Abstract
Identification of the binary interactions between viral and host proteins has become a valuable tool for investigating viral tropism and pathogenesis. Here, we present the first systematic protein interaction screening of the unique variola virus proteome by using yeast 2-hybrid screening against a variety of human cDNA libraries. Several protein–protein interactions were identified, including an interaction between variola G1R, an ankryin/F-box containing protein, and human nuclear factor kappa-B1 (NF-κB1)/p105. This represents the first direct interaction between a pathogen-encoded protein and NF-κB1/p105. Orthologs of G1R are present in a variety of pathogenic orthopoxviruses, but not in vaccinia virus, and expression of any one of these viral proteins blocks NF-κB signaling in human cells. Thus, proteomic screening of variola virus has the potential to uncover modulators of the human innate antiviral responses.
Keywords: ankyrin repeats, Skp1, yeast 2-hybrid
Smallpox, a devastating infectious disease dreaded throughout much of recorded history, is caused by variola virus, a member of the Poxviridae family of DNA viruses that replicate entirely in the cytoplasm (1). Currently approved live smallpox vaccines are associated with significant safety risks and, consequently, the deliberate release of variola virus would have catastrophic consequences on global public health (2). However, the mechanisms that contribute to smallpox pathogenesis in man are poorly understood at the molecular level (3). Public health concerns regarding the possible reemergence of variola virus have led to renewed interest in the pathogenesis of smallpox and in the development of therapies and safer vaccines (2, 4, 5).
Analysis of the 186,102 bp that constitute the entire DNA genome of a highly virulent variola virus clone, isolated from Bangladesh (VAR-BSH) in 1975, revealed 187 closely spaced major ORFs specifying putative major proteins containing ≥65 aa (6). The majority of these proteins were markedly similar (>90% identity) to major gene products encoded by vaccinia virus, the smallpox vaccine and the most extensively studied orthopoxvirus, whereas the remaining proteins reflected variola-specific sequences or ORF divergences for variant proteins, which are often truncated or elongated compared with their vaccinia counterparts (6). ORFs that cluster near the genomic termini encode many proteins that are predicted to mediate variola virus transmissibility and virulence for its only natural host, humans (6).
Understanding viral pathogen interactions with the host is key to understanding how complex parameters like virus tropism and virulence are regulated. Identification of virus-host protein interactions have the potential to reveal important insights into how viruses infect and cause disease in specific hosts and why they occasionally leap into host species. The introduction of an in vivo yeast genetic screening method provided a means to accelerate the rate of progress in the identification of protein–protein interactions (7, 8). This approach has been successfully used in a genome-wide analysis of vaccinia virus protein–protein interactions (9) and, recently, a directed yeast 2-hybrid screening was conducted to identify human protein binding partners of the entire predicted proteome of vaccinia virus (http://proteinbank.vbi.vt.edu/ProteinBank/p/search/landingpage.dll). Because of the marked similarity between the majority of variola and vaccinia proteins (6), a selected yeast 2 hybrid search was also conducted for this report that focused exclusively on identifying variola specific human protein interactions that would have no direct counterparts from vaccinia virus. Our results revealed some interactions between distinctive variola and human proteins and identified a class of NF-κB inhibitors that is highly conserved among pathogenic orthopoxviruses.
Results and Discussion
It has been reported that the terminal regions of variola virus encode several proteins and variants of other poxvirus proteins that potentially augment variola virus transmissibility and virulence for its only natural host, humans (6). To better understand the potential role of these proteins and to help assess their possible involvement in modulating the human innate antiviral responses, after World Health Organization (WHO) approval, we initiated a selected yeast 2-hybrid screenings of all of the unique variola specific ORFs that are absent, truncated, or elongated compared with their vaccinia counterparts. These screens were carried out using 4 independent cDNA libraries prepared from various human tissues, cancer cell lines or a collection of clones [human tongue/tonsil, human spleen, a combination of breast tumor and prostate tumor cell lines, and the National Institutes of Health sponsored Mammalian Gene Collection (MGC; http://mgc.nci.nih.gov) respectively]. Bait constructs expressing either full length (VAR-BSH genes: C18L, A27L, A39L, B4L, B9R, B10R, B11R, B14L, B19R, B20R, and B22R) or fragments (VAR-BSH genes: D8L, C18L, B22R, G1R, and G2R) representing various variola virus protein domains were used (supporting information (SI) Table S1). At least 5 million yeast colonies were screened for each library. Of the 14 different variola ORFs examined, only 3 (B22R, G1R, and G2R) revealed consistent interactions with known human proteins (Table S2). However, the lack of any significant hits for the other variola ORFs examined does not necessarily reflect a lack of host-interacting partners because it may be a consequence of one or more of the inherent caveats associated with the yeast 2-hybrid system.
Screening using a bait construct that expresses residues 300–650 from VAR-BSH B22R protein, a surface glycoprotein of unknown function, identified 3 candidate human partners, including the C-terminal region of the cytokine inducible kinase (CNK; residues 513–608) [AKA; polo-like kinase 3 (PLK3)], the C-terminal region of mutated in colorectal cancer isoform 2 (MCC; residues 634–825) and the C-terminal region of spinophilin, protein phosphatase 1 regulatory subunit 9B (PPP1R9B; residues 676–817) (10) (Table S2). Both CNK, a putative serine/threonine kinase, and MCC, a candidate colorectal tumor suppressor gene, have been suggested to play a role in regulation of cell cycle progression and tumorigenesis (11, 12), implying a potential role for B22R in modulation of cell cycle progression during viral infection.
Variola G2R, a tumor necrosis factor receptor (TNFR) homolog, called CrmB, functions not only as a soluble decoy TNFR but also as a highly specific binding protein for chemokines that mediate recruitment of immune cells to mucosal surfaces and the skin, sites of virus entry and viral replication at late stages of smallpox (3). Screening using a bait construct that expresses residues 1–150 of VAR-BSH G2R protein, containing the TNFR domain and the preligand assembly domain (PLAD), identified 3 candidate interacting proteins, including the N-terminal half of calcium modulating cyclophilin ligand (CAML; residues 18–184) (13, 14), the mucin, oligomeric mucus/gel-forming (MUC5B; several fragments) (15) and the N-terminal region of solute carrier family 39, member 7 (SLC39A7; between residues 34–152) (16) (Table S2). However, further work is needed to verify these interactions and help understand their physiological relevance during viral infection.
Screening using a bait construct that expresses residues 200–585 of VAR-BSH G1R protein, containing several ankyrin (ANK) repeats and a C-terminal F-box like domain, identified 2 candidate interacting human proteins, including the C-terminal region of nuclear factor kappa-B, subunit 1 (NF-κB1/p105; between residues 577–714) and S-phase kinase-associated protein 1, isoform b (SKP1A; between residues 30–152) (Table S2). NF-κB1 belongs to the NF-κB family of transcription factors that regulate the expression of numerous genes encoding proteins that are crucial elements of innate and acquired immune responses (reviewed in ref. 17). NF-κB1 is synthesized as a large precursor of 105 (p105) kDa that is proteolytically processed by the proteasome, which removes its C-terminal half, to produce the active NF-κB1 p50 (18). SKP1A is a component of the SCF complexes involved in the regulated ubiquitination of specific protein substrates that targets them for degradation by the proteasome (reviewed in refs. 19, 20). Poxviruses, like many other viruses, express proteins to interfere with NF-κB signaling pathways, preventing the production of proinflammatory molecules and enhancing virus survival (21, 22). However, to date, no pathogen-encoded protein has been shown to interact directly with NF-κB1/p105, making G1R an attractive target for further characterization.
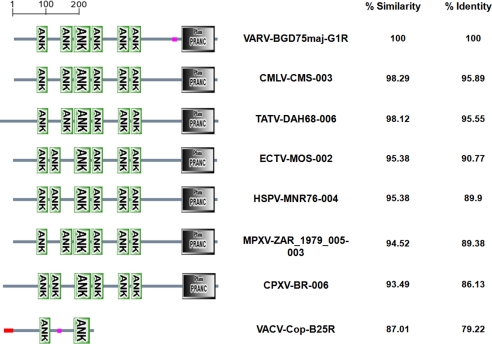
G1R belongs to a larger poxvirus family of ANK repeat proteins that also contain a C-terminal F-box like domain [AKA; Pox proteins Repeats of ANkyrin - C-terminal (PRANC)] known to be necessary for interaction with components of the SCF (Skp1, culin-1 and F-box) complex (23, 24). An ortholog search among the Poxvirus Bioinformatics Resource (PBR) proteins indicated that although G1R protein is substantially divergent from any vaccinia counterparts; it is highly conserved among several other pathogenic orthopoxvirus (Fig. 1). Analysis of the amino acid sequences of variola G1R or any of its orthologs using the simple modular architecture research tool (SMART) for protein domain identification and analysis of protein domain architectures, predicted 6 ANK domains, spanning the N-terminal two-thirds of the protein, and a C-terminal F-box like domain (PRANC) (Fig. 1).
Fig. 1.
Sequence and domain architecture conservation of Variola G1R with orthologs from other orthopoxviruses. SMART for protein domain identification and analysis of protein domain architectures (49) was used for the analysis of the amino acid sequences of variola G1R, camelpox 003, taterapox 006, ectromelia 002, horsepox 004, monkeypox 003, cowpox 006, and vaccinia B25R. The percentage similarity and percentage identity between the various viral proteins were obtained from the PBR. ANK, ankyrin repeat; PRANC, Pox proteins Repeats of ANkyrin - C-terminal.
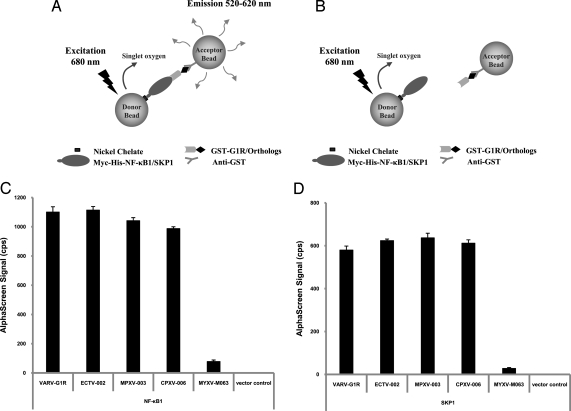
As with any yeast 2-hybrid screen, false positives need to be excluded because some of the identified interactions from the screen may not interact in mammalian cells in vivo. To verify whether G1R and either NF-κB1 p105 or SKP1A can interact in mammalian cells, their association was further analyzed in transfected cells, using full-length constructs and the AlphaScreen technology (PerkinElmer) (25, 26). The AlphaScreen assay system utilizes 2 different bead types, described as donor beads and acceptor beads. In the present study, we established a protein interaction assay by using a Myc-His tagged host protein and a GST-fused viral protein, immobilized on Nickel-coated donor and anti-GST-coated acceptor beads via Nickel and anti-GST, respectively. Only when the beads are brought into close proximity by a protein–protein interaction, do they produce an AlphaScreen emission signal (Fig. 2A). In contrast, a severe reduction or a complete lack of an AlphaScreen signal is interpreted as a lack of interaction (Fig. 2B).
Fig. 2.
Variola G1R interaction with both NF-κB1 and SKP1A is conserved among orthologs from other pathogenic orthopoxviruses. (A) Nickel-coated donor and anti-GST-coated acceptor beads are brought into close proximity when the Myc-His-host-GST–viral protein complex conjugates to the respective beads. On laser light excitation, singlet oxygen is produced by the photosensitizer in the donor bead and can diffuse to the acceptor bead, giving rise to a chemiluminescent/fluorescent signal, i.e., the AlphaScreen signal. (B) In the absence of the Myc-His-host–GST–viral protein complex formation, the acceptor and donor beads are no longer in close proximity and no AlphaScreen signal is detected. BSR-T7/5 cells were cotransfected with the pcDNA3.1MycHis plasmid expressing either NF-κB1 (C) or SKP1A (D) along with the pANT7_cGST plasmid expressing variola G1R, ECTV-002, MPXV-003, CPXV-006, MYXV-M063, or an empty pANT7_cGST plasmid. After transfection, cell lysates were subjected to the AlphaScreen assay. The presented results represent the average of 3 independent transfections.
To test for G1R interaction with NF-κB1, BSR-T7/5 cells were cotransfected with the pcDNA3.1MycHis plasmid expressing NF-κB1 along with the pANT7_cGST plasmid expressing VAR-BSH G1R. For control purposes, cells were also cotransfected with the NF-κB1 expression plasmid along with either an empty pANT7_cGST plasmid or a control pANT7_cGST plasmid expressing an irrelevant control protein, M063 (MYXV-M063) from myxoma virus (27). As seen in Fig. 2C, a GST fusion of VAR-BSH G1R protein, VARV-G1R, was able to interact with a Myc-His-tagged NF-κB1, confirming their interaction as revealed by the yeast 2-hybrid screen and demonstrating that the interaction occurs between full-length proteins. In contrast, both empty vector and MYXV-M063 controls failed to support any significant AlphaScreen signal (Fig. 2C). Next, to assess how conserved this interaction is among G1R orthologs, we examined the ability of selected G1R orthologs (ECTV-002 from ectromelia virus strain Moscow, MPXV-003 from monkeypox virus strain Zaire_1979–005 and CPXV-006 from cowpox virus strain Brighton Red) to interact with NF-κB1, using the AlphaScreen system. As shown in Fig. 2C, GST fusions of G1R orthologs (ECTV-002, MPXV-003, and CPXV-006) were able to also interact with Myc-His-tagged NF-κB1, demonstrating the conservation of this interaction among other pathogenic orthopoxviruses.
As for the interaction with SKP1A (Fig. 2D), similar to NF-κB1 interaction results, VARV-G1R was also able to interact with SKP1A, as compared with both vector and MYXV-M063 controls, confirming their interaction as revealed by our yeast 2-hybrid screen and demonstrating that the interaction occurs between full-length proteins in mammalian cells. Furthermore, G1R orthologs (ECTV-002, MPXV-003, and CPXV-006) were also able to interact with SKP1A, demonstrating the conservation of this interaction among pathogenic orthopoxviruses (Fig. 2D). Interestingly, a recent report demonstrated that ECTV-002 interacts with both SKP1A and conjugated ubiquitin (28), substantiating these interaction results with SKP1A.
To evaluate their impact on NF-κB function in human cells, the effect of the transient expression of each of the viral proteins (VARV-G1R, ECTV-002, MPXV-003, and CPXV-006) upon the expression of the firefly luciferase gene under the control of an NF-κB-regulated promoter was examined. For control purposes, the expression of a cotransfected renilla luciferase gene, under the control of a constitutively active promoter, was also examined. Treatment of HEK293 cells with TNF resulted in an 11.57-fold relative increase in luciferase activity over that in untreated cells (Fig. 3A). However, HEK293 cells transfected with VARV-G1R expression plasmid showed a reduction in the relative fold increase in luciferase activity in response to TNFα treatment (6.75-fold, Fig. 3A). Likewise, HEK293 cells transfected with expression plasmids for ECTV-002, MPXV-003, or CPXV-006 showed varying degrees of reduction (4.92-, 4.87-, and 8.44-fold, respectively) in the relative fold increase in luciferase activity in response to TNFα treatment (Fig. 3A). The reduction in luciferase activity represented a uniformly significant inhibition of TNFα-induced NF-κB activation as a result of the expression of VARV-G1R, ECTV-002, MPXV-003, or CPXV-006 (Fig. 3B). These results demonstrate that VARV-G1R and its orthologs specifically inhibit inducible NF-κB-regulated gene expression in human cells.
Fig. 3.
Variola G1R family expression inhibits NF-κB activity. (A) HEK293 cells were cotransfected with a 1:1:1 mix of plasmids, pNFκB-Luc (gives inducible NF-κB-dependent expression of firefly luciferase gene), pRL-TK (gives low-level constitutive expression of sea pansy luciferase gene), and pcDNA3.1MycHis (either empty or expressing VARV-G1R, ECTV-002, MPXV-003, or CPXV-006). At 48-h posttransfection cells were either left untreated or treated with TNFα (20 ng/mL) for 6 h and then assayed for both sea pansy and firefly luciferase activity. The presented values represent the average of 3 independent transfections. (B) The percentage inhibition of TNFα-induced NF-κB activation was obtained by calculating the percentage reduction in the relative fold luciferase activity in cells transfected with viral expression plasmid versus empty plasmid, after TNFα treatment. (C) Indirect immunofluorescence and confocal microscopy analysis demonstrating the effect of ECTV-002 on TNFα-induced nuclear translocation of NF-κB/p65 in HeLa cells. Mock transfected HeLa cells were labeled in the absence (i–iv) or presence (v–viii) of TNFα treatment. HeLa cells transfected with Flag-ECTV-002 expression plasmid were labeled in the absence (ix–xii) or presence (xiii–xvi) of TNFα treatment. (D) A graphical representation of the percentage of cells displaying a nuclear localization of p65. (E) HEK 293 cells were either mock transfected or transfected with plasmids constitutively expressing VARV-G1R, ECTV-002, MPXV-003, CPXV-006, or MYXV-M063. At 24-h posttransfection cells were either left untreated or treated with TNFα (20 ng/mL) for 30 min and then analyzed for NF-κB1 p105/p50 or β-actin by Western blot.
Given the ability of this class of viral Ankyrin-repeat proteins to inhibit inducible NF-κB-regulated gene expression, we next evaluated its effect on the nuclear translocation of NF-κB p65 (RelA) in response to TNF treatment, using ECTV-002 as the test protein (28). HeLa cells were either mock transfected or transiently transfected with a Flag-tagged ECTV-002 expression plasmid. Activation of mock-transfected HeLa cells with TNF (Fig. 3C, v–viii) induced the translocation of NF-κB p65 to the nucleus compared with the untreated mock-transfected cells (Fig. 3C, i–iv). In contrast, expression of the Flag-tagged ECTV-002 protein blocked the TNF-induced nuclear translocation of NF-κB p65 (Fig. 3C, xiii–xvi), confirming the inhibitory effect of the G1R family of proteins on NF-κB activity at the cellular level. Similar results were obtained by individually transfecting VARV-G1R, MPXV-003, or CPXV-006 (Fig. S1). Fig. 3D shows a graphical representation of the percentage of cells demonstrating NF-κB p65 nuclear translocation, expressed as the percentage of total cells examined.
The continuously selected interactions between host and pathogens during their coevolution have shaped not only the immune system but also the countermeasures exploited by pathogens. Viruses of many families actively inhibit a diverse array of molecular targets to elude immune detection and destruction (reviewed in refs. 29–35). Among these, the poxviruses are especially adept at interfering with immune pathways, such as those mediated by NF-κB (reviewed by refs. 36–42). However, no viral protein has been shown to interact directly with NF-κB1/p105, categorizing the G1R family of proteins as unique NF-κB inhibitors. Given that the ANK repeat domain has not been reported to occur within cellular F-box proteins, it has been suggested that poxviruses use these ANK/F-box proteins to dictate target specificity to SCF1 ubiquitin ligases and thereby exploit the cell's ubiquitin-proteasome machinery to target specific host proteins for elimination (24, 28). However, the ability of a transiently expressed G1R family protein to inhibit the nuclear translocation of NF-κB p65 suggests that the interaction between G1R and NF-κB1/p105 might instead be interfering with the processing/degradation of NF-κB1/p105, therefore preventing the release of active NF-κB p50 and the formation of the active NF-κB p65/p50 heterodimer that subsequently translocates to the nucleus. To assess the effect of these proteins on the stability of NF-κB1/p105, HEK293 cells were transfected with plasmids expressing G1R family member proteins, each individually, and then treated in the presence or absence of TNFα. For control purposes, HEK293 cells were also either mock transfected or transfected with an expression plasmid for the control protein MYXV-M063. In the absence of TNFα, no degradation of NF-κB1/p105 was observed (Fig. 3E, lane 7). However, treatment of cells with TNFα, in the absence of any plasmid transfection, induced the complete degradation of NF-κB1/p105 (Fig. 3E, lane 6). In contrast to the transfection of MYXV-M063 expression plasmid that failed to block the degradation of NF-κB1/p105 (Fig. 3E, lane 5), transfection of plasmids expressing any of the G1R family member proteins blocked the degradation of NF-κB1/p105 (Fig. 3E, lanes 1–4). Because NF-κB1/p105, like IκBs, bind to mature NF-κBs and sequester them in the cytoplasm (43), these results show, for the first time, that the G1R family proteins inhibit NF-κB activation by binding and stabilizing NF-κB1/p105 and preventing its degradation. However, at this point, it is not yet clear whether the demonstrated inhibitory effect of G1R against NF-κB is a reflection of its ability to interact with both NF-κB1/p105 and SKP1A simultaneously, or whether these are separate events.
In conclusion, proteomic information on virus–host interactions continues to reveal insights into basic biological processes in mammalian cells. The variola G1R protein family interactions with NF-κB1 and SKP1 provide a framework for hypotheses of how the NF-κB signaling pathways can be strategically controlled, not only during infection with pathogenic orthopoxviruses but likely other viral pathogens as well. Further studies of these interactions should improve our ability to manipulate NF-κB signaling during other proinflammatory response scenarios, and provide insights into many other aspects of virus tropism and pathogenesis.
Materials and Methods
Cell Culture.
Human embryonic kidney 293 (HEK293) cells (ATCC CRL1573) and HeLa cells (ATCC CCL2) were maintained in Dulbecco's modified Eagle's medium (DMEM; Gibco BRL) supplemented with 100 units/mL penicillin, 100 μg/mL streptomycin, 2 mM l-glutamine, and 10% FBS (Gibco BRL). BSR T7/5 cells [stably expressing T7 RNA polymerase under control of the cytomegalovirus (CMV) promoter] (44) were maintained in DMEM supplemented with 100 units/mL penicillin, 100 μg/mL streptomycin, 2 mM l-glutamine, and 10% FBS with the addition of geneticin (200 μg/mL) to the cell culture medium every second passage.
Reagents and Antibodies.
Lyophilized recombinant human tumor necrosis factor alpha (TNFα; R&D Systems Inc.) was reconstituted at a concentration of 20 μg/mL in a filter-sterilized solution of PBS containing 0.1% BSA (Sigma); aliquots were stored at −80 °C. Rabbit polyclonal antibody for NF-κB1 p105/p50, was purchased from Cell Signaling Technology and mouse monoclonal antibody for β-actin was from Ambion. HRP-conjugated goat anti-rabbit and anti-mouse IgG antibodies were purchased from Jackson Laboratory.
Plasmid Constructs.
Permission was requested of, and granted by, the WHO to receive a gel-purified PCR amplicon containing the VARV-G1R coding region. Cloning was done by using the Gateway cloning technology (Invitrogen). The Gateway technology is based on the bacteriophage lambda site-specific recombination system that facilitates the integration of lambda into the Escherichia coli chromosome (45). In the Gateway technology, the components of the lambda recombination system are modified to improve the specificity and efficiency of the system (46). Coding regions for viral (VARV-G1R, ECTV-002, MPXV-003, CPXV-006, and MYXV-M063) and human (NF-κB1 and SKP1A) proteins were PCR amplified, using primers containing gene-specific sequences flanked by sequences representing modified components of the lambda recombination system (Table S3) and incorporated into pDNOR222 plasmid (Invitrogen). The resultant entry clones were then subjected to LR recombination reactions with destination vectors pcDNA3.1MycHis (Invitrogen), pcDNA-DEST40 (Invitrogen), and/or pANT7_cGST (Harvard Institute of Proteomics) (47) to generate corresponding expression clones. Cloning strategies used resulted in expressed proteins that were tagged at the C terminus with MycHis, V5, or GST tags, respectively. A codon-optimized version of Flag-ECTV-002 was generated by GeneArt and subsequently subcloned into pcDNA3 using the BamHI and HindIII restriction sites.
Yeast 2-Hybrid Screening.
Yeast 2-hybrid screening was carried out at Myriad Genetics using the various variola virus proteins (Table S1) as baits with a mating based method. The drop out media and plates are prepared according to Guthrie and Fink (SI Text) (48).
Bioinformatics Tools.
SMART is an online resource (http://smart.embl.de/) that was used for protein domain identification and the analysis of protein domain architectures (49). The ortholog search among other poxviruses was done through the PBR (http://www.poxvirus.org/).
Alphascreen Assays.
The amplified luminescent proximity homogeneous assay (AlphaScreen; PerkinElmer) (25, 26) is a bead-based technology for screening biomolecular interactions in a microplate format (50). The assay utilizes 2 different bead types, described as donor beads and acceptor beads, and the surfaces of these beads are coated with reactive aldehyde groups to which biomolecules can be covalently conjugated. A specific biomolecular interaction brings donor and acceptor beads in close proximity, and a photosensitizer in the donor beads then excites the acceptor beads that deliver output photons. On excitation at 680 nm, the photosensitizer converts ambient oxygen into a hyperexcited singlet oxygen state that can diffuse up to 200 nm in solution and interact with chemiluminescent groups on a proximate acceptor bead. This transfer of energy ultimately emits light at 520–620 nm, which constitutes the AlphaScreen signal (Fig. 2A).
BSR T7/5 cells were seeded at 50% confluence onto 24-well plates the day before being transfected. Cells were transfected according to the manufacturer's protocol using 2 μL of Lipofectamine 2000 reagent (Gibco BRL) and 0.8 μg of DNA, in serum-free MEM, per each well. The DNA was a 1:1 mix of host protein (NF-κB1 or SKP1) expression vector to viral protein (VARV-G1R, ECTV-002, MPXV-003, CPXV-006, or MYXV-M063) expression vector. The expression vectors used were either the pcDNA3.1MycHis, for the host proteins, or the pANT7_cGST (47), for the viral proteins. Expression from the pcDNA3.1MycHis plasmid is under the control of either CMV or T7 RNA polymerase promoter and expressed proteins are fused at their C-terminal with both Myc and His tags. However, expression from the pANT7_cGST plasmid is under the control of T7 RNA polymerase promoter and expressed proteins are fused at their C-terminal with a GST tag. Forty-eight hours posttransfection, cells were collected in 500 μL of PBS, pelleted and then lysed in 20 μL of lysis buffer (100 mM NaCl, 100 mM Tris, pH 8.0, 0.5% Nonidet P-40, containing 25 μL/mL Roche complete protease inhibitor). The cellular extracts were then separated after centrifugation for 5 min at 13,000 rpm.
All assays were carried out in 384-well white opaque plates (PerkinElmer). Unless otherwise stated, 5 μL of anti-GST acceptor beads (PerkinElmer, 6760603C), diluted 50-fold (20 μg/mL final concentration of beads in assay) in PBS (PBS; 3.2 mM Na2HPO4, 0.5 mM KH2PO4, 1.3 mM KCl, 135 mM NaCl, pH 7.4) containing 0.1% BSA, and 5 μl of cell extract were mixed to a final volume of 20 μL of with 0.1% BSA/PBS assay buffer, and incubated at room temperature for 1.5 h. Subsequently, 5 μL of Nickel Chelate donor beads (PerkinElmer, AS101D), diluted 50-fold in 0.1% BSA/PBS assay buffer (20 μg/mL final concentration of beads in assay), were added to a final volume of 25 μL of and incubation was continued for another 1.5 h at room temperature. The plates were then read on an EnVision multiplate reader (PerkinElmer). Owing to the light-sensitivity of the beads, all assay steps were performed under subdued lighting, and the incubation steps were carried out in the dark.
Transfections and Luciferase Assays.
HEK 293 cells (1 × 104) were seeded at 50% confluence onto 96-well plates the day before being transfected. Cells were transfected according to the manufacturer's protocol using 0.5 μL of Lipofectamine 2000 reagent (Gibco BRL) and 0.2 μg of DNA, in serum-free MEM, per each well. The DNA was a 1:1:1 mix of NF-κB reporter vector to constitutive expression vector to viral protein expression vector. The NF-κB reporter vector used was the Stratagene pNFκB-Luc plasmid, which gives inducible NF-κB-dependent expression of firefly (Photinus pyralis) luciferase driven by a synthetic promoter comprising a TATA box preceded by 5 direct repeats of the sequence containing the NF-κB binding element (51). The constitutive expression vector used was the Promega pUCbased pRL-TK vector, which gives low level constitutive expression of sea pansy (Renilla reniformis) luciferase from the promoter of the herpes virus thymidine kinase gene. The viral protein expression vector was the Invitrogen pcDNA3.1MycHis vector that gives a constitutive level of expression of each of the viral proteins (VARV-G1R, ECTV-002, MPXV-003, or CPXV-006), independently, from the CMV promoter. At 6-h posttransfection, FBS was added to the medium for a final concentration of 10%. At 48-h posttransfection, the cells were treated with 20 ng/mL TNFα for 6 h. To determine luciferase values, the Promega Dual-Luciferase Reporter Assay System was used according to the manufacturer's instructions, The lysates were analyzed for firefly and sea pansy luciferase activity using the Appliskan multimode microplate reader (Thermo Scientific). The relative fold changes were determined by normalizing the ratios of firefly to sea pansy luciferase activities in each transfected cell group to the value obtained for nontransfected cells.
Western Blot Analysis of NF-κB1 p105/p50.
HEK 293 cells were seeded at 50% confluence onto a 24-well plate the day before being transfected. Cells were either mock transfected or transfected with plasmids constitutively expressing the viral proteins VARV-G1R, ECTV-002, MPXV-003, CPXV-006, or MYXV-M063. At 24-h posttransfection, the cells were treated for 30 min in the presence or absence of 20 ng/mL TNFα. Cells were harvested and lysed with RIPA buffer [50 mM Tris, 150 mM NaCl, 0.1% SDS, 0.5% sodium deoxycholate, 1% Nonidet P-40, 1 mM PMSF, protease inhibitor mixture (Roche)]. Protein samples were analyzed by SDS/PAGE and transferred to PVDF membranes (GE Healthcare). Membranes were blocked with 5% nonfat milk in Tris-buffered saline/0.05% Tween-20 (TBST; 20 mM Tris, 150 mM NaCl, 0.05% Tween-20 pH 7.6) and probed for NF-κB1 p105/p50 or β-actin proteins with primary antibody diluted (1:2000 or 1:5000, respectively) in 5% nonfat milk/TBST followed by a secondary incubation with HRP-conjugated goat anti-rabbit or anti-mouse antiserum, diluted 1:5000 in 5% nonfat milk/TBST. The membrane was then incubated in enhanced chemiluminescence substrate (Pierce), and autoradiography was used to detect the specific proteins.
Immunofluorescence Confocal Microscopy.
HeLa cells (3 × 104) plated on coverslips were either mock transfected or transiently transfected with 2 μg of pcDNA3-Flag-ECTV-002. Cells were mock treated or treated with 10 ng/mL TNFα (Roche) for 20 min at 37 °C at 12-h posttransfection. Coverslips were then washed with PBS and fixed with 2% paraformaldehyde (Sigma) for 10 min, permeabilized with 1% Nonidet P-40 (Sigma) and blocked with 30% goat serum (Invitrogen). Coverslips were incubated with both rabbit anti-p65 (Santa Cruz) and mouse anti-Flag (Sigma) diluted 1:200 in PBS containing 1% FBS for 1 h. Coverslips were incubated in these secondary antibodies: goat anti-rabbit-Alexa Fluor 546 and goat anti-mouse-Alexa Fluor 488 (Invitrogen) diluted at 1:400 in PBS containing 1% FBS for 1 h in the dark. After staining, coverslips were mounted on microscope slides with 7.5 μL of 50% glycerol containing N-propyl-gallate and 250 μg/mL 4′,6-diamino-2-phenylindole (DAPI) (Invitrogen) to visualize the nuclei. Cells were visualized using the 40X oil-immersion objective of a Zeiss Axiovert 200M fluorescent microscope outfitted with an ApoTome 10 optical sectioning device (Zeiss). To quantify the number of cells displaying a nuclear localization of p65 > 75 cells were counted.
Supplementary Material
Acknowledgments.
We thank Dr. Richard Moyer for providing cowpox DNA and Dr. Klaus Frueh for providing monkeypox DNA. This work was supported by contract DHHSN266200400057C to Myriad Genetics, Inc.
Footnotes
Conflict of interest statement: Myriad authors hold stock and or stock options in Myriad Genetics, Inc.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0900452106/DCSupplemental.
References
- 1.Moss B. Poxviridiae: The viruses and their replication. In: Fields BN, Knipe DM, PM H, editors. Fields' Virology. 5th Ed. Vol 2. Philadelphia: Wolters Kluwer Health/Lippincott Williams and Wilkins; 2007. pp. 2905–2946. [Google Scholar]
- 2.Grosenbach DW, et al. Immune responses to the smallpox vaccine given in combination with ST-246, a small-molecule inhibitor of poxvirus dissemination. Vaccine. 2008;26:933–946. doi: 10.1016/j.vaccine.2007.11.095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Alejo A, et al. A chemokine-binding domain in the tumor necrosis factor receptor from variola (smallpox) virus. Proc Natl Acad Sci USA. 2006;103:5995–6000. doi: 10.1073/pnas.0510462103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Henderson DA. The looming threat of bioterrorism. Science. 1999;283:1279–1282. doi: 10.1126/science.283.5406.1279. [DOI] [PubMed] [Google Scholar]
- 5.Smith GL, McFadden G. Smallpox: Anything to declare? Nat Rev Immunol. 2002;2(7):521–527. doi: 10.1038/nri845. [DOI] [PubMed] [Google Scholar]
- 6.Massung RF, et al. Analysis of the complete genome of smallpox variola major virus strain Bangladesh-1975. Virology. 1994;201:215–240. doi: 10.1006/viro.1994.1288. [DOI] [PubMed] [Google Scholar]
- 7.Chien CT, Bartel PL, Sternglanz R, Fields S. The two-hybrid system: A method to identify and clone genes for proteins that interact with a protein of interest. Proc Natl Acad Sci USA. 1991;88:9578–9582. doi: 10.1073/pnas.88.21.9578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fields S, Song O. A novel genetic system to detect protein–protein interactions. Nature. 1989;340:245–246. doi: 10.1038/340245a0. [DOI] [PubMed] [Google Scholar]
- 9.McCraith S, Holtzman T, Moss B, Fields S. Genome-wide analysis of vaccinia virus protein–protein interactions. Proc Natl Acad Sci USA. 2000;97:4879–4884. doi: 10.1073/pnas.080078197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Allen PB, Ouimet CC, Greengard P. Spinophilin, a novel protein phosphatase 1 binding protein localized to dendritic spines. Proc Natl Acad Sci USA. 1997;94:9956–9961. doi: 10.1073/pnas.94.18.9956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Matsumine A, et al. MCC, a cytoplasmic protein that blocks cell cycle progression from the G0/G1 to S phase. J Biol Chem. 1996;271:10341–10346. doi: 10.1074/jbc.271.17.10341. [DOI] [PubMed] [Google Scholar]
- 12.Zimmerman WC, Erikson RL. Polo-like kinase 3 is required for entry into S phase. Proc Natl Acad Sci USA. 2007;104:1847–1852. doi: 10.1073/pnas.0610856104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bram RJ, Crabtree GR. Calcium signalling in T cells stimulated by a cyclophilin B-binding protein. Nature. 1994;371:355–358. doi: 10.1038/371355a0. [DOI] [PubMed] [Google Scholar]
- 14.Feng P, et al. Kaposi's sarcoma-associated herpesvirus mitochondrial K7 protein targets a cellular calcium-modulating cyclophilin ligand to modulate intracellular calcium concentration and inhibit apoptosis. J Virol. 2002;76:11491–11504. doi: 10.1128/JVI.76.22.11491-11504.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Troxler RF, Offner GD, Zhang F, Iontcheva I, Oppenheim FG. Molecular cloning of a novel high molecular weight mucin (MG1) from human sublingual gland. Biochem Biophys Res Commun. 1995;217:1112–1119. doi: 10.1006/bbrc.1995.2884. [DOI] [PubMed] [Google Scholar]
- 16.Huang L, Kirschke CP, Zhang Y, Yu YY. The ZIP7 gene (Slc39a7) encodes a zinc transporter involved in zinc homeostasis of the Golgi apparatus. J Biol Chem. 2005;280:15456–15463. doi: 10.1074/jbc.M412188200. [DOI] [PubMed] [Google Scholar]
- 17.Sun SC, Ley SC. New insights into NF-kappaB regulation and function. Trends Immunol. 2008;29:469–478. doi: 10.1016/j.it.2008.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Beinke S, Ley SC. Functions of NF-kappaB1 and NF-kappaB2 in immune cell biology. Biochem J. 2004;382:393–409. doi: 10.1042/BJ20040544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cardozo T, Pagano M. The SCF ubiquitin ligase: Insights into a molecular machine. Nat Rev Mol Cell Biol. 2004;5:739–751. doi: 10.1038/nrm1471. [DOI] [PubMed] [Google Scholar]
- 20.Hershko A. The ubiquitin system for protein degradation and some of its roles in the control of the cell-division cycle (Nobel lecture) Angew Chem, Int Ed. 2005;44:5932–5943. doi: 10.1002/anie.200501724. [DOI] [PubMed] [Google Scholar]
- 21.Gedey R, Jin XL, Hinthong O, Shisler JL. Poxviral regulation of the host NF-kappaB response: The vaccinia virus M2L protein inhibits induction of NF-kappaB activation via an ERK2 pathway in virus-infected human embryonic kidney cells. J Virol. 2006;80:8676–8685. doi: 10.1128/JVI.00935-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shisler JL, Jin XL. The vaccinia virus K1L gene product inhibits host NF-kappaB activation by preventing IkappaBalpha degradation. J Virol. 2004;78:3553–3560. doi: 10.1128/JVI.78.7.3553-3560.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mercer AA, Fleming SB, Ueda N. F-box-like domains are present in most poxvirus ankyrin repeat proteins. Virus Genes. 2005;31:127–133. doi: 10.1007/s11262-005-1784-z. [DOI] [PubMed] [Google Scholar]
- 24.Sonnberg S, Seet BT, Pawson T, Fleming SB, Mercer AA. Poxvirus ankyrin repeat proteins are a unique class of F-box proteins that associate with cellular SCF1 ubiquitin ligase complexes. Proc Natl Acad Sci USA. 2008;105:10955–10960. doi: 10.1073/pnas.0802042105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ullman EF, et al. Luminescent oxygen channeling immunoassay: Measurement of particle binding kinetics by chemiluminescence. Proc Natl Acad Sci USA. 1994;91:5426–5430. doi: 10.1073/pnas.91.12.5426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ullman EF, et al. Luminescent oxygen channeling assay (LOCI): Sensitive, broadly applicable homogeneous immunoassay method. Clin Chem. 1996;42:1518–1526. [PubMed] [Google Scholar]
- 27.Barrett JW, et al. Myxoma virus M063R is a host range gene essential for virus replication in rabbit cells. Virology. 2007;361:123–132. doi: 10.1016/j.virol.2006.11.015. [DOI] [PubMed] [Google Scholar]
- 28.van Buuren N, Couturier B, Xiong Y, Barry M. Ectromelia virus encodes a novel family of F-box proteins that interact with the SCF complex. J Virol. 2008;82:9917–9927. doi: 10.1128/JVI.00953-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Alcami A, Koszinowski UH. Viral mechanisms of immune evasion. Trends Microbiol. 2000;8:410–418. doi: 10.1016/S0966-842X(00)01830-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Haig DM. Subversion and piracy: DNA viruses and immune evasion. Res Vet Sci. 2001;70:205–219. doi: 10.1053/rvsc.2001.0462. [DOI] [PubMed] [Google Scholar]
- 31.Loo YM, Gale M., Jr Viral regulation and evasion of the host response. Curr Top Microbiol Immunol. 2007;316:295–313. doi: 10.1007/978-3-540-71329-6_14. [DOI] [PubMed] [Google Scholar]
- 32.McFadden G, Murphy PM. Host-related immunomodulators encoded by poxviruses and herpesviruses. Curr Opin Microbiol. 2000;3:371–378. doi: 10.1016/s1369-5274(00)00107-7. [DOI] [PubMed] [Google Scholar]
- 33.Schroder M, Bowie AG. An arms race: Innate antiviral responses and counteracting viral strategies. Biochem Soc Trans. 2007;35:1512–1514. doi: 10.1042/BST0351512. [DOI] [PubMed] [Google Scholar]
- 34.Tortorella D, Gewurz BE, Furman MH, Schust DJ, Ploegh HL. Viral subversion of the immune system. Annu Rev Immunol. 2000;18:861–926. doi: 10.1146/annurev.immunol.18.1.861. [DOI] [PubMed] [Google Scholar]
- 35.Unterholzner L, Bowie AG. The interplay between viruses and innate immune signaling: Recent insights and therapeutic opportunities. Biochem Pharmacol. 2008;75:589–602. doi: 10.1016/j.bcp.2007.07.043. [DOI] [PubMed] [Google Scholar]
- 36.Alcami A. New insights into the subversion of the chemokine system by poxviruses. Eur J Immunol. 2007;37:880–883. doi: 10.1002/eji.200737215. [DOI] [PubMed] [Google Scholar]
- 37.Barrett JW, Cao JX, Hota-Mitchell S, McFadden G. Immunomodulatory proteins of myxoma virus. Semin Immunol. 2001;13:73–84. doi: 10.1006/smim.2000.0298. [DOI] [PubMed] [Google Scholar]
- 38.Dunlop LR, Oehlberg KA, Reid JJ, Avci D, Rosengard AM. Variola virus immune evasion proteins. Microbes Infect. 2003;5:1049–1056. doi: 10.1016/s1286-4579(03)00194-1. [DOI] [PubMed] [Google Scholar]
- 39.Haga IR, Bowie AG. Evasion of innate immunity by vaccinia virus. Parasitology. 2005;130:S11–S25. doi: 10.1017/S0031182005008127. [DOI] [PubMed] [Google Scholar]
- 40.Moss B, Shisler JL. Immunology 101 at poxvirus U: Immune evasion genes. Semin Immunol. 2001;13:59–66. doi: 10.1006/smim.2000.0296. [DOI] [PubMed] [Google Scholar]
- 41.Seet BT, et al. Poxviruses and immune evasion. Annu Rev Immunol. 2003;21:377–423. doi: 10.1146/annurev.immunol.21.120601.141049. [DOI] [PubMed] [Google Scholar]
- 42.Smith GL. Vaccinia virus immune evasion. Immunol Lett. 1999;65(1–2):55–62. doi: 10.1016/s0165-2478(98)00125-4. [DOI] [PubMed] [Google Scholar]
- 43.Hayden MS, Ghosh S. Shared principles in NF-kappaB signaling. Cell. 2008;132:344–362. doi: 10.1016/j.cell.2008.01.020. [DOI] [PubMed] [Google Scholar]
- 44.Buchholz UJ, Finke S, Conzelmann KK. Generation of bovine respiratory syncytial virus (BRSV) from cDNA: BRSV NS2 is not essential for virus replication in tissue culture, and the human RSV leader region acts as a functional BRSV genome promoter. J Virol. 1999;73:251–259. doi: 10.1128/jvi.73.1.251-259.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ptashne M. A Genetic Switch: Phage λ and Higher Organisms. 2nd Ed. Cambridge, MA: Cell Press : Blackwell Scientific Publications; 1992. [Google Scholar]
- 46.Bushman W, Thompson JF, Vargas L, Landy A. Control of directionality in lambda site specific recombination. Science. 1985;230:906–911. doi: 10.1126/science.2932798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ramachandran N, et al. Self-assembling protein microarrays. Science. 2004;305:86–90. doi: 10.1126/science.1097639. [DOI] [PubMed] [Google Scholar]
- 48.Guthrie C, Fink GR. Methods in Enzymology. Vol 194. San Diego: Academic, Inc.; 1991. Guide to yeast genetics and molecular biology; p. 933. [PubMed] [Google Scholar]
- 49.Letunic I, et al. SMART 5: Domains in the context of genomes and networks. Nucleic Acids Res. 2006;34:D257–D260. doi: 10.1093/nar/gkj079. Database issue. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Gesellchen F, Prinz A, Zimmermann B, Herberg FW. Quantification of cAMP antagonist action in vitro and in living cells. Eur J Cell Biol. 2006;85:663–672. doi: 10.1016/j.ejcb.2006.01.009. [DOI] [PubMed] [Google Scholar]
- 51.Sen R, Baltimore D. Multiple nuclear factors interact with the immunoglobulin enhancer sequences. Cell. 1986;46:705–716. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.