Abstract
α-Synuclein (αSyn) is a small cytosolic protein of unknown function, which is highly enriched in the brain. It is genetically linked to Parkinson’s disease (PD) in that missense mutations or multiplication of the gene encoding αSyn cause early-onset familial PD. Furthermore, the neuropathological hallmarks of both sporadic and familial PD, Lewy bodies and Lewy neurites, contain insoluble aggregates of αSyn. Several studies have reported evidence that αSyn can inhibit phospholipase D (PLD), which hydrolyzes phosphatidylcholine to form phosphatidic acid and choline. Although various hypotheses exist regarding the roles of αSyn in health and disease, no other specific biochemical function for this protein has been reported to date. Because PLD inhibition could represent an important function of αSyn, we sought to extend existing reports on this interaction. Using purified proteins, we tested the ability of αSyn to inhibit PLD activity in cell-free assays. We also examined several cell lines and transfection conditions to assess whether αSyn inhibits endogenous or overexpressed PLD in cultured mammalian cells. In yeast, we extended our previous report of an interaction between αSyn and PLD-dependent phenotypes, for which PLD activity is absolutely necessary. Despite testing a range of experimental conditions, including those previously published, we observed no significant inhibition of PLD by αSyn in any of these systems. We propose that the previously reported effects of αSyn on PLD activity could be due to increased endoplasmic reticulum-related stress associated with αSyn overexpression in cells, but are not likely due to a specific and direct interaction between αSyn and PLD.
Parkinson’s disease (PD1) is a debilitating neurodegenerative disorder that affects over one million people in the United States (for review, see (1)). Although more than 90% of PD cases are believed to occur sporadically, several genes are known to cause familial forms of PD. Among these, point mutations or increased dosage of the α-synuclein (αSyn) gene are known to cause rare cases of early-onset familial PD (2-7). The αSyn protein is also the major component of Lewy bodies and Lewy neurites, the insoluble aggregates that are neuropathological hallmarks of both familial and sporadic PD (8). Thus, dysregulation of αSyn at either the genetic or protein level can contribute to PD-type neurodegeneration.
αSyn is a small, highly conserved cytosolic protein of unknown function. It is enriched in the brain, particularly at presynaptic terminals. Proposed functions of αSyn include lipid binding, regulation of membrane composition, and regulation of neurotransmitter release and/or of the reserve pool of synaptic vesicles (9-15). Given the hypothesis that loss of axonal terminals in the striatum may precede the death of nigral neurons in PD (1, 16), these observations suggest that the dysfunction of αSyn at the synapse could be an early event in the pathogenesis of PD.
Phospholipase D (PLD) is a membrane-associated enzyme that hydrolyzes phosphatidyl-choline to form phosphatidic acid (PA) and choline. PA is an essential metabolic intermediate and an intracellular signaling molecule that can be further hydrolyzed into diacylglycerol, another important signaling molecule (for review, see (17)). Mammalian cells express two PLD isoforms, PLD1 and PLD2. They are regulated by distinct cellular mechanisms but perform a similar hydrolytic reaction. In cell-free systems, PLD1 requires an activator such as Rho or ARF GTPases, while PLD2 is highly active in these systems (18, 19). The precise function of PLD in the cell has not yet been determined, though proposed functions include roles in exocytosis, endocytosis, and intracellular signaling (16, 20, 21).
In 1998, in vitro studies suggested that αSyn and its close family member, β-synuclein (βSyn), could act as inhibitors of PLD2 (22). Three other labs extended these findings over the next six years, suggesting that αSyn can inhibit PLD activity in mammalian cells (23), in yeast (24), and in cell-free assays (25). Although several hypotheses exist regarding the physiological and pathological roles of αSyn in the brain, no other specific biochemical function for αSyn has been proposed to date. Furthermore, if PLD regulation is a physiological function of αSyn, then PLD dysregulation due to gain or loss of αSyn function could be an early event in the pathogenesis of PD and other synucleinopathies.
Here, we sought to further explore the mechanism of the interaction between αSyn and PLD. We used cell-free and cell-based systems to directly assay PLD activity in the presence or absence of αSyn. In yeast, we extended our previously published genetic findings on αSyn inhibition of PLD. These various approaches did not yield evidence that αSyn significantly inhibits PLD. Based on our data, as well as previous work showing that αSyn expression can induce cytotoxicity associated with endoplasmic reticulum (ER) stress, we suggest that the previously published effects of αSyn on PLD activity may be nonspecific or attributable to general ER stress, rather than a direct and physiological inhibition of PLD by the αSyn protein.
EXPERIMENTAL PROCEDURES
Materials
Chromatographically enriched human PLD1b (hPLD1b) and 6-his-tagged human PLD2a (hPLD2a) were generated as previously described (26, 27) and tested for activity using an exogenous substrate assay (17, 28). Recombinant αSyn protein was generously provided by P.T. Lansbury (29) and by J.M. George (25). Following resuspension in phosphate buffered saline, the protein was filtered through a 0.22 μm syringe filter followed by a 100,000 MWCO centrifugal filter. The filtered solution was probed by SDS-PAGE and Coomassie stain to confirm the monomeric nature of the recombinant protein preparation. Lipids, including phosphatidyl-butanol, were purchased from Avanti Polar Lipids (Alabaster, AL). 3H-Oleic acid was purchased from PerkinElmer Life Science Products (Newtown, CT). The αSyn expression plasmid was provided by R. Sharon. Unless otherwise stated, all other reagents were purchased from Sigma-Aldrich (St. Louis, MO).
Cell-Based PLD Activity Assay
Endogenous PLD activity was measured as described previously (30). Briefly, 24 hours before the assay, HEK 293, HeLa, or undifferentiated PC12 cells were transfected as indicated using LipofectAMINE 2000 (Invitrogen, Carlsbad, CA). Cells were labeled with 3H-oleic acid (10 μCi/mL) overnight in serum-free medium supplemented with 0.25 mg/mL fatty acid free bovine serum albumin (BSA). On the day of the assay, some cells were pre-incubated for 5 minutes in serum-free medium containing the PLD inhibitor VU0155056 (2 μM) (27). Cells were then stimulated for 30 min with serum-free medium containing 1 μM phorbol-12-myristate-13-acetate (PMA) with 0.3% butanol and/or VU0155056, as indicated. Lipids were extracted using chloroform : methanol : HCl 0.1 N (1:1:1), then spotted onto silica-coated TLC plates. The plates were developed in chloroform : methanol : acetic acid : acetone : water (50:10:10:20:5), and the radioactivity was visualized on autoradiographic film with a Kodak Transcreen LE enhancing screen, scanned, and quantified using Quantity One software (BioRad Laboratories, Hercules, CA). A phosphatidylbutanol standard was run on each plate and visualized with sublimed iodine (17).
Cell-Free PLD Activity Assay
PLD activity was assayed as described previously (28). Briefly, purified hPLD2a (15 nM final concentration) was added to reaction buffer (50 mM HEPES pH 7.5, 80 mM KCl, 3 mM EGTA, 0.1 mM DTT, 4.5 mM MgCl2, 4.5 mM CaCl2, 10 μM GTPγS) containing lipid vesicles and 3H-PC (total DPPC : POPE : PI(4,5)P2 : cholesterol at a molar ratio of 10:100:6.2:1.4) in a total volume of 60 μl per assay tube. Various concentrations of αSyn were added to some reactions as indicated, and the PLD inhibitor VU0155056 (20 μM) was added to other reactions as a control. Reactions were incubated at 37°C for 30 min, then terminated with 200 μl trichloroacetic acid (10% v/v) and 100 μl BSA (10% w/v). Free 3H-choline released by hPLD2a activity was measured by scintillation counting. Similar assays were performed using purified hPLD1b.
Yeast Plasmids and Strains
The sec14-1, cki1Δ, and sec14-1 cki1Δ strains were described previously (24). Here, we generated the sec14-1 cki1Δ spo14Δ strain by replacing SPO14 with a KanMX cassette by homologous recombination in the sec14-1 cki1Δ strain. Colony PCR was used to verify the gene disruption. A SPO14 entry clone (pDONR221) and the Advanced Gateway destination vectors pAG413GPD-ccdB or pAG416GPD-ccdB (31) were used in a Gateway® LR reaction (Invitrogen) to construct pAG413GPD-SPO14 and pAG416GPD-SPO14, respectively. To construct the pAG413GPD-αSyn plasmid, an αSyn entry clone was used in a Gateway® LR reaction with the pAG413GPD-ccdB destination vector.
Yeast Transformation and Spotting Assays
Yeast cultures were maintained according to standard protocols (32). We used the PEG/lithium acetate method to transform yeast with DNA (33). Yeast cells carrying the αSyn and/or SPO14 plasmids were grown overnight at 30°C in liquid media containing glucose until they reached log or mid-log phase. Cultures were then normalized for OD600, serially diluted and spotted onto synthetic solid media containing glucose and lacking either uracil (for pAG416GPD plasmids) or histidine (for pAG413GPD plasmids) and were grown at 23°C or 37°C for 2-3 days.
Statistical Analyses
For each assay, at least three independent experiments were conducted, each in triplicate. Results were analyzed by one-way ANOVA with Tukey’s post-hoc tests, as indicated in the figure legends.
RESULTS
αSyn Does Not Inhibit PLD Activity in Living Cells
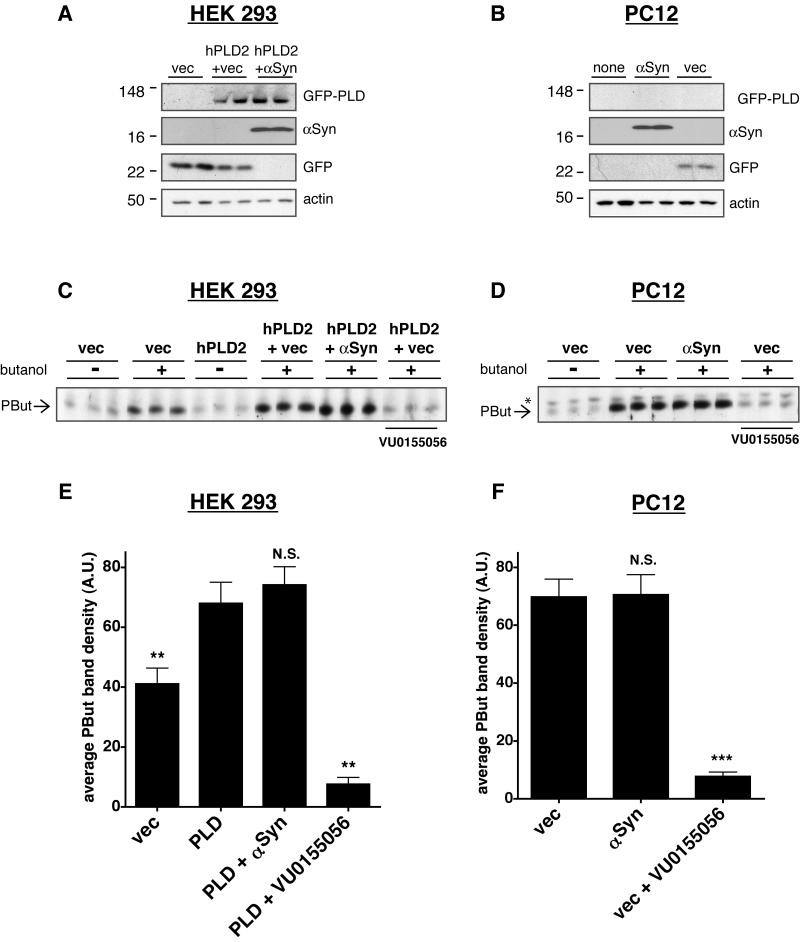
A previous report suggested that αSyn can inhibit PLD activity in living cells (23). We sought to repeat and extend these findings to determine the extent to which such inhibition could be relevant to PD pathogenesis. We used two different cell lines: HEK 293 and PC12 cells. Both cell lines express low or undetectable levels of αSyn (Figures 1A and 1B), and both show moderate levels of endogenous PLD activity (Figures 1C and 1D). The HEK 293T cells are similar to the HEK 293 cells that were used in the previously published work (23). Cells were transfected with αSyn and/or EGFP-hPLD2, or with empty EGFP vector as a negative control. PC12 cells were not transfected with EGFP-hPLD2 because these cells are known to have high levels of endogenous PLD2 activity (34). Transfection efficiency for both cell lines was monitored in two ways: under a fluorescent microscope, >50% of the cells in GFP-transfected wells fluoresced green (data not shown), and Western blot results showed high levels of expression (Figures 1A and 1B).
Figure 1. αSyn does not inhibit PLD activity in mammalian cells.
Cells were transiently transfected with empty EGFP vector as a control (vec), αSyn, or GFP-hPLD2, and PLD activity was then assayed. Some culture wells were used for immunoblots; representative results show αSyn and GFP-hPLD2 protein expression levels in (A) HEK 293 cells and (B) PC12 cells. First panel: GFP-hPLD2 visualized using polyclonal rabbit anti-GFP antibody (Invitrogen); PC12 cells were not transfected with GFP-hPLD2. Second panel: monoclonal mouse anti-αSyn (LB509, Santa Cruz Biotechnology); Third panel: EGFP vector control visualized using anti-GFP antibody. Fourth panel: monoclonal mouse anti-actin (Abcam) was used as a loading control. Twenty-four hours following transient transfection as indicated, PLD activity was assayed. The PLD inhibitor VU0155056 (2 μM; (27)) was used as a positive control for inhibition. Representative images are shown from (C) HEK 293 cells and (D) PC12 cells; signal from the PLD product phosphatidylbutanol is indicated with an arrow and a background band is indicated with an asterisk. Results from three independent experiments, each conducted in triplicate, were quantified and graphed (E and F). Graphs represent means + SEM. Phosphatidylbutanol signal was quantified in each lane, and background-subtracted results were compared by one-way ANOVA with Tukey’s post-hoc tests. (E) Quantification of results from HEK 293 cells. **, p < 0.01 compared with hPLD2-transfected cells. (F) Quantification of results from PC12 cells. ***, p < 0.001 compared with vector-transfected cells.
Because 1-butanol can replace water in the PLD-catalyzed hydrolysis of PC and is favored 1000-fold over water in this reaction, the addition of a small amount of butanol to the culture medium allows for the production of the PLD-specific product phosphatidylbutanol (PBut), which cannot be metabolized by the cells. Quantification of accumulated PBut levels, therefore, serves as a sensitive and specific indicator of the total PLD activity in the cells within the given time frame. Cells were activated with phorbol-12-myristate-13-acetate (PMA), an inducer of the protein kinase C pathway and, indirectly, of PLD (30). PMA was added to the cells in the presence or absence of 1-butanol, then lipids were extracted in chloroform/methanol and run on a TLC plate to separate the phospholipids. In both cell lines, αSyn did not inhibit the activity of either endogenous or overexpressed PLD (Figure 1). In contrast, the small molecule PLD inhibitor VU0155056 (27) dramatically reduced PLD activity. We conclude that αSyn does not detectably inhibit PLD activity in these cell types.
αSyn Does Not Inhibit PLD Activity in Cell-Free Systems
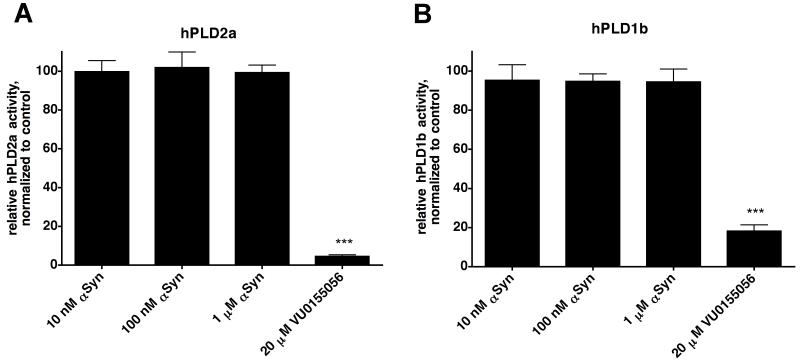
Previous work suggested that αSyn can inhibit PLD activity in cell-free assays (22, 25). Before extending these findings, we first sought to replicate them. We used protocols similar to those used in the published work to assay the activity of hPLD2a (15 nM) in the presence or absence of αSyn at different concentrations. Based on the published findings, we expected to see near-maximal inhibition of hPLD2a activity with 100nM αSyn (22, 25). However, αSyn did not inhibit hPLD2a in our in vitro assays at concentrations up to 1 μM (Figure 2A). For completeness, we also tested the effect of αSyn on the activity of hPLD1b (3 nM), although the published work exploring the inhibition of PLD1 by αSyn indicated that such inhibition was incomplete at best (22, 23). As with hPLD2a, hPLD1b activity was not affected by αSyn in our cell-free assays (Figure 2B).
Figure 2. αSyn does not inhibit PLD in a cell-free assay.
(A) The activity of hPLD2a was assayed in the presence or absence of wild-type human αSyn. Differences between these conditions were not significant, whereas the addition of VU0155056 resulted in a significant decrease in hPLD2a activity. (B) The activity of hPLD1b was assayed with or without wild-type human αSyn. Differences between these conditions were not significant. For all of these experiments, reactions were incubated at 37°C for 30 min, terminated, and free 3H-choline release was quantified by scintillation counting (see Experimental Procedures). The small-molecule PLD inhibitor VU0155056 (20 μM) was used as a control for inhibition in some reactions, as indicated. Background-subtracted values were normalized to the averaged control values and compared by one-way ANOVA with Tukey’s post-hoc tests. Graphs represent means + SEM from 3 independent experiments, total n=9 per condition. ***, p < 0.001 in pairwise comparisons with each αSyn concentration.
Two of the PD-causing mutations in αSyn affect the protein’s lipid binding properties and were shown in one report to have differential effects on αSyn inhibition of hPLD2 (25). Specifically, the authors reported that the A53T mutation increased hPLD2 inhibition, while the A30P mutation did not differ significantly from wild-type protein. In our assays, however, neither A30P nor A53T showed significant inhibition of hPLD2a at the concentrations tested (data not shown), consistent with our findings with the wild-type protein. Thus, in our hands, αSyn did not inhibit hPLD1b or hPLD2a in a cell-free system despite our best efforts to replicate the previously published conditions. These results were unanticipated, and possible explanations are explored in the Discussion.
Effect of αSyn on Yeast PLD
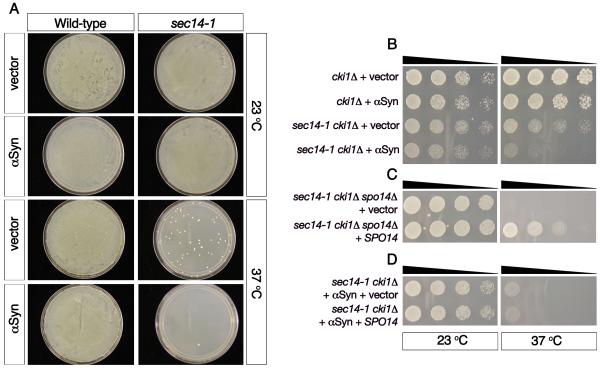
Following the unexpected lack of inhibition of PLD by αSyn in mammalian cells and cell-free assays, we returned to the yeast system in which this inhibition has previously been inferred from the results of genetic interactions. In the yeast Saccharomyces cerevisiae, the SPO14 gene encodes PLD activity (35-37). We first investigated the effect of αSyn overexpression in yeast carrying a mutation in the SEC14 gene, which encodes an essential phosphatidylinositol and phosphatidylcholine transfer protein (38, 39). Yeast cells with a temperature-sensitive mutation in SEC14 (sec14-1) grow normally at the permissive temperature (23°C) but are unable to grow at the restrictive temperature (37°C) (40, 41). Occasionally, sec14-1 cells can acquire “bypass mutations” in one of at least 6 different pathways, conferring the ability to grow at the restrictive temperature (42). In agreement with previously published work (24), expression of low levels of αSyn abolished the sec14-1 bypass mutants’ ability to grow at the restrictive temperature (Figure 3A).
Figure 3. αSyn effects on yeast growth are not rescued by overexpression of PLD.
(A) Yeast cells with a temperature-sensitive mutation in SEC14 (sec14-1) grow normally at 23°C, but are unable to grow at 37°C, unless they acquire various “bypass mutations” (sec14-1, vector, 37°C). As reported previously (24), αSyn expression prevents the ability of the sec14-1 bypass mutants to grow at the restrictive temperature (sec14-1, αSyn, 37°C). (B) Deletion of the choline kinase 1 gene (cki1Δ), allows the sec14-1 mutants to grow at the restrictive temperature in a PLD-dependent manner, and αSyn expression in these cells blocks this rescue effect. (C) Deletion of SPO14 (spo14Δ) in the sec14-1/cki1Δ mutants abolishes their ability to grow at the restrictive temperature. Overexpression of functional SPO14 restores growth to sec14-/cki1Δ/spo14Δ cells (C) but is unable to rescue the αSyn-induced growth defect (D). (n=3)
To determine whether the observed inhibition was PLD-dependent, we focused on the well-characterized sec14 bypass mutant, cki1Δ. Deletion of the choline kinase, CKI1, allows sec14-1 mutants to grow at the restrictive temperature (Figure 3B) in a PLD-dependent manner (Figure 3C) (42), and expressing αSyn in these cells blocked this effect (Figure 3B). We hypothesized that if the effect of αSyn on sec14-1/cki1Δ cells was caused by PLD inhibition, then expressing saturating levels of ectopic PLD should overcome this growth defect. To this end, we created a plasmid to overexpress SPO14 (which encodes the yeast PLD homolog) and verified its functionality by virtue of its ability to restore growth to sec14-1/cki1Δ/spo14 Δ cells (Figure 3C), which express the sec14-1 and cki1Δ mutations and have an additional genomic deletion of SPO14. However, overexpressing this functional SPO14 in the sec14-1/cki1Δ cells was not sufficient to rescue the αSyn-induced growth defect (Figure 3D).
DISCUSSION
To date, the only specific biochemical function that has been proposed for αSyn is a potential inhibition of PLD. Because the normal function of αSyn may be related to its role in PD, we sought to repeat and extend the reported findings regarding this interaction and to explore how it might relate to PD pathogenesis. However, in spite of our efforts to use several independent but complimentary assay systems and to include conditions similar to those previously reported, we were unable to repeat many of the published findings.
In mammalian cells, we observed no significant inhibition of PLD activity by overexpression of human αSyn. We used both the neuronal rat PC12 cell line and the non-neuronal human HEK 293 cell line in order to ensure that any effects we observed were not specific to one particular cell line or species. Furthermore, the HEK 293 cells are similar to the cells used in the previously published work showing αSyn inhibition of cellular PLD (23). Similar to those experiments, we overexpressed both αSyn and hPLD2 in these cells by transient transfection. However, unlike the earlier work, we controlled for possible titration effects by co-expressing two plasmids in all of the experimental conditions. Thus, cells were transfected with αSyn and GFP, or GFP-hPLD2 and GFP, or αSyn and GFP-hPLD2. However, αSyn did not inhibit PLD activity (Figure 1E) in these experiments. We also overexpressed αSyn in PC12 and HeLa cells to determine whether the protein could inhibit the activity of the highly expressed endogenous PLD in these two cell types. As with the HEK 293 cells, we observed no inhibition of PLD by αSyn in either PC12 (Figure 1F) or HeLa cells (data not shown).
Based on one report of a possible direct interaction between αSyn and PLD (23), we attempted to co-immunoprecipitate these two proteins from HeLa cells using antibodies against αSyn or the HA tag on our PLD2 construct. However, we were unable to obtain evidence of co-immunoprecipitation of these proteins (data not shown). In all of our experiments we included a small-molecule PLD inhibitor, VU0155056, to establish the maximal level of inhibition that we could expect to see in these assays. While the PLD inhibitor consistently reduced PLD activity in our assays, overexpression of αSyn did not.
In cell-free assays, we did not observe inhibition of PLD activity upon the addition of recombinant αSyn protein, although we achieved dramatic inhibition with the small-molecule inhibitor VU0155056. Thus, the activity of our hPLD1b and hPLD2a enzymes was sensitive to inhibition under these conditions, in spite of the failure of recombinant αSyn to affect activity levels. We used up to 67-fold molar excess of αSyn compared with our PLD isoenzymes. Based on the published data, we expected this excess of αSyn to yield at least 50% inhibition of hPLD2a in the cell-free assays. However, in our hands αSyn had no effect on hPLD2a or hPLD1b activity. Furthermore, an inhibition that requires greater molar excess would not appear to be physiologically relevant, nor can it be immediately understood with regard to protein-protein interactions in a living cell.
We then turned to experiments in yeast as a tool to explore the physiological relevance of the reported inhibition of PLD by αSyn. Using the sec14-1 temperature-sensitive mutant yeast strain, we showed that αSyn prevented the acquisition of bypass mutations at the restrictive temperature, as previously published (24). Such bypass mutations can be acquired through 6 different pathways (42), among which the well-characterized CKI1 pathway is PLD-dependent (43). Inhibition by αSyn overexpression of all possible bypass mutation pathways implies that this effect is not likely to depend on PLD activity. In the sec14-1 cki1Δ double-mutant strain in which bypass mutations are acquired in a PLD-dependent manner, αSyn prevented growth at the restrictive temperature but this deficit could not be rescued by co-expression of functional Spo14. This result further strengthens our conclusion that αSyn does not directly affect PLD in this system.
In separate work, some of us (A.D.G., S.L.) have recently reported that αSyn accumulation causes endoplasmic reticulum (ER) stress and activation of the unfolded protein response (UPR) secondary to a severe block in vesicular trafficking between the ER and Golgi (44). Intriguingly, it has been found that sec14 bypass mutants are particularly sensitive to ER stress and require an intact UPR for survival (45). Inducing ER stress, either genetically (by mutating the UPR pathway components IRE1 or HAC1) or chemically (by treatment with the N-linked glycosylation inhibitor tunicamycin) abolishes the ability of sec14-1/cki1Δ cells to grow at the restrictive temperature (45), and this deleterious effect is strictly independent of PLD activity. Thus, we propose a new interpretation for the confirmed ability of αSyn to inhibit sec14 bypass mutant growth (24). Rather than a direct inhibition of PLD activity by αSyn, the increased ER stress present in αSyn-expressing cells may be sufficient to prevent the sec14 bypass mutants from growing. This interpretation is consistent with our failure to suppress the effect by overexpressing SPO14 (i.e., PLD).
In light of the variety of assays and conditions we used, we conclude that αSyn is not a robust inhibitor of PLD. From the experiments in cell-free systems we conclude that αSyn does not directly inhibit PLD activity, and from the experiments in yeast and mammalian cells we conclude that αSyn does not inhibit PLD through either a direct or indirect interaction. It may be that previous findings implicating αSyn in PLD inhibition in cells could instead be interpreted as effects of general αSyn cytotoxicity, because overexpression of PLD in these systems does not rescue the effects of αSyn. We further conclude that dysregulation of PLD inhibition is not likely to be involved in the role of αSyn in PD pathogenesis.
ACKNOWLEDGEMENT
We thank Michelle Armstrong, Matthew Hemming, and Tracy Young-Pearse for their advice and assistance.
Footnotes
This work was supported by the Brigham and Women’s Hospital Udall Center for Excellence in Parkinson’s Disease, NIH/NINDS grant NS038375 (DJS).
- αSyn
- α-Synuclein
- βSyn
- β-synuclein
- BSA
- bovine serum albumin
- ER
- endoplasmic reticulum
- hPLD1b
- human phospholipase D 1b
- hPLD2a
- human phospholipase D 2a
- PD
- Parkinson’s Disease
- PA
- phosphatidic acid
- PBut
- phosphatidylbutanol
- PLD
- phospholipase D
- PMA
- phorbol-12-myristate-13-acetate
REFERENCES
- (1).Dauer W, Przedborski S. Parkinson’s disease: mechanisms and models. Neuron. 2003;39:889–909. doi: 10.1016/s0896-6273(03)00568-3. [DOI] [PubMed] [Google Scholar]
- (2).Kruger R, Kuhn W, Muller T, Woitalla D, Graeber M, Kosel S, Przuntek H, Epplen JT, Schols L, Riess O. Ala30Pro mutation in the gene encoding alpha-synuclein in Parkinson’s disease. Nat Genet. 1998;18:106–108. doi: 10.1038/ng0298-106. [DOI] [PubMed] [Google Scholar]
- (3).Singleton AB, Farrer M, Johnson J, Singleton A, Hague S, Kachergus J, Hulihan M, Peuralinna T, Dutra A, Nussbaum R, Lincoln S, Crawley A, Hanson M, Maraganore D, Adler C, Cookson MR, Muenter M, Baptista M, Miller D, Blancato J, Hardy J, Gwinn-Hardy K. alpha-Synuclein locus triplication causes Parkinson’s disease. Science. 2003;302:841. doi: 10.1126/science.1090278. [DOI] [PubMed] [Google Scholar]
- (4).Polymeropoulos MH, Lavedan C, Leroy E, Ide SE, Dehejia A, Dutra A, Pike B, Root H, Rubenstein J, Boyer R, Stenroos ES, Chandrasekharappa S, Athanassiadou A, Papapetropoulos T, Johnson WG, Lazzarini AM, Duvoisin RC, Di Iorio G, Golbe LI, Nussbaum RL. Mutation in the alpha-synuclein gene identified in families with Parkinson’s disease. Science. 1997;276:2045–2047. doi: 10.1126/science.276.5321.2045. [DOI] [PubMed] [Google Scholar]
- (5).Zarranz JJ, Alegre J, Gomez-Esteban JC, Lezcano E, Ros R, Ampuero I, Vidal L, Hoenicka J, Rodriguez O, Atares B, Llorens V, Gomez Tortosa E, del Ser T, Munoz DG, de Yebenes JG. The new mutation, E46K, of alpha-synuclein causes Parkinson and Lewy body dementia. Ann Neurol. 2004;55:164–173. doi: 10.1002/ana.10795. [DOI] [PubMed] [Google Scholar]
- (6).Chartier-Harlin MC, Kachergus J, Roumier C, Mouroux V, Douay X, Lincoln S, Levecque C, Larvor L, Andrieux J, Hulihan M, Waucquier N, Defebvre L, Amouyel P, Farrer M, Destee A. Alpha-synuclein locus duplication as a cause of familial Parkinson’s disease. Lancet. 2004;364:1167–1169. doi: 10.1016/S0140-6736(04)17103-1. [DOI] [PubMed] [Google Scholar]
- (7).Ibanez P, Bonnet AM, Debarges B, Lohmann E, Tison F, Pollak P, Agid Y, Durr A, Brice A. Causal relation between alpha-synuclein gene duplication and familial Parkinson’s disease. Lancet. 2004;364:1169–1171. doi: 10.1016/S0140-6736(04)17104-3. [DOI] [PubMed] [Google Scholar]
- (8).Spillantini MG, Schmidt ML, Lee VM, Trojanowski JQ, Jakes R, Goedert M. Alpha-synuclein in Lewy bodies. Nature. 1997;388:839–840. doi: 10.1038/42166. [DOI] [PubMed] [Google Scholar]
- (9).Jo E, McLaurin J, Yip CM, St George-Hyslop P, Fraser PE. alpha-Synuclein membrane interactions and lipid specificity. J Biol Chem. 2000;275:34328–34334. doi: 10.1074/jbc.M004345200. [DOI] [PubMed] [Google Scholar]
- (10).Murphy DD, Rueter SM, Trojanowski JQ, Lee VM. Synucleins are developmentally expressed, and alpha-synuclein regulates the size of the presynaptic vesicular pool in primary hippocampal neurons. J Neurosci. 2000;20:3214–3220. doi: 10.1523/JNEUROSCI.20-09-03214.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (11).Cabin DE, Shimazu K, Murphy D, Cole NB, Gottschalk W, McIlwain KL, Orrison B, Chen A, Ellis CE, Paylor R, Lu B, Nussbaum RL. Synaptic vesicle depletion correlates with attenuated synaptic responses to prolonged repetitive stimulation in mice lacking alpha-synuclein. J Neurosci. 2002;22:8797–8807. doi: 10.1523/JNEUROSCI.22-20-08797.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (12).Abeliovich A, Schmitz Y, Farinas I, Choi-Lundberg D, Ho WH, Castillo PE, Shinsky N, Verdugo JM, Armanini M, Ryan A, Hynes M, Phillips H, Sulzer D, Rosenthal A. Mice lacking alpha-synuclein display functional deficits in the nigrostriatal dopamine system. Neuron. 2000;25:239–252. doi: 10.1016/s0896-6273(00)80886-7. [DOI] [PubMed] [Google Scholar]
- (13).Gitler AD, Shorter J. Prime time for alpha-synuclein. J Neurosci. 2007;27:2433–2434. doi: 10.1523/JNEUROSCI.0094-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (14).Larsen KE, Schmitz Y, Troyer MD, Mosharov E, Dietrich P, Quazi AZ, Savalle M, Nemani V, Chaudhry FA, Edwards RH, Stefanis L, Sulzer D. Alpha-synuclein overexpression in PC12 and chromaffin cells impairs catecholamine release by interfering with a late step in exocytosis. J Neurosci. 2006;26:11915–11922. doi: 10.1523/JNEUROSCI.3821-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (15).Kubo S, Nemani VM, Chalkley RJ, Anthony MD, Hattori N, Mizuno Y, Edwards RH, Fortin DL. A combinatorial code for the interaction of alpha-synuclein with membranes. J Biol Chem. 2005;280:31664–31672. doi: 10.1074/jbc.M504894200. [DOI] [PubMed] [Google Scholar]
- (16).Bernheimer H, Birkmayer W, Hornykiewicz O, Jellinger K, Seitelberger F. Brain dopamine and the syndromes of Parkinson and Huntington. Clinical, morphological and neurochemical correlations. J Neurol Sci. 1973;20:415–455. doi: 10.1016/0022-510x(73)90175-5. [DOI] [PubMed] [Google Scholar]
- (17).Brown HA, Henage LG, Preininger AM, Xiang Y, Exton JH. Biochemical analysis of phospholipase D. Methods Enzymol. 2007;434:49–87. doi: 10.1016/S0076-6879(07)34004-4. [DOI] [PubMed] [Google Scholar]
- (18).Exton JH. Phospholipase D. Ann N Y Acad Sci. 2000;905:61–68. doi: 10.1111/j.1749-6632.2000.tb06538.x. [DOI] [PubMed] [Google Scholar]
- (19).Klein J. Functions and pathophysiological roles of phospholipase D in the brain. J Neurochem. 2005;94:1473–1487. doi: 10.1111/j.1471-4159.2005.03315.x. [DOI] [PubMed] [Google Scholar]
- (20).Levy BD, Fokin VV, Clark JM, Wakelam MJ, Petasis NA, Serhan CN. Polyisoprenyl phosphate (PIPP) signaling regulates phospholipase D activity: a ‘stop’ signaling switch for aspirin-triggered lipoxin A4. Faseb J. 1999;13:903–911. doi: 10.1096/fasebj.13.8.903. [DOI] [PubMed] [Google Scholar]
- (21).Oude Weernink PA, Lopez de Jesus M, Schmidt M. Phospholipase D signaling: orchestration by PIP2 and small GTPases. Naunyn Schmiedebergs Arch Pharmacol. 2007;374:399–411. doi: 10.1007/s00210-007-0131-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (22).Jenco JM, Rawlingson A, Daniels B, Morris AJ. Regulation of phospholipase D2: selective inhibition of mammalian phospholipase D isoenzymes by alpha- and beta-synucleins. Biochemistry. 1998;37:4901–4909. doi: 10.1021/bi972776r. [DOI] [PubMed] [Google Scholar]
- (23).Ahn BH, Rhim H, Kim SY, Sung YM, Lee MY, Choi JY, Wolozin B, Chang JS, Lee YH, Kwon TK, Chung KC, Yoon SH, Hahn SJ, Kim MS, Jo YH, Min do S. alpha-Synuclein interacts with phospholipase D isozymes and inhibits pervanadate-induced phospholipase D activation in human embryonic kidney-293 cells. J Biol Chem. 2002;277:12334–12342. doi: 10.1074/jbc.M110414200. [DOI] [PubMed] [Google Scholar]
- (24).Outeiro TF, Lindquist S. Yeast cells provide insight into alpha-synuclein biology and pathobiology. Science. 2003;302:1772–1775. doi: 10.1126/science.1090439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (25).Payton JE, Perrin RJ, Woods WS, George JM. Structural determinants of PLD2 inhibition by alpha-synuclein. J Mol Biol. 2004;337:1001–1009. doi: 10.1016/j.jmb.2004.02.014. [DOI] [PubMed] [Google Scholar]
- (26).Henage LG, Exton JH, Brown HA. Kinetic analysis of a mammalian phospholipase D: allosteric modulation by monomeric GTPases, protein kinase C, and polyphosphoinositides. J Biol Chem. 2006;281:3408–3417. doi: 10.1074/jbc.M508800200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (27).Scott SA, Selvy PE, Buck J, Cho H, Criswell TL, Armstrong MD, Arteaga CL, Lindsley C, Brown HA. Isoform selective phospholipase D inhibitors modulate lipid signaling species and cancer cell invasiveness. Nature Chem Biol. 2009;5:108–117. doi: 10.1038/nchembio.140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (28).Brown HA, Gutowski S, Moomaw CR, Slaughter C, Sternweis PC. ADP-ribosylation factor, a small GTP-dependent regulatory protein, stimulates phospholipase D activity. Cell. 1993;75:1137–1144. doi: 10.1016/0092-8674(93)90323-i. [DOI] [PubMed] [Google Scholar]
- (29).Volles MJ, Lansbury PT., Jr. Relationships between the sequence of alpha-synuclein and its membrane affinity, fibrillization propensity, and yeast toxicity. J Mol Biol. 2007;366:1510–1522. doi: 10.1016/j.jmb.2006.12.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (30).Walker SJ, Brown HA. Measurement of G protein-coupled receptor-stimulated phospholipase D activity in intact cells. Methods Mol Biol. 2004;237:89–97. doi: 10.1385/1-59259-430-1:89. [DOI] [PubMed] [Google Scholar]
- (31).Alberti S, Gitler AD, Lindquist S. A suite of Gateway((R)) cloning vectors for high-throughput genetic analysis in Saccharomyces cerevisiae. Yeast. 2007;24:913–919. doi: 10.1002/yea.1502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (32).Guthrie C, Fink AE, editors. Methods in Enzymology. Academic Press; San Fransisco: 2002. Guide to yeast genetics and molecular and cell biology. [Google Scholar]
- (33).Ito H, Fukuda Y, Murata K, Kimura A. Transformation of intact yeast cells treated with alkali cations. J Bacteriol. 1983;153:163–168. doi: 10.1128/jb.153.1.163-168.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (34).Gibbs TC, Meier KE. Expression and regulation of phospholipase D isoforms in mammalian cell lines. J Cell Physiol. 2000;182:77–87. doi: 10.1002/(SICI)1097-4652(200001)182:1<77::AID-JCP9>3.0.CO;2-B. [DOI] [PubMed] [Google Scholar]
- (35).Rose K, Rudge SA, Frohman MA, Morris AJ, Engebrecht J. Phospholipase D signaling is essential for meiosis. Proc Natl Acad Sci U S A. 1995;92:12151–12155. doi: 10.1073/pnas.92.26.12151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (36).Waksman M, Eli Y, Liscovitch M, Gerst JE. Identification and characterization of a gene encoding phospholipase D activity in yeast. J Biol Chem. 1996;271:2361–2364. doi: 10.1074/jbc.271.5.2361. [DOI] [PubMed] [Google Scholar]
- (37).Ella KM, Dolan JW, Qi C, Meier KE. Characterization of Saccharomyces cerevisiae deficient in expression of phospholipase D. Biochem J. 1996;314(Pt 1):15–19. doi: 10.1042/bj3140015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (38).Bankaitis VA, Aitken JR, Cleves AE, Dowhan W. An essential role for a phospholipid transfer protein in yeast Golgi function. Nature. 1990;347:561–562. doi: 10.1038/347561a0. [DOI] [PubMed] [Google Scholar]
- (39).Bankaitis VA, Malehorn DE, Emr SD, Greene R. The Saccharomyces cerevisiae SEC14 gene encodes a cytosolic factor that is required for transport of secretory proteins from the yeast Golgi complex. J Cell Biol. 1989;108:1271–1281. doi: 10.1083/jcb.108.4.1271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (40).Novick P, Ferro S, Schekman R. Order of events in the yeast secretory pathway. Cell. 1981;25:461–469. doi: 10.1016/0092-8674(81)90064-7. [DOI] [PubMed] [Google Scholar]
- (41).Novick P, Field C, Schekman R. Identification of 23 complementation groups required for post-translational events in the yeast secretory pathway. Cell. 1980;21:205–215. doi: 10.1016/0092-8674(80)90128-2. [DOI] [PubMed] [Google Scholar]
- (42).Cleves AE, McGee TP, Whitters EA, Champion KM, Aitken JR, Dowhan W, Goebl M, Bankaitis VA. Mutations in the CDP-choline pathway for phospholipid biosynthesis bypass the requirement for an essential phospholipid transfer protein. Cell. 1991;64:789–800. doi: 10.1016/0092-8674(91)90508-v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (43).Sreenivas A, Patton-Vogt JL, Bruno V, Griac P, Henry SA. A role for phospholipase D (Pld1p) in growth, secretion, and regulation of membrane lipid synthesis in yeast. J Biol Chem. 1998;273:16635–16638. doi: 10.1074/jbc.273.27.16635. [DOI] [PubMed] [Google Scholar]
- (44).Cooper AA, Gitler AD, Cashikar A, Haynes CM, Hill KJ, Bhullar B, Liu K, Xu K, Strathearn KE, Liu F, Cao S, Caldwell KA, Caldwell GA, Marsischky G, Kolodner RD, Labaer J, Rochet JC, Bonini NM, Lindquist S. Alpha-synuclein blocks ER-Golgi traffic and Rab1 rescues neuron loss in Parkinson’s models. Science. 2006;313:324–328. doi: 10.1126/science.1129462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (45).Chang HJ, Jones EW, Henry SA. Role of the unfolded protein response pathway in regulation of INO1 and in the sec14 bypass mechanism in Saccharomyces cerevisiae. Genetics. 2002;162:29–43. doi: 10.1093/genetics/162.1.29. [DOI] [PMC free article] [PubMed] [Google Scholar]