Abstract
Amyotrophic lateral sclerosis is a degenerative disorder of motor neurons that typically develops in the 6th decade and is uniformly fatal, usually within 5 years. To identify genetic variants associated with susceptibility and phenotypes in sporadic ALS, we performed a genome-wide SNP analysis in sporadic ALS cases and controls. A total of 288,357 SNPs were screened in a set of 1,821 sporadic ALS cases and 2,258 controls from the U.S. and Europe. Survival analysis was performed using 1,014 deceased sporadic cases. Top results for susceptibility were further screened in an independent sample set of 538 ALS cases and 556 controls. SNP rs1541160 within the KIFAP3 gene (encoding a kinesin-associated protein) yielded a genome-wide significant result (P = 1.84 × 10−8) that withstood Bonferroni correction for association with survival. Homozygosity for the favorable allele (CC) conferred a 14.0 months survival advantage. Sequence, genotypic and functional analyses revealed that there is linkage disequilibrium between rs1541160 and SNP rs522444 within the KIFAP3 promoter and that the favorable alleles of rs1541160 and rs522444 correlate with reduced KIFAP3 expression. No SNPs were associated with risk of sporadic ALS, site of onset, or age of onset. We have identified a variant within the KIFAP3 gene that is associated with decreased KIFAP3 expression and increased survival in sporadic ALS. These findings support the view that genetic factors modify phenotypes in this disease and that cellular motor proteins are determinants of motor neuron viability.
Keywords: genome-wide association study, single nucleotide polymorphism
Amyotrophic lateral sclerosis (ALS) is an age-dependent, degenerative disorder of motor neurons (1) that typically develops in the 6th decade and is uniformly fatal, usually within 5 years (2). Approximately 10% of ALS cases are dominantly inherited (3); 20% of these are caused by mutations in the gene encoding copper/zinc superoxide dismutase 1 (SOD1) (4); mutations in the TARDBP gene (5, 6) account for ≈5% of cases. Rare familial cases arise from mutations in genes encoding the vesicle-associated membrane associated protein B (7), alsin (a RAB5-guanine nucleotide exchange factor) (8, 9), senataxin (10) or dynactin (11). Recently, we reported that ≈5% of familial ALS cases are due to mutations in the FUS/TLS gene (12, 13) whose product binds DNA and RNA, as does TARDBP. The cause of sporadic ALS is thought to be multifactorial, with environmental, infectious and genetic etiologies. Reported associations with variants in diverse genes (14–25) have proven difficult to replicate. Advances in the technology for large-scale genotyping of single nucleotide polymorphisms (SNPs) have facilitated unbiased, genome-wide association studies. Examples include the identification of IL2RA and IL7RA variants as risk factors for multiple sclerosis (26–28) and the recent reports of 6 new gene regions associated with type 2 diabetes (29–35). To test the hypothesis that commonly occurring genetic variants modify susceptibility, survival, site of onset or age of onset in sporadic ALS, we have undertaken a multicenter genetic analysis of 1,821 sporadic ALS cases (SALS) and 2,258 controls.
Results
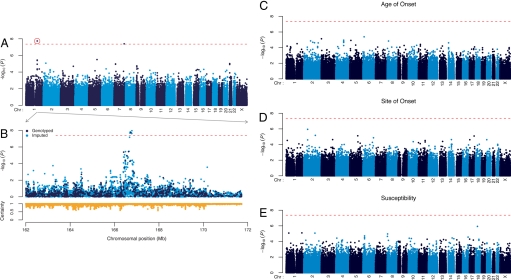
Genotypes were obtained from 3 sources in the U.S. (Boston, Atlanta, National Institute of Neurological Disorders and Stroke) and 3 in Europe (London, France, Netherlands), using Illumina BeadArrays. Survival information was not available for samples from the NINDS and France. No SNPs generated significant P values for association with susceptibility, site of onset, and age of onset of disease after Bonferroni correction (288,357 SNPs × 4 phenotypes) (Fig. 1 C–E, Table 1, see also Table S1). In a further attempt to reveal any SNPs that were associated with susceptibility of sporadic ALS, we elected to genotype those SNPs that yielded a P < 5.0 × 10−4 (153 in total) in a confirmatory (Stage 2) panel consisting of 538 ALS cases and 556 controls. Survival information was not available for most of the samples. Successful genotypes were obtained for 139 (90.8%) of the SNPs; none of the variants yielded a significant P value after Bonferroni multiple test correction (Table S1). Although our study failed to confirm recent reports that susceptibility to sporadic ALS may be mediated by variants in the inositol-triphosphate receptor (ITPR2) (21), DPP6 (22, 23) or a novel, brain-expressed gene (FLJ10986) (24), these discrepancies may reflect differences in methodology or case populations (Table S2).
Fig. 1.
Plot of −log10(P) for survival, age of onset, site of onset and susceptibility of sporadic ALS. Analysis for survival, age of onset, site of onset and susceptibility was performed for 288,357 SNPs and the results for the entire genome were plotted as shown in A and (C–E). The x axis represents the chromosomal position and the y axis represents the −log10 of the P value for each SNP. The dotted line represents the cutoff for Bonferroni significance. (A) P values from linear regression analysis of survival. SNPs rs1541160 (circled) and rs855913 were significant after Bonferroni correction. (B) A closer view of the rs1541160 region is shown. Dark points represent SNPs typed in the study, and light points represent SNPs whose genotypes were imputed. (Lower) Imputation certainty for each imputed SNP, defined as the average maximum posterior genotype call probability. The chromosomal region spans 5 Mb on either side of SNP rs1541160. Positions are in National Center for Biotechnology Information build-35 coordinates.
Table 1.
SNPs yielding best P-values in four tested categories
| Category | SNP | Chr | Position |
P |
Nearest gene | Freq. |
Regr. coeff. | SE | OR | R2 | Alleles | ||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Raw | Corrected | Case | Cont. | Limb | Bulbar | ||||||||||
| Susceptibility | rs10438933 | 18 | 27527127 | 1.18 × 10−6 | 1 | B4GALT6 | 0.155 | 0.124 | 1.296 | A → G | |||||
| rs16856202 | 1 | 228461886 | 7.98 × 10−6 | 1 | DISC1 | 0.020 | 0.038 | 0.499 | A → C | ||||||
| rs873917 | 1 | 39801888 | 8.37 × 10−6 | 1 | NT5C1A | 0.316 | 0.285 | 1.162 | G → T | ||||||
| rs10192369 | 2 | 161206395 | 8.53 × 10−6 | 1 | RBMS1 | 0.512 | 0.473 | 1.170 | C → T | ||||||
| rs10503354 | 8 | 5940352 | 1.09 × 10−5 | 1 | MCPH1 | 0.134 | 0.166 | 0.773 | G → A | ||||||
| Site of Onset | rs7577894 | 2 | 55920555 | 1.13 × 10−6 | 1 | EFEMP1 | 0.386 | 0.471 | 0.708 | T → C | |||||
| rs13015447 | 2 | 167203485 | 6.71 × 10−6 | 1 | SCN7A | 0.344 | 0.425 | 0.709 | T → G | ||||||
| rs7702057 | 5 | 115755737 | 7.60 × 10−6 | 1 | SEMA6A | 0.033 | 0.066 | 0.488 | C → A | ||||||
| rs8066857 | 17 | 68207698 | 8.11 × 10−6 | 1 | SLC39A11 | 0.172 | 0.235 | 0.676 | C → T | ||||||
| rs3734803 | 6 | 151961759 | 1.37 × 10−5 | 1 | C6orf97 | 0.177 | 0.243 | 0.669 | C → T | ||||||
| Age of Onset | rs697739 | 6 | 16850012 | 4.14 × 10−6 | 1 | ATXN1 | −2.036 | 0.4409 | 0.0097 | G → A | |||||
| rs2619566 | 3 | 2599938 | 7.42 × 10−6 | 1 | CNTN4 | −3.032 | 0.6746 | 0.0107 | A → G | ||||||
| rs1486860 | 4 | 113751047 | 1.12 × 10−5 | 1 | ALPK1 | 1.833 | 0.4161 | 0.0082 | T → C | ||||||
| rs2451852 | 5 | 16642040 | 1.20 × 10−5 | 1 | FLJ20152 | −2.503 | 0.5702 | 0.0097 | T → C | ||||||
| rs2055593 | 2 | 98450631 | 1.39 × 10−5 | 1 | CNGA3 | −2.136 | 0.4903 | 0.0103 | G → A | ||||||
| Survival | rs1541160 | 1 | 166727460 | 1.84 × 10−8 | 0.021 | KIFAP3 | 0.582 | 0.1026 | 0.0293 | T → C | |||||
| rs855913 | 7 | 148641310 | 4.02 × 10−8 | 0.046 | ZNF746 | 1.079 | 0.1951 | 0.0279 | C → A | ||||||
| rs11241713 | 5 | 123147464 | 3.17 × 10−6 | 1 | CSNK1G3 | 0.786 | 0.1677 | 0.0202 | G → T | ||||||
| rs648576 | 1 | 166631085 | 3.63 × 10−6 | 1 | KIFAP3 | 0.490 | 0.1052 | 0.0198 | C → T | ||||||
| rs3177980 | 1 | 166408144 | 4.10 × 10−6 | 1 | SELL | 0.509 | 0.1098 | 0.0248 | A → G | ||||||
In contrast to susceptibility, site of onset, and age of onset, SNPs rs1541160 and rs855913 generated significant P values after Bonferroni correction (288,357 SNPs × 4 phenotypes) for association with disease survival, using linear regression (Fig. 1A and Table 1). For SNP rs1541160, the nominal and Bonferroni-corrected P values were 1.84 × 10−8 and 0.021. Within the region of rs1541160, several SNPs (including imputed SNP alleles) yielded a cluster of positive values; 4 of the imputed SNPs were significant after Bonferroni correction (Fig. 1B). SNP rs1541160 maps within intron 8 of the KIFAP3 gene (encoding a kinesin-associated protein) on chromosome 1. For SNP rs855913, the nominal and Bonferroni-corrected P values were 4.02 × 10−8 and 0.046. This SNP lies ≈10 kb upstream of the ZNF746 gene. This gene was not further characterized for 3 reasons. First, a sensitivity analysis of this SNP revealed that it does not replicate within the individual Boston population (P = 0.264). Second, in our sensitivity analyses, had we analyzed the U.S. as the Stage 1 population, we would not have identified this variant due to its relatively high P value (0.0073) and low ranking (2169th). This is in contrast with SNP rs1541160 that emerges as significant in our study, whether considering the aggregate of all cases or each individual population. Finally, for the ZNF746 gene variant in question, the homozygotes for the minor allele are rare (0.7%) so that it is difficult to ascertain the reliability of the results (despite having >1,821 ALS cases in our screening study).
The genotype frequencies of rs1541160 are 9.9% (CC), 39.7% (CT) and 50.4% (TT); the minor allele frequency is 29.7% (Table S3). The rate of genotyping rs1541160 was 100%. Hardy–Weinberg testing revealed that rs1541160 is in equilibrium (controls P = 0.541, cases P = 0.527, all P = 0.970). Haplotypes defined by 3 SNPs, rs2750014, rs4656729 and rs12123693, but excluding rs1541160, yielded association with survival comparable to that of rs1541160 (P = 1.35 × 10−9), indicating that genotyping artifacts specific to rs1541160 are not generating the association. Further tests confirmed that this association is not biased by population stratification (SI Methods). Pairwise linkage disequilibrium (LD) analysis for ≈50 SNPs distributed across the locus defined by KIFAP3 and 5 neighboring genes (SCYL3, C1orf156, Clorf112, and Selectins E and L) revealed disequilibrium that spanned ≈155 kb from marker rs2750014 to rs1216443 but was centered on rs1541160 within the gene KIFAP3 (Fig. S1).
Our approach to identify variants associated with increased survival was based on a joint analysis of 4 DNA sets. This approach is more powerful than a 2-staged method in which a set of SNPs within an initial population below a cutoff P value is verified within a secondary confirmation population (36). However, because several genome-wide association studies (GWAS) have used 2-stage analyses (21, 22, 24, 30, 32, 33), we have investigated how such an approach would influence our results. We performed a sensitivity analysis in which we dropped each of the 4 populations in turn from the study and computed the P value associated with both the remaining populations (i.e., simulating a Stage 1 study) and the removed population (i.e., simulating a Stage 2 study). In each case, this sensitivity testing (Table 2) revealed that rs1541160 remained in the top 5 survival-associated SNPs. The largest increase in the P value (to 5.50 × 10−6) was observed after removal of the Boston DNA set, which contains the most samples (n = 398). Furthermore, the P values for rs1541160 from each individual site (the simulated Stage 2 study) also yielded significant P values, with the exception of that from Atlanta (P = 0.079), which contains the lowest number of samples (n = 90). Interestingly, the median survival increased at a minimum of 20.51% (Netherlands) and a maximum of 78.54% (London) in individuals harboring a CC genotype for rs1541160 as compared with a TT genotype (Table 2). We also performed the sensitivity analysis by grouping the population into U.S. (Atlanta and Boston) and Europe (London and Netherlands). By this approach, both the U.S. and Europe yielded a high ranking for rs1541160 if used as a Stage 1 population (20th and 31st, respectively). Furthermore, both the U.S. and Europe yielded significant individual P values (5.55 × 10−5 and 7.70 × 10−5, respectively) (Table 2). These results also confirm that the observed association is not due to population stratification, which would not be expected to yield a significant P value for each individual population.
Table 2.
Sensitivity analysis of rs1541160 within four populations. Median Survival is represented in years
| Stage 2 population | Stage 1 P | Stage 1 rank | Stage 2 P | Stage 1 sample size | Stage 2sample size | Stage 2 median surv. CC | Stage 2 median Surv. TT | Survival increase (CC vs. TT) |
|---|---|---|---|---|---|---|---|---|
| Atlanta | 2.89E-07 | 1st | 0.079 | 924 | 90 | 4.00 | 2.77 | 44.40% |
| Boston | 5.50E-06 | 5th | 8.86E-04 | 616 | 398 | 4.27 | 2.93 | 45.73% |
| London | 4.00E-06 | 5th | 1.42E-03 | 804 | 210 | 4.66 | 2.61 | 78.54% |
| Netherlands | 2.60E-07 | 1st | 0.022 | 698 | 316 | 2.82 | 2.34 | 20.51% |
| United States | 7.70E-05 | 31st | 5.55E-05 | 526 | 488 | 4.23 | 2.85 | 48.42% |
| Europe | 5.55E-05 | 20th | 7.70E-05 | 488 | 526 | 3.39 | 2.42 | 40.08% |
| Total | 1.84 × 10−8 | 1,014 | 3.96 | 2.67 | 46.44% | |||
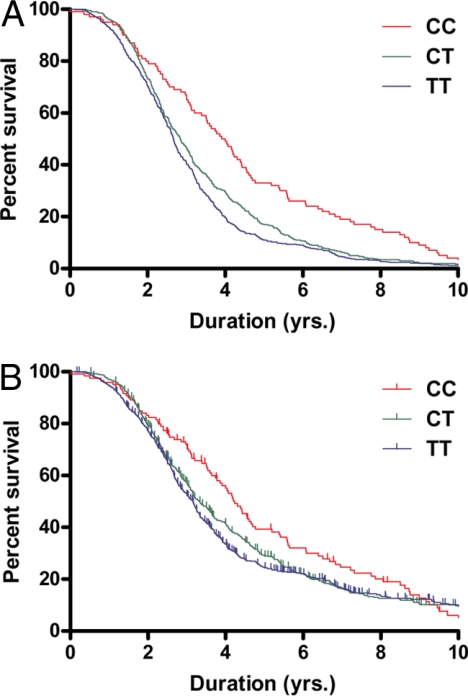
The absolute median survival for the CC, CT, and TT genotypes were 3.96, 2.84 and 2.67, respectively. The absolute mean survival for the CC, CT, and TT genotypes were 4.60, 3.40, and 3.07, respectively. As assessed by linear regression analysis, the mean and median survival increments for the CC genotype were 14.0 and 14.9 months, respectively, compared with the TT genotype, based on the analysis with SNP rs1541160 alone. With genotypic-based survival curve analysis using the Peto-Prentice generalized Wilcoxon text and deceased ALS cases (Fig. 2A), the P value for the rs1541160 SNP is 3.87 × 10−6 (n = 1,014). A censored analysis considering all of the cases (Fig. 2B) yielded a P value for rs1541160 of 1.82 × 10−3 (n = 1,321).
Fig. 2.
Influence of alleles of rs1541160 in KIFAP3 on survival in sporadic ALS. Survival curves were generated from sporadic ALS cases with different rs1541160 genotypes and analyzed using the Peto-Prentice generalized Wilcoxon test; each curve plots percentage survival versus duration for each of the 3 genotypes observed for rs1541160. To better visualize the differences between the genotypes, the curves were truncated at 10 years. As shown, individuals harboring the CC genotype (red) display an increased survival as compared with individuals with the CT (green) and TT (blue) genotypes. (A) uncensored data; (B) censored data. In B, small vertical marks superimposed on the survival curve show censored points at which individuals were lost to further assessment.
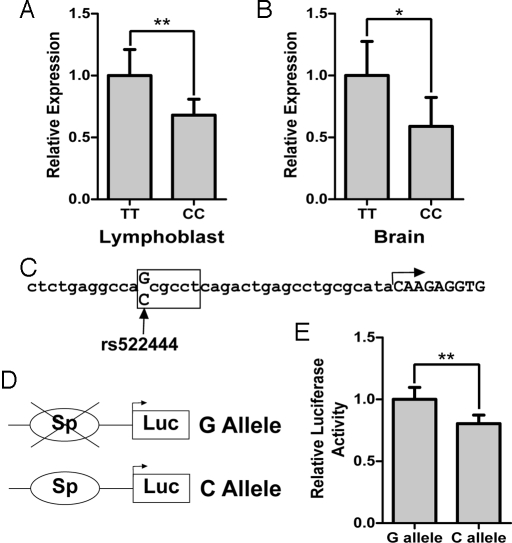
Sequence analysis of the KIFAP3 coding region and exon/intron boundaries of 8 individuals homozygous for the CC and 4 for the TT rs1541160 genotype (12 individuals) did not reveal variants in strong LD with rs1541160, suggesting that the KIFAP3-mediated increase in survival is not due to an alteration in its protein sequence. To determine whether the expression of KIFAP3 is modified by the genotype of rs1541160, we performed real-time PCR on lymphoblastoid cell lines harboring either a CC (n = 38) or TT (n = 40) genotype for rs1541160. KIFAP3 expression in the CC genotypes was 31.9% less than that in the TT genotypes (Fig. 3A) (P = 0.0084, Wilcoxon 2-sample test). A comparison of occipital lobe brain samples homozygous for either the C (n = 9) or T (n = 17) alleles again revealed a decrease in expression of KIFAP3 (41.1%) in the CC as compared with TT samples (Fig. 3B) (P = 0.025, Wilcoxon 2-sample test). A second real-time PCR probe for KIFAP3 confirmed the findings for both lymphoblast and brain samples. Western blotting of the brain samples confirmed a decrease in KIFAP3 protein (69.8%) in CC samples compared with TT samples (Fig. S2) (P = 0.014, Wilcoxon 2-sample test).
Fig. 3.
Association of rs1541160/rs522444 with expression of KIFAP3. (A and B) Total RNA was isolated from lymphoblastoid cell lines (A) or occipital cortex brain tissue harboring (B) either a CC or TT genotype for rs1541160. Relative expression of KIFAP3 was determined by real-time PCR. As shown, individuals harboring the CC genotype display decreased expression compared with individuals with the TT genotype. Error bars represent the 95% C.I. (C) The sequence of the KIFAP3 promoter region is shown. The arrow indicates the transcriptional start site. SNP rs522444 is indicated at the −25 position. The box represents the location of the putative Sp1 binding site. (D) The KIFAP3 promoter region and 5′ UTR (633 bp) were amplified from individuals harboring either the CC or TT genotype and subcloned upstream of the firefly luciferase gene. The schematic (not drawn to scale) represents the resultant constructs, which differ only at a single base pair located at rs522444. (E) The resultant constructs were transfected into SKN-AS cells and relative luciferase activity was measured. The error bars represent the 95% C.I. A promoterless vector yielded <1% relative activity. The construct containing the G allele displays higher luciferase activity relative to the C allele. *, P < 0.05; **, P < 0.01.
Given that rs1541160 is located in the eighth intron of KIFAP3, we decided to examine SNPs within the promoter region which may influence gene regulation to determine whether any were in linkage disequilibrium with rs1541160. Sequencing of ≈1.6 kb of this promoter within 8 individuals homozygous for the CC and 4 for the TT rs1541160 genotype revealed a variant (C/G, previously identified as rs522444) located at −25 bp relative to the transcription start site (Fig. 3C). This variant is in complete linkage disequilibrium with rs1541160 for all 24 chromosomes (r2 = 1.00). Genotypes derived from the HapMap project for rs1541160 and rs522444 further documented that these 2 SNPs are in complete LD (r2 = 1.00) in Caucasian and SubSaharan African populations (evident through analysis of 116 and 118 chromosomes, respectively). We further confirmed this high level of LD by genotyping an additional 1,017 individuals (r2 = 0.99).
Analysis of the KIFAP3 promoter using transcription element search system (TESS) (37) revealed that the KIFAP3 gene lacks a TATA box and that rs522444 lies within a putative Sp1 binding site (log-likelihood score = 12) (Fig. 3C). Moreover, the C allele of rs522444, which is in linkage disequilibrium with the lower expressing C allele of rs1541160, creates the putative Sp1 binding site, whereas the G allele at rs522444 destroys this site. Comparison with the promoter region in other primates demonstrates that the G allele is evolutionarily conserved suggesting this is the ancestral allele and that the KIFAP3 gene is not normally regulated by a Sp1 binding site. Because Sp1 family members binding to cognate Sp1 binding sites can both repress and activate gene expression (38), we sought to define the influence of variants of rs522444 on KIFAP3 promoter activity by subcloning the promoter region and 5′ UTR of KIFAP3 upstream of the firefly luciferase gene. The constructs (Fig. 3D), differing only at the variant position, were transfected into the neuroblastoma cell line SKN-AS and the resulting luciferase activity was measured. This revealed that the promoter harboring the C allele for rs522444 displayed significantly decreased transcriptional activity (19.6%, P = 0.0044, Wilcoxon 2-sample test) (Fig. 3E). This result is analogous to the decreased expression of KIFAP3 in association with the rs1541160 C allele in both brain and lymphoblast tissues. We conclude that rs522444 C variant alone suffices to attenuate expression of the KIFAP3 gene.
Discussion
In a set of 4,079 DNA samples from sporadic ALS cases and controls we have completed an unbiased analysis of ≈288,000 SNPs distributed across the genome and identified a SNP (rs1541160) within the KIFAP3 gene that is associated with reduced KIFAP3 expression and longer survival. Other SNPs in the region of rs1541160 trended toward association (Fig. 1B); the 4th and 10th highest SNPs in the survival analysis were also within the KIFAP3 gene (Table S1). The failure to detect other significant SNP genotypes is subject to multiple interpretations. Perhaps most importantly, this suggests that there is not a single, readily detectable genetic variant that exerts a preponderant influence on either the risk of developing sporadic ALS or ALS phenotypes other than survival. In the present study, the absence of strongly associated SNPs other than rs1541160 may reflect other factors including inherent heterogeneity in the populations studied, locus and allelic heterogeneity, the inability of our present study design to detect underlying epistatic interactions of multiple gene variants, the effect of a microdeletions or insertions or inadequacies in the power of our study to detect genes of small effect. (For susceptibility studies, assuming conservatively a genome-wide significance level of 5.0 × 10−8 and a minor allele frequency of 0.3, for genotypic relative risks of 1.3 and 1.5 the corresponding powers are 53% and 100%). (Table S4).
That homozygosity for SNP rs1541160 confers survival variation of ≈14 months is of clinical importance in a disorder with a mean survival of only 3–5 years. Why attenuation of KIFAP3 expression should slow progression in ALS is unclear. With the kinesin motor proteins KIF3A and KIF3B, KIFAP3 forms a trimeric motor complex, KIF3, that mediates binding between the motor proteins and their cargoes, serving multiple functions such as chromosomal cytokinesis and anterograde transport (39, 40). Presumably, reduced levels of KIFAP3 modulate survival by favorably affecting both the stoichiometry of KIFAP3 and the KIF3 complex and one or more transport functions, such that the CC genotype of rs522444 is beneficial. Heightened expression of KIFAP3 is reportedly an early marker of disease in transgenic mutant SOD1 mice (41), suggesting that levels of KIFAP3 reflect adverse events within motor neurons. In human sporadic ALS, expression of 2 other kinesin-related proteins (KIF3Ab and KIF1Bb) is reportedly reduced (42). A recent report documents that KIFAP3 (also described as KAP3) binds mutant SOD1 protein, slowing axonal transport of choline acetyl transferase in motor neurons; moreover, KIFAP3/KAP3 colocalizes with mutant SOD1 in human motor neurons at autopsy (43). It is conceivable that diminished expression of KIFAP3/KAP3 has a beneficial impact on the SOD1-motor protein interaction in ALS. More generally, mutations in motor proteins are implicated in multiple motor neuron degenerative disorders in both humans (11, 44) and mice (45).
It is encouraging that investigations employing complex genetics may provide fresh insight into sporadic ALS (21–24, 46). Few genetic factors that modify ALS survival are reported (19, 20, 47); none were identified in the previous ALS genome analyses (21–24, 46). The identification of KIFAP3 as a determinant of progression rate of sporadic ALS is therefore promising; insights into this pathway may provide new targets for therapies to slow this devastating disease, for example, by reducing levels of KIFAP3 expression or modifying its interactions with 1 or more protein binding partners.
Materials and Methods
Genotypes were obtained from 3 sources in the U.S. (917 ALS, 912 controls) and 3 in Europe (904 ALS, 1,346 controls) (Table S5). To maximize the power of this study, we combined these into a single set of cases and controls (36). Duration information was obtained from 4 of the sites. A set of 307,776 SNPs common to all sites was used for this analysis. Multiple quality control measures were applied to the set of DNAs and SNPs. 10,360 SNPs were eliminated because they were not in Hardy–Weinberg equilibrium (P < 10−6 in controls) or demonstrated call rates <0.95 or minor allele frequencies <0.001. An additional 9,059 SNPs were eliminated by tests for divergence of cases and control call rates and for nonrandom missing genotype data (to determine whether genotypes are missing with respect to the true genotype as defined by the observed genotypes of nearby SNPs). Samples were excluded if their call rates were <95%; if genotypes revealed duplicate samples, relatedness (proportion of genome IBD >0.2), or excess homozygosity or heterozygosity (inbreeding coefficient >0.05 or <-0.025); or if reported gender did not match the genotypically-assessed gender. The sample set was additionally subjected to stratification analysis; based on the distribution of pairwise genome-wide identity-by-state distances, we applied complete linkage hierarchical cluster analysis and classical multidimensional scaling. As a result, 72 outlier samples, defined as 3 standard deviations from the group mean, were eliminated, leaving the cases and controls as in Table S5. For the final analysis, 288,357 SNPs were evaluated (Table S6). After applying quality control metrics to the full set of DNAs and SNPs (SI Methods), 288,357 SNPs remained that were evaluated in 4,079 DNA samples, yielding 1,176,208,203 genotypes (Table S6). Genotypic and phenotypic data are available through the dbGaP database at the National Center for Biotechnology Information web site (www.ncbi.nlm.nih.gov/UH). Genotypes were used for the analysis of 4 SALS phenotypes: susceptibility, site of onset, age of onset and survival of disease. Multiple analyses failed to detect biases introduced by stratification of our case-control cohorts (see SI Methods).
Supplementary Material
Acknowledgments.
We dedicate this article to the memories of Frieda R. Abraham, Donna Roberts, and Jennifer Estess. We thank Daniel Mirel (Broad Institute) for all of his assistance, all of the clinicians who assisted in recruiting cases and controls, particularly to clients with ALS and their families, and Jerry Xu and Dennis Gurgul of the Enterprise Research IS group at Partners Healthcare for support and use of the Hewlett-Packard computing facilities. The Boston-based investigations were supported by the ALS Therapy Alliance, Project ALS, the Angel Fund, the Pierre L. de Bourgknecht ALS Research Foundation, the Al-Athel ALS Research Foundation (R.H.B., J.E.L., D.M.M.-Y., I.R.-L, T.J.K.), the ALS Family Charitable Foundation, the American Academy of Neurology Foundation/ALS Association (A.-M.W.), the National Institute of Neurological Disorders and Stroke (R.H.B., T.J.K.), the Harvard Business School Class of 2007, the Yamner family, Avichai Kremer, Guy Yamen, Nate Boaz, and Professor Robert S. Kaplan. London-based investigations were supported by the Motor Neurone Disease Association of Great Britain and Northern Ireland (A.A.-C.), the Medical Research Council (A.A.-C.), the Wellcome Trust (P.N.L. and C.E.S.), and Psychiatry Research Trust (Tim Perkins Fund) (P.N.L., A.A.-.C., and C.E.S.). The investigations in France were supported by Assistance Publique, Hopitaux de Paris, Institut National de la Santé et de la Recherche Médicale, GIS-Institut des Maladies Rares and the Ministere de l'Enseignement Superieur et de la Recherche. V.S. was partially suported by a donation of the Peviani Family. We thank DNA banks maintained by the Coriell Institute (with support from the National Institute of Neurological Disorders and Stroke) and Genethon. This study used data from the SNP Database at the National Institute of Neurological Disorders and Stroke Human Genetics Resource Center DNA and Cell Line Repository (http://ccr.coriell.org/ninds). Investigations in The Netherlands were supported by The Netherlands Organisation for Scientific Research, the “Prinses Beatrix Fonds,” the Kersten Foundation, the Adessium Foundation, J. R. van Dijk and VSB Fonds (L.H.v.d.B.). This work was supported by Howard Hughes Medical Institute (P.S. and H.R.H.), the Packard Center for ALS Research (J.D.G. and M.P.), The Muscular Dystrophy Association (U.S.) (S.C. and O.H.), and National Center for Research Resources Grant U54 RR020278–01 (to The Broad Institute Center for Genotyping and Analysis). H.R.H. is Howard Hughes Medical Institute Investigator.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0812937106/DCSupplemental.
References
- 1.Tandan R, Bradley WG. Amyotrophic lateral sclerosis: Part 1. Clinical features, pathology and ethical issues in management. Ann Neurol. 1985;18:271–280. doi: 10.1002/ana.410180302. [DOI] [PubMed] [Google Scholar]
- 2.Tandan R, Bradley WG. Amyotrophic lateral sclerosis: Part 2. Etiopathogenesis. Ann Neurol. 1985;18:419–431. doi: 10.1002/ana.410180402. [DOI] [PubMed] [Google Scholar]
- 3.Mulder D, Kurland L, Offord K, Beard C. Familial adult motor neuron disease: Amyotrophic lateral sclerosis. Neurology. 1986;36:511–517. doi: 10.1212/wnl.36.4.511. [DOI] [PubMed] [Google Scholar]
- 4.Rosen DR, et al. Mutations in Cu/Zn superoxide dismutase gene are associated with familial amyotrophic lateral sclerosis. Nature. 1993;362:59–62. doi: 10.1038/362059a0. [DOI] [PubMed] [Google Scholar]
- 5.Sreedharan J, et al. TDP-43 mutations in familial and sporadic amyotrophic lateral sclerosis. Science. 2008;319:1668–1672. doi: 10.1126/science.1154584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kabashi E, et al. TARDBP mutations in individuals with sporadic and familial amyotrophic lateral sclerosis. Nat Genet. 2008;40:572–574. doi: 10.1038/ng.132. [DOI] [PubMed] [Google Scholar]
- 7.Nishimura AL, et al. A mutation in the vesicle-trafficking protein VAPB causes late-onset spinal muscular atrophy and amyotrophic lateral sclerosis. Am J Hum Genet. 2004;75:822–831. doi: 10.1086/425287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hadano S, et al. A gene encoding a putative GTPase regulator is mutated in familial amyotrophic lateral sclerosis 2. Nat Genet. 2001;29:166–173. doi: 10.1038/ng1001-166. [DOI] [PubMed] [Google Scholar]
- 9.Yang Y, et al. The gene encoding alsin, a protein with three guanine-nucleotide exchange factor domains, is mutated in a form of recessive amyotrophic lateral sclerosis. Nat Genet. 2001;29:160–165. doi: 10.1038/ng1001-160. [DOI] [PubMed] [Google Scholar]
- 10.Chen YZ, et al. DNA/RNA helicase gene mutations in a form of juvenile amyotrophic lateral sclerosis (ALS4) Am J Hum Genet. 2004;74:1128–1135. doi: 10.1086/421054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Puls I, et al. Mutant dynactin in motor neuron disease. Nat Genet. 2003;33:455–456. doi: 10.1038/ng1123. [DOI] [PubMed] [Google Scholar]
- 12.Kwiatkowski TJ, Jr, et al. Mutations in the FUS/TLS gene on chromosome 16 cause familial amyotrophic lateral sclerosis. Science. 2009;323:1205–1208. doi: 10.1126/science.1166066. [DOI] [PubMed] [Google Scholar]
- 13.Vance C, et al. Mutations in FUS, an RNA processing protein, cause familial amyotrophic lateral sclerosis type 6. Science. 2009;323:1208–1211. doi: 10.1126/science.1165942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Greenway MJ, et al. ANG mutations segregate with familial and “sporadic” amyotrophic lateral sclerosis. Nat Genet. 2006;38:411–413. doi: 10.1038/ng1742. [DOI] [PubMed] [Google Scholar]
- 15.Lambrechts D, et al. VEGF is a modifier of amyotrophic lateral sclerosis in mice and humans and protects motoneurons against ischemic death. Nat Genet. 2003;34:383–394. doi: 10.1038/ng1211. [DOI] [PubMed] [Google Scholar]
- 16.Wang XS, et al. Increased incidence of the Hfe mutation in amyotrophic lateral sclerosis and related cellular consequences. J Neurol Sci. 2004;227:27–33. doi: 10.1016/j.jns.2004.08.003. [DOI] [PubMed] [Google Scholar]
- 17.Saeed M, et al. Paraoxonase cluster polymorphisms are associated with sporadic ALS. Neurology. 2006;67:771–776. doi: 10.1212/01.wnl.0000227187.52002.88. [DOI] [PubMed] [Google Scholar]
- 18.Slowik A, et al. Paraoxonase gene polymorphisms and sporadic ALS. Neurology. 2006;67:766–770. doi: 10.1212/01.wnl.0000219565.32247.11. [DOI] [PubMed] [Google Scholar]
- 19.Moulard B, et al. Association between centromeric deletions of the SMN gene and sporadic adult-onset lower motor neuron disease. Ann Neurol. 1998;43:640–644. doi: 10.1002/ana.410430513. [DOI] [PubMed] [Google Scholar]
- 20.Veldink JH, et al. Homozygous deletion of the survival motor neuron 2 gene is a prognostic factor in sporadic ALS. Neurology. 2001;56:749–752. doi: 10.1212/wnl.56.6.749. [DOI] [PubMed] [Google Scholar]
- 21.van Es MA, et al. ITPR2 as a susceptibility gene in sporadic amyotrophic lateral sclerosis: A genome-wide association study. Lancet Neurol. 2007;6:869–877. doi: 10.1016/S1474-4422(07)70222-3. [DOI] [PubMed] [Google Scholar]
- 22.van Es MA, et al. Genetic variation in DPP6 is associated with susceptibility to amyotrophic lateral sclerosis. Nat Genet. 2008;40:29–31. doi: 10.1038/ng.2007.52. [DOI] [PubMed] [Google Scholar]
- 23.Cronin S, et al. A genome-wide association study of sporadic ALS in a homogenous Irish population. Hum Mol Genet. 2008;17:768–774. doi: 10.1093/hmg/ddm361. [DOI] [PubMed] [Google Scholar]
- 24.Dunckley T, et al. Whole-genome analysis of sporadic amyotrophic lateral sclerosis. N Engl J Med. 2007;357:775–788. doi: 10.1056/NEJMoa070174. [DOI] [PubMed] [Google Scholar]
- 25.Simpson CL, et al. Variants of the elongator protein 3 (ELP3) gene are associated with motor neuron degeneration. Hum Mol Genet. 2009;18:472–481. doi: 10.1093/hmg/ddn375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gregory SG, et al. Interleukin 7 receptor alpha chain (IL7R) shows allelic and functional association with multiple sclerosis. Nat Genet. 2007;39:1083–1091. doi: 10.1038/ng2103. [DOI] [PubMed] [Google Scholar]
- 27.Lundmark F, et al. Variation in interleukin 7 receptor alpha chain (IL7R) influences risk of multiple sclerosis. Nat Genet. 2007;39:1108–1113. doi: 10.1038/ng2106. [DOI] [PubMed] [Google Scholar]
- 28.International Multiple Sclerosis Genetics Consortium. Risk alleles for multiple sclerosis identified by a genomewide study. N Engl J Med. 2007;357:851–862. doi: 10.1056/NEJMoa073493. [DOI] [PubMed] [Google Scholar]
- 29.Frayling TM. Genome-wide association studies provide new insights into type 2 diabetes aetiology. Nat Rev Genet. 2007;8:657–662. doi: 10.1038/nrg2178. [DOI] [PubMed] [Google Scholar]
- 30.Sladek R, et al. A genome-wide association study identifies novel risk loci for type 2 diabetes. Nature. 2007;445:881–885. doi: 10.1038/nature05616. [DOI] [PubMed] [Google Scholar]
- 31.Steinthorsdottir V, et al. A variant in CDKAL1 influences insulin response and risk of type 2 diabetes. Nat Genet. 2007;39:770–775. doi: 10.1038/ng2043. [DOI] [PubMed] [Google Scholar]
- 32.Saxena R, et al. Genome-wide association analysis identifies loci for type 2 diabetes and triglyceride levels. Science. 2007;316:1331–1336. doi: 10.1126/science.1142358. [DOI] [PubMed] [Google Scholar]
- 33.Scott LJ, et al. A genome-wide association study of type 2 diabetes in Finns detects multiple susceptibility variants. Science. 2007;316:1341–1345. doi: 10.1126/science.1142382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zeggini E, et al. Replication of genome-wide association signals in UK samples reveals risk loci for type 2 diabetes. Science. 2007;316:1336–1341. doi: 10.1126/science.1142364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wellcome Trust Case Control Consortium. Genome-wide association study of 14,000 cases of seven common diseases and 3,000 shared controls. Nature. 2007;447:661–678. doi: 10.1038/nature05911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Skol AD, Scott LJ, Abecasis GR, Boehnke M. Joint analysis is more efficient than replication-based analysis for two-stage genome-wide association studies. Nat Genet. 2006;38:209–213. doi: 10.1038/ng1706. [DOI] [PubMed] [Google Scholar]
- 37.Schug J. Using TESS to predict transcription factor binding sites in DNA sequence. In: Baxevanis A, editor. Current Protocols in Bioinformatics. Hoboken, NJ: J. Wiley and Sons; 2003. [DOI] [PubMed] [Google Scholar]
- 38.Kaczynski J, Cook T, Urrutia R. Sp1- and Kruppel-like transcription factors. Genome Biol. 2003;4:206. doi: 10.1186/gb-2003-4-2-206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gottardi CJ, den Elzen NR, Yap AS. When domestiques rebel: Kinesins, cadherins and neuronal proliferation. Nat Cell Biol. 2005;7:445–447. doi: 10.1038/ncb0505-445. [DOI] [PubMed] [Google Scholar]
- 40.Hirokawa N. Stirring up development with the heterotrimeric kinesin KIF3. Traffic. 2000;1:29–34. doi: 10.1034/j.1600-0854.2000.010105.x. [DOI] [PubMed] [Google Scholar]
- 41.Dupuis L, et al. Differential screening of mutated SOD1 transgenic mice reveals early up-regulation of a fast axonal transport component in spinal cord motor neurons. Neurobiol Dis. 2000;7:274–285. doi: 10.1006/nbdi.2000.0292. [DOI] [PubMed] [Google Scholar]
- 42.Pantelidou M, et al. Differential expression of molecular motors in the motor cortex of sporadic ALS. Neurobiol Dis. 2007;26:577–589. doi: 10.1016/j.nbd.2007.02.005. [DOI] [PubMed] [Google Scholar]
- 43.Tateno M, et al. Mutant SOD1 impairs axonal transport of choline acetyltransferase and acetylcholine release by sequestering KAP3. Hum Mol Genet. 2009;18:942–955. doi: 10.1093/hmg/ddn422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhao C, et al. Charcot–Marie–Tooth disease type 2A caused by mutation in a microtubule motor KIF1Bbeta. Cell. 2001;105:587–597. doi: 10.1016/s0092-8674(01)00363-4. [DOI] [PubMed] [Google Scholar]
- 45.Hafezparast M, et al. Mutations in dynein link motor neuron degeneration to defects in retrograde transport. Science. 2003;300:808–812. doi: 10.1126/science.1083129. [DOI] [PubMed] [Google Scholar]
- 46.Schymick JC, et al. Genome-wide genotyping in amyotrophic lateral sclerosis and neurologically normal controls: First stage analysis and public release of data. Lancet Neurol. 2007;6:322–328. doi: 10.1016/S1474-4422(07)70037-6. [DOI] [PubMed] [Google Scholar]
- 47.Cudkowicz M, et al. Epidemiology of SOD1mutations in amyotrophic lateral sclerosis. Ann Neurol. 1997;41:210–212. doi: 10.1002/ana.410410212. [DOI] [PubMed] [Google Scholar]
- 48.Simon-Sanchez J, et al. Genome-wide SNP assay reveals structural genomic variation, extended homozygosity and cell-line induced alterations in normal individuals. Hum Mol Genet. 2007;16:1–14. doi: 10.1093/hmg/ddl436. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.