Abstract
High affinity monoamine transporters efficiently terminate neurotransmission through synaptic reuptake of released neurotransmitter. We recently cloned and characterized a novel low-affinity, high capacity plasma membrane monoamine transporter (PMAT) that is strongly expressed in the human brain and efficiently transports 5-HT and dopamine (DA). In efforts to understand the physiological function of PMAT and its relevance in monoaminergic pathways, we cloned the PMAT homolog from the mouse brain, demonstrated its capability for transporting 5-HT and DA, and determined the regional and cellular localization of mouse plasma membrane monoamine transporter (mPMAT) in adult mouse brain by reverse-transcription polymerase chain reaction, non-radioactive in situ hybridization, and immunohistochemical methods. Our results showed that mPMAT mRNA and protein are broadly expressed in the mouse brain and are particularly abundant in forebrain cortex, olfactory tubercle, hippocampus, cerebellum and epithelial cells of the choroid plexus. Dual-immunofluorescence histochemistry with established phenotypic markers microtubule-associated protein (MAP2) and glial fibrillary acidic protein (GFAP) revealed that mPMAT is expressed in neuronal cells but not in astrocytes. mPMAT is co-expressed in many brain regions with the high affinity 5-HT transporter (SERT) and the dopamine transporter (DAT), but is also found in certain sites that receive monoamine innervation but lack significant expression of SERT or DAT. These findings suggest that mPMAT is a widely distributed, neuronally-expressed transporter, which may support the role of 5-HT and DA uptake under certain conditions.
Keywords: uptake2, ENT4, brain localization, in situ hybridization, 5-HT, dopamine
In the CNS, the monoamine neurotransmitters 5-HT, dopamine (DA), and norepinephrine (NE) control a variety of physiological, behavioral, and endocrine functions (Carlsson, 1987; Greengard, 2001). Monoamines are also critically involved in the pathophysiology and pharmacotherapy of a number of brain disorders, including Parkinson’s disease, depression, schizophrenia, and drug addiction (Carlsson, 1987; Greengard, 2001; Gainetdinov et al., 2002). The plasma membrane–expressed transporters for 5-HT, DA, and NE play a key role in determining the intensity and duration of monoamine signaling by removing these transmitters from the extracellular space (Iversen, 1971; Blakely et al., 1994; Torres et al., 2003). Uptake of released monoamines into presynaptic neurons is mainly carried out by a family of Na+- and Cl−-dependent high affinity plasma membrane transporters, which include the dopamine transporter (DAT), the 5-HT transporter (SERT), and the norepinephrine transporter (NET). These high affinity transporters, also known as uptake1, share high sequence similarity and belong to the solute carrier 6 (SLC6) family. In the CNS, DAT, SERT and NET are expressed almost exclusively in the nerve terminals of monoaminergic neurons, and they represent the major targets of many psychotropic drugs and neurotoxins (Amara and Kuhar, 1993; Blakely et al., 1994; Torres et al., 2003).
In addition to uptake1, a Na+- and Cl−-independent, low-affinity, high-capacity transport system, termed uptake2, was recognized and first characterized in peripheral tissues (Lightman and Iversen, 1969; Iversen, 1971; Eisenhofer, 2001). Uptake2 displays different pharmacological profiles from uptake1, and is thought to play a role in catecholamine removal in peripheral tissues with sympathetic innervation (Lightman and Iversen, 1969; Iversen, 1971; Eisenhofer, 2001). In the CNS, a number of studies have described uptake2-like monoamine transport activities in tissue preparations from various brain areas including the cortex, striatum and accumbal tissues (Hendley et al., 1970; Burrows et al., 1981; Mireylees et al., 1986; Inazu et al., 1999). Recent studies in brain tissues from DAT and SERT knock-out mice also provided evidence of the existence of monoamine uptake activities that cannot be fully explained by the presence of high affinity transporters (Mundorf et al., 2001; Ravary et al., 2001; Sora et al., 2001; Wayment et al., 2001). However, in contrast to the well-characterized high affinity transporters, the role of the low affinity transporters in CNS neurotransmission is still unknown.
Recent molecular cloning work has identified that organic cation transporter 3 (OCT3) in the SLC22 family represents one of the uptake2 transporters for biogenic amines (Grundemann et al., 1998; Wu et al., 1998). Human OCT3 favors epinephrine and NE over 5-HT and DA (Grundemann et al., 1998; Amphoux et al., 2006), and is broadly expressed in a number of tissues, including the liver, heart, placenta, skeletal muscle, kidney, and brain (Grundemann et al., 1998; Wu et al., 2000). In 2004, our laboratory reported the cloning and characterization of a novel uptake2-type transporter in the human brain, which we termed plasma membrane monoamine transporter (PMAT) (Engel et al., 2004). The 530-residue human plasma membrane monoamine transporter (hPMAT) protein is not homologous to uptake1 transporters or the OCTs, but exhibits low sequence homology (~20% identity) to members in the equilibrative nucleoside transporter family (SLC29). We previously demonstrated that PMAT, also alternatively termed equilibrative nucleoside transporter 4 (ENT4), exhibits typical uptake2 characteristics, mediating Na+ and Cl− independent, low affinity and high capacity transport of monoamine neurotransmitters (Engel et al., 2004). Among the monoamines, PMAT favors 5-HT and DA over NE, epinephrine and histamine (Engel et al., 2004; Engel and Wang, 2005). The transport efficiency (Vmax/Km) of PMAT toward 5-HT and DA in cell expression systems is nearly comparable to that of SERT and DAT (Engel et al., 2004). PMAT also transports the dopaminergic neurotoxin 1-methyl-4-phenylpyridinium (MPP+) with comparable affinity to other biogenic amine transporters. Northern analysis using multiple human tissues showed that PMAT mRNA is highly expressed in the brain and present in several gross regions of human brain (Engel et al., 2004). However, the regional distribution and cellular localization of PMAT in mammalian brains have not been determined. It is not known whether there are any preferential sites of PMAT expression in the brain, nor in which cell types the transporter is expressed.
In this study, we cloned the mouse plasma membrane monoamine transporter (mPMAT) isoform and mapped the mRNA and protein expression patterns of mPMAT in adult mouse brain. Our results suggest that PMAT is broadly expressed in various neuronal cells of many brain regions and may contribute to 5-HT and DA uptake in areas where the high-affinity transporters are absent or expressed at low levels.
EXPERIMENTAL PROCEDURES
Cloning of mPMAT cDNA
A cDNA clone (BC025599) isolated from mouse mammary tumor tissues was identified as the homolog of human SLC29A4 gene by BLAST search of the NCBI database. Primers (upstream: 5′-tgccatgggctctatcggaagccagc-3′; downstream: 5′-tctagaccctgggcg gtgaagacaagttccagc-3′) flanking the open reading frame were used to amplify a 1.6 kb cDNA fragment by polymerase chain reaction (PCR) from mouse brain cDNA (Clontech, Mountain View, CA, USA). PCR was conducted for 35 cycles at 94° C for 1 min, 56° C for 1 min and 72° C for 3 min. The cDNA fragment was then subcloned into the pCR-II-TOPO vector (Invitrogen, Carlsbad, CA, USA). Three clones from independent PCR reactions were isolated and sequenced at the UW Biochemistry Sequencing Center. One clone, which showed 100% sequence identity to BC025599, was selected for further studies.
Expression of mPMAT and hPMAT in Madin-Darby canine kidney (MDCK) cells
MDCK cells were maintained in minimum essential medium (MEM) supplemented with 10% fetal bovine serum (FBS), 2 mM L-glutamine, 100 μg/ml penicillin/streptomycin at 37 °C in a 5% CO2 incubator. To facilitate the establishment of a cell line stably expressing mPMAT, the mPMAT cDNA was subcloned in-frame into the C-terminal of an enhanced yellow fluorescent protein (YFP) vector pEYFP-C1 (Invitrogen) using the EcoRI and XbaI sites. The construct was then transfected into MDCK cells using Lipofectamine2000 according to manufacturer’s instruction (Invitrogen). The pEYFP-C1 vector DNA was transfected into MDCK cells as a control. Transfected cells were selected with 1000 μg/ml G-418 for 2 weeks and YFP positive cells were purified using a FACScan Vantage SE flow cytometry sorter (BD Biosciences, San Jose, CA, USA). Sorted cells were maintained in MEM medium containing 10% FBS and G418 for transport and immunoblot assays. In addition, mPMAT subcloned into pcDNA3 vector (Invitrogen) was transiently transfected into MDCK cells for immunoblot and immunocytochemistry (ICC) studies. For hPMAT, previously established cell lines stably expressing hPMAT and YFP-tagged hPMAT (Xia et al., 2007) were used for immunoblot and ICC studies.
Transport assays
Transport studies were performed as described previously (Engel and Wang, 2005). Briefly, MDCK cells stably transfected with pEYFP-C1 vector or YFP-tagged mPMAT were plated in 24-well plates and allowed to grow for 3 days. Transport assays were performed at 37 °C in Krebs-Ringer-Henseleit (KRH) buffer (5.6 mM glucose, 125 mM NaCl, 4.8 mM KCl, 1.2 mM KH2PO4, 1.2 mM CaCl2, 1.2 mM MgSO4, and 25 mM Hepes, pH 7.4) containing 1 μM of [3H]5-HT (27.1 Ci/mmol) or [3H]DA (59.7 Ci/mmol). Uptake was performed in the presence or absence of a PMAT inhibitor decynium 22 (1 μM). At the end of incubation, uptake was terminated by washing the cells three times with ice-cold KRH buffer. The cells were solubilized and radioactivity was quantified by liquid scintillation counting. Protein content in each uptake well was measured using a Bio-Rad Protein Assay Kit (Pierce, Rockford, IL, USA), and the uptake in each well was normalized to its protein content.
Reverse-transcription polymerase chain reaction (RT-PCR) analyses of tissue and brain distributions of mPMAT
An RT-PCR strategy was used to determine the tissue distribution of mPMAT mRNA transcripts using a commercially available mouse multi-tissue cDNA panel (OriGene Technologies, Rockville, MD, USA). PCR primers (upstream: 5′-CCTCCTCGCCTT-GGGTCCCTTGCTC-3′; downstream: 5′-CTGATGCCCATTAG-CAGCGAGAAGAC-3′) were designed to amplify a 941 bp fragment of mPMAT. Amplification of mouse GAPDH was used as an internal control. To determine the distribution of mPMAT mRNA in mouse brain, a cDNA panel (OriGene) containing purified, lyophilized cDNAs from various regions of adult mouse brain was used. Amplicons of mPMAT, mouse serotonin transporter (mSERT), mouse norepinephrine transporter (mNET) and mouse dopamine transporter (mDAT) were obtained by single-gene (for mPMAT) or multiplex (for mNET, mSERT and mDAT) PCR. The β-actin gene was also amplified as a reaction control. The PCR products were analyzed by agarose gel electrophoresis in the presence of 0.05 μg/ml ethidium bromide, and the identity of each gene product was verified by molecular size, restriction analysis or direct sequencing of purified cDNA bands.
Localization of mPMAT mRNA in brain sections using non-isotopic in situ hybridization (ISH)
For ISH, sense and antisense digoxigenin (DIG)-labeled cRNA probes were prepared from a 392-bp PCR-amplified fragment of the mPMAT 3′ untranslated region. A BLASTn search of querying this fragment against the NCBI database showed that there is no alignment of this region with any other mouse DNA sequences. For in vitro transcription, the fragment was cloned into pCR-II-TOPO vector (Invitrogen). The construct was linearized in sense and antisense directions, and in vitro transcribed using the DIG-UTP labeling kit according to manufacturer’s recommendations (Roche Applied Sciences, Indianapolis, IN, USA). The quality and purity of transcribed sense and antisense probes were confirmed for correct size by RNA denaturing gel electrophoresis in the presence of 0.05 μg/ml ethidium bromide. The purified cRNA probes were then stored at −80 °C until use. For all ISH studies, experiments were performed using commercially-obtained frozen sections of mouse brain from CD1 mice (Zyagen, San Diego, CA, USA). Non-radioactive ISH was performed using a previously described procedure (Hevner et al., 2001). Handling and treatment of slides was performed under RNAse-free conditions. All solutions used in the pre-hybridization and hybridization procedures were DEPC-treated and autoclaved prior to use. To prepare tissues for hybridization, frozen tissue sections were removed from −80 °C, air-dried 10 min at room temperature and fixed in 4% (w/v) paraformaldehyde (pH 8.0) in 1× phosphate-buffered saline (PBS). Fixed sections were washed in PBS (2×10 min), permeabilized in 0.1 M HCl (10 min) and acetylated in stirring 0.1 M triethanolamine (pH 8.0). Acetic anhydride (0.25%, v/v) was added dropwise for 1 min and the reaction was continued for 15 min. Sections were then washed in PBS (2×10 min) and incubated in 50% deionized formamide/2× SSC (1× SSC is 0.15 M sodium chloride and 1.5 mM trisodium citrate) for 30 min at 42 °C to inhibit nonspecific binding of probes. Hybridization solution (20% dextran sulfate in formamide, 40×Denhardt’s solution, 20×SSC, 0.5 M EDTA, 500 μg/ml yeast tRNA, 500 μg/ml salmon sperm DNA) (Roche Applied Science) containing DIG-labeled probes (1 μg/ml) was incubated at 85 °C for 3 min to denature the probes, and then immediately placed on ice until use. Approximately 100 μl of hybridization solution was added to each section to cover the tissue. Hybridization took place for over 16 h at 42 °C inside an RNAse-free hybridization chamber (Sigma Genosys, St. Louis, MO, USA) humidified with 50% formamide/2× SSC. After hybridization, the slides were washed under high-stringency conditions. Briefly, slides were rinsed in 50% formamide/2× SSC at 42 °C, 20 min × three times followed by washing with NTE buffer (0.5 M sodium chloride, 10 mM Tris, 1 mM EDTA; pH 8.0) at 37 °C for 5 min. The sections were treated with RNase A (Roche Applied Science) in NTE buffer (20 μg/ml) at 37 °C for 30 min followed by rinsing in NTE buffer for 5 min and washing in 0.1×SSC at 42 °C, 20 min × three times. For immunodetection of the DIG antigen, the slides were equilibrated in 0.1 M Tris buffer (pH 7.4) for 10 min blocked with diluted normal goat serum (Vector Laboratories, Burlingame, CA, USA) in PBS for 60 min and incubated with anti-DIG-AP Fab Fragments (Roche Applied Science) in blocking serum for 2 h at room temperature. Slides were washed in PBS (3×10 min) developed with the alkaline phosphatase substrate NBT/BCIP (Roche Applied Science) (200 μl in 10 ml 0.1 M Tris buffer containing 0.5 M magnesium chloride, pH 9.5) for 2–24 h and evaluated by photomicroscopy using a Nikon Eclipse E600 microscope with a QImaging Retigia EX CCD Camera (Nikon Instruments, Melville, NY, USA). Brain regions were identified using the Franklin and Paxinos (1997) adult mouse brain atlas.
Production and purification of PMAT antibody
Comparative analysis of human, mouse and rat PMAT protein sequences identified two peptide sequences correspond to amino acids 90 – 103 (TDVDYLHHKYPGTS) and 469–482 (ILAAGKVSPKQREL) in hPMAT that are 100% conserved across human and rodent PMATs but divergent with other SLC29 family members. The two peptides plus an additional C-terminal cysteine for carrier conjugation were chemically synthesized and conjugated to keyhole limpet hemocyanin (KLH). The synthetic peptide-KLH conjugates were purified to >95% homogeneity and used to immunize New Zealand White rabbits. The polyclonal rabbit antisera were commercially prepared using standard protocols by Sigma Genosys. Antisera with the highest ELISA titer toward the purified antigen were further tested by immunoblotting using whole cell lysates prepared from MDCK cells expressing hPMAT or mPMAT. The antibody directed toward the amino acids 469–482 was highly reactive and specific toward human and mPMAT as determined by Western blot. This antibody, designated P469, was affinity purified by chromatography on a sepharose 4B column (ProSci Inc., Poway, CA, USA) and was used in subsequent immunoblotting and immunolocalization studies. The pre-immune sera and P469 antisera pre-absorbed with the antigenic peptide were included as controls in immunohistochemistry (IHC) and ICC experiments.
Immunoblotting
For protein extraction, non-transfected, vector-, hPMAT-, YFP-hPMAT-, mPMAT-, or YFP-mPMAT-transfected MDCK cells were harvested and washed three times with cold Dulbecco’s PBS at 4 °C. Cell pellets were resuspended and incubated on ice for 1 h in a lysis buffer (50 mM Tris–HCl, pH 7.4) containing 1% Nonidet P-40, 20 μg/ml phenylmethylsulfonyl fluoride and protease inhibitor cocktail (Roche Applied Science). The lysates were centrifuged at 13,000×g at 4 °C for 10 min to remove cell debris. The protein contents from the supernatant were quantified using a BCA protein assay (Pierce). An aliquot of the supernatant (20 μg protein) was boiled for 5 min, separated on 7.5% SDS polyacrylamide gels, and electrophoretically transferred to a PVDF membrane (Millipore, Billerica, MA, USA). The membrane was blocked with 5% (w/v) non-fat dry milk powder (BioRad Laboratories, Hercules, CA, USA) in PBS for 1 h and incubated with the P469 primary antibody diluted 1:1600 in 1% milk in PBS for 2 h following three times washing with PBS containing 0.1% Tween-20. The blot was then incubated with horseradish peroxidase– conjugated goat anti-rabbit IgG (1:20,000 dilution). Immunoreactive proteins were detected by chemiluminescence using Supersignal West Pico Reagent (Pierce) followed by autoradiography.
ICC
ICC was performed on MDCK cell lines and primary cultures of neurons prepared from rat ventral mesencephalon as described by (Hsuan et al., 2006). Cultures of primary neurons or cell lines (stably transfected with hPMAT or transiently transfected with mPMAT) were fixed in 4% paraformaldehyde and permeabilized in 0.2% Triton X-100 in 1× PBS. The permeabilized cells were blocked using 10% FBS for 90 min at room temperature, with gentle shaking, followed by incubation with P469 primary antibody (diluted 1:200 in blocking buffer) for 1 h at room temperature. Cells were washed in 1× phosphate-buffered saline with 0.05% Tween-20 (PBST) over 45 min, and incubated with the Alexa-488 fluorescent secondary antibody (1: 1000) (Molecular Probes, Carlsbad, CA, USA) in the dark for 30 min. The cells were washed in PBST and treated with Topo3 dye (Molecular Probes) for 5 min and then analyzed by confocal microscopy as previously described (Engel et al., 2004). The pcDNA3-transfected MDCK cells were included as a control. For neuronal culture ICC, primary cultures were treated with the pre-absorbed P469 antisera as a control.
Localization of PMAT protein in brain sections by IHC
For all IHC studies, experiments were performed using commercially-obtained cryosections of mouse brain to minimize the use of animals. Frozen sections of mouse brain tissues (Zyagen) were fixed in ice cold acetone for 5 min, washed briefly in PBS and incubated in 0.3% hydrogen peroxide in methanol for 20 min. After washing once in PBS for 5 min, the sections were blocked in diluted normal goat serum (Vector Laboratories) in PBS followed by incubation with P469 (diluted 1:500 in blocking buffer) overnight at room temperature. Control incubations consisted of diluted blocking serum without antibody or diluted blocking serum containing pre-immune sera or peptide antigen pre-absorbed P469 antibody. The slides were then washed in PBS (3×10 min) at room temperature and incubated with biotinylated secondary antibody followed by treatment with the Vectastain ABC Reagent, according to manufacturer’s instructions (Vector Laboratories). The slides were developed with DAB peroxidase substrate solution (Vector Laboratories) for 2–5 min and examined by photomicroscopy as described above. Mouse brain regions were identified using a mouse brain atlas (Franklin and Paxinos, 1997). For immunofluorescence studies in human brain tissues, frozen human cerebellum slides (Analytical Biological Service, Wilmington, DE, USA) fixed in cold acetone were thawed at room temperature, briefly washed with PBST (0.05% Tween-20), blocked with 10% FBS in PBST for 1 h, and incubated with the primary P469 antibody (1:200) diluted in 10% FBS in PBST for 2 h. After washing three times for 15 min with PBST, the sections were stained with AlexaFluor 488-conjugated goat anti-rabbit IgG (1:1000 in blocking buffer) (Molecular Probes) for 30 min and washed three times with PBST. The nuclei were counterstained with Topro-3 dye (Molecular Probes) in PBS for 5 min, washed twice in PBS and mounted. The sections were observed and photographed by confocal microscopy using a Zeiss LSM 510 META confocal microscope at the University of Washington Keck Imaging Center (Seattle, WA, USA).
Co-localization of PMAT with neuronal or astroglial markers by double-label immunofluorescence
Mouse brain cryosections were prepared for IHC as described above. Chicken anti-mouse polyclonal antibodies against glial fibrillary acidic protein (GFAP) (Chemicon, Temecula, CA, USA) or all isoforms of microtubule-associated protein 2 (MAP2) (Abcam, Inc., Cambridge, MA, USA) were diluted 1:1000 and 1:10,000, respectively, in PBS containing 3% normal goat serum. Tissue sections were incubated with diluted primary antibody markers containing either P469 antibody (1:1000) or peptide antigen pre-adsorbed P469 (1:1000) (control) for 24 h at 4 °C. Tissue sections were washed three times in PBS for over 2.5 h (at room temperature), and then incubated with a mixture of Alexa Fluor 488- and Alexa Fluor 555-conjugated goat anti-chicken IgG (H+L) secondary antibodies (diluted 1:1000 in PBS) (Molecular Probes) for 60 min at room temperature. Tissue sections were washed again three times in PBS over 90 min (at room temperature), and then prepared for analysis by fluorescence microscopy as described above.
RESULTS
Cloning and functional expression of mPMAT
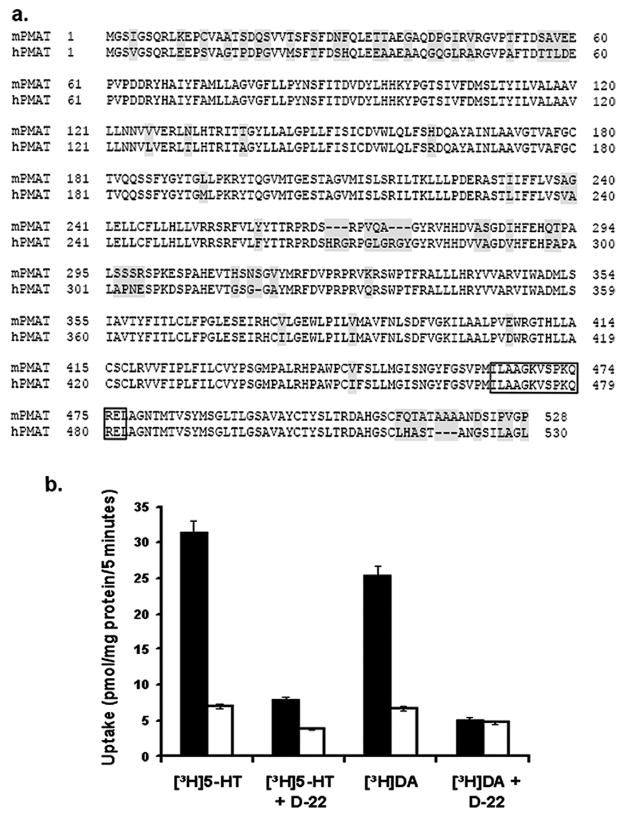
Using PCR, we isolated the full length mPMAT cDNA from a mouse brain cDNA library. The mPMAT cDNA encodes a protein of 528 amino acid residues and is 86% identical to hPMAT (hPMAT, 530 a.a.) (Fig. 1a). Expression of YFP-tagged mPMAT in MDCK cells resulted in a significant increase in cellular accumulation of 5-HT (1 μM), which was completely abolished in the presence of 1 μM decynium22 (Fig. 1b), a potent inhibitor of hPMAT (Engel et al., 2004). Similarly, mPMAT-specific, decynium22-sensitive uptake was also observed for DA at 1 μM (Fig. 1b). These data demonstrated that, like hPMAT, the mouse isoform also transports biogenic amines and is sensitive to inhibition by decynium22. These data suggest that the key functional features of hPMAT are preserved in mPMAT.
Fig. 1.
Cloning and functional characterization of mPMAT. (a) Amino acid sequence alignment of mPMAT and hPMAT. Shaded areas correspond to non-conserved regions in the mPMAT and hPMAT amino acid sequences. The boxed region indicates the peptide sequence used to generate the p469 antibody. (b) Transport of DA and 5-HT (1 μM each) by MDCK cells stably transfected with either pEYFP-C1 vector (open bars) or mPMAT-pEYFP (filled bars), in the presence and absence of decynium-22 (D-22) (1 μM), a specific inhibitor of hPMAT.
Detection of mPMAT mRNA in various mouse tissues and brain regions by RT-PCR
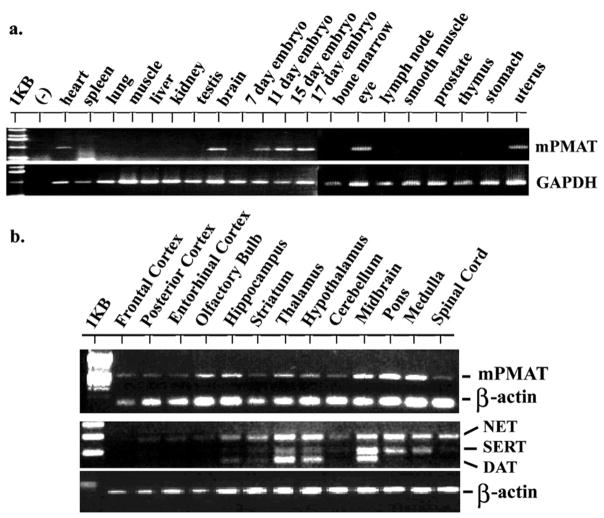
The distribution of mPMAT in various mouse tissues was analyzed by PCR using a multiple tissue cDNA panel. mPMAT transcripts were detected in whole brain, eye, heart, uterus and cDNAs prepared from whole embryos at gestational days 11–17 (Fig. 2a). No amplification was found in other tissues including spleen, lung, skeletal muscle, liver, kidney, testis, bone marrow, lymph nodes, smooth muscle, prostate, thymus, stomach and 7-day old embryos. As an initial step to characterize the mPMAT expression in the brain, we PCR amplified mPMAT from a cDNA panel containing multiple regions representative of the entire adult mouse brains (Fig. 2b). Widespread expression of mPMAT transcripts was detected in nearly all brain regions tested (Fig. 2b). In contrast, expression of the high affinity transporters, especially mDAT and mSERT, was localized to the midbrain and brain stem regions (Fig. 2b). These data are in agreement with our previous Northern blot studies in human brains (Engel et al., 2004), which also showed a broad expression of hPMAT in the adult brain.
Fig. 2.
Tissue and brain regional distribution of mPMAT mRNA analyzed by RT-PCR. (a) Tissue distribution of mPMAT analyzed by PCR using a mouse multi-tissue cDNA panel. (b) Top panel shows RT-PCR amplification of mPMAT transcripts from different brain regions. The bottom panel shows multiplex PCR analysis of mouse NET, SERT and DAT in various regions of the mouse brain. In all samples, amplification of GAPDH or β-actin was performed as an internal control.
Brain distribution of mPMAT mRNA revealed by non-isotopic ISH
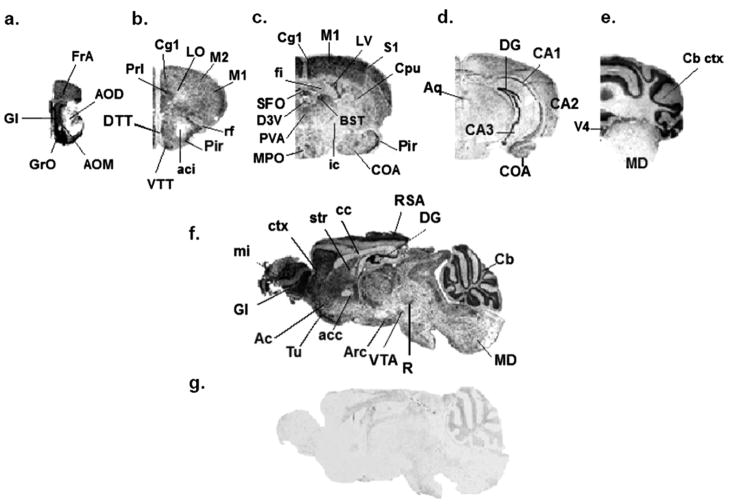
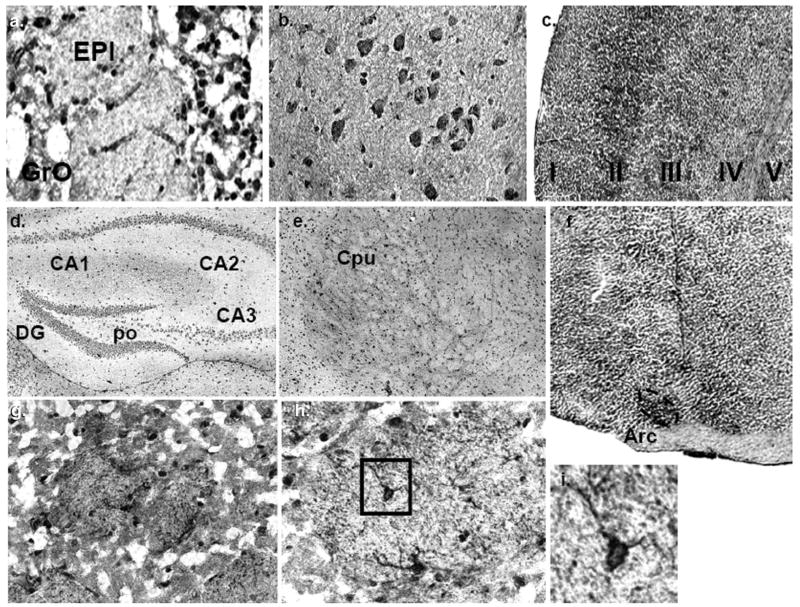
The distribution of mPMAT mRNA on sagittal and coronal cryosections of brains from adult mice was investigated by non-isotopic ISH using DIG-labeled cRNA probes followed by DIG-alkaline phosphatase IHC. ISH with mPMAT antisense probes revealed widespread ISH signals in multiple brain regions (Fig. 3). The observed signals were restricted to the cytoplasm of cells, as was expected for mRNA hybridization. Depleting mRNA by RNAase-treatment of tissues prior to hybridization resulted in no signal in antisense-treated tissue sections (data not shown). Further, ISH of adjacent sections with mPMAT sense probes showed little to no specific labeling (Fig. 3g). As determined by non-isotopic ISH with the antisense probe, mPMAT mRNA was widely expressed in multiple regions of the forebrain, midbrain and hindbrain (Figs. 3–5). In the forebrain, intense mRNA labeling was found in the olfactory bulb, cortex, olfactory tubercle, hippocampal field (CA1-3) areas and dentate gyrus (Figs. 3 and 4). The olfactory bulb contained strong hybridization signals, with the greatest signal intensity appearing in the mitral and glomerular cell layers (Fig. 4a). The forebrain cortex showed high mRNA labeling (Fig. 3a–c and f; Fig. 4b–d). The greatest intensity of hybridization signal in the cortex was found in the piriform cortex (Fig. 4d). High levels of expression were also observed in the frontal association area (Fig. 3a), motor (Fig. 3b and c), somatosensory (Fig. 3c), cingulate, prelimbic and visual (Fig. 3b and c) cortices. mRNA expression was consistently dense in the neurons of layer II throughout the forebrain (Fig. 4b and c). Cortical areas with lower mPMAT expression included the perirhinal cortex and temporal cortex association area (data not shown). The lateral orbital cortex showed much lower levels of mRNA labeling (Fig. 3b). The dorsal and ventral taenia tecta showed a prominent hybridization signal (Fig. 3b). In the hippocampus, the granule cell layer of the dentate gyrus displayed the greatest ISH signals of the forebrain regions (Fig. 3d and f; Fig. 4e–g). The CA1-3 regions of the hippocampus also showed dense hybridization signals (Fig. 4e). In the amygdala, we observed light to moderate hybridization signals in neurons in the cortical amygdaloid areas (Fig. 3c and d). Moderate to dense mRNA labeling in many neurons of the caudate putamen/striatum was observed (Fig. 3c and 4h). The bed nucleus of the stria terminalis, supracapsular part, showed a strong hybridization signal (Fig. 3c). We did not observe strong labeling in the internal capsule or globus pallidus (Fig. 3c). In the thalamus, a strong hybridization signal was observed in neurons of the anterior paraventricular thalamic nucleus (Fig. 3c) and the medial geniculate complex (Fig. 5e). Specific regions of the hypothalamus showed intense mRNA labeling. Strong mRNA hybridization was observed in the medial preoptic area (Fig. 3c) as well as the arcuate hypothalamic nucleus (Fig. 3f and Fig. 4i–j).
Fig. 3.
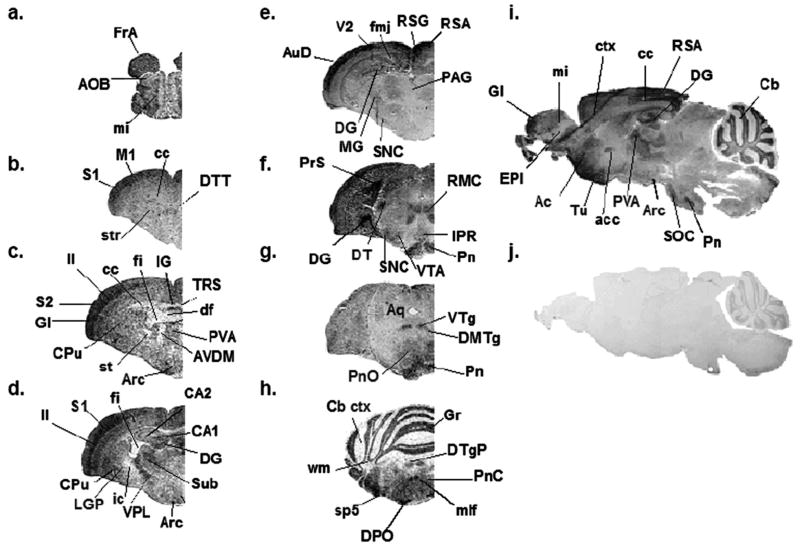
Regional distribution of mPMAT mRNA in the mouse brain analyzed by non-radioactive ISH. Coronal (a–e) or sagittal (f–g) cryosections of mouse brain were subjected to non-radioactive DIG ISH using cRNA probes specific for mPMAT. Panels a–f show ISH labeling with the antisense probe whereas panel g represents the sense probe control. Images were obtained at 2× magnification.
Fig. 5.
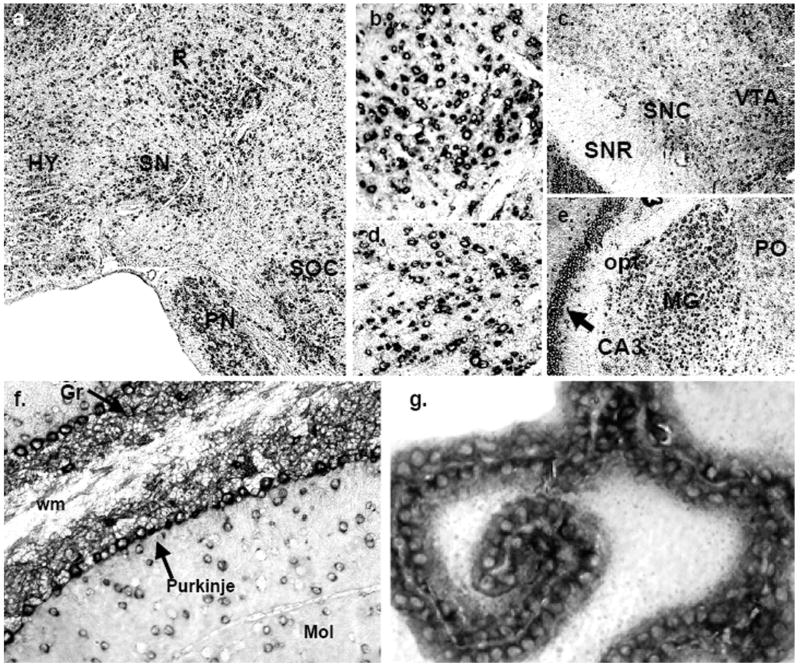
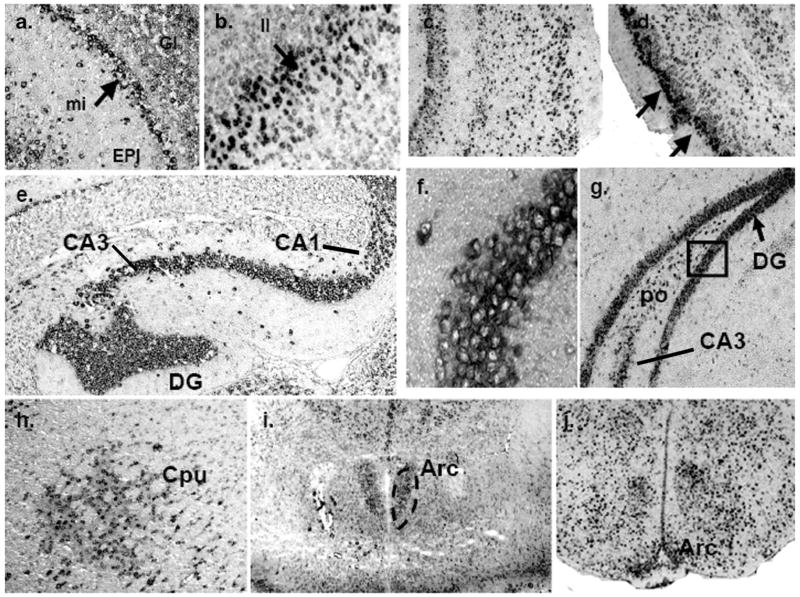
Localization of mPMAT mRNA in midbrain and hindbrain as revealed by non-radioactive ISH. (a) mPMAT mRNA is observed in the midbrain substantia nigra (SN) and red nucleus (R), pons (PN and SOC) and hypothalamus (HY). The regions in panels b and d correspond to high-magnification (40×) images of the R and SN in the section from panel a. (c) mPMAT mRNA expression in the substantia nigra compact part (SNC), substantia nigra reticular part (SNR), and the ventral tegmental area (VTA) (e) Expression of mPMAT in the midbrain hippocampal CA3 area and thalamic areas (MG and PO). (f) mPMAT-expressing neurons in the molecular (Mol) and granular layers (Gr, arrow) of the cerebellum, and in Purkinje neurons (arrows). The white matter tract (wm) is not labeled. (g) High-magnification (60×) of mPMAT expression in the choroid plexus (third ventricle).
Fig. 4.
Localization of mPMAT mRNA in forebrain regions as revealed by non-radioactive ISH. (a) mPMAT antisense cRNA probes densely label the mitral (mi) and glomerular layers (Gl) of the olfactory bulb. (b–d) Layers of the frontal cortex (b and c) and piriform cortex (d) are strongly labeled. (e–g) Intense labeling of mPMAT mRNA in the granular layers of the dentate gyrus (DG), CA1 and CA3 regions of the hippocampus. (f) High magnification (60×) of boxed area in panel g showing cytoplasmic staining of hippocampus granule cells. (h) Expression of mPMAT in the caudate putamen (striatum). (i, j) Expression of mPMAT in the arcuate nucleus (area surrounded by dashed line) of the hypothalamus.
In contrast to the forebrain and interbrain regions, the midbrain appeared to express relatively lower levels of mPMAT (Fig. 3d and f). However, several midbrain structures—red nucleus, substantia nigra and ventral tegmental area— displayed strong ISH signals (Fig. 3f and Fig. 5a–d). The red nucleus was populated by many neurons containing dense cytoplasmic labeling (Fig. 5b). The substantia nigra compact part, but not the reticular part, also contained many neurons with a strong cytoplasmic hybridization signals (Fig. 5c and d). Additionally, the ventral tegmental area showed dense mRNA labeling (Fig. 5a). In the hindbrain, the pontine nuclei were densely labeled (Fig. 5a). Within the pontine nuclei, very light staining was observed in the pedunculopontine tegmental nucleus, interstitial nucleus of the medial longitudinal fasciculus and hypoglossal nucleus (data not shown). There was intense staining of the superior olivary complex (Fig. 5a).
In the cerebellum, mPMAT expression was very intense (Fig. 3e and f; Fig. 5f). The granular cell layer in particular showed very dense hybridization signal, as did the Purkinje cell layer, and there was moderate labeling of many neurons in the molecular layer (Fig. 5f). In the medulla, the overall staining was light to moderate; however, there were scattered cells with intense staining (Fig. 3e and f).
In the circumventricular system, the subfornical organ (Fig. 3c) contained dense mPMAT hybridization signals. We also observed dense hybridization within the epithelial cells of all ventricular choroid plexuses (Fig. 3c and e; Fig. 5g). We did not observe significant mPMAT mRNA expression in the white matter or fiber tracts (Fig. 3 and Fig. 5e and f).
With regard to cell type specificity, all cells, with the exception of the choroidal epithelial cells, exhibited morphology consistent with that of neuronal cells. No specific ISH signals in astroglial cells, based upon morphology, were observed.
Generation and characterization of PMAT antibodies
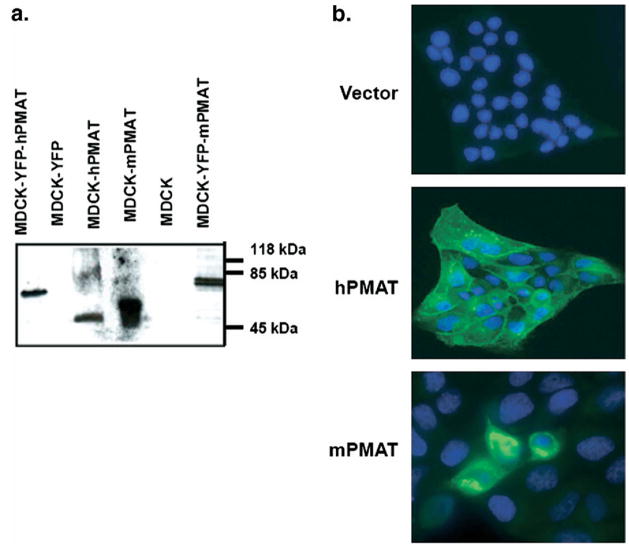
Comparative analysis of human, mouse and rat PMAT sequences identified two peptide sequences that are 100% conserved across human and rodent PMATs, but divergent with other SLC29 family members. The two peptides were synthesized, conjugated to KLH, and polyclonal antisera were prepared and affinity purified. One antibody, P469, generated toward 469 – 482 a.a. in hPMAT (or 464 – 477 a.a. in mPMAT) (Fig. 1a) specifically reacted with human and mPMAT proteins by immunoblot and ICC analysis of transfected MDCK cells (Fig. 6). Specifically, Western analysis showed that the P469 antibody detected the 58 kDa untagged mPMAT or hPMAT proteins as well as the 83 kDa YFP-tagged mPMAT or hPMAT proteins (Fig. 6a). Immunoreactive products were absent from cell lysates prepared from untransfected or vector-transfected MDCK cells (Fig. 6a), or when PMAT-expressing cell lysates were blotted with pre-immune sera or peptide antigen pre-absorbed antibody (data not shown). Additionally, when blotted with a commercial monoclonal anti-YFP antibody, the same 83 kDa bands appeared in cells expressing YFP-tagged mPMAT or YFP-tagged hPMAT (data not shown). ICC studies showed that in hPMAT stably transfected MDCK cells and mPMAT transiently transfected MDCK cells, the P469 antibody showed specific staining of plasma membranes and cell bodies (Fig. 6b). In contrast, there was no specific staining of MDCK cells transfected with an empty vector. Together, these data demonstrated the P469 peptide antibody is highly reactive to human and mPMAT isoforms. This antibody was used in subsequent immunolocalization studies.
Fig. 6.
Validation of the P469 polyclonal antibody. (a) Western blot analysis of untagged (58 kDa) and YFP-tagged (83 kDa) mPMAT or hPMAT expressed in MDCK cells. Untransfected or YFP-transfected cells were used as controls. (b). Immunocytochemical analysis of hPMAT (stable transfection) and mPMAT (transient transfection) in MDCK cells. Cells were stained with the P469 anti-PMAT antibody (1:200) and the nuclei were counterstained with Topro-3 dye.
Localization of mPMAT protein in various mouse brain regions and cell groups
Consistent with our ISH data, immunostaining of sagittal and coronal mouse brain sections with P469 antibody revealed widespread expression of mPMAT protein throughout the adult mouse brain (Figs. 7–9). The staining was particularly abundant in the forebrain cortex, olfactory tubercle, ventral striatum, hippocampal CA1-3 fields, dentate gyrus, pontine and tegmentum nuclei, and cerebellum. In the olfactory system, immunoreactivity toward P469 appeared most intense in the mitral cells, with comparatively weaker signals prevailing in the accessory olfactory bulb (Fig. 7a) and the external plexiform layer (Fig. 7i and Fig. 8a). Of all forebrain regions, mPMAT immunoreactivity was the highest in the cortex (Fig. 7b–e and i). In particular, the cell bodies and processes of neurons in layer II (Fig. 7c and d; Fig. 8c) of the cortex contained the densest mPMAT immunoreactivity. Furthermore, all other cortices were densely labeled (Fig. 7b–e). Within the cortex, we observed immunolabeling of various large and small neurons with their cell bodies and processes stained, as well as labeling of surrounding neuropil (Fig. 8b). In contrast, immunolabeling of glia or astroglial-like cells was not typically observed. In the hippocampus, the granular cells of the dentate gyrus contained dense mPMAT labeling, which was among the highest in the forebrain (Fig. 7i and Fig. 8d). The hippocampal CA1-3 fields also exhibited significant immunoreactivity (Fig. 8d). The fimbria of the hippocampus was not labeled (Fig. 7c). Intense staining of the presubiculum was present in sections of anterior hippocampus (Fig. 7f). mPMAT labeling was moderate throughout the dorsal taenia tecta (Fig. 7b). The caudate putamen and striatum contained dense immunolabeling (Fig. 7b–d); however, the stria terminalis did not display significant immunoreactivity to P469. Moderate labeling of some of the striosomes was observed (Fig. 7b and Fig. 8e). At higher magnification, significant labeling of striatal cells was apparent (Fig. 8g and h); striatal neurons displayed P469-immunoreactive cell bodies and processes (Fig. 8h and i). In the ventral striatum, the accumbens shell was densely immunoreactive (Fig. 7i); however, olfactory tubercle was among the most densely immunoreactive areas of the brain (Fig. 7i). The dorsal fornix was not labeled, but the indusium and triangular nucleus of the septum showed high immunoreactivity (Fig. 7c). The lateral septal nucleus also showed dense staining. The lateral globus pallidus and ventral pallidum displayed pronounced expression (Fig. 7d). Most thalamic areas, including cell bodies and neuropil, contained moderate to dense immunoreactivity (Fig. 7c, d and i). The ventral posterolateral nucleus, submedius thalamic nucleus and ventral posteromedial nucleus and medial geniculate nucleus of the thalamus were densely labeled (Fig. 7d, e and i). In contrast, the anterioventral nucleus showed very low immunoreactivity (Fig. 7d). Many hypothalamic areas were moderately labeled (several structures shown in Fig. 7c). The arcuate nucleus also exhibited dense mPMAT immunoreactive staining (Figs. 7c and 8f).
Fig. 7.
Regional distribution of mPMAT protein in the mouse brain as revealed by IHC. Immunohistochemical labeling of mPMAT was obtained by incubating adjacent mouse brain coronal sections (a–h) or sagittal sections (i–j) with affinity-purified P469 anti-PMAT antibody (a–i) or a control antisera containing the antigen preabsorbed primary antibody (j). Images are shown at 2× magnification.
Fig. 9.
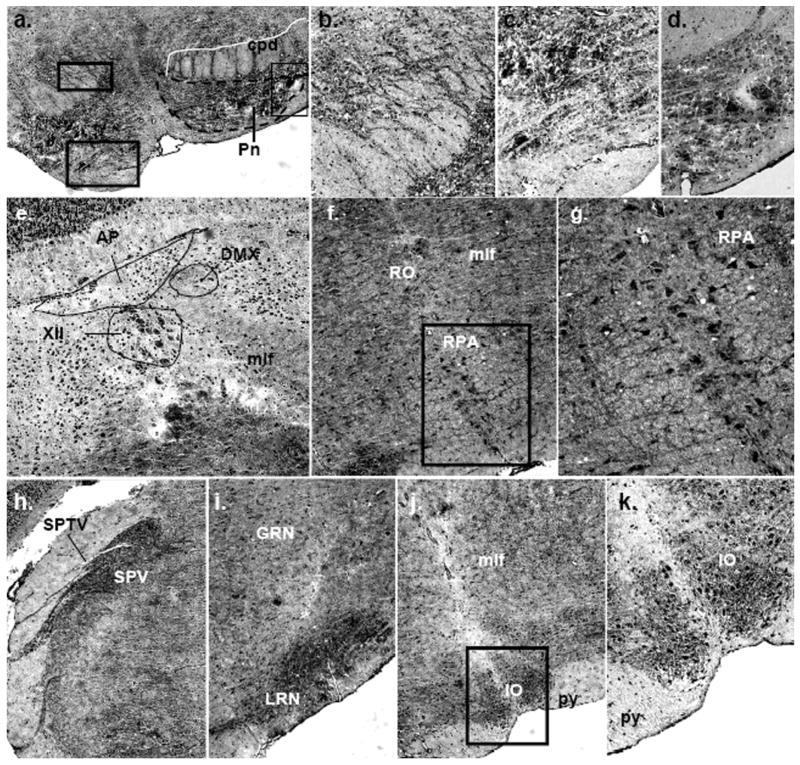
Immunolocalization of mPMAT protein in the midbrain and hindbrain. (a–d) Dense mPMAT staining in the pontine nuclei cell bodies (a, c, d) and fibers (a, b). (e) Expression of mPMAT in the medulla showing labeling of cells in the area postrema, dorsal motor nucleus of the vagus nerve, hypoglossal nucleus and medial longitudinal fasciculus. (f, g) mPMAT immunoreactivity in neurons of the nucleus raphe obscurus and nucleus raphe pallidus. Panel g contains the boxed region in panel f. (h–k) Immunolabeling of the spinal trigeminal nerve tract and nucleus, gigantoreticular and lateral reticular nucleus and inferior olivary complex. Images were obtained at 20× (a, e, h–j) and 40× (b–d, f, g, k) magnification.
Fig. 8.
Immunolocalization of mPMAT protein in the forebrain. (a) mPMAT expression in the granular neurons of the olfactory bulb. (b, c) mPMAT expression n neurons of the neocortex. (d) mPMAT expression in CA1-3 fields and dentate gyrus (DG) of the hippocampus. (e, g–i) Expression of mPMAT protein in the caudate putamen (striatum) at low magnification (e) (4×) and at higher magnification (g–i) (20×–60×). The striosomes are shown in panels g and h. Panel h shows a high-magnification (40×) image of a striosome containing densely-labeled neurons (example shown in boxed area). The plasma membrane and projections of striatal neurons are strongly labeled (panel i, boxed area from panel h). (f) Dense expression of mPMAT protein in the arcuate nucleus of the hypothalamus.
In the midbrain, mPMAT immunoreactivity was pronounced in the tegmentum. Immunoreactive cells were detected in the midbrain substantia nigra pars compacta and ventral tegmental area (Fig. 7f). The red nucleus was also very densely labeled (Fig. 7f). Of the hindbrain, the pontine nuclei were the most densely immunostained regions containing strong labeling of both cell bodies and fibers (Fig. 7f–i; Fig. 9a–d).
In the cerebellum, mPMAT immunoreactivity was very dense in the cerebellar cortex, particularly within the granule cell layer (Fig. 7h). Low to intermediate labeling was observed in Purkinje and molecular cell layers. In contrast to our ISH data, which showed low to intermediate labeling of the medulla, mPMAT protein immunoreactivity in the neuropil and fibers of the medulla was very intense (Fig. 7h; Fig. 9f–k). Additionally, there were many clearly labeled cell bodies in the area postrema, dorsal motor nucleus of the vagus nerve and hypoglossal nucleus (Fig. 9e). In a more caudal coronal section of the medulla, we observed very dense labeling of the neuropil and cell bodies of the inferior olivary complex, lateral reticular nucleus and gigantocellular reticular nucleus (Fig. 9i–k). Overall, mPMAT immunostaining in the hindbrain was strongest in the cerebellar granular layers, pontine nuclei and medulla cortex.
In the fiber tracts, we observed light mPMAT immunoreactivity in the corpus callosum, consistent with neuronal fiber staining (Fig. 7i). However, other white matter structures including the stria terminalis, internal capsule and optic chiasm were not significantly stained (Fig. 7c and d). The anterior commissure displayed dense immunoreactivity compared with other white matter structures (Fig. 7i). The white matter of the cerebellum showed low to moderate labeling (Fig. 7h). The pyramid of the medulla was not strongly labeled (Fig. 9j and k) and the spinal tract of the trigeminal nerve showed very faint staining compared with surrounding cells (Fig. 9h).
PMAT expression in neuronal and astroglial cells
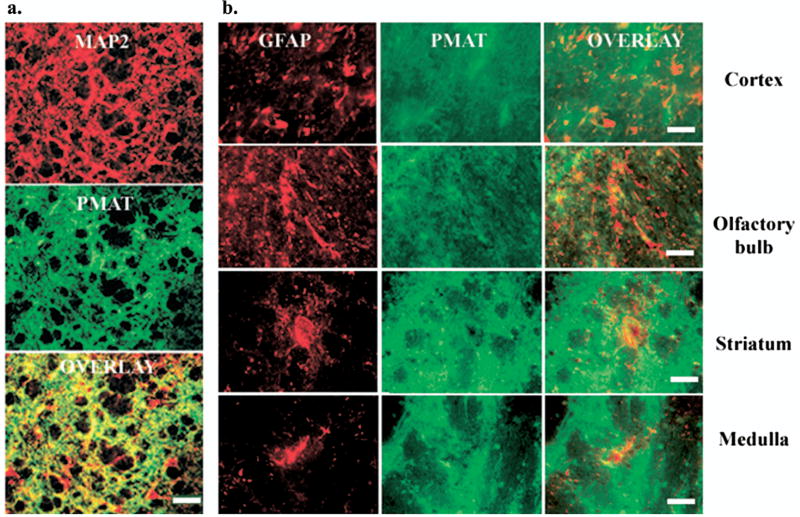
Our ISH and IHC detected mPMAT expression in diverse groups of neurons including pyramidal neurons, interneurons, granular neurons and Purkinje cells. In contrast, we did not observe significant expression in astroglial cells by morphology. To confirm the cellular localization of mPMAT, we performed double-immunofluorescence IHC on mouse brain sections using the P469 antibody and specific markers for neurons and astrocytes (Fig. 10). Antibodies against MAP2 or GFAP were co-incubated with either the P469 antibody or peptide antigen preadsorbed control antisera followed by fluorescence IHC with Alexa-fluor-488 or -555-conjugated secondary antibodies. The P469-antibody showed staining (green) patterns similar to MAP2 (red) and the signals overlapped (orange) extensively throughout the brain. An example is shown in Fig. 10b, where intense anti-MAP2 and P469 co-labeling of neuronal cell bodies and processes was observed in striatum. A number of neurons immunoreactive to anti-MAP2 but not to P469 were also observed scattering throughout the sections, indicating that mPMAT is not expressed in all MAP2-positive cells. In contrast, expression of GFAP was restricted to astrocytes and displayed a localization pattern that was distinct from that of mPMAT in all brain regions. Fig. 10a shows individual staining (green for P469 and red for anti-GFAP) and their overlays in fore-brain cortex, olfactory bulb, striatum and medulla.
Fig. 10.
PMAT co-localizes with neuronal but not astroglial cell markers. Dual-immunofluorescence histochemistry was performed to establish the localization of mPMAT in relation to prototypical markers for astrocytes and neurons. (a) Immunofluorescence of a section from striatum incubated with antibodies against the MAP2 (top panel) and mPMAT (middle panel). Bottom panel is an overlay of top and middle images. (b) Immunofluorescence staining of GFAP (images in the left column) and mPMAT (center column) in representative brain regions. Image overlays (right column) reveal distinct expression patterns for GFAP and mPMAT. Scale bars=100 μm (a), 50 μm (b).
PMAT expression in primarily cultured rat neurons
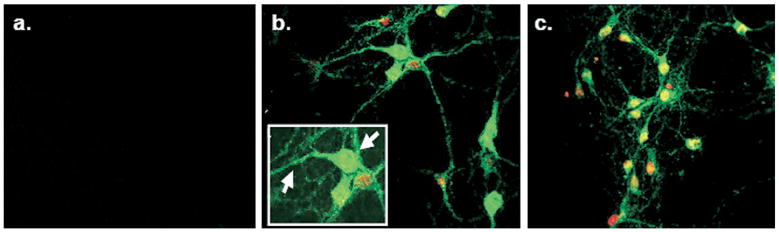
Our ISH and IHC studies showed high levels of PMAT expression in various neurons in the mouse brain. To provide additional evidence, we obtained and treated primary-cultured neurons dissected from E14 rat ventral mesencephalon (prepared as described by Hsuan et al., 2006) with the P469 anti-PMAT antibody or peptide antigen preadsorbed control antisera. The P469 antibody was directed toward a 14 a.a. region absolutely conserved among rat, mouse and hPMAT isoforms. Significant PMAT expression was observed in neuronal cultures incubated with the P469 antibody (Fig. 11b and c), whereas no fluorescent labeling was observed in the cultures treated with the pre-absorbed control antibody (Fig. 11a). Immunoreactive signals were present in the cell bodies, dendrites and axons, where fluorescent labeling appeared punctate throughout the axon (Fig. 11b and inset, arrows). These data are consistent with our observation in mouse brain that PMAT is expressed in neuronal cells.
Fig. 11.
PMAT immunofluorescence in primary ventral mesencephalic cultures prepared from E14 rats. Panel a illustrates neuron cultures stained with peptide preabsorbed P469 antisera. Panels b and c contain cultures labeled with the P469 antibody. Arrows indicate PMAT expression in cell in neural processes and cell bodies. The cell nuclei were counterstained with Topro-3 (red).
Immunolocalization of hPMAT in human cerebellum
Immunofluorescence histochemistry was also performed on sections of human cerebellum with the P469 antibody. We observed extensive staining of neuronal fibers in the cerebellar cortex, but not in the control antisera-treated section (Fig. 12). Immunostaining was particularly prominent in axons and dendrites projecting from the nerve cell bodies. Somatic staining was relatively light and did not overlap with the counterstaining of the cell nuclei. No PMAT-immunoreactive cells with the typical morphology of glia were observed.
Fig. 12.
Fluorescence immunolocalization of hPMAT in human cerebellar cortex. Immunofluorescence histochemistry was performed on sections of human cerebellum cortex incubated with preimmune antisera (1:200 dilution) (a) or P469 anti-PMAT antibody (1:200 dilution) (b). The nuclei were counterstained with Topro-3 (red). Images were obtained at 20× magnification.
DISCUSSION
This study accomplished several objectives toward characterizing PMAT, a low affinity, high capacity monoamine transporter recently discovered in our laboratory. Firstly, we determined the regional distribution of mPMAT in the mouse brain using RT-PCR, non-radioactive ISH and immunohistochemical methods. Secondly, we showed that mPMAT protein is present in neuronal rather than astroglial cell types. Finally, we cloned the mPMAT isoform, and demonstrated that mPMAT transports monoamine neurotransmitters in a manner similar to hPMAT. In addition, we also developed a polyclonal antibody capable of cross-reacting with human and rodent PMAT proteins. Ultimately, the information and reagents generated by our study will assist efforts to further clarify the role of low-affinity monoamine transport mechanisms in the brain.
mPMAT mRNA and protein are widely distributed across the brain
Data generated from our studies unanimously showed that mPMAT mRNA and protein are widely expressed in the mouse brain. The highest expression levels of both PMAT mRNA and protein are observed in forebrain cortex, olfactory tubercle, hippocampus, cerebellum and choroid plexus. With regard to mRNA expression, the distributional patterns of mPMAT observed in our ISH study are in general agreement with ISH data for mPMAT (Slc29a4) recently released by the Allen Brain Atlas (http://brainatlas.com/aba/), which also revealed widespread expression of mPMAT across the brain. There was also a general agreement between mPMAT mRNA distribution as detected by ISH and RT-PCR, and mPMAT protein distribution in our IHC and ICC studies. Protein and mRNA detection methods consistently showed high expression levels in the forebrain (dentate gyrus, cortex and olfactory areas) and cerebellum, with relatively lower levels in the midbrain. However, some differences in mRNA and protein distribution were noted in several areas, including medulla, the corpus callosum and anterior commissure. Such discordance may be explained as proteins can be delivered to a distance along neural projections whereas mRNA expression is generally confined to the cytoplasm in the cell body of neuronal cells.
PMAT is localized in neurons
Our ISH and IHC studies consistently detected mPMAT signals in neurons of diverse populations, including morphologically distinct cell types such as pyramidal neurons, interneurons, granular neurons and Purkinje cells. IHC and ICC studies revealed staining of both cell bodies and neuropil (Figs. 8–11). In addition, the ventricular epithelial cells of the choroid plexus also highly express mPMAT. Notably, we did not detect any specific labeling of astrocytes in ISH or IHC that could be differentiated from background staining. Using antibodies directed against the established astrocyte marker GFAP and all three isoforms of the neuronally-expressed MAP2 protein, our data further confirmed that mPMAT expression is confined to neuronal rather than astroglial cells (Fig. 10). While the neurochemical properties of the PMAT-expressing neurons need further investigation, the observation that PMAT-immunoreactive cell bodies are found throughout multiple brain regions and in multiple neuronal subtypes which do not synthesize monoamines (e.g. interneurons and Purkinje cells), suggests that, in contrast to SERT and DAT, PMAT is not confined to neurons that predominantly produce monoamines.
Comparison of mPMAT immunoreactivity with DAT and SERT
Like its human counterpart, mPMAT also transports 5-HT and DA (Fig. 1b). We have previously hypothesized that PMAT may represent an additional mechanism for monoamine clearance in the brain, especially for 5-HT and DA (Engel et al., 2004). Using an in vitro hybrid depletion approach, we recently provided indirect evidence that PMAT may contribute significantly to total brain 5-HT uptake (Zhou et al., 2007). While detailed neurochemical and functional testing in animal models is needed to investigate this mechanism, a comparison of the brain expression patterns of PMAT with that of SERT and DAT may provide initial clues to the physiological function of PMAT. To this end, we have compiled literature data on DAT and SERT protein expression (Garris et al., 1993; Ciliax et al., 1995; Freed et al., 1995; Qian et al., 1995; Revay et al., 1996; Sur et al., 1996; Ase et al., 2000b; Zhou et al., 2002; Delis et al., 2004), and compared with the regional brain distribution of mPMAT protein (Tables 1 and 2). Because the distributional patterns for SERT and DAT are very similar in rat and mouse (Blakely et al., 1994; Qian et al., 1995), we included data from both species. For some brain regions, little information is available for SERT and DAT by IHC studies. For these regions, we also included data from quantitative autoradiography studies, which have been shown to correlate well with protein expression data (Blakely et al., 1994; Qian et al., 1995; Torres et al., 2003).
Table 1.
Comparison of PMAT expression with SERT and DAT expression in the forebrain
| Brain area, Telencephalon | Protein density |
Reference | ||
|---|---|---|---|---|
| PMAT | SERT | DAT | ||
| Olfactory areas | ||||
| Accessory olfactory bulb | +/− to + | + | Ase (2000b) | |
| Anterior olfactory nucleus | + to ++ | + | ++ | Ase (2000b), Delis (2004) |
| Glomerular layer | +++ | ++to +++ | ++ | Sur (1996), Revay (1996) |
| Granular cell layer | +++ | ++ to +++ | +/− | Sur (1996), Ase (2000b), Revay (1996) |
| Mitral cell layer | +++ | ++ | Sur (1996), Ase (2000b) | |
| Cortex | ||||
| Agranular insular cortex | ++ to +++ | + | +/− to + | Ase (2000b), Freed (1995) |
| Auditory cortex | +++ | + | +/− to + | Ase (2000b), Freed (1995), Delis (2004) |
| Cingulate cortex, area 1 | + to ++ | +/− | +/−; ++ | Ase (2000b), Garris (1993), Ciliax (1995), Freed (1995) |
| Cingulate cortex, area 2 | + to ++ | +/− | +/−; ++ | Ase (2000b), Garris (1993), Ciliax (1995), Freed (1995) |
| Claustrum | +/− | ++ | ++ | Ase (2000b), Ciliax (1995), Freed (1995), Delis (2004) |
| Frontal association area | ++ to +++ | +/− | Garris (1993) | |
| Infralimbic cortex | + to ++ | +/− | Garris (1993) | |
| Entorhinal cortex | +++ | +++ | +/− to + | Qian (1995), Freed (1995), Delis (2004) |
| Orbital cortex | + to ++ | + to ++ | +/− to + | Ase (2000b), Freed (1995) |
| Piriform cortex | +++ | + | +/− to + | Ase (2000b), Freed (1995), Delis (2004) |
| Prelimbic cortex | ++ to +++ | + | +/− to + | Ase (2000b), Freed (1995) |
| Primary motor cortex | ++ to +++ | +/− | +/− to + | Ase (2000b), Freed (1995), Delis (2004) |
| Secondary motor cortex | ++ to +++ | +/− | +/− to + | Ase (2000b), Freed (1995), Delis (2004) |
| Primary visual cortex | +++ | +/− | +/− to + | Ase (2000b), Freed (1995), Delis (2004) |
| Secondary visual cortex | +++ | +/− | +/− to + | Ase (2000b), Freed (1995), Delis (2004) |
| Primary somatosensory cortex | +++ | + | +/− to + | Ase (2000b), Freed (1995), Delis (2004) |
| Secondary somatosensory cortex | +++ | + | +/− to + | Ase (2000b), Freed (1995), Delis (2004) |
| Retrosplenial agranular cortex | ++ to +++ | +/− | +/− to + | Ase (2000b), Freed (1995), Delis (2004) |
| Retrosplenial granular cortex | ++ to +++ | +/− | +/− to + | Ase (2000b), Freed (1995), Delis (2004) |
| Hippocampus | ||||
| CA1 field | ++ | + to ++ | + | Sur (1996), Qian (1995), Delis (2004) |
| CA2 field | ++ | +; ++ | + | Ase (2000b), Zhou (2002), Delis (2004) |
| CA3 field | ++ | +; +++ | + | Ase (2000b), Sur (1996), Qian (1995), Delis (2004) |
| Dentate gyrus | +++ | +/−; + | + | Ase (2000b), Sur (1996), Delis (2004) |
| Amygdala | ||||
| Basolateral amygdaloid nucleus | ++ | ++ | + | Ase (2000b), Sur (1996), Revay (1996) |
| Cortical amygdaloid nucleus | ++ | + | + | Sur (1996), Revay (1996) |
| Lateral amygdaloid nucleus | ++ | + | +/−; ++ | Sur (1996), Revay (1996), Ciliax (1996) |
| Striatum | ||||
| Caudate putamen | ++ | +++ | +++ | Sur (1996), Ciliax (1996), Freed (1995) |
| Islands of Cajella | ++ | +++ | Freed (1995) | |
| Lateral septal nucleus | ++ | ++ to +++ | + to ++ | Sur (1996), Freed (1995) |
| Nucleus accumbens, core | ++ | +; +++ | +++ | Ase (2000b), Sur (1996), Ciliax (1996), Freed (1995), Delis (2004) |
| Nucleus accumbens, shell | ++ | +/− | +++ | Sur (1996), Ciliax (1996), Freed (1995), Delis (2004) |
| Olfactory tubercle | +++ | ++ | +++ | Ase (2000b), Ciliax (1996), Freed (1995), Delis (2004) |
| Pallidum | ||||
| Bed nucleus of the stria terminalis | +/− | +/− | Ciliax (1996) | |
| Globus pallidus | ++ | ++ to +++ | +++ | Ase (2000b), Ciliax (1996) |
| Medial septal nucleus | ++ | +; +++ | + | Ase (2000b), Sur (1996), Delis (2004) |
| Triangular septal nucleus | ++ to +++ | + | + | Ase (2000b), Delis (2004) |
| Ventral pallidum | +++ | +++ | +++ | Ase (2000b), Sur (1996), Delis (2004) |
Rating scale for protein immunoreactivity density: +/−, very low/absent; +, low; ++, intermediate to high; +++, very high.
Marks separated by a semicolon (e.g. +, ++) indicate differences in reported values between studies. Blank areas indicate regions for which specific expression data are not available.
Table 2.
Comparison of PMAT expression with SERT and DAT expression in the midbrain and hindbrain
| Brain area | Protein density |
Reference | ||
|---|---|---|---|---|
| PMAT | SERT | DAT | ||
| Diencephalon | ||||
| Thalamus | ||||
| Medial geniculate nucleus | +++ | + | + | Ase (2000b), Delis (2004) |
| Paraventricular thalamic nucleus | ++ | +++ | ++ | Ase (2000b), Delis (2004) |
| Hypothalamus | ||||
| Arcuate hypothalamic nucleus | +++ | +; +/− | Garris (1993), Ciliax (1996), Freed (1995) | |
| Lateral hypothalamic area | ++ | ++ | +/+; − | Ase (2000b), Ciliax (1996), Delis (2004) |
| Preoptic area | +/− to + | ++ | +/− | Ase (2000b), Ciliax (1996) |
| Subfornical organ | +++ | +/− | Ciliax (1996) | |
| Substantia innominata | +/− to + | +/− | Ciliax (1996) | |
| Suprachiasmatic nucleus | +++ | +/− | Ciliax (1996) | |
| Supraoptic nucleus | +/− to + | +/− | Ciliax (1996) | |
| Ventrolateral preoptic nucleus | ++ | +/− | Ciliax (1996) | |
| Zona incerta | + to ++ | + | + | Ase (2000b), Revay (1996), Delis (2004) |
| Mesencephalon | ||||
| Caudal linear nucleus of the raphe | +/− to + | +++ | +/− | Ase (2000b), Sur (1996), Qian (1995), Ciliax (1996), Freed (1995) |
| Dorsal raphe nucleus | +/− | +++ | +/− | Ase (2000b), Sur (1996), Qian (1995), Zhou (2002), Ciliax (1996), Freed (1995) |
| Deep gray layer of the superior colliculus | + | + | + | Ase (2000b), Delis (2004) |
| Deep white layer of the superior colliculus | + | +++ | + | Ase (2000b), Delis (2004) |
| Intermediate gray layer of the superior colliculus | +++ | + | + | Ase (2000b), Delis (2004) |
| Superficial gray layer of the superior colliculus | +++ | + | + | Ase (2000b), Delis (2004) |
| External cortex of the inferior colliculus | +/− | + | + | Ase (2000b), Delis (2004) |
| Nucleus of the brachium of the inferior colliculus | ++ | ++ | + | Ase (2000b), Delis (2004) |
| Periaqueductal gray | +/− to + | +++; ++to +++ | +/−; + | Ase (2000b), Sur (1996), Ciliax (1996), Delis (2004) |
| Deep mesencephalic nucleus | + | ++ | Ase (2000b) | |
| Interpeduncular nucleus | ++ | ++ | Ase (2000b) | |
| Red nucleus | +++ | + | Ase (2000b) | |
| Substantia nigra pars compacta | + to ++ | +++ | +++ | Ase (2000b), Sur (1996), Qian (1995), Zhou (2002), Ciliax (1996), Freed (1995) |
| Substantia nigra pars reticularis | + | ++ | Ciliax (1996), Freed (1995) | |
| Ventral tegmental area | + to ++ | +++ | +++ | Ase (2000b), Zhou (2002), Ciliax (1996), Freed (1995) |
| Locus coeruleus | +/− to + | ++; +++ | +/− | Ase (2000b), Sur (1996), Qian (1995), Zhou (2002), Ciliax (1996), Freed (1995) |
| Rhombencephalon | ||||
| Pontine reticular nucleus | ++ to +++ | + | Ase (2000b) | |
| Cerebellar granule cell layers | +++ | +/− | +/− to + | Sur (1996), Delis (2004) |
| Cerebellar molecular cell layers | + to ++ | +/− | + | Sur (1996), Delis (2004) |
| Purkinje cell layers | + to ++ | +/− | Sur (1996) | |
| Inferior olive | +++ | + | Ase (2000b) | |
| Medial vestibular nucleus, medial caudal part | +/− | +/− | Sur (1996) | |
| Medial vestibular nucleus, parvicellular part | +++ | +/− | Sur (1996) | |
| Pontine nuclei | +++ | + | Ase (2000b) | |
| Raphe magnus nucleus | +++ | +++ | Qian (1995) | |
| Reticulotegmental nucleus of the pons | ++ | ++ to +++ | Qian (1995) | |
Rating scale for protein immunoreactivity density: +/−, very low/absent; +, low; ++, intermediate to high; +++, very high.
Marks separated by a semicolon (e.g. +, ++) indicate differences in reported values between studies. Blank areas indicate regions for which specific expression data are not available.
Although 5HT and DA-producing cell bodies are restricted to the midbrain and brain stem regions, projections of the serotonergic and dopaminergic neurons virtually innervate the entire CNS. As seen in Tables 1 and 2, PMAT protein is co-expressed in many brain regions with SERT and/or DAT. However, several sites that lack significant expression of the high affinity transporters also have high expression levels of PMAT. Notably, PMAT is expressed with high intensity in several sites that reportedly lack or express SERT only at very low levels, including the cerebellum, accumbens shell, dentate gyrus of the hippocampus and many structures of the forebrain cortex (Tables 1 and 2). In these areas, PMAT may play a significant role in 5-HT clearance. For instance, in the nucleus accumbens, which is innervated by serotonergic fibers and contains high tissue levels of 5-HT (8.17 ng/g in mouse) (Ase et al., 2000a), two distinct types of 5-HT axons exist (Mamounas et al., 1991; Brown and Molliver, 2000). The accumbens core is innervated by 5-HT axons that express SERT, but in the shell region nearly all 5-HT axons lack SERT (Brown and Molliver, 2000). The intense staining of PMAT in the nucleus accumbens shell indicates that this transporter may represent a possible candidate for terminating 5-HT neurotransmission in this area. PMAT protein is also highly expressed in cerebellum, an area that has the lowest SERT expression and is often used as a “SERT-free” region (Kish et al., 2005). 5-HT is known to play important neuromodulatory roles in the cerebellum, which contains low but detectable levels of 5-HT (0.94 ng/g in mouse) (Ase et al., 2000a) and expresses a number of 5-HT receptors (Schweighofer et al., 2004). The intense PMAT expression in both mouse and human cerebellum (Table 2 and Fig. 12) suggests that this transporter may play an important role in modulating 5-HT signaling in this structure. Likewise, PMAT may also play a role in dopaminergic neurotransmission in areas (e.g. medial prefrontal cortex) that receive DA input but poorly express DAT (Wayment et al., 2001).
It is also possible that the broad expression of PMAT mRNA and protein is related to a more general role in maintaining low levels of monoamine neurotransmitters throughout the brain. It is well established that released monoamine neurotransmitters can escape high affinity uptake1 transporters and diffuse away (i.e. spill over) from the synaptic cleft (Garris et al., 1994; Bunin and Wightman, 1999). Because the brain is an isolated compartment (due to the presence of the blood– brain and blood–CSF barriers), the existence of a widely available uptake system, such as PMAT, may help to prevent the buildup of diffused amines in the extracellular space of the brain. In this regard, other uptake2 transporters, such as OCT3, could also contribute. OCT3 is highly expressed in the circumventricular organs such as area postrema and subfornical organ, but low levels of expression are also found throughout the brain (Vialou et al., 2004; Amphoux et al., 2006). hPMAT and OCT3 seem to exhibit complementary substrate preferences toward monoamines; PMAT favors 5HT and DA whereas OCT3 favors NE and epinephrine (Grundemann et al., 1998; Engel et al., 2004; Amphoux et al., 2006). As most metabolizing enzymes, such as the monoamine oxidases A and B, are intracellular enzymes widely expressed in the various brain cells (Saura et al., 1996; Jahng et al., 1997), cellular uptake of monoamines by PMAT and OCT3 is a prerequisite for these enzymes to gain assess to their substrates. PMAT and OCTs may thus work together with monoamine metabolizing enzymes to maintain a low background level of these signaling molecules.
CONCLUSION
In summary, our studies suggest that PMAT is a widely distributed, neuronally-expressed monoamine transporter in the mammalian brain. The specificity of PMAT toward biogenic amines and its wide expression in the CNS indicate a role of this transporter in safeguarding a low baseline level of extracellular monoamines in the brain. Furthermore, in areas that receive monoamine innervations but lack high-affinity transporters, PMAT may take an active role in terminating monoaminergic neurotransmission. While more studies are necessary to uncover the function of this new biogenic amine transporter in the CNS, the work presented here represents a necessary step toward understanding the physiological roles of PMAT in the mammalian brain.
Acknowledgments
We thank Ms. Heather Klintworth and Dr. Zhengui Xia for providing primary cultures of rat neurons for this study. This work was supported in part by NIH grant GM066233 and NIH Pharmacological Sciences Training grant T32 GM 07750.
Abbreviations
- CA
field of the hippocampus
- DA
dopamine
- DAT
dopamine transporter
- DIG
digoxigenin
- FBS
fetal bovine serum
- GFAP
glial fibrillary acidic protein
- hPMAT
human plasma membrane monoamine transporter
- ICC
immunocytochemistry
- IHC
immunohistochemistry
- ISH
in situ hybridization
- KLH
keyhole limpet hemocyanin
- KRH
Krebs-Ringer-Henseleit
- MAP2
microtubule-associated protein
- mDAT
mouse dopamine transporter
- MDCK
Madin-Darby canine kidney
- MEM
minimum essential medium
- mNET
mouse nor-epinephrine transporter
- mPMAT
mouse plasma membrane monoamine transporter
- mSERT
mouse serotonin transporter
- NE
norepinephrine
- NET
norepinephrine transporter
- OCT
organic cation transporter
- PBS
phosphate-buffered saline
- PBST
phosphate-buffered saline with 0.05% Tween-20
- PCR
polymerase chain reaction
- PMAT
plasma membrane monoamine transporter
- RT-PCR
reverse-transcription polymerase chain reaction
- SERT
serotonin transporter
- YFP
yellow fluorescent protein
Abbreviations in the figures
- Ac
accumbens nucleus
- acc
anterior commissure
- aci
anterior commissure, intrabulbar part
- AOB
anterior olfactory bulb
- AOD
anterior olfactory nucleus, dorsal part
- AOM
anterior olfactory nucleus, medial part
- AP
area postrema
- Aq
aqueduct
- Arc
arcuate nucleus of the hypothalamus
- AuD
auditory cortex, dorsal part
- AuDM
auditory cortex, dorsomedial part
- BST
bed nucleus of the stria terminalis
- Cb
cerebellum
- cc
corpus collosum
- Cg1
cingulate cortex, area 1
- COA
cortical amygdalar area
- cpd
cerebral peduncle
- Cpu
caudate putamen
- ctx
cerebral cortex
- df
dorsal fornix
- DG
dentate gyrus
- DMTg
dorsomedial tegmental area
- DMX
dorsal motor nucleus of the vagus nerve
- DPO
dorsal periolivary region
- DT
dorsal terminal nucleus of the accessory optic tract
- DTgP
dorsal tegmental nucleus, pericentral part
- DTT
dorsal tenia tecta
- D3V
dorsal third ventricle
- EP1
external plexiform layer of the olfactory bulb
- fi
fimbria hippocampus
- fmi
forceps minor of the corpus callosum
- FrA
frontal association cortex
- Gl
glomerular layer of the olfactory bulb
- Gr
granular layer of the cerebellum
- GRN
gigantocellular reticular nucleus
- GrO
granular layer of the olfactory bulb
- HY
hypothalamus
- Ic
internal capsule
- IG
induseum griseum
- IO
inferior olivary complex
- IPR
interpenduncular nucleus, rostral subnucleus
- LGP
lateral globus pallidus
- LO
lateral orbital cortex
- LRN
lateral reticular nucleus
- LV
lateral ventricle
- MD
medulla
- MG
medial geniculate nucleus
- mi
mitral cell layer of the olfactory bulb
- mlf
medial longitudinal fasciculus
- Mol
cerebellar molecular layer
- MPO
medial preoptic nucleus
- mv
medial vestibular nucleus
- M1
primary motor cortex
- M2
secondary motor cortex
- opt
optic tract
- PAG
periaqueductal gray
- Pir
piriform cortex
- PnC
pontine reticular nucleus, caudal
- PnO
pontine reticular nucleus, oral
- PN/Pn
pontine nucleus
- PO/po
posterior thalamic nuclear group
- Prl
prelimbic cortex
- PrS
presubiculum
- PVA
paraventricular thalamic nucleus, anterior part
- PVH
paraventricular hypothalamic nucleus
- py
pyramid
- R
red nucleus
- rf
rhinal fissure
- RMC
red nucleus, magnocellular part
- RO
nucleus raphe obscurus
- RPA
nucleus raphe pallidus
- RSA
retrosplenial agranular cortex
- RSG
retrosplenial granular cortex
- SFO
subfornical organ
- SN
substantia nigra
- SNC
substantia nigra, compact part
- SNR
substantia nigra, reticular part
- SOC
superior olivary complex
- SPTV
spinal tract of the trigeminal nerve
- SPV
spinal nucleus of the trigeminal nerve
- sp5
spinal trigeminal tract
- str
striatum
- Sub
submedius thalamic nucleus
- S1
primary somatosensory cortex
- S2
secondary somatosensory cortex
- TRS
triangular nucleus of the septum
- Tu
olfactory tubercle
- VPL
ventral posterolateral thalamic nucleus
- VTA
ventral tegmental area
- VTg
ventral tegmental nucleus
- VTT
ventral tenia tecta
- V2
secondary visual cortex
- V4
fourth ventricle
- wm
white matter
- XII
hypoglossal nucleus
- I–VI
layers 1–6 of the cerebral cortex
References
- Amara SG, Kuhar MJ. Neurotransmitter transporters: recent progress. Annu Rev Neurosci. 1993;16:73–93. doi: 10.1146/annurev.ne.16.030193.000445. [DOI] [PubMed] [Google Scholar]
- Amphoux A, Vialou V, Drescher E, Bruss M, Mannoury La Cour C, Rochat C, Millan MJ, Giros B, Bonisch H, Gautron S. Differential pharmacological in vitro properties of organic cation transporters and regional distribution in rat brain. Neuropharmacology. 2006;50:941–952. doi: 10.1016/j.neuropharm.2006.01.005. [DOI] [PubMed] [Google Scholar]
- Ase AR, Reader TA, Hen R, Riad M, Descarries L. Altered serotonin and dopamine metabolism in the CNS of serotonin 5-HT(1A) or 5-HT(1B) receptor knockout mice. J Neurochem. 2000a;75:2415–2426. doi: 10.1046/j.1471-4159.2000.0752415.x. [DOI] [PubMed] [Google Scholar]
- Ase AR, Strazielle C, Hebert C, Botez MI, LaLonde R, Descarries L, Reader TA. Central serotonin system in Dystonia musculorum mutant mice: biochemical, autoradiographic and immunocytochemical data. Synapse. 2000b;37:179–193. doi: 10.1002/1098-2396(20000901)37:3<179::AID-SYN2>3.0.CO;2-0. [DOI] [PubMed] [Google Scholar]
- Blakely RD, De Felice LJ, Hartzell HC. Molecular physiology of norepinephrine and serotonin transporters. J Exp Biol. 1994;196:263–281. doi: 10.1242/jeb.196.1.263. [DOI] [PubMed] [Google Scholar]
- Brown P, Molliver ME. Dual serotonin (5-HT) projections to the nucleus accumbens core and shell: relation of the 5-HT transporter to amphetamine-induced neurotoxicity. J Neurosci. 2000;20:1952–1963. doi: 10.1523/JNEUROSCI.20-05-01952.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bunin MA, Wightman RM. Paracrine neurotransmission in the CNS: involvement of 5-HT. Trends Neurosci. 1999;22:377–382. doi: 10.1016/s0166-2236(99)01410-1. [DOI] [PubMed] [Google Scholar]
- Burrows GH, Myers MM, Whittemore SR, Hendley ED. Calcium-sensitive accumulation of norepinephrine in rat cerebral cortex. Eur J Pharmacol. 1981;69:301–312. doi: 10.1016/0014-2999(81)90476-3. [DOI] [PubMed] [Google Scholar]
- Carlsson A. Perspectives on the discovery of central monoaminergic neurotransmission. Annu Rev Neurosci. 1987;10:19–40. doi: 10.1146/annurev.ne.10.030187.000315. [DOI] [PubMed] [Google Scholar]
- Ciliax BJ, Heilman C, Demchyshyn LL, Pristupa ZB, Ince E, Hersch SM, Niznik HB, Levey AI. The dopamine transporter: immunochemical characterization and localization in brain. J Neurosci. 1995;15:1714–1723. doi: 10.1523/JNEUROSCI.15-03-01714.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delis F, Mitsacos A, Giompres P. Dopamine receptor and transporter levels are altered in the brain of Purkinje cell degeneration mutant mice. Neuroscience. 2004;125:255–268. doi: 10.1016/j.neuroscience.2004.01.020. [DOI] [PubMed] [Google Scholar]
- Eisenhofer G. The role of neuronal and extraneuronal plasma membrane transporters in the inactivation of peripheral catecholamines. Pharmacol Ther. 2001;91:35–62. doi: 10.1016/s0163-7258(01)00144-9. [DOI] [PubMed] [Google Scholar]
- Engel K, Wang J. Interaction of organic cations with a newly identified plasma membrane monoamine transporter. Mol Pharmacol. 2005;68:1397–1407. doi: 10.1124/mol.105.016832. [DOI] [PubMed] [Google Scholar]
- Engel K, Zhou M, Wang J. Identification and characterization of a novel monoamine transporter in the human brain. J Biol Chem. 2004;279:50042–50049. doi: 10.1074/jbc.M407913200. [DOI] [PubMed] [Google Scholar]
- Franklin KBJ, Paxinos G. The mouse brain in stereotaxic coordinates. San Diego, CA: Academic Press; 1997. [Google Scholar]
- Freed C, Revay R, Vaughan RA, Kriek E, Grant S, Uhl GR, Kuhar MJ. Dopamine transporter immunoreactivity in rat brain. J Comp Neurol. 1995;359:340–349. doi: 10.1002/cne.903590211. [DOI] [PubMed] [Google Scholar]
- Gainetdinov RR, Sotnikova TD, Caron MG. Monoamine transporter pharmacology and mutant mice. Trends Pharmacol Sci. 2002;23:367–373. doi: 10.1016/s0165-6147(02)02044-8. [DOI] [PubMed] [Google Scholar]
- Garris PA, Ciolkowski EL, Pastore P, Wightman RM. Efflux of dopamine from the synaptic cleft in the nucleus accumbens of the rat brain. J Neurosci. 1994;14:6084–6093. doi: 10.1523/JNEUROSCI.14-10-06084.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garris PA, Collins LB, Jones SR, Wightman RM. Evoked extracellular dopamine in vivo in the medial prefrontal cortex. J Neurochem. 1993;61:637–647. doi: 10.1111/j.1471-4159.1993.tb02168.x. [DOI] [PubMed] [Google Scholar]
- Greengard P. The neurobiology of slow synaptic transmission. Science. 2001;294:1024–1030. doi: 10.1126/science.294.5544.1024. [DOI] [PubMed] [Google Scholar]
- Grundemann D, Schechinger B, Rappold GA, Schomig E. Molecular identification of the corticosterone-sensitive extraneuronal catecholamine transporter. Nat Neurosci. 1998;1:349–351. doi: 10.1038/1557. [DOI] [PubMed] [Google Scholar]
- Hendley ED, Taylor KM, Snyder SH. 3H-Normetanephrine uptake in rat brain slices. Relationship to extraneuronal accumulation of norepinephrine. Eur J Pharmacol. 1970;12:167–179. doi: 10.1016/0014-2999(70)90062-2. [DOI] [PubMed] [Google Scholar]
- Hevner RF, Shi L, Justice N, Hsueh Y, Sheng M, Smiga S, Bulfone A, Goffinet AM, Campagnoni AT, Rubenstein JL. Tbr1 regulates differentiation of the preplate and layer 6. Neuron. 2001;29:353–366. doi: 10.1016/s0896-6273(01)00211-2. [DOI] [PubMed] [Google Scholar]
- Hsuan SL, Klintworth HM, Xia Z. Basic fibroblast growth factor protects against rotenone-induced dopaminergic cell death through activation of extracellular signal-regulated kinases 1/2 and phosphatidylinositol-3 kinase pathways. J Neurosci. 2006;26:4481– 4491. doi: 10.1523/JNEUROSCI.4922-05.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inazu M, Kubota N, Takeda H, Zhang J, Kiuchi Y, Oguchi K, Matsumiya T. Pharmacological characterization of dopamine transport in cultured rat astrocytes. Life Sci. 1999;64:2239–2245. doi: 10.1016/s0024-3205(99)00175-7. [DOI] [PubMed] [Google Scholar]
- Iversen LL. Role of transmitter uptake mechanisms in synaptic neurotransmission. Br J Pharmacol. 1971;41:571–591. doi: 10.1111/j.1476-5381.1971.tb07066.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jahng JW, Houpt TA, Wessel TC, Chen K, Shih JC, Joh TH. Localization of monoamine oxidase A and B mRNA in the rat brain by in situ hybridization. Synapse. 1997;25:30–36. doi: 10.1002/(SICI)1098-2396(199701)25:1<30::AID-SYN4>3.0.CO;2-G. [DOI] [PubMed] [Google Scholar]
- Kish SJ, Furukawa Y, Chang LJ, Tong J, Ginovart N, Wilson A, Houle S, Meyer JH. Regional distribution of serotonin transporter protein in postmortem human brain: is the cerebellum a SERT-free brain region? Nucl Med Biol. 2005;32:123–128. doi: 10.1016/j.nucmedbio.2004.10.001. [DOI] [PubMed] [Google Scholar]
- Lightman SL, Iversen LL. The role of uptake2 in the extraneuronal metabolism of catecholamines in the isolated rat heart. Br J Pharmacol. 1969;37:638–649. doi: 10.1111/j.1476-5381.1969.tb08502.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mamounas LA, Mullen CA, O’Hearn E, Molliver ME. Dual serotoninergic projections to forebrain in the rat: morphologically distinct 5-HT axon terminals exhibit differential vulnerability to neurotoxic amphetamine derivatives. J Comp Neurol. 1991;314:558–586. doi: 10.1002/cne.903140312. [DOI] [PubMed] [Google Scholar]
- Mireylees SE, Brammer NT, Buckley GA. A kinetic study of the in vitro uptake of [3H]dopamine over a wide range of concentrations by rat striatal preparations. Biochem Pharmacol. 1986;35:4065–4071. doi: 10.1016/0006-2952(86)90029-8. [DOI] [PubMed] [Google Scholar]
- Mundorf ML, Joseph JD, Austin CM, Caron MG, Wightman RM. Catecholamine release and uptake in the mouse prefrontal cortex. J Neurochem. 2001;79:130–142. doi: 10.1046/j.1471-4159.2001.00554.x. [DOI] [PubMed] [Google Scholar]
- Qian Y, Melikian HE, Rye DB, Levey AI, Blakely RD. Identification and characterization of antidepressant-sensitive serotonin transporter proteins using site-specific antibodies. J Neurosci. 1995;15:1261–1274. doi: 10.1523/JNEUROSCI.15-02-01261.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ravary A, Muzerelle A, Darmon M, Murphy DL, Moessner R, Lesch KP, Gaspar P. Abnormal trafficking and subcellular localization of an N-terminally truncated serotonin transporter protein. Eur J Neurosci. 2001;13:1349–1362. doi: 10.1046/j.0953-816x.2001.1511.x. [DOI] [PubMed] [Google Scholar]
- Revay R, Vaughan R, Grant S, Kuhar MJ. Dopamine transporter immunohistochemistry in median eminence, amygdala, and other areas of the rat brain. Synapse. 1996;22:93–99. doi: 10.1002/(SICI)1098-2396(199602)22:2<93::AID-SYN1>3.0.CO;2-C. [DOI] [PubMed] [Google Scholar]
- Saura J, Bleuel Z, Ulrich J, Mendelowitsch A, Chen K, Shih JC, Malherbe P, Da Prada M, Richards JG. Molecular neuroanatomy of human monoamine oxidases A and B revealed by quantitative enzyme radioautography and in situ hybridization histochemistry. Neuroscience. 1996;70:755–774. doi: 10.1016/s0306-4522(96)83013-2. [DOI] [PubMed] [Google Scholar]
- Schweighofer N, Doya K, Kuroda S. Cerebellar aminergic neuromodulation: towards a functional understanding. Brain Res Brain Res Rev. 2004;44:103–116. doi: 10.1016/j.brainresrev.2003.10.004. [DOI] [PubMed] [Google Scholar]
- Sora I, Hall FS, Andrews AM, Itokawa M, Li XF, Wei HB, Wichems C, Lesch KP, Murphy DL, Uhl GR. Molecular mechanisms of cocaine reward: combined dopamine and serotonin transporter knockouts eliminate cocaine place preference. Proc Natl Acad Sci U S A. 2001;98:5300–5305. doi: 10.1073/pnas.091039298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sur C, Betz H, Schloss P. Immunocytochemical detection of the serotonin transporter in rat brain. Neuroscience. 1996;73:217–231. doi: 10.1016/0306-4522(96)00030-9. [DOI] [PubMed] [Google Scholar]
- Torres GE, Gainetdinov RR, Caron MG. Plasma membrane monoamine transporters: structure, regulation and function. Nat Rev Neurosci. 2003;4:13–25. doi: 10.1038/nrn1008. [DOI] [PubMed] [Google Scholar]
- Vialou V, Amphoux A, Zwart R, Giros B, Gautron S. Organic cation transporter 3 (Slc22a3) is implicated in salt-intake regulation. J Neurosci. 2004;24:2846–2851. doi: 10.1523/JNEUROSCI.5147-03.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wayment HK, Schenk JO, Sorg BA. Characterization of extracellular dopamine clearance in the medial prefrontal cortex: role of monoamine uptake and monoamine oxidase inhibition. J Neurosci. 2001;21:35–44. doi: 10.1523/JNEUROSCI.21-01-00035.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu X, Huang W, Ganapathy ME, Wang H, Kekuda R, Conway SJ, Leibach FH, Ganapathy V. Structure, function, and regional distribution of the organic cation transporter OCT3 in the kidney. Am J Physiol Renal Physiol. 2000;279:F449–F458. doi: 10.1152/ajprenal.2000.279.3.F449. [DOI] [PubMed] [Google Scholar]
- Wu X, Kekuda R, Huang W, Fei YJ, Leibach FH, Chen J, Conway SJ, Ganapathy V. Identity of the organic cation transporter OCT3 as the extraneuronal monoamine transporter (uptake2) and evidence for the expression of the transporter in the brain. J Biol Chem. 1998;273:32776–32786. doi: 10.1074/jbc.273.49.32776. [DOI] [PubMed] [Google Scholar]
- Xia L, Engel K, Zhou M, Wang J. Membrane localization and pH-dependent transport of a newly cloned organic cation transporter (PMAT) in kidney cells. Am J Physiol Renal Physiol. 2007;292:F682–F690. doi: 10.1152/ajprenal.00302.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou FC, Lesch KP, Murphy DL. Serotonin uptake into dopamine neurons via dopamine transporters: a compensatory alternative. Brain Res. 2002;942:109–119. doi: 10.1016/s0006-8993(02)02709-9. [DOI] [PubMed] [Google Scholar]
- Zhou M, Engel K, Wang J. Evidence for significant contribution of a newly identified monoamine transporter (PMAT) to serotonin uptake in the human brain. Biochem Pharmacol. 2007;73:147–154. doi: 10.1016/j.bcp.2006.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]