchliffe (2006) Science 311, 1430). Location of the protein-bound QNf (which is shown here in reference to cluster N2) was calculated based on our spin-spin interaction data [35].
Mitchell identified ubiquinone as one of the key redox components involved in the coupling of electron transfer to vectorial proton translocation in his original formulation of the chemiosmotic hypothesis [10, 11]. Many of the membrane-bound protein complexes in the respiratory and photosynthetic systems are known to react with quinone. X-ray crystallographic information clearly confirmed the general functional roles of quinones, for example, at the Qo and Qi sites of mitochondrial complex III [4,8] as well as at the QA and QB sites of the bacterial photosynthetic reaction center [23,36]. Various research groups detected protein-bound forms of quinone species by their semiquinone signals using EPR spectroscopy.
Early in 1983, Suzuki and King detected SQ signals in NADH-reduced isolated bovine heart complex I using a high frequency Q-band EPR (34 GHz) [29]. These authors showed that the SQ signals would be derived from protein-bound species as judged from their anisotropic line shape with gz,y,x = 2.0060, 2.0051, 2.0022. They also resolved spectra into two distinct SQ species based on their different sensitivity to rotenone and sulfhydryl reagent, but both species had extremely slow spin relaxation (the signals were detectable even at 23°C). Subsequently in 1989, Vinogradov’s group discovered the presence of extremely fast-relaxing SQ signals in the complex I segment of the respiratory chain in the tightly coupled bovine heart submitochondrial particles (SMP). This SQ species was found to be uncoupler- and rotenone-sensitive. These investigators suggested a possible spin-spin interaction between SQ and cluster N2 [2]. Our following collaborative studies [9,30] revealed the existence of two distinct protein-bound SQ species. They can be distinguished as fast relaxing SQNf (PI/2 > 150 mW at 40 K) and slow relaxing SQNS (PI/2 = 1 ~ 10 mW at 40 K) species. These SQ species also differ in their sensitivity to both uncouplers and rotenone, which suggested that their locations and functional roles are different from each other. It was proposed that SQNf is involved in the proton pumping (as described below) while SQNs functions as a converter between one and two electron transfer pathways in complex I [20]. Both SQNf and SQNs species were shown to be anionic forms at a pH close to neutral [34].
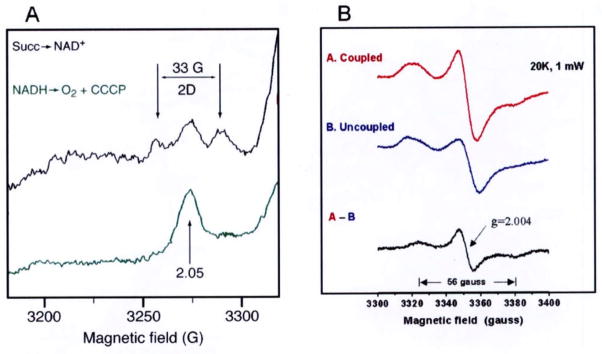
Because of the 33 gauss splitting of the gz peak of cluster N2 and very fast spin relaxation of the , the spin-coupling between SQNf and N2 was suggested. However, without crystallographic information, experimental proof for their direct spin-spin interaction could not be obtained for some time. A detailed profile of SQNf spectra was obtained as the difference between coupled and uncoupled submitochondrial particles under the aerobic steady state with added NADH. Finally, we detected a 56 gauss splitting of the g = 2.004 SQNf signal < 25 K (Fig. 2B) [35] in addition to the 33 gauss splitting of the g// = 2.05 peak of cluster N2 observed < 16 K (Fig. 2A) [9,30]. We analyzed these spin-spin interaction spectra with a computer simulation program which includes contribution from both exchange and dipolar-couplings as well as the g-strain effect. We arrived at the following conclusions: (i) cluster N2 and SQNf are directly spin-coupled, and their center-to-center distance was calculated to be 12Å; (ii) The vector connecting cluster N2 and SQNf is close to the membrane plane direction, and SQNf is located only 5Å deeper than N2 in the membrane normal direction [17,35].
Fig. 2.
(A) The 33 gauss splitting of the g// = 2.05 signal of cluster N2 [9] and (B) the 56 gauss splitting of the gx,y,z = 2.004 signal of the protein-bound QNf [35].
All of these results indicated that SQNf is directly involved in the energy coupling mechanism in complex I, but this mechanism could not be readily explained by the classic Mitchell’s chemiosmotic loop mechanism [10,11]. Thus, Ohnishi and Salerno proposed a novel complex I proton-pump mechanism [17]. In this hypothesis, the equilibrium between two conformational states of the QNf-binding protein is uniquely dependent on the . Since ΔpH < 0.5 in the respiring mitochondria, the membrane potential (Δψ) is the dominant component of [16]. Therefore we focused only on Δψ for the simplicity of the model. The physical shift between the two conformational states changes the thermodynamic stability of SQNf, thereby gating the proton pump. This proton pump is connected to the positive side of the membrane via a proton-well, as originally proposed by Mitchell [11].
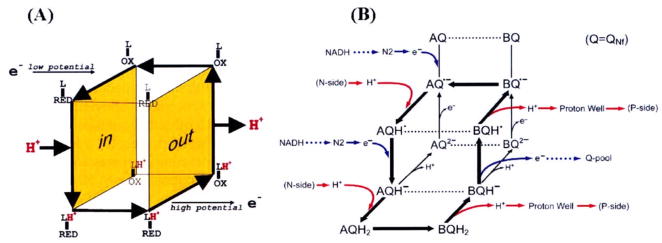
A simpler expression of “cubic model” for the proton pump in cytochrome c oxidase [31] and our conceptually-related “open-box model” of complex I proton pump [17] are shown side by side in Fig. 3(A) and (B). The latter was developed as reported [18,21,22], independent from the cytochrome c oxidase model. Since there is no proton carrier such as Q in cytochrome c oxidase, it has 2 redox states (oxidized and reduced) the redox-cofactor (electron carrier) is linked with a proton accepting pump-site (L). In our model for complex I, the redox component (quinone) functions simultaneously as an electron and proton carrier. The SQ stability responds critically to the transmembrane Δψ, thereby playing a role as a gating component for the proton pump in complex I. In Fig. 3B, the “A state” represents the input-state conformation, where electrons equilibrate with the low potential side of the coupling site and protons equilibrate with the N-side of the membrane. The “B state” signifies the output-state conformation, where electrons equilibrate with the high potential Q-pool (via QNs) and protons equilibrate with the P-side of the membrane via a proton well.
Fig 3.
(A) Simple cubic model of the proton pump in cytochrome c oxidase (2 redox states of the gating component; oxidized and reduced state) and (B) Open box model of complex I (3 redox states; oxidized, intermediate (SQ), and fully reduced state).
In the priming step of this pump, Q is reduced to by cluster N2, and serves as a “gate” as well as a carrier for the pump. It accepts one electron from the low potential side and 2 protons from the N-side, becoming AQH2. Then a comformational change occurs and AQH2 is converted to BQH2, which is the reduced output state. Then, it releases 2 protons to the P side via the proton well, and donates 1 electron to Q-pool via QNs (which is a n = 1 → n = 2 converter). Our experimental data indicated that the EPR signal of in the presence of Δψ is larger than that of . The latter does not accumulate during turnover, and returns directly to without going through the BQ and AQ states. The return process to the A state must be rather fast. Subsequent cyclic processes involve only 2 redox states, semiquinone ( ) and fully-reduced quinone (QH2) states of the QNf species via QNfH· and QNfH−. They both carry electrons and protons in both the input state A and the output state B (they follow along the route shown by thick arrows). For the oxidation of one molecule of NADH, this cycle turns twice for one NADH molecule (2 electron substrate), 4 protons are vectorially transported by the 2 cycles in this “open-box model”. In both cytochrome c oxidase and complex I systems, thick arrows denote a possible route of redox-linked proton translocation. More detailed mechanism is discussed in [17].
A key assumption in this hypothesis is that the SQ state is stabilized in the presence of Δψ. Using purified bovine heart complex I prepared at Yoshikawa’s laboratory [15], and preparing tightly coupled reconstituted proteoliposomes from phospholipids, we tested this hypothesis. We wanted to confirm that (i) an inside-positive membrane potential (Δψ), is created by electron transfer from NADH to Q, (ii) it would induce stabilization of the SQ EPR signals, and (iii) an uncoupler would diminish the signal.
2. Materials and methods
2.1. Chemicals
DiBAC4(5) (bis-(1, 3-dibutylbarbituric acid) pentamethine oxonol) was purchased from Molecular Probes (Eugene, OR). Sodium cholate and Calbiosorb™ Adsorbent were bought from Calbiochem (La Jolla, CA). All phospholipids were obtained from Avanti Polar Lipids (Alabaster, AL). All other chemicals were purchased from Sigma-Aldrich (St. Louis, MO).
2.2. Purification of complex I
In brief: A mitochondrial preparation was obtained from 1.1 Kg of minced bovine heart muscle. It was homogenized at 11,000 rpm for 5 min in 6 liter of 20 mM sodium phosphate buffer (pH 7.4) using a large scale blender (Nihon Seiki Kaisha Ltd. Japan) followed by centrifugation at 2,230 × g for 20 min. The supernatant was centrifuged at 18,480 × g for 30 minutes. The precipitate was suspended in 40 mM HEPES buffer (pH 7.8) containing 300 mM sucrose. The mitochondrial preparation (21 mg/ml) was solubilized with deoxycholate. The solubilized fraction (the supernatant obtained by centrifugation of the solubilized mitochondrial preparation) was subjected to density gradient centrifugation (40,000 rpm for 12 hours). The complex I fraction was further purified by Poros 20 HQ chromatography. The purified fraction was dialyzed against 40 mM HEPES (pH 7.8) containing 300 mM sucrose.
2.3. Preparation of proteoliposomes
We used a cholate-dialysis method developed by Ragan et al. [25,26] with a slight modification. Octylglucoside, which was used for the reconstitution of proteoliposomes of Yarrowia lipolytica complex I [5], could not be used because it denatured bovine heart complex I (Yoshikawa et al. unpublished result). We evaporated a total of 100 mg of phospholipids (phosphatidylcholine 75%, phosphatidylethanolamine 20% and cardiolipin 5%; all in chloroform) and suspended it with a 10 ml buffer solution containing 2.5% sodium cholate, 15 mM Na2SO4 and 5 mM HEPES (pH 7.5). After sonicating for a few minutes in a bath sonifier (until the suspension became clear), 2 mg bovine heart complex I was added. Then we dialyzed the mixture at 4°C to remove cholate with a 2 liter dialysis solution (15 mM Na2SO4, and 5 mM HEPES; pH 7.5). After several hours, we exchanged the dialysis solution, and added 50 ml of the Calbiosorb™ Adsorbent suspension to the 2 liter dialysis solution (to facilitate the removal of cholate) and dialyzed it overnight. We could tell whether cholate was removed or not by shaking the dialysis solution. When it was removed, the dialysis solution did not form soapy bubbles.
2.4. Measurement of membrane potential
A fluorescent dye, DiBAC4(5), which is sensitive to the membrane potential was used at 2 μM concentration. A Hitachi/Perkin-Elmer fluorescence spectrophotometer Model 650 was used with the excitation at 595 nm and emission at 615 nm. When the membrane potential was established, the fluorescence increased, but when the potential was dissipated, the fluorescence decreased.
2.5. Assay for NADH oxidation and the measurement of respiratory control
The experiments were performed in a 1 ml cuvette which was placed in a Hitachi Model 557 dual-beam spectrophotometer (with a built-in magnetic stirring device) set at the two wavelength mode (340–420 nm) for assaying NADH oxidation. Temperature was kept at 30 ± 0.1°C by circulating water from a thermostatic water bath. First, proteoliposomes and 100 μM decylubiquinone (DBQ) or Q1 were mixed in the cuvette. Then, 50 μM NADH was added and the decrease in NADH concentration was monitored. The activity of NADH oxidation was determined from the slope of the recording. Then, the uncoupler (CCCP) was added. From the ratio between the slopes after and before the CCCP addition, the “respiratory control” ratio was calculated.
2.6. EPR samples for SQ signal measurement in the presence of the NADH-induced membrane potential
We added 0.3 ml of proteoliposome suspension to a EPR quartz tube (O.D. diameter 4 mm). First, the electron accepter (final concentration of 400 μM Q1) was added and the mixture was incubated for 2 minutes at 20°C. Then, 200 μM NADH was added. After 30 seconds, the tube was frozen with a 2-methylbutane solution cooled in dry ice.
2.7. EPR measurements
Semiquinone EPR signals were recorded by Bruker EMXmicro spectrometer (X-band; 9.44 GHz). The cavity temperature was kept at 180K using a nitrogen flow cryostat. Other EPR conditions: microwave power, 2 mW; scanning range, 100 gauss, modulation amplitude, 6 gauss; conversion time and time constant, 81.92 ms. Semiquinone signals were detected at around g = 2.004 with a peak-to-peak linewidth of 7.4 gauss.
3. Results and discussion
3.1. Measurements of NADH oxidation and uncoupling by CCCP
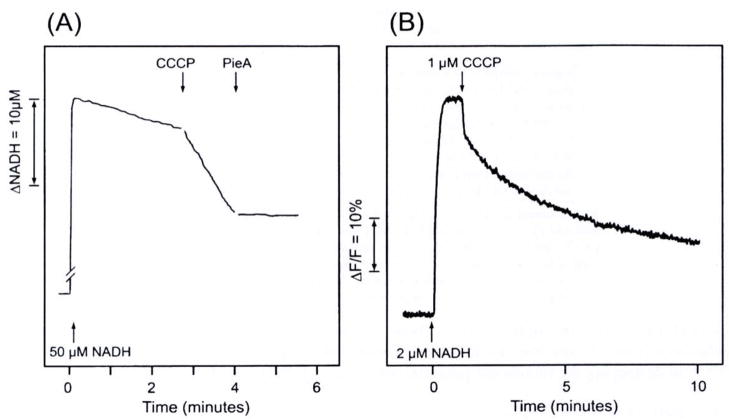
Figure 4(A) shows NADH oxidation by proteoliposomes in the presence of DBQ (an analogue of coenzyme Q10). Since coenzyme Q10 is insoluble in aqueous solution, we used its analogue for in vitro experiments. The respiration was enhanced 5.8 times by the addition of an uncoupler (0.3 μM CCCP). This indicates that electron transfer and proton transport was tightly coupled, and therefore, when uncoupled by CCCP, the rate of electron transfer was increased. In analogy with the mitochondrial respiratory control, this ratio may be regarded as the “Respiratory Control” ratio. The uncoupled respiration was inhibited by 0.5 μM piericidin A by 99%. These results demonstrated that in our reconstituted proteoliposomes, electron transfer and proton transport are tightly coupled, and in that respect, the system is similar to mitochondria or coupled submitochondrial particles. The tightly coupled proteoliposomes would serve as an excellent model system to study the function and mechanism of complex I.
Fig. 4.
(A) The measurement of NADH oxidation (340–420nm). Proteoliposomes (containing 0.003 mg complex I and 0.15 mg phospholipids) were suspended in an 1 ml buffer solution (15 mM Na2SO4, and 5 mM HEPES; pH 7.5) and the suspension was incubated with 100 μM DBQ for 3 minutes. Concentrations of added substances: 50 μM NADH, 0.4 μM CCCP, and 0.5 μM piericidin A (Pie. A). Temperature: 30°C. (B) Fluorescence measurement (excitation 595 nm, emission 615 nm) of the formation of an inside-positive membrane potential with electron transport. The potential was formed with the addition of 2 μM NADH, but it was destroyed by 1 μM gCCCP. The suspension contained 0.003 mg complex I/ml, 2 μM DBQ, 2μM DiBAC4,(5), 15mM Na2SO4, and 5 mM HEPES (pH 7.5). Temperature: 25°C.
3.2. Formation of membrane potential was detected by a potential-sensitive dye
Figure 4(B) shows that the membrane potential formation by the electron transport from NADH to DBQ was detected by a fluorescent dye, DiBAC4(5). As shown in the figure, the potential was dissipated by the addition of the uncoupler, CCCP. These results confirmed that the inside-positive membrane potential was formed by the inward translocation of protons coupled with NADH oxidation. The efficacy of CCCP in destroying the membrane potential was clearly demonstrated.
3.3. Comparison between DBQ andQ1
Table 1 shows differences between DBQ and Q1 as electron acceptors. Although Q1 was found to be a good electron acceptor, DBQ was slightly better than Q1 in that the former provided a higher rate of NADH oxidation and a higher respiratory control ratio with CCCP. The degree of inhibition by piericidin A was also higher with DBQ. However, we found that the size of SQ EPR signals in the proteoliposome system was greater with Q1 than with DBQ (almost doubled; data not shown). Therefore, for the following EPR study, we used Q1.
Table 1.
Comparison between Q1 and DBQ.
| Q1 | DBQ | |
|---|---|---|
| (A) Original complex I suspension. | ||
| (a) Rate of NADH oxidation (μmoles mg−1 min−1) | 5.0 | 7.9 |
| (b) Inhibition (%) by piericidin A | 98.5 | 99.0 |
| (B) Proteoliposomes suspension. | ||
| (a) Rate of NADH oxidation | 0.32 | 0.43 |
| (b) Rate after CCCP addition | 0.83 | 2.4 |
| (c) Respiratory control ratio | 2.6 | 5.7 |
| (d) Inhibition (%) by piericidin A | 88.5 | 99.0 |
Experimental conditions: The buffer solution contains 50 μM NADH, 100 μM Q, or DBQ, 15 mM Na2SO4 and 5 mM HEPES (pH. 7.5), 30°C. (A) 3 μg/ml Complex I, 0.1 mg/ml asolectin and 0.1 % dodecyl maltoside were added to the buffer. (B) Reconstituted proteoliposomes (protein concentration: 1.4 mg/ml; phospholipids: 70 mg/ml) was suspended in the same buffer solution as described before.
3.4. Δψ dependent enhancement of semiquinone EPR signals during the NADH-induced respiration
Figure 5 shows experiments with 200 μM NADH and 400 μM Q1. The black trace indicates the experiment without uncoupler (CCCP), and the red trace the experiment with 30 μM CCCP. The semiquinone EPR signal was the result of NADH oxidation by Q1. The result also showed that with the addition of CCCP, the SQ signal decreased by about 60%.
Fig. 5.
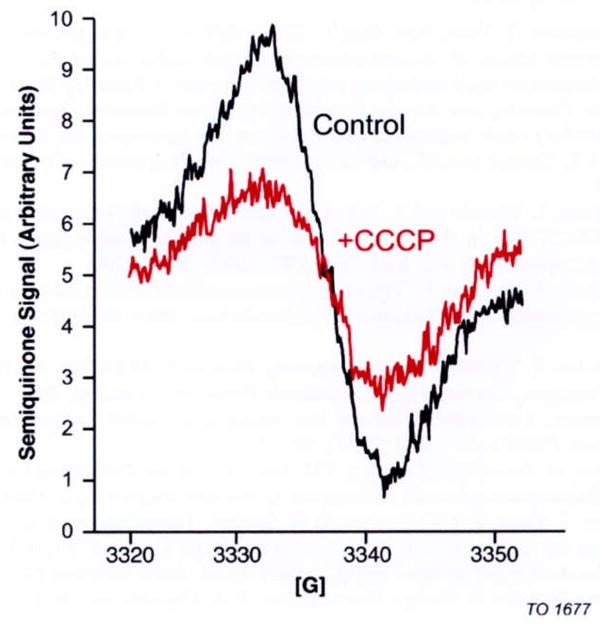
EPR signals of SQ in the steady state of complex I proteoliposomes (1.4 mg complex I/ml). EPR spectra were recorded at the sample temperature of 180 K. The black trace shows the SQ signal in the coupled state and the red trace in the uncoupled state (+30 μM CCCP). The proteoliposomes were suspended in the 15 mM solution of Na2SO4, 5 mM HEPES buffer (pH 7,5) and 400 μM Q1 at 20°C. Then, 200 μM NADH was added, and 30 seconds later, the mixture was frozen at −80°C See text for the EPR conditions.
4. Conclusions
Our proteoliposomes could build up an inside-positive membrane potential coupled with NADH → DBQ electron transport as evidenced by the measurement with a membrane potential dye, DiBAC4(5). The tightness of the coupling was shown by a high “Respiratory Control” ratio of 5.8. The formation of the semiquinone signals during steady state electron transfer from NADH to Q1 were detected by EPR measurement at 180 K. When the membrane potential was destroyed with the uncoupler (CCCP), the SQ signals were decreased. These data strongly support the reaction mechanism proposed by Ohnishi and Salerno, in which the SQ state would be stabilized by an inside-positive membrane potential [17]. It is of special note that each step of this hypothetical mechanism can be experimentally tested. We are currently conducting thermodynamic studies of the bound Q species in this model system. To further refine this hypothesis, crystallographic structural information of the whole complex I is needed.
Acknowledgments
This study was supported in part by NIH Grant GM30736 to T.O. and by Grant-in-aid for Scientific Research on Priority Areas and for 21 st Century Center of Excellence Program from the Ministry of Education, Culture, Sports, Science and Technology, Japan to S. Y. The authors thank Dr. J.C. Salerno for stimulating discussions.
Abbreviations
- CCCP
carbonyl cyanide 3-chrolophenylhydrazone
- DBQ
decylubiquinone
- DiBAC4(5)
bis-(1, 3-dibutylbarbituric acid) pentamethine oxonol
- EPR
electron paramagnetic resonance
- Fe/S
iron-sulfur
References
- 1.Brandt U. Energy Converting NADH: Quinone Oxidoreductase (Complex I) Annu Rev Biochem. 2006;75:69–92. doi: 10.1146/annurev.biochem.75.103004.142539. [DOI] [PubMed] [Google Scholar]
- 2.Burbaev DS, Moroz IA, Kotlyar AB, Sled VD, Vinogradov AD. Ubisemiquinone in the NADH-ubiquinone reductase region of the mitochondrial respiratory chain. FEBS Lett. 1989;254:47–51. [Google Scholar]
- 3.Carroll J, Fearnley IM, Shannon RJ, Hirst J, Walker JE. Analysis of the subunit composition of complex I from bovine heart mitochondria. Mol Cell Proteomics. 2003;2:117–126. doi: 10.1074/mcp.M300014-MCP200. [DOI] [PubMed] [Google Scholar]
- 4.Cramer WA, Zhang H, Yan J, Kurisu G, Smith JL. Evolution of photosynthesis: Time-independent structure of the cytochrome b6f complex. Biochemistry. 2004;43:5921–5929. doi: 10.1021/bi049444o. [DOI] [PubMed] [Google Scholar]
- 5.Dröse S, Galkin AS, Brandt U. Proton pumping by complex I (NADH: ubiquinone oxidoreductase) from Yarrowia lipolytic a reconstituted into proteoliposomes. Biochim Biophys Acta. 2005;1710:87–95. doi: 10.1016/j.bbabio.2005.10.001. [DOI] [PubMed] [Google Scholar]
- 6.Galkin AS, Grivennikova VG, Vinogradov AD. H+/2e stoichiometry in NADH-quinone reductase reactions catalyzed by bovine heart submitochondrial particles. FEBS Lett. 1999;451:157–161. doi: 10.1016/s0014-5793(99)00575-x. [DOI] [PubMed] [Google Scholar]
- 7.Hirst J, Carroll J, Fearnley IM, Shannon RJ, Walker JE. The nuclear encoded subunits of complex I from bovine heart mitochondria. Biochim Biophys Acta. 2003;1604:135–150. doi: 10.1016/s0005-2728(03)00059-8. [DOI] [PubMed] [Google Scholar]
- 8.Hunte C, Palsdottir H, Trumpower BL. Proton motive pathways and mechanisms in the cytochrome bc1 complex. FEBS Letters. 2003;545:39–46. doi: 10.1016/s0014-5793(03)00391-0. [DOI] [PubMed] [Google Scholar]
- 9.Magnitsky S, Toulokhonova L, Yano T, Sled VD, Hägerhälll C, Grivennikova VG, Burbaev DS, Vinogradov AD, Ohnishi T. EPR characterization of ubisemiquinones and iron-sulfur cluster N2, central components of the energy coupling in the NADH-ubiquinone oxidoreductase (complex I) in situ. J Bioenerg Biomembr. 2002;34:193–208. doi: 10.1023/a:1016083419979. [DOI] [PubMed] [Google Scholar]
- 10.Mitchell P. Chemiosmotic Coupling and Energy Transduction. Glynn Research; Bodmin: 1968. [Google Scholar]
- 11.Mitchell P. Keilin’s respiratory chain concept and its chemiosmotic consequences. Science. 1979;206:1148–1159. doi: 10.1126/science.388618. [DOI] [PubMed] [Google Scholar]
- 12.Moser CC, Farid TA, Chobot SE, Dutton PL. Electron tunneling chains of mitochondria. Biochim Biophys Acta. 2006;1757:1096–1109. doi: 10.1016/j.bbabio.2006.04.015. [DOI] [PubMed] [Google Scholar]
- 13.Nakamaru-Ogiso E, Yano T, Ohnishi T, Yagi T. Characterization of the iron-sulfur cluster coordinated by a cysteine cluster motif (CXXCXXXCX27C) in the Nqo3 subunit in the proton- translocating NADH-quinone oxidoreductase (NDH-1) of Thermus thermophilus HB-8. J Biol Chem. 2002;277:1680–1688. doi: 10.1074/jbc.M108796200. [DOI] [PubMed] [Google Scholar]
- 14.Nakamaru-Ogiso E, Yano T, Yagi T, Ohnishi T. Characterization of the Iron-Sulfur Cluster N7 (N1 c) in the Subunit NuoG of the Proton-translocating NADH-quinone Oxidoreductase from Escherichia coli. J Biol Chem. 2005;280:301–307. doi: 10.1074/jbc.M410377200. [DOI] [PubMed] [Google Scholar]
- 15.Shinzawa-Ito Nakashima K, Yoshikawa S, Bioenerg J. Biomembr. 2002;34:11–19. doi: 10.1023/a:1013862502185. [DOI] [PubMed] [Google Scholar]
- 16.Nicholls DG, Ferguson SJ. Bioenergetics. Vol. 3. Academic Press, Inc; London: 2002. [Google Scholar]
- 17.Ohnishi T, Salerno JC. Conformation-driven and semiquinone-gated proton-pump mechanism in the NADH-ubiquinone oxidoreductase. FEBS Letters. 2005;579:45–55. doi: 10.1016/j.febslet.2005.06.086. [DOI] [PubMed] [Google Scholar]
- 18.Ohnishi T, Salerno JC. In: Iron-Sulfur Proteins. Spiro TG, editor. Wiley Publishing Co; New York: 1982. pp. 285–327. [Google Scholar]
- 19.Ohnishi T. Iron-sulfur clusters/semiquinones in Complex I. Biochim Biophys Acta. 1998;1364:186–206. doi: 10.1016/s0005-2728(98)00027-9. [DOI] [PubMed] [Google Scholar]
- 20.Ohnishi T, Johnson JJE, Yano T, LoBrutto R, Widger RW. Thermodynamic and EPR studies of slowly relaxing ubisemiquinone species in the isolated bovine heart complex I. FEBS Letters. 2005;579:500–506. doi: 10.1016/j.febslet.2004.11.107. [DOI] [PubMed] [Google Scholar]
- 21.Ohnishi T. Studies on the mechanism of site I energy conservation. Eur J Biochem. 1976;64:91–103. doi: 10.1111/j.1432-1033.1976.tb10277.x. [DOI] [PubMed] [Google Scholar]
- 22.Ohnishi T. In: Membrane Proteins in Energy Transduction. Capaldi RA, editor. Marcel Dekker, Inc; New York: 1979. pp. 1–87. [Google Scholar]
- 23.Okamura MY, Paddock ML, Graige MS, Feher G. Proton and electron transfer in bacterial reaction centers. Biochim Biophys Acta. 2000;1458:148–163. doi: 10.1016/s0005-2728(00)00065-7. [DOI] [PubMed] [Google Scholar]
- 24.Pohl T, Uhlmann M, Kaufenstein M, Friedrich T. Lambda Red-mediated mutagenesis and efficient large scale affinity purification of the Escherichia coli NADH:ubiquinone oxidoreductase (complex I) Biochemistry. 2007;46:10694–10702. doi: 10.1021/bi701057t. [DOI] [PubMed] [Google Scholar]
- 25.Ragan CI, Racker E. resolution and Reconstitution of the mitochondrial electron transport system. J Biol Chem. 1973;248:6876–6884. [PubMed] [Google Scholar]
- 26.Ragan CI, Hinkle PC. Ion transport and respiratory control in vesicles formed from reduced nicotine amide adenine dinucleotide coenzyrne Q reductase and phospholipids. J Biol Chem. 1975;250:8472–8476. [PubMed] [Google Scholar]
- 27.Sazanov LA, Hinchliffe P. Structure of the Hydrophilic Domain of Respiratory Complex I from Thermus thermophilus. Science. 2006;311:1430–1436. doi: 10.1126/science.1123809. [DOI] [PubMed] [Google Scholar]
- 28.Sazanov LA. Respiratory complex I: mechanistic and structural insights provided by the crystal structure of the hydrophilic domain. Biochemistry. 2007;46:2275–2288. doi: 10.1021/bi602508x. [DOI] [PubMed] [Google Scholar]
- 29.Suzuki H, King TE. Evidence of an ubisemiquinone radical(s) from the NADH-ubiquinone reductase of the mitochondrial respiratory chain. J Biol Chem. 1983;258:352–358. [PubMed] [Google Scholar]
- 30.Vinogradov AD, Sled VD, Burbaev DS, Grivennikova VG, Moroz IA, Ohnishi T. Energy-dependent complex I-associated ubisemiquinones in submitochondrial particles. FEBS Lett. 1995;370:83–87. doi: 10.1016/0014-5793(95)00803-h. [DOI] [PubMed] [Google Scholar]
- 31.Wikström M. Cytochrome c oxidase: 25 years of the elusive proton pump. Biochim Biophys Acta. 2004;12:241–247. doi: 10.1016/j.bbabio.2003.07.013. [DOI] [PubMed] [Google Scholar]
- 32.Wikström M. Two protons are pumped from the mitochondrial matrix per electron transferred between NADH and ubiquinone. FEBS Lett. 1984;169:300–304. doi: 10.1016/0014-5793(84)80338-5. [DOI] [PubMed] [Google Scholar]
- 33.Yagi T, Matsuno-Yagi A. The Proton-Translocating NADH-Quinone Oxidoreductase in the Respiratory Chain: The Secret Unlocked. Biochemistry. 2003;42:2266–2274. doi: 10.1021/bi027158b. [DOI] [PubMed] [Google Scholar]
- 34.Yano T, Magnitsky S, Ohnishi T. Characterization of the complex I-associated ubisemiquinone species: toward the understanding of their functional roles in the electron/proton transfer reaction. Biochim Biophys Acta. 2000;1459:299–304. doi: 10.1016/s0005-2728(00)00164-x. [DOI] [PubMed] [Google Scholar]
- 35.Yano T, Dunham WR, Ohnishi T. Characterization of the Δ -sensitive ubisemiquinone species (SQNf) and the interaction with cluster N2: new insight into the energy-coupled electron transfer in complex I. Biochemistry. 2005;44:1744–1754. doi: 10.1021/bi048132i. [DOI] [PubMed] [Google Scholar]
- 36.Zhu Z, Gunner MR. Energetics of Quinone-Dependent Electron and Proton Transfers in Rhodobacter spheroides, Photosynthetic Reaction Centers. Biochemistry. 2005;44:82–96. doi: 10.1021/bi048348k. [DOI] [PubMed] [Google Scholar]