Abstract
Acyl-coenzyme A (CoA) thioesterases are a large family of enzymes that hydrolyze acyl-CoA esters to the free fatty acid and CoA and thereby regulate essential cellular functions such as lipid metabolism, membrane synthesis, signal transduction, and gene transcription. To better understand the virulence mechanisms of Campylobacter jejuni, and its possible link to membrane lipid biosynthesis, we have investigated C. jejuni thioesterases, annotated as putative proteins. While little is known about fatty acid biosynthesis and regulation in C. jejuni, remarkable differences in the genome and its organization from Escherichia coli, the paradigm system, raise questions as to the functions of these putative proteins. Here we present the crystal structure and biochemical analysis of Cj0915, defining the first functional thioesterase from C. jejuni. The structure of Cj0915 reveals the hot dog fold with an YciA-type hexameric assembly. Enzymatic assays performed with the purified protein show that Cj0915 is an efficient thioesterase with a broad specificity toward acyl-CoA substrates. This study provides a framework for investigation on roles of the Cj0915 thioesterase in virulence, and functional activities associated with the Cj0915 thioesterase in vivo.
Keywords: crystal structure, hot dog fold, thioesterase, membrane lipid biosynthesis, Campylobacter jejuni
Introduction
Thioesterases are a large family of enzymes that catalyze the hydrolysis of thioesters to free fatty acids and thiols including coenzyme A (CoA) and the acyl carrier proteins. Apart from the established function for acyl-CoA thioesterases in termination of fatty acid synthesis, the cellular functions of these enzymes remain largely unknown. Nonetheless, it is conceivable that they regulate lipid metabolism by maintaining proper levels of acyl-CoA, CoA, and free fatty acids [1]. The importance of thioesterase function can be appreciated when one examines many different biochemical processes involving long-chain acyl-CoA esters and free fatty acids, such as membrane synthesis, signal transduction, and gene regulation [1]. Thioesterases appear to be mainly evolved within the α/β fold hydrolase superfamily and the hot dog fold enzyme superfamily [2-4]. A typical α/β fold thioesterase is an eight-stranded mostly parallel α/β structure, and contains a catalytic triad Ser-His-Asp. The hot dog fold thioesterase, first observed with the structure of the Escherichia coli β-hydroxydecanoyl thiol ester dehydratase (FabA) [5], is a mixed α+β structure, which adopts a five- to seven-stranded antiparallel β-sheet as the ‘bun’ wrapping around a central α-helix ‘sausage’.
Campylobacter jejuni, from the epsilon group of proteobacteria, is a Gram-negative, microaerophilic, motile, spiral-shaped bacterium, like its related gastric pathogen Helicobacter pylori [6,7]. Despite being a major food-borne pathogen and the availability of multiple genome sequences, remarkably little is understood about the details of molecular pathogenesis of C. jejuni. Interestingly, the genome of C. jejuni NCTC11168 contains 1,641,481 base pairs in length [6], approximately one third of the Escherichia coli genome in size, and represents one of the most compact genomes among proteobacteria sequenced to date. Another intriguing attribute of the genome includes scarce organization of genes into operons or clusters [6], which raises fundamental issues concerning transcriptional regulation in Campylobacter. These observations raise important questions as to the mechanisms of fundamental cellular processes in this organism, which mostly derive from knowledge acquired from the E. coli system as the paradigm. For example, while structural and functional studies of enzymes catalyzing lipid metabolic pathways have been performed for the E. coli system and a few other pathogens, the corresponding enzymatic attributes are totally absent for C. jejuni.
In a recent study, we found that Cj0977, a σ28-regulated non-flagellar virulence protein from C. jejuni, adopts a homodimeric hot dog fold [8]. Moreover, the putative binding site for an acyl-CoA, identified by structural comparison with other hot dog fold proteins, is indeed required for the Cj0977 virulence function, suggesting a possible function of Cj0977 as an acyl-CoA binding regulatory protein [8]. In an effort to gain insight into the structural basis of C. jejuni virulence, we launched a long-term study on the link between membrane lipid biosynthesis and virulence in C. jejuni. Since Cj0977 did not show any detectable thioesterase activity, we first focused on identifying a functional hot dog fold thioesterase from C. jejuni. Of note, the structures of as many as seven different hot dog thioesterases have been determined from the E. coli genome: PaaI (PDB code, 2fs2), YciA (PDB code, 1yli), YbgC (PDB code, 1s5u), YbaW (PDB code, 1njk), YbdB (PDB code, 1vh9), YdiI (PDB code, 1vi8), and thioesterase II (PDB code, 1c8u). Many of theses structures are contributed by the structural genomics projects, and functional assignment and characterization remain elusive for many of these structures [9-11]. By contrast, the C. jejuni genome appears to contain only two hot dog fold thioesterases Cj0915 and Cj0965c, according to the genome sequence of C. jejuni NCTC11168, reflecting the compact genome structure. The Cj0915 protein is a homolog of E. coli YciA, and the Cj0965c protein is a homolog of E. coli YbgC, encoded by the ybgC gene of the tol-pal gene cluster [12].
One major challenge in this post-genomics era is to assign the biological function of numerous genes encoding proteins of unknown functions. In this study, we identified and purified two putative thioesterases, Cj0915 and Cj0965c, from the C. jejuni genome sequence. We report the crystal structure of Cj0915, revealing the hot dog fold assembled into a hexameric quaternary structure. To link the structural information with a function, we investigated the activity of the recombinant enzymes. We show that the Cj0915 protein functions as a major acyl-CoA thioesterase of C. jejuni, whereas Cj0965c displays negligible thioesterase activity. The structure and biochemical analysis of Cj0915 define the first functional thioesterase from C. jejuni.
MATERIALS AND METHODS
Chemicals
Restriction enzymes were purchased from New England Biolabs. Taq DNA polymerase and T4 DNA ligase were purchased from Promega. Primers were synthesized by IDT. DNA sequencing was performed by SeqWright. All acyl-CoA compounds were purchased from Sigma.
Cloning
The genes corresponding to Cj0915 and Cj0965c of NCTC 11168 were amplified by PCR using Taq DNA polymerase (Promega) and C. jejuni strain 81-176 genomic DNA. Primers p915_F1 (5’- ccgggatccATG AGAGATATGGGAGAACC-3’) and p915_R137 (5’- ccgctcgagTTAAGCATTT AAAAAGCCATG-3’) were used for Cj0915, and primers p965_F1 (5’-ccgggatccATGAAAATGCGTGTGTATTATG AAG –3’) p965_R124 (5’-ccgctcgagTTAAAATAA TTC TTCAAAGAGC-3’) were used for Cj0965c. For the cloning purpose, two restriction sites, BamHI and XhoI, were introduced to the forward and reverse primers, respectively. PCR was performed in a Sprint Thermal Cycler (Thermo Fisher Scientific) as follows: 95°C for 5 min, 25 cycles of denaturation / annealing / elongation (94°C for 45 sec / 55°C for 43 sec / 72°C for 2 min), and finally 72°C for 10 min. The PCR fragments were then inserted into the pGEX-6p-1 vector (GE Healthcare) using the BamHI and XhoI restriction sites. The ligation reaction was used to transform E. coli DH5α strain. Following the presence of the cloned fragments was confirmed by colony PCR and by double digestion of plasmids, all plasmids were purified using QIAprep® Spin Miniprep Kit (Qiagen). The cloned gene sequences were confirmed by DNA sequencing.
Expression and Purification
For expression of Cj0915, the recombinant plasmid pGEX∷Cj0915 was transformed into BL21(DE3) competent cells. Typically, the cells were grown at 37°C in 1 L of Luria-Bertani media with 100 μg/ml ampicillin. Once OD600nm reached ~0.8, isopropyl β-D-thiogalactopyranoside (IPTG) was added to a final concentration of 0.2 mM, and the culture was incubated at room temperature for 3 hrs. For expression of Cj0965c, the same procedures were applied, except that the protein expression was induced for 90 min. Cells were harvested at 4,000g for 20 min, resuspended in 80 mL of PBS containing 1 % Triton X-100 (Sigma) and Complete™ EDTA-free protease inhibitor (Roche Diagnostics), and then stored at -20°C until use.
Following cell lysis using sonication, cell lysate was centrifuged at 40,000g for 20 min. GST-fusion proteins were isolated by affinity chromatography using Glutathione-Sepharose 4B beads (GE Healthcare). After binding for 2 hrs at 4°C and extensive washing with PBS in the presence of 0.05 % Triton X-100 and 1mM DTT, the Cj0915 or Cj0965c portions of the fusion proteins were cleaved by adding 80 units PreScission protease (GE Healthcare) in 3 mL of cleavage buffer containing 50mM Tris-HCl pH 7.0, 150 mM NaCl, 1 mM EDTA, and 1 mM DTT) and incubated overnight 4°C. The Cj0915 or Cj0965c moiety was separated and collected by centrifugation at 1000g for 5 min. The protein was next purified by gel filtration using Sephacryl-S100 16/60 column (GE Healthcare) equilibrated with buffer containing 20 mM Tris-HCl (pH 8.0) and 100 mM NaCl. The peak fractions were combined, concentrated, and further purified using a high resolution gel filtration column (Superdex 75 10/300 GL) equilibrated with buffer containing 50 mM HEPES (pH 7.0), 200 mM NaCl, 0.1 mM EDTA and 5% glycerol. The highly pure peak fractions were then concentrated to ~4 mg/mL.
Crystallization and Data collection
Crystals of Cj0915 were formed in a 1:1 mixture (2 μl of droplet) of protein (4 mg/mL in buffer of 50mM HEPES (pH 7.0) / 200 mM NaCl / 0.1 mM EDTA / 5% glycerol) and reservoir solution containing 0.7 – 0.8 M NaCl / 0.1 M Na-citrate (pH 5) using the hanging drop vapor diffusion method at 17°C. Tiny cubic crystals appeared within 1-2 days and slowly grew to typical dimensions of ~0.05 × 0.05 × 0.05 mm. For data collection, crystals were flash cooled in liquid N2 after a brief incubation in 1.2 M NaCl / 0.1 M Na-citrate (pH 5) / 20% glycerol or 20% ethylene glycol as cryo protectant.
Cj0915 diffraction data were collected using single crystals at 100K on beamline 19BM at the Advanced Photon Source (APS), Argonne National Laboratory. The data sets were indexed and integrated using HKL2000 and scaled with SCALEPACK to 2.6Å [13]. Cj0915 crystal belonged to space group P4132 with unit cell dimensions of a = b = c = 103.018 Å with a monomeric protomer per asymmetric unit.
Structure determination and refinement
The structure of Cj0915 was solved by molecular replacement using the program MOLREP [14] and the structure of Hi0827 from Haemophilus influenzae (PDB code: 1YLI) as a search model. The top solution had a correlation coefficient of 34.3% and an R-factor of 59.1%. Manual model building was performed using COOT [15]. Refinement was carried out using CNS1.1 and Refmac5 [16,17]. The first refinement cycle by Refmac5 gave an R-factor of 34.8% and Rfree of 49.4%. Solvent molecules were placed at positions where spherical electron density peaks were found above 1.5σ in the |2Fo-Fc| map and above 3.5σ in the |Fo-Fc| map, and where stereo-chemically reasonable hydrogen bonds were allowed. The final model has an R-factor of 20.1% and Rfree of 25.3%. Structural analysis of the final model using the Protein Data Bank validation suite indicated almost all residues are in the most favored regions of the Ramachandran plot. The summary of data collection and refinement statistics is given in Table 1. The coordinates and structure factors of the Cj0915 have been deposited at Protein Data bank (PDB id code 3D6L).
Table 1.
Crystallographic data and refinement statistics.
| Data Collection | |
| Wavelength (Å) | 0.97915 |
| Space group | P4132 |
| Cell parameters (Å) | a = b = c =103.018 |
| Resolution (Å) | 50-2.6 (2.69-2.60) |
| Reflections (Total / Unique) | 119,675 / 6,240 |
| Completeness (%)a | 100 (100) |
| Rmerge (%)a,b | 10.4(33.9) |
| <I/σ(I)>a | 5.6 (3.6) |
| Redundancya | 19.1 (13.9) |
| Structure Refinement | |
| Resolution (Å) | 50-2.6 |
| No. reflections (working/test) | 5,920 / 286 |
| |F|/σ(|F|) | >0 |
| R (%)a,c/Rfreed(%) | 20.1/25.3 |
| Total number of atomse | 1,024 |
| Asymmetric unit content | 1 protomer |
| Average B factor (Å2) | 23.4 |
| rms from ideality | |
| Bonds (Å) | 0.013 |
| Angles (°) | 1.484 |
Overall data, high-resolution shell in parentheses.
Rmerge = Σ | Ii - 〈I〉|/Σ|Ii|, where Ii is the intensity of an observation. 〈I〉 the mean value for that reflection, and the summations are over all equivalents.
R-factor = Σh∥Fo(h)| - |Fc(h)∥/ΣhFo(h), where Fo and Fc are the observed and calculated structure factor amplitudes, respectively.
Rfree was calculated with 5% of the data excluded from the refinement.
includes 20 water molecules and 2 chloride ions.
Acyl-CoA thioesterase Assays
Acyl-CoA thioesterase activity was measured by using a DTNB [5,5’-dithio-bis(2-nitrobenzoic acid)]-based continuous spectrophotometric assay, as described previously [18]. Briefly, the enzyme activity was measured from the formation of 5-thio-2-nitrobenzoate produced from DTNB reacting with CoA released by hydrolysis from the substrate and was monitored through its absorbance at 412 nm (molar extinction coefficient of 13,600 M-1·cm-1). In the standard assay condition, the reaction mixture contained 100 mM HEPES (pH 7.5), 200 mM KCl, 1.8 mM DTNB and 100 μM substrate. The reaction mixture was pre-incubated at room temperature for 5 min in a quartz cuvette with a 1-cm light path length. The reaction was initiated by adding the enzyme (0.55 μg) to reaction mixture (final volume, 500 μL) at room temperature. The absorbance of 5-thio-2-nitrobenzoate was measured at 412 nm using UV-Visible Cary 50 Bio Spectrophotometer (Varian, Palo Alto, CA) for 5 min. The kinetic parameters of Vmax and Km were determined by nonlinear regression (Eq. 1) using the SigmaPlot version 10.1 software (Systat Software, Chicago, USA).
| (1) |
where x is the substrate concentration, y is the initial velocity, Vmax is the maximum velocity, n is the curve slope factor and K is Michaelis constant.
The kcat (s-1) was calculated from the ratio of Vmax (μmol·min-1·mg-1) and the total enzyme concentration. The enzyme concentration was determined by Bradford assay following the manufacturer’s instruction for the Bradford reagent (Bio-Rad, Hercules, USA). One unit (U) of activity was defined as the amount of enzyme releasing 1 μmol of CoA per minute under the standard condition.
RESULTS
Identification of an acyl CoA thioesterase from Campylobacter jejuni
In our quest to find a functional hot dog fold thioesterase of C. jejuni, two gene products, Cj0915 and Cj0965c, which were annotated as putative thioesterases, were selected from the genome sequence of C. jejuni strain NCTC 11168 (GenBank#AL111168) [6]. The 137–amino acid Cj0915 (Mr = 14757 Da) is 37% identical to the Escherichia coli thioesterase YciA (132 amino acids). In addition, Cj0915 appears to be similar to eukaryotic acyl-CoA hydrolases. For example, Cj0915 and human cytoplasmic acyl-CoA thioesterases share over 30% identical residues within the hot dog fold domain (GenBank#AAB61211). The 124-amino acid Cj0965c (Mr = 14584 Da) is 31% identical to the E. coli YbgC (134 amino acids) protein. Multiple sequence alignments using ClustalW clearly indicated that these thioesterases are highly conserved across different bacterial species (data not shown).
Both Cj0915 and Cj0965c are predicted to be cytoplasmic soluble proteins. However, whereas the recombinant Cj0915 could be purified as a highly soluble protein, the recombinant Cj0965c was only partially soluble (~10% of total expression). Yet, once the soluble fraction of Cj0965c was isolated from the total lysate, the protein remained soluble and stable for the further purification steps. Accordingly, highly pure recombinant Cj0915 and Cj0965c were produced as described in Material and Methods. The quaternary structure of the native proteins was assessed by high-resolution gel filtration. While Cj0915 is a hexamer (~90 kDa) in solution, Cj0965c forms tetramers (~60 kDa) in solution (Fig. 1A).
Figure 1.
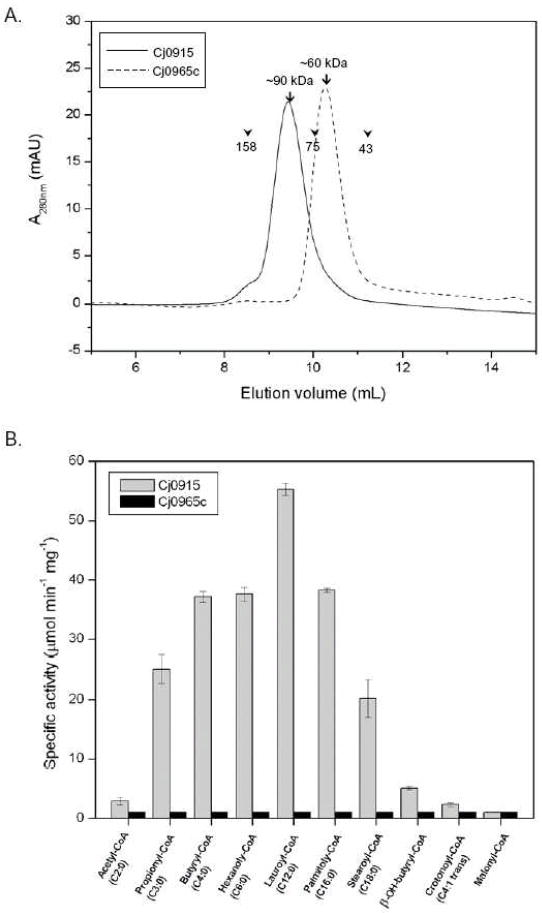
Purification and enzyme activity of Cj0915 and Cj0965c. (A) Cj0915 and Cj0965c proteins were purified as described in Material and Methods. After removing the GST-tag, the recombinant proteins were applied to a high-resolution gel filtration column, Superdex 75 10/300 GL. Size exclusion chromatograms of each protein were overlaid for comparison. The apparent molecular masses of Cj0915 and Cj0965c were estimated to be 90 kDa and 60 kDa, respectively. The elution peak positions of molecular standards are indicated as arrowheads (Aldolase, 158 kDa; Conalbumin, 75 kDa; and Ovalbumin, 43 kDa). (B) Observed rates of acyl-CoA hydrolysis catalyzed by purified Cj0915 and Cj0965c proteins. Following high-resolution gel filtration of each protein, highly pure fractions were combined and concentrated to ~4 mg/ml for crystallization and enzyme assays. Enzyme activities on various CoA thioesters were measured under the standard assay condition at a fixed substrate concentration of 100 μM at pH 7.5 and at 25°C. For each substrate, the enzyme activities are shown as bars (left, Cj0915; right, Cj0965c).
The two purified recombinant proteins were subjected to crystallization, yielding cubic crystals for Cj0915, and plate and needle shaped crystals for Cj0965c. Concomitantly, these protein solutions were tested for thioesterase activity by using aliphatic acyl CoA compounds with various chain lengths. While Cj0965c did not show any measurable activity, Cj0915 was very active toward a wide range of acyl moiety chain lengths (Fig. 1B), as described below. Therefore, this study further focused on investigating the structure and function of Cj0195, a C. jejuni thioesterase.
Structure of Cj0915
The native Cj0915 protein crystallized in the space group P4132 with one molecule per asymmetric unit and diffracted to a resolution of 2.6 Å. (Table I). The structure was determined by molecular replacement using the crystal structure of Haemophilus influenzae YciA homolog (Hi0827: PDB code, 1yli) [19]. The final structure is well ordered, and electron densities for all residues, but the first and last three residues, are clearly interpretable. Accordingly, the refined model includes 133 of the 137 total residues, two chloride ions, and 20 ordered water molecules.
As expected from the sequence conservation of these proteins, Cj0915 monomer adopts a typical β+α ‘hot dog’ fold, which was first observed in the structure of E. coli β-hydroxydecanoyl thiol ester dehydratase (FabA) [5]. Since then, this characteristic fold has been found in numerous other enzymes, notably, thioesterases catalyzing diverse acyl-CoA substrates as well as many putative proteins [4, 20-26]. The protomer of Cj0915 (overall dimensions of 30 Å × 25 Å × 52 Å) consists of a five-stranded β-sheet with topology β1/β3/β4/β5/β2 that wraps around a central five-turn α helix, α1, positioned between β1 and β2 (Fig. 2A). A short α helix formed at the C-terminus extends over the open side of the β sheet, whereas a 310 helix located at the opposite side is involved in the formation of hexamer.
Figure 2.

Crystal structure of Cj0915. (A) Ribbon diagram of the Cj0915 protomer. The secondary structures are labeled. N and C denote the N and C termini of Cj0915, respectively. The central helix (α1) is highlighted in red. (B) Ribbon diagram of the Cj0915 dimer harboring two symmetrical active site tunnels at the dimer interface. (C) Ribbon diagram of the Cj0915 hexamer (trimer of dimers) is viewed approximately down the 3-fold axis, showing the biological quaternary architecture of Cj0915. Each subunit is shown in different color.
While the Cj0915 crystals contained a monomeric protomer per asymmetric unit, the quaternary structure of Cj0915 hexamer could be easily identified and assembled using crystallographic symmetry. The structure of Cj0915 is a trimer of dimers, consistent with the gel filtration chromatography result (Fig. 1A). The Cj0915 monomers dimerize through the β2 strands forming a ten-stranded antiparallel β sheet (Fig. 2B). The dimer formation buries ~1620 Å2 of solvent-accessible surface area per protomer. The major driving forces for the dimerization of two subunits are the hydrogen-bonding interaction between two anti-parallel β2 strands from each subunit, and interaction between the first two turns of the symmetrical central α-helix by packing against each other in an antiparallel way. Of note, this anti-parallel β strand hydrogen-bonding interaction is strictly conserved among hot dog fold proteins with various quaternary structures. The enzyme active site is formed at the dimer interface, generating two symmetrical active sites.
Three identical dimers joined together to form a compact hexamer with its 3-fold rotational axis coincident to a crystallographic triad (Fig. 2C). Three dimers are juxtaposed such that five strands from each subunit form a large twisted β barrel containing 30 β strands with overall dimensions of 70 Å × 70 Å × 52 Å. Contacts between dimers are extensive with the total buried area of ~11,278 Å2, as one subunit interacts with four other subunits, where subunit A contacts B (dimer pair), C, E, and F. The intricate intermolecular contacts within the hexamer are mediated by amino acid residues from β1, 310-helix, the loop between α1 and β2, and the loop between β4 and β5. This hexameric assembly is very similar to that of H. influenzae YciA (HiYciA), the sole structure reported to date [19]. Indeed, the number of structures with this quaternary structural arrangement is increasing, mainly contributed by the structural genomics centers, while the functional assignment of these proteins remains mostly absent (PDB codes: 1VPM, 1Y7U, 2GVH, 2QQ2, 2V1O, 2Q2B, 2EIS, 3B7K, 2BI0, 1Q6W).
Acyl-CoA thioesterase activity of Cj0915
Following structural evidence that Cj0915 is similar to the known thioesterase H. influenzae YciA, we next sought to assign the biochemical function of the protein. To assess the specificity of Cj0915 in hydrolyzing the acyl moieties of acyl-CoA derivatives, we tested Cj0915 with following aliphatic acyl-CoA thioesters: acetyl-CoA (C2:0), propionyl-CoA (C3:0), butyryl-CoA (C4:0), hexanoyl-CoA (C6:0), lauroyl-CoA (C12:0), and palmitoyl-CoA (C16:0), stearoyl-CoA (18:0), β-OH-butyryl-CoA, crotonoyl (4:1 trans) and malonyl-CoA. The acyl-CoA substrates with chain length longer than C18 were not studied because of the activity inhibition attributed to the detergent-like effect of long chain acyl-CoA [27]. The initial velocity of hydrolysis of these acyl-CoA substrates at fixed concentration (100 μM) was determined by a continuous spectrophotometric assay as described in Materials & Methods (Fig. 1B). Cj0915 hydrolyzed a wide range of saturated aliphatic acyl-CoA compounds, except for acetyl-CoA. For the substrates with the same chain length, Cj0915 clearly preferred saturated acyl-CoA substrates to unsaturated substrates (C4:0 vs. C4:1 trans). Hydrophilic functional groups such as hydroxyl and carboxyl groups in the acyl moiety also decreased Cj0915 activity (C4:0 vs. β-OH-butyryl-CoA and malonyl-CoA). The kinetic parameters were determined for the four most active substrates, butyryl-CoA, hexanoyl-CoA, lauroyl-CoA, and palmitoyl-CoA, using steady-state kinetic methods (Figure 3; Table 2). The kcat values for these active substrates range from 9.6 to 15.6 s-1, while the Km values range from 10.6 to 40.8 μM. The catalytic efficiency (kcat/Km) of Cj0915 for these active substrates varied from 3.3×105 to 1.1×106 M-1•s-1 (Table 2). In considering that the kcat/Km values of most of enzymes are ~ 1×106 M-1•s-1, although for the enzymes with catalytic perfection the value is over ~ 1×108 M-1•s-1, the measured activity of Cj0915 is physiologically relevant, and therefore Cj0915 is a functional acyl-CoA thioesterase of C. jejuni. In a recent commendable work, the substrate specificities of E. coli YciA and H. influenzae YciA were examined in depth [10]. Together with our study, three common substrates examined for all three enzymes are n-butyryl-CoA, lauroyl-CoA, and palmitoyl-CoA. The kcat/Km values for n-butyryl-CoA / lauroyl-CoA / palmitoyl-CoA were determined as 3.3×105 /1.1× 106/9.1×105 M-1•s-1, 9.9×104 / 4.9×105 / 1.1×105 M-1•s-1, 6.2×106 / 1.4×106 / 1.6×105 M-1•s-1 for Cj0915, E. coli YciA, and H. influenzae YciA, respectively. Apparently, all three enzymes displayed broad substrate specificities for the chain length of the aliphatic acyl group. Therefore, Cj0915 is an efficient enzyme, comparable to the known YciA proteins.
Figure 3.

Substrate saturation curves of Cj0915. Thioesterase activity was measured under the standard assay condition at various concentrations of substrates: butyryl-CoA, hexanoyl-CoA, lauroyl-CoA, and palmitoly-CoA. Curves were the best-fit curves fitted by the Hill rate equation (Eq. 1) as described in Materials and Methods. The average of triplicate data points is plotted.
Table 2.
Steady-state kinetic parameters for Cj0915-catalyzed hydrolysis of acyl-CoA thioesters at pH 7.5 and at 25°C determined as described in Materials and Methods.
| Substrate | Vmax (μmol·min-1·mg-1 | kcat (s-1) | Km (μM) | kcat/Km (M-1 s-1) |
|---|---|---|---|---|
| Butyryl-CoA | 55.0 ± 5.3 | 13.6 ± 1.3 | 40.8 ± 10.3 | 3.3×105 |
| Hexanoly-CoA | 54.3 ± 8.2 | 13.4 ± 2.0 | 39.3 ± 15.2 | 3.4×105 |
| Lauroyl-CoA | 63.3 ± 2.9 | 15.6 ± 0.7 | 13.9 ± 1.4 | 1.1×106 |
| Palmitoly-CoA | 38.9 ± 1.3 | 9.6 ± 0.3 | 10.6 ± 0.6 | 9.1×105 |
Acyl-CoA binding and catalysis by Cj0915
Regardless of their quaternary structures, all the hot dog fold superfamily enzymes studied to date contain two symmetrical active site tunnels at the dimer interface. The acyl-CoA molecule is bound at this subunit-subunit interface with the acyl moiety buried at the core of the dimer and the CoA moiety rested on the surface of the dimer. While the present crystal structure of Cj0915 does not contain a ligand, the active sites in the Cj0915 dimer are readily identified by superposition of structures of hot dog fold thioesterases in complex with various CoA derivatives (Fig. 4A). The acyl-CoA ligands can be divided into three structural moieties: adenosine 3’, 5’-diphosphate, 4-phosphopantetheine, and specificity determining acyl chain (Fig. 4B). For each moiety, mainly two different subunits (referred to as A and B) contribute key residues in forming the recognition site or interacting with the ligands. Interestingly, while the recognition pocket of the 4-phosphopantetheine and acyl moieties is formed by the hot dog fold dimer pair, the contact sites of the adenosine 3’, 5’-diphosphate moiety involve different subunits, depending on the quaternary structure assembly.
Figure 4.

Proposed active site of Cj0915. (A) The Cj0915 dimer (green shades) overlaid with the H. influenzae YciA (gray shades). The CoA ligand (shown in stick model) is fitted onto the cleft after superimposing the Cj0915 structure with HiYciA bound with CoA (PDB code 1YL1). (B) Schematic presentation of ligand recognition sites. Shown are conserved residues critical for recognition of the 4-phosphopantetheine moiety. The corresponding residues in Cj0915 are indicated in green (subunit A) and pale green (subunit B). The corresponding amino acids to HiYciA are indicated in gray. (C) Closeup view of the active site of Cj0915 superimposed with HiYciA-CoA complex. As in Fig.4A, Cj0915 and HiYciA are in green and gray ribbons. Residues defining the proposed active site are labeled.
From various tetrameric structures bound with ligands, it is interesting to observe that the placement of the adenosine 3’, 5’-diphosphate moiety is dictated by the mode of quaternary structure assembly. In YbgC and Pseudomonas sp. β-hydroxybenzoic acid thioesterases (PsHBT) structures (both belong to a distinct group of εγ-tetramers as discussed below), the polar adenosine 3’,5’-diphosphate moiety interacts with the same hot dog dimer pair [23]. On the other hand, in both Thermus thermophilus PaaI (TtPaaI) and Arthrobacter β-hydroxybenzoic acid thioesterases (ArHBT) structures (both belong to a distinct group of βγ-tetramers as discussed below), molecular contacts of the adenosine 3’, 5’-diphosphate moiety involve one subunit from one dimer and another subunit from the second dimer of the tetramer (therefore, three subunits are involved in ligand binding) [24,25]. The HiYciA structure in complex with CoA revealed that the polar adenosine 3’,5’-diphosphate moiety interacts with the hot dog pair dimer in the same way as εγ-tetramers [19]. Based on the HiYciA structure bound to CoA, it is obvious that Cj0915 would form elaborated electrostatic interactions with the 3’-phosphoryl group of the CoA ribose through residues Asn80, Thr81, and Ser82 of subunit A, which correspond to Arg90, Ser91, and Ser92 in HiYciA, respectively (Fig. 4B & 4C).
Comparison of the recognition sites of 4-phosphopantetheine clearly indicates that key recognition residues and/or sequence motifs include Val50, Thr51, Ile52, Ser53, Val113, and Gly119 (from subunit A), and Ile24, Phe25, Gly26, Phe59, Lys60, Glu61, and Pro62 (from subunit B) (Fig. 4B & 4C). These residues are mostly located at the beginning of the central helix α1, the beginning and the end of β2, and the loop between β3 and β4 (Fig. 5A). Cleary, the ligand binding site is conserved, albeit not identical, suggesting a very similar binding pocket structure for the 4-phosphopantetheine moiety (Fig. 4C), with conserved sequence motifs such as (I)-(F)-(G) at the N terminus of α1, (V)-(T)-(I/V)-X at the N terminus of β2, and (G)-X-(S/T)-S at the connecting loop between β3 and β4 (Fig. 5A).
Figure 5.

(A) Structure-based sequence alignment of three distinct quaternary structures of hot dog fold thioesterases: hexamers, βγ-tetramers, and εγ-tetramers. While the protomer of both Cj0915 (YciA) and Cj0965c (YbgC) homologs adopts 5 stranded β-sheet hot dog fold, that of Thermus thermophilus PaaI (Arthrobacter HBT) consists of 6-stranded β-sheet. Catalytic residues Asp/Glu are highlighted in red. The secondary structure elements are shown above primary sequences of each class based on the Cj0915, EcYbgC, and ArHBT structures. For each group, identical residues are highlighted in gold, pale blue, and silver. C, Campylobacter jejuni; H, Haemophilus influenzae, E, Escherichia coli, Ps, Pseudomonas sp.; T, Thermus thermophilus; Ar, Arthrobacter sp. (B) Distinct quaternary assemblies with hot dog fold acyl-CoA thioesterases determined by crystallography. Ribbons diagrams were made using coordinates of Cj0915 (PDB code 3D6L), EcYbgC (PDB code 1S5U), and TtPaaI (PDB code 1WN3).
The crystal structures of Cj0915 homologs (i.e. hexameric thioesterases) solved to date only contain CoA. As for the specificity determining acyl moiety, however, structural insights can be gained from other thioesterases such as TtPaaI bound with the ligand hexanoyl-CoA and ArHBT bound with the ligand 4-hydroxyphenacyl-CoA. The ligand pocket that adapts the 4-hydroxyphenacyl moiety in the crystal structure is inaccessible to solvent. In the absence of the acyl moiety, the hollow corresponding the pocket of HiYciA bound with CoA is now only partially buried by a salt bridge between Asp33 of one subunit and Arg58 of second subunit [19]. By analogy, structural comparison between one HiYciA dimer and the corresponding crystallographic dimer of Cj0915 suggests the same might be true for Cj0915 involving Asn23 of one subunit and Arg48 of the second subunit (Fig. 5A). However, without ligand binding in Cj0915, the loop containing Asn23 is flexible and not well superimposed with the HiYciA structure.
Structurally and mechanistically, hot dog fold thioesterases possess intriguing attributes. For example, β-hydroxybenzoic acid thioesterases from Pseudomonas sp. and from Arthrobacter adopt two distinct quaternary assemblies, βγ- and εγ-tetramers (see below) [20,23,24]. Mechanistically, these two orthologs employ two distinct sites of the catalytic carboxylate residue [20,23,24]. While the catalytic carboxylate residue of PsHBT is Asp17 located at the connecting loop between 310 helix and the central α-helix, the catalytic carboxylate residue of ArtHBT is Glu73 located in the middle of the central α-helix (Fig. 5A). Therefore, it is interesting to observe that the catalytic carboxylate residue of E. coli YciA is Asp37 (Asp44 of HiYciA and Asp34 of Cj0915) located in the central α-helix, the equivalent position to Glu73 of ArtHBT, although Cj0915/YciA-like thioesterases are structurally reminiscent of PsHBT, including the quaternary structure assembly mode and the binding position of the adenosine 3’, 5’-diphosphate moiety as discussed above (Fig. 5A & 5B).
DISCUSSION
In an effort to understand the fatty acid biosynthesis pathway of Campylobacter jejuni and its possible link to virulence of this organism, we initiated a long-term investigation on structural and functional characterization of enzymes catalyzing C. jejuni lipid metabolism. Because there is no information available on this fundamental biological process in this important pathogen, genome sequence data banks were employed to search for relevant putative enzymes. We identified two candidate thioesterases, Cj0915 and Cj0965c, which showed strong sequence identities to their homologs from γ-proteobacteria such as Escherichia coli and Haemophilus influenzae.
The Cj0915 protein is a homolog of YciA proteins from E. coli (EcYciA; b1253 of K12 strain) and H. influenzae. (HiYciA, HI0827 of Rd KW20 strain). Whereas EcYciA and HiYciA share over 60% primary sequence identity, the Cj0915 is 37% and 35% identical to EcYciA and HiYciA, respectively. Unlike many other C. jejuni proteins showing strong identity to corresponding Helicobacter pylori proteins, reflecting that both organisms belong to ε-proteobacteria, the Cj0915 protein is only 25% identical to its H. pylori homolog HP0891. Interestingly, however, Cj0915 shows over 30% identity to eukaryotic thioesterase. To explore how the function of YciA proteins might support the cell, a recent study examined the yciA gene context for many γ-proteobacteria containing homologs of EcYciA or HiYciA [10]. While the YciA protein appears to be in a diverse gene context, one emerging common point of YciA proteins suggested the involvement of YciA in the biogenesis of the inner membrane for division or transport protein [10]. To extend this observation, we analyzed the gene context of Cj0915. Despite the structural and functional conservation among YciA proteins as demonstrated in this study, the gene locus of Cj0915 does not suggest any similar gene organization observed in many γ-proteobacteria. In considering that C. jejuni genome rarely contains gene clusters or operons, this observation is not surprising. Nonetheless, we note that a neighboring gene Cj0914c corresponds to CiaB, one of the secreted Campylobacter invasion antigens and a virulence factor protein [28]. It would be interesting to know whether or not the function of Cj0915 links with CiaB secretion and virulence in C. jejuni.
The Cj0965c protein is a homolog of the E. coli YbgC protein, another E. coli hot dog fold thioesterase encoded by the ybgC gene of the tol-pal gene cluster [12]. The YbgC homologs are highly conserved proteins across the Tol–Pal system, which is required for maintaining outer membrane integrity and is well conserved in Gram-negative bacteria [12]. The system consists of (at least) five proteins: TolA, TolQ, TolR, TolB, and Pal (peptidoglycan-associated lipoprotein). In E. coli, the ybgC gene is part of the operon involving ybgC-tolQ-tolR-tolA [12]. Previous studies reported that YbgC exhibits a significant level of thioesterase activity toward short chain acyl-CoA thioesters showing the kcat/Km values only up to 4×10 M-1•s-1 [29]. However, this magnitude of enzyme efficiency is not considered as physiologically relevant. In this study, Cj0915 showed efficient thioesterase activity toward aliphatic acyl-CoA, comparable to HiYciA or EcYciA, whereas we could not detect measurable activity with Cj0965c. This (very) low activity of YbgC or Cj0965c toward aliphatic acyl-CoA appears to be an inherent property of the enzyme. Interestingly, the Tol-Pal system is almost conserved even in C. jejuni with the gene locus cj0109 - cj0110 - cj0111 - cj0112 - cj0113 - cj0114 corresponding to TolQ –TolR - a putative protein –TolB –Pal -YbgF. However, unlike this region in other well-conserved organisms, tolA and cj0965c (ybgC homolog) in C. jejuni are not part of the operon. While all these proteins remain putative proteins of unknown function, our observation raises an interesting question as to whether or not this quasi-conserved tol-pal locus plays a role in C. jejuni virulence.
Although all known hot dog thioesterases share the common fold, it is remarkable to observe different ways of oligomer formation as well as enzyme activity for various substrates. Structurally, at least three distinct quaternary assemblies have been identified: βγ-tetrameric, εγ-tetrameric, and hexameric thioesterases. Crystal structures and enzymatic characterization have been reported for many tetrameric thioesterases, where two dimers can associate on the β-sheet side (referred to as βγ-mode) or on the helical side (referred to as εγ-mode) (Fig. 5B) [25]. One of the remarkable findings from the previous structural studies is exemplified by two orthologs: Arthrobacter HBT (βγ-mode) and Pseudomonas HBT (εγ-mode), suggesting evolving nature of these quaternary structures. The Cj0915 thioesterase is related to YciA proteins in sequence and adopts a hexamer like HiYciA [19]. Therefore, this quaternary structure appears to be preferred to this group of thioesterases. In the Cj0915 hexamer three dimers are associated on the helical side, similar to εγ-mode tetramers. However, as one hexamer contains three dimers, the individual five-stranded anti-parallel β–sheets of six protomers form a continuous and twisted 30-stranded β-barrel (Fig. 5B). It would be interesting to know if other YciA homologs can adopt other than the quaternary structure revealed by Cj0915 and HiYciA.
Mechanistically, one of the remarkable observations with the tetrameric hot dog thioesterase family is that its catalytic carboxylate site is located at one of two conserved positions of the enzyme. This divergence of the catalytic residue and mechanism may define two evolutionary distinct classes. Based on the crystal structures and their functional attributes, it is now reasonable to rationalize this mechanistic diversity. From the quaternary structural viewpoint, the Cj0915 (YciA) thioesterase is rather similar to YbgC proteins, as the formation of the tetramer or the hexamer is via helical side (the β sheet outside) (Fig. 5B). However, from the mechanistic viewpoint, the Cj0915 thioesterase shares the same catalytic carboxylate site, the conserved amino acid Asp/Glu located in the center of the signature α-helix of the hot dog fold (Fig. 5A). Although YbgC proteins adopt the typical hot dog fold and also contain the conserved Asp/Glu residues in the central α-helix, this carboxylate residue is not at the geometrically ideal position for a nuclephile or catalytic base. This is due to the inclination of the central helix. In Cj0915 (YciA) and TtPaaI-like thioesterases, the angle between the central helix and the perpendicular plane to hot dog fold β sheet is about 45° (Fig. 6). By contrast, in the YbgC-like thioesterases the central helix is tilted 30° from the perpendicular plane to β sheet, which renders the Asp/Glu position further away from the attacking carbonyl center (Fig. 6). Therefore, for this group of enzyme a new catalytic site appears to be evolved to adapt a more productive catalytic efficiency of the enzyme.
Figure 6.

Variable geometry among hot dog fold dimers. The observed angles between the central helix (cylinder) and the perpendicular plane to the hot dog fold β sheet (Cα chain) are indicated.
Oligomerization of proteins represents a fundamental strategy for producing protein structural and functional complexity [30]. Plausible advantages for higher order structures would include genetic economy, functional gain, structural stability, and increased potential for regulation [31]. Although some arguments have been proposed to rationalize the selection pressures to evolve and maintain specific dimerization interfaces, few direct tests have been performed to address specific selection benefits of the quaternary structures observed. In this regard, the hot dog fold thioesterases represent an excellent system to investigate, as we start to collect a significant body of structural and functional data. Why some primary structures, such as Cj0915 (YciA) homologs, form hexamers, and some other primary structures, Cj0965c (YbgC) homologs, form tetramers? What is the driving force for the evolution of specific hot dog fold thioesterases? While these fundamental questions await experimental evidence, the structure-based sequence alignment of the three acyl-CoA thioesterases clearly shows specific sequence conservation on the regions involved in the higher order quaternary structure in each class (Fig. 5A).
In conclusion, the crystal structure and the functional identification of Cj0915 delineate the first functional acyl-CoA thioesterase of C. jejuni. Our study contributes to establishing a novel class of hexameric thioesterases. Together with previous structural studies, it should be possible to investigate the molecular evolution of quaternary structures in the hot dog fold thioesterases. Finally, this study provides a framework for investigation on roles of the Cj0915 thioesterase in virulence, and functional activities associated with the Cj0915 thioesterase in vivo.
Acknowledgments
We thank the staff of beamline 19BM of the Structural Biology Center at APS (Argonne National Laboratory) for help during data collection, Dr. Pat Guerry for C. jejuni 81-117 genomic DNA of exceptional quality and Drs. Joe Eichberg and Tim Cooper for helpful discussions.
Abbreviations
- CoA
coenzyme A
- IPTG
isopropyl β-D-thiogalactopyranoside
- PBS
Phosphate Buffered Saline
- GST
Glutathione S-transferase
- DTT
dithiothreitol
- DTNB
5,5’-dithio-bis(2-nitrobenzoic acid)
- HEPES
N-(2-hydroxyethyl)piperazine-N’-2-ethanesulfonate
- HBT
β-hydroxybenzoic acid thioesterases
Footnotes
This work was supported in part by National Institutes of Health Grant RO1 AI068943 (to H.J.Y.) and by the R. Welch Foundation grant E-1616 (to H.J.Y.)
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Hunt MC, Alexson SE. The role Acyl-CoA thioesterases play in mediating intracellular lipid metabolism. Prog Lipid Res. 2002;41:99–130. doi: 10.1016/s0163-7827(01)00017-0. [DOI] [PubMed] [Google Scholar]
- 2.Nardini M, Dijkstra BW. Alpha/beta hydrolase fold enzymes: the family keeps growing. Curr Opin Struct Biol. 1999;9:732–737. doi: 10.1016/s0959-440x(99)00037-8. [DOI] [PubMed] [Google Scholar]
- 3.Li C, Hassler M, Bugg TD. Catalytic promiscuity in the alpha/beta-hydrolase superfamily: hydroxamic acid formation, C--C bond formation, ester and thioester hydrolysis in the C--C hydrolase family. Chembiochem. 2008;9:71–76. doi: 10.1002/cbic.200700428. [DOI] [PubMed] [Google Scholar]
- 4.Dillon SC, Bateman A. The Hotdog fold: wrapping up a superfamily of thioesterases and dehydratases. BMC Bioinformatics. 2004;5:109. doi: 10.1186/1471-2105-5-109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Leesong M, Henderson BS, Gillig JR, Schwab JM, Smith JL. Structure of a dehydratase-isomerase from the bacterial pathway for biosynthesis of unsaturated fatty acids: two catalytic activities in one active site. Structure. 1996;4:253–264. doi: 10.1016/s0969-2126(96)00030-5. [DOI] [PubMed] [Google Scholar]
- 6.Parkhill J, Wren BW, Mungall K, Ketley JM, Churcher C, Basham D, Chillingworth T, Davies RM, Feltwell T, Holroyd S, Jagels K, Karlyshev AV, Moule S, Pallen MJ, Penn CW, Quail MA, Rajandream MA, Rutherford KM, van Vliet AH, Whitehead S, Barrell BG. The genome sequence of the food-borne pathogen Campylobacter jejuni reveals hypervariable sequences. Nature. 2000;403:665–668. doi: 10.1038/35001088. [DOI] [PubMed] [Google Scholar]
- 7.Guerry P. Campylobacter flagella: not just for motility. Trends Microbiol. 2007;15:456–461. doi: 10.1016/j.tim.2007.09.006. [DOI] [PubMed] [Google Scholar]
- 8.Yokoyama T, Paek S, Ewing CP, Guerry P, Yeo HJ. Structure of a σ28-Regulated Nonflagellar Virulence Protein from Campylobacter jejuni. J Mol Biol. 2008;384:364–376. doi: 10.1016/j.jmb.2008.09.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Song F, Zhuang Z, Finci L, Dunaway-Mariano D, Kniewel R, Buglino JA, Solorzano V, Wu J, Lima CD. Structure, function, and mechanism of the phenylacetate pathway hot dog-fold thioesterase PaaI. J Biol Chem. 2006;281:11028–11038. doi: 10.1074/jbc.M513896200. [DOI] [PubMed] [Google Scholar]
- 10.Zhuang Z, Song F, Zhao H, Li L, Cao J, Eisenstein E, Herzberg O, Dunaway-Mariano D. Divergence of function in the hot dog fold enzyme superfamily: the bacterial thioesterase YciA. Biochemistry. 2008;47:2789–2796. doi: 10.1021/bi702334h. [DOI] [PubMed] [Google Scholar]
- 11.Li J, Derewenda U, Dauter Z, Smith S, Derewenda ZS. Crystal structure of the Escherichia coli thioesterase II, a homolog of the human Nef binding enzyme. Nat Struct Biol. 2000;7:555–559. doi: 10.1038/76776. [DOI] [PubMed] [Google Scholar]
- 12.Sturgis JN. Organisation and evolution of the tol-pal gene cluster. J Mol Microbiol Biotechnol. 2001;3:113–122. [PubMed] [Google Scholar]
- 13.Otwinowski Z, Minor W. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 14.Vagin A, Teplyakov A. An approach to multi-copy search in molecular replacement. Acta Crystallogr D. 2000;56:1622–1624. doi: 10.1107/s0907444900013780. [DOI] [PubMed] [Google Scholar]
- 15.Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr D. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 16.Brunger AT, Adams PD, Clore GM, DeLano WL, Gros P, Grosse-Kunstleve RW, Jiang JS, Kuszewski J, Nilges M, Pannu NS, Read RJ, Rice LM, Simonson T, Warren GL. Crystallography & NMR system: A new software suite for macromolecular structure determination. Acta Crystallogr D. 1998;54:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- 17.Murshudov GN, Vagin AA, Lebedev A, Wilson KS, Dodson EJ. Efficient anisotropic refinement of macromolecular structures using FFT. Acta Crystallogr D. 1999;55:247–255. doi: 10.1107/S090744499801405X. [DOI] [PubMed] [Google Scholar]
- 18.Alexson SE, Nedergaard J. A novel type of short- and medium-chain acyl-CoA hydrolases in brown adipose tissue mitochondria. J Biol Chem. 1988;263:13564–13571. [PubMed] [Google Scholar]
- 19.Willis MA, Zhuang Z, Song F, Howard A, Dunaway-Mariano D, Herzberg O. Structure of YciA from Haemophilus influenzae (HI0827), a hexameric broad specificity acyl-coenzyme A thioesterase. Biochemistry. 2008;47:2797–2805. doi: 10.1021/bi702336d. [DOI] [PubMed] [Google Scholar]
- 20.Benning MM, Wesenberg G, Liu R, Taylor KL, Dunaway-Mariano D, Holden HM. The three-dimensional structure of 4-hydroxybenzoyl-CoA thioesterase from Pseudomonas sp. Strain CBS-3. J Biol Chem. 1998;273:33572–33579. doi: 10.1074/jbc.273.50.33572. [DOI] [PubMed] [Google Scholar]
- 21.Forwood JK, Thakur AS, Guncar G, Marfori M, Mouradov D, Meng W, Robinson J, Huber T, Kellie S, Martin JL, Hume DA, Kobe B. Structural basis for recruitment of tandem hotdog domains in acyl-CoA thioesterase 7 and its role in inflammation. Proc Natl Acad Sci U S A. 2007;104:10382–10387. doi: 10.1073/pnas.0700974104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kimber MS, Martin F, Lu Y, Houston S, Vedadi M, Dharamsi A, Fiebig KM, Schmid M, Rock CO. The structure of (3R)-hydroxyacyl-acyl carrier protein dehydratase (FabZ) from Pseudomonas aeruginosa. J Biol Chem. 2004;279:52593–52602. doi: 10.1074/jbc.M408105200. [DOI] [PubMed] [Google Scholar]
- 23.Thoden JB, Holden HM, Zhuang Z, Dunaway-Mariano D. X-ray crystallographic analyses of inhibitor and substrate complexes of wild-type and mutant 4-hydroxybenzoyl-CoA thioesterase. J Biol Chem. 2002;277:27468–27476. doi: 10.1074/jbc.M203904200. [DOI] [PubMed] [Google Scholar]
- 24.Thoden JB, Zhuang Z, Dunaway-Mariano D, Holden HM. The structure of 4-hydroxybenzoyl-CoA thioesterase from Arthrobacter sp. strain SU. J Biol Chem. 2003;278:43709–43716. doi: 10.1074/jbc.M308198200. [DOI] [PubMed] [Google Scholar]
- 25.Kunishima N, Asada Y, Sugahara M, Ishijima J, Nodake Y, Sugahara M, Miyano M, Kuramitsu S, Yokoyama S, Sugahara M. A novel induced-fit reaction mechanism of asymmetric hot dog thioesterase PaaI. J Mol Biol. 2005;352:212–228. doi: 10.1016/j.jmb.2005.07.008. [DOI] [PubMed] [Google Scholar]
- 26.Holm L, Sander C. Dali: a network tool for protein structure comparison. Trends Biochem Sci. 1995;20:478–480. doi: 10.1016/s0968-0004(00)89105-7. [DOI] [PubMed] [Google Scholar]
- 27.Tilton GB, Shockey JM, Browse J. Biochemical and molecular characterization of ACH2, an acyl-CoA thioesterase from Arabidopsis thaliana. J Biol Chem. 2004;279:7487–7494. doi: 10.1074/jbc.M309532200. [DOI] [PubMed] [Google Scholar]
- 28.Konkel ME, Klena JD, Rivera-Amill V, Monteville MR, Biswas D, Raphael B, Mickelson J. Secretion of virulence proteins from Campylobacter jejuni is dependent on a functional flagellar export apparatus. J Bacteriol. 2004;186:3296–3303. doi: 10.1128/JB.186.11.3296-3303.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhuang Z, Song F, Martin BM, Dunaway-Mariano D. The YbgC protein encoded by the ybgC gene of the tol-pal gene cluster of Haemophilus influenzae catalyzes acyl-coenzyme A thioester hydrolysis. FEBS Lett. 2002;516:161–163. doi: 10.1016/s0014-5793(02)02533-4. [DOI] [PubMed] [Google Scholar]
- 30.Ali MH, Peisach E, Allen KN, Imperiali B. X-ray structure analysis of a designed oligomeric miniprotein reveals a discrete quaternary architecture. Proc Natl Acad Sci U S A. 2004;101:12183–12188. doi: 10.1073/pnas.0401245101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Marianayagam NJ, Sunde M, Matthews JM. The power of two: protein dimerization in biology. Trends Biochem Sci. 2004;29:618–625. doi: 10.1016/j.tibs.2004.09.006. [DOI] [PubMed] [Google Scholar]


