Abstract
The apoptotic effects of docosahexaenoic acid (DHA) and other ω-3 polyunsaturated fatty acids (PUFAs) have been documented in cell and animal studies. The molecular mechanism by which DHA induces apoptosis is unclear. Although there is no direct evidence, some studies have suggested that DNA damage generated through lipid peroxidation may be involved. Our previous studies showed that DHA, because it is high degree of unsaturation, can give rise to the acrolein-derived 1,N2-propanodeoxyguanosine (Acr-dG) as a major class of DNA adducts via lipid oxidation. As a first step to investigate the possible role of oxidative DNA damage in apoptosis induced by DHA, we examined the relationships between oxidative DNA damage and apoptosis caused by DHA in human colon cancer HT-29 cells. The apoptosis and oxidative DNA damage, including Acr-dG and 8-oxo-deoxyguanosine (8-oxo-dG) formation, in cells treated with DHA and ω-6 PUFAs, including arachidonic acid (AA) and linoleic acid (LA), were measured. DHA induced apoptosis in a dose- and time-dependent manner with a concentration range from 0 to 300 µM as indicated by increased caspase-3 activity and PARP cleavage. In contrast, AA and LA had little or no effect at these concentrations. The Acr-dG levels were increased in HT-29 cells treated with DHA at 240 and 300µM, and the increases were correlated with the induction of apoptosis at these concentrations, while no significant changes were observed for 8-oxo-dG. Because proteins may compete with DNA to react with Acr, we then examined the effects of BSA on the DHA induced apoptosis and oxidative DNA damage. The addition of BSA to HT-29 cell culture media significantly decreases Acr-dG levels with a concomitant decrease in the apoptosis induced by DHA. The reduced Acr-dG formation is attributed to the reaction of BSA with acrolein as indicated by increased levels of total protein carbonyls. Similar correlations between Acr-dG formation and apoptosis were observed in HT-29 cells directly incubated with 0 to 200µM of acrolein. Additionally, DHA treatment increased level of DNA strand breaks and caused cell cycle arrested at G1 phase. Taken together, these results demonstrate the parallel relationships between the Acr-dG level and apoptosis in HT-29 cells, suggesting that the formation of Acr-dG in cellular DNA may contribute to apoptosis induced by DHA.
Keywords: polyunsaturated fatty acids, apoptosis, chemoprevention, colon cancer, docosahexaenoic acid (DHA), arachidonic acid (AA), linoleic (LA), acrolein, 4-hydroxy-2-nonenal, cyclic deoxyguanosine adducts, oxidative DNA damage, 32P-postlabeling
Introduction
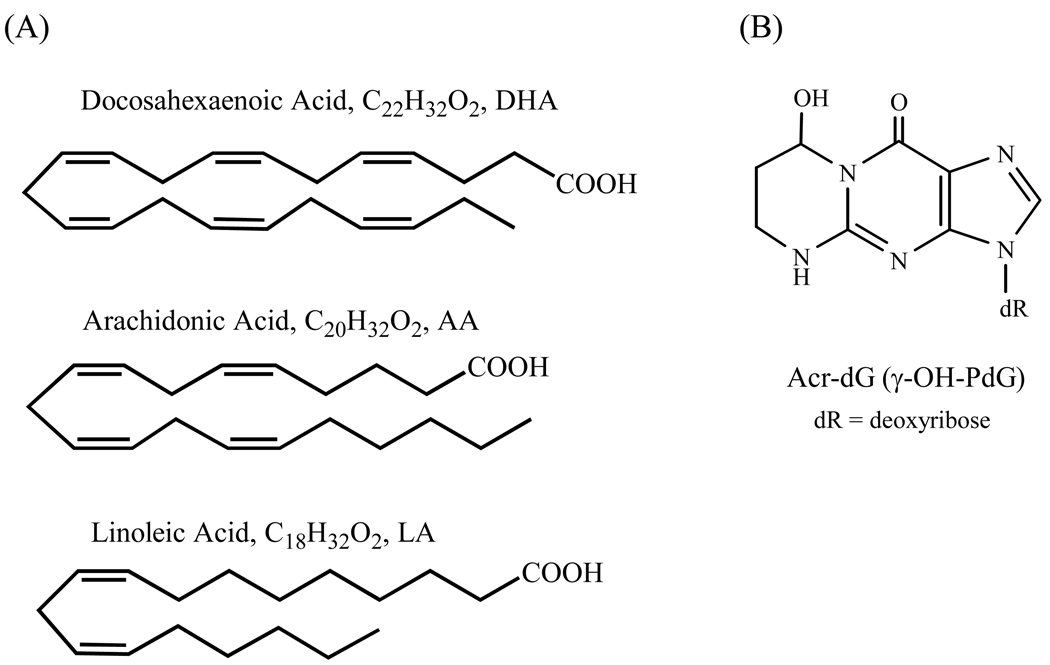
The opposite effects of ω-3 and ω-6 polyunsaturated fatty acids (PUFAs) on colon carcinogenesis have been well-documented (1–5); ω-3 PUFAs, including docosahexaenoic acid (DHA) and eicosapentaenoic acid (EPA), are protective against colon cancer, whereas ω-6 PUFAs, such as linoleic acid (LA) and arachidonic acid (AA), promote it. DHA, abundant in fish oil, is one of the ω-3 PUFAs which has been widely studied. Despite the collective evidence has shown that DHA and other ω-3 PUFAs possess anti-carcinogenic activity, implicating their roles in cancer prevention, the underlying molecular mechanisms and its application in cancer prevention and therapy remain unclear and are under debate (6,7). The key question is why relatively subtle structural variations between ω-3 and ω-6 fatty acids result in such a dramatic difference in the modulation of carcinogenesis (Figure 1A). Here, we report a study on the role of oxidative DNA damage in apoptosis caused by DHA in human colon cancer cells. Our hypothesis is that the formation of specific oxidative DNA damage induced by DHA and possibly other ω-3 PUFAs could trigger apoptotic response that may underlie a mechanism by which ω-3 PUFAs protect against colon tumorigenesis.
Fig. 1.
Structures of PUFAs (A) and Acr-dG (B).
Lipid peroxidation is an important endogenous source for the formation of a variety of DNA adducts (8–12). PUFAs can be oxidized by reactive oxygen species with the generation of electrophilic compounds, such as α,β-unsaturated aldehydes (enals), capable of forming DNA or protein adducts both of which could trigger apoptotic responses from cells. The cyclic 1,N2 -propanodeoxyguanosine (PdG) adducts are reaction products of enals with deoxyguanosine (dG). Our previous studies showed a distinct pattern of the cyclic adduct formation from ω-3 vs. ω-6 PUFAs under oxidative conditions (13). The shorter chain acrolein (Acr)-, crotonaldehyde- and pentenal-dG adducts are the major products from oxidized ω-3 PUFAs, whereas the longer chain heptenal- and 4-hydroxy-2-nonenal (HNE)-dG adducts are exclusively formed by ω-6 PUFAs. We found that the formation of Acr adducts is correlated with the number of double bonds in PUFAs, thus DHA and EPA are the main sources of their formation and the order of the Acr adduct formation is DHA>EPA>AA>LA. Among the cyclic dG adducts identified in vitro, so far only Acr and HNE adducts are ubiquitously detected in tissues of rodents and humans (8,14,15). Crotonaldehyde-dG adducts are also present in DNA, however, the in vivo levels are often too low to be consistently detected. For reasons still unclear, among the Acr-dG stereoisomers (α-OH- and γ-OH-PdG, originally designated as Acr-dG 1, 2 and 3, respectively, see reference 8), γ-OH-PdG (Acr-dG 3) has been detected as the major isomer in vivo (Figure 1B) by an HPLC-based 32P-postlabeling assay (8,10). However, a recent study using LC-mass spectrometry showed that both α- and γ-OH-Acr-dG are present almost in equal amount in DNA from human lung cells (15). The reason for the differences in these studies is unclear at present. The basal levels of Acr adducts in tissues usually are in the range of adducts per 107 bases, whereas HNE adducts are in adducts per 109 bases (16). The nearly two orders of magnitude higher levels of Acr adducts than HNE adducts could be attributed to either the greater reactivity and more abundance of Acr than HNE or more efficient repair of HNE than Acr adducts or both. As a major lipid peroxdation-derived DNA adduct in vivo, Acr-dG adducts have been extensively studied for their roles in mutagenicity in vitro, but the results are mixed depending on the assay conditions (17–21).
As a defense mechanism, cells respond to various forms of DNA damage by undergoing apoptosis through different sensing and signaling pathways. One of the most studied DNA damages is the radiation induced DNA strand breaks which cause replication stress which could initiate the apoptotic pathways (22–23). DNA damage can also cause transcriptional stress generating the transcription-coupled DNA strand breaks that have also been shown to activate apoptosis responses (24). In addition, DNA repair systems can also be engaged and play important roles in cell cycle checkpoint recognition and apoptosis initiation (25–27). Except for DNA strand breaks, relatively few DNA adducts have been shown to trigger apoptosis (25, 28, 29). In this study, we examined, as a first step to investigate the roles of the Acr-derived cyclic dG adduct and 8-oxo-dG in apoptosis, the relationships between the oxidative DNA damages and apoptosis induced by DHA in human colon HT-29 cells.
Materials and methods
Chemicals
Acr, calf thymus DNA and micrococcal nuclease were obtained from Sigma-Aldrich Co. (St. Louis, MO). Spleen phosphodiesterase was purchased from Worthington Biochemical (Lakewood, NJ), and [γ-32P]-ATP and T4 polynucleotide kinase were from Amersham (Piscataway, NJ). Nuclease P1 was obtained from Yamasa Shoyu Co. (Choshi, Japan). All other reagents, unless otherwise stated, were from Sigma-Aldrich Co. and Fisher Chemical (Fair Lawn, NJ). The 3’-monophosphate of Acr-dG was prepared as previously described (30). DHA, LA and AA were purchased from Cayman Chemical Company (Ann Arbor, MI).
Cell culture and PUFA treatments
The human colonic carcinoma HT-29 cells were routinely cultured at 37°C with 5% carbon dioxide in the DMEM medium with 10% FBS, 50 I.U./ml penicillin and 50 µg/ml streptomycin (all from Mediatech Inc, Herndon, VA). DHA, AA and LA were freshly prepared from the stock solutions before each treatment. All the treatments were applied to cells when they reached ~40% confluence. Cells were treated with different PUFAs in the concentration range from 0 to 300 µM and with indicated concentrations for 0, 4, 8, 12, and 24 h. Unless otherwise indicated, the fatty acids were dissolved in small volume of ethanol (<0.5% of total medium volume) as a vehicle and cells treated with vehicle alone were used as controls. In experiments where the effects of BSA on apoptosis were examined, a routinely used method was used to deliver the PUFAs in the form of BSA-PUFA complex (31, 32). Briefly, media containing certain concentrations of PUFAs and BSA were first incubated in a 37°C shaker for 1 h before being added to the cells. After each treatment, cells were collected from the dishes and processed for assays described below.
Apoptosis Assays
For caspase-3 activities, the harvested cells were washed once with ice-cold PBS. The cell pellets were then suspended in assay buffer (50 mM HEPES pH7.4, 100 mM NaCl, 10 mM DTT, 1 mM EDTA, 0.1% CHAPS) and lysed by freezing and thawing three times. The lysates were centrifuged at 10000×g for 15 min, and the supernatants, postnuclear cell lysates, were collected. To measure caspase-3 activity in the supernatants, the Ac-DEVD-R110 (Roche Applied Science, Indianapolis, IN), the rhodamine 110-labeled peptide, was added as substrate, and the release of rhodamine 110 from the substrate was monitored by measuring the fluorescent every 10 min for 1 h with a 485±20 nm excitation filter and a 528±20 nm emission filter on a spectrophotometer microplate reader. The amount of R110 released per min was determined, and then normalized with protein amount, so the final caspase-3 activities were expressed as nmol R110/min/mg cellular protein.
The standard procedure to measure the PARP cleavage was used. Briefly, the cells will be harvested, washed and lysed. Proteins were separated by SDS-PAGE gel electrophoresis and then transferred to a nitrocellulose membrane. The blot was blocked with 5% milk in 1×TBST solution and incubated with the anti-PARP antibody (BD Biosciences, Franklin Lakes, NJ) overnight. After incubating with secondary antibody, the blot was developed in SuperSignal West Chemiluminescent Substrate (Pierce Biotechnology, Inc, Rockford, IL) for 5min and visualized on a Syngene Bio Imaging System.
For Annexin V-FITC staining, cells treated with Acr were washed twice with PBS and resuspended in binding buffer (10 mM Hepes/NaOH, pH 7.5, 140 mM NaCl, and 2.5 mM CaCl2). Cell suspension was then incubated with annexin V–FITC and propidium iodide (PI) for 10 min at room temperature in the dark. The population of annexin V-positive cells was evaluated by flow cytometry.
Detection of Acr-dG and 8-oxo-dG adducts in DNA
DNA was isolated from PBS washed cells by a previously reported method (33), quantified by absorbance at 260 nm, and its purity was checked with A260/A230 (>1.8) and A260/A280 (>1.8). About 20~50µg of DNA were obtained from approximately 106 cells and used for the assays.
The Acr-dG levels in DNA were determined by an improved 32P-Postlabeling/Solid Phase Extraction (SPE)/HPLC method (34). Briefly, DNA was first digested with micrococcal nuclease and spleen phosphodiesterase, then applied to a preconditioned SPE column (C18, 200 mg, 1 ml volume; Varian, Harrbor City, CA). The column was then washed with 1.7 ml of 5 mM ammonium formate pH 3.5 containing 2% methanol to remove most of the unmodified nucleotides, the eluted solution was collected, dried and analyzed on HPLC system-1 to quantify the amount of dG 3’-monophosphate (3’-mP) in the DNA sample. The Acr-dG 3’-mP was then eluted with 0.7 ml of 30% methanol in water and collected in tubes with 0.05 mM of GSH, which has been shown to prevent artificial adduct formation. The eluted adduct fraction was dried, incubated with nuclease P1 to further hydrolyze residual unmodified nucleotides, and then labeled and converted to 32P labeled Acr-dG 5’-monophosphate (5’-mP) by T4 polynucleotide kinase in the presence of [γ- 32P]ATP. After labeling, the 32P-labeled Acr-dG 5’-mP was separated with a second SPE, spiked with the Acr-dG 5’-mP as UV marker and purified with reverse-phase HPLC System 2 followed by ion-pairing HPLC System 3. Finally, the collected Acr-dG 5’-mP fraction was converted into ring-opened Acr-dG 5’-mP and analyzed on a reverse phase HPLC System 4. The amount of the Acr-dG adducts was quantified by comparing its radioactivity with that of the Acr-dG standard that was gone through the same analysis procedure. The adduct level was obtained by normalizing the adduct amount with the amount of the normal dG in that same sample.
All HPLC systems consist of Shimadzu or Waters solvent delivering components with photodiode array or UV/VIS detector. The columns used in each system is a Prodigy ODS 3 C18 reverse-phase column (5 µm, 250 mm × 4.6 mm) from Phenomenex (Torrance, CA). The solvents and gradient program used in each system are: System 1, A: 50 mM NaH2PO4 (pH 5.8), B: 50% methanol with 0~30% A in 15 min 0.7 ml/min; System 2, A: 5mM sodium citrate (pH 8.1), B: methanol/water 50:50 with 0→30% B in 40 min at 0.6 ml/min; Systems 3, A: 50mM triethylamine phosphate (pH 6.4), B: methanol/water 50:50 with 0→40% B in 40 min at 0.6 ml/min; Systems 4 uses a β-Ram radio-flow detector (IN/US systems, Inc., Fairfield, NJ, USA) to measure radioactivity, with A: 5 mM sodium citrate (pH 5), B: methanol/water 50:50 with 0→50% B in 50 min at 0.6 ml/min.
The 8-oxo-dG was analyzed with an HPLC-Electrochemical assay described before (35).
Protein carbonyl formation assay
To measure the protein oxidation level in the cell culture media and in cells, the traditional 2,4-dinitrophenylhydrazine (DNPH) assay was used (36). Briefly, 200µl of cell culture medium or cell lysate was reacted with 800µl DNPH dissolved in 2.5M HCl by incubating at room temperature at dark for 1 h. At the end of the reaction, 1ml 20% TCA was added to precipitate the proteins, the sample was vortexed and put on ice for 5 min. After centrifugation, the protein pellet was first washed by vortexing with 1ml ethanol/ethyl acetate (1:1) for two to three times, then resuspended into 500ul guanidine hydrochloride by vortexing. After removing any debris by centrifugation, absorbance at 370nm was measured.
Cell cycle analysis
About 1~2×106 cells were trypsinized and washed with PBS. Then 0.5ml cold PBS was added and the cells were gently suspended by pipetting to obtain single cell suspension. To fix the cells, 0.5ml ice-cold absolute ethanol was added three times followed by mixing and suspending cells well after each addition of ethanol. The final suspension, 1~2×106 cells in 2.0ml 75% ethanol, was kept on ice for at least 20min, then stained with 50 µg/ml PI, 50 µg/ml RNase A, and 0.1% Triton X-100 by incubating at room temperature for 30min followed by at least 30min on ice. The FACS assay was done on a Becton Dickinson FACSort system and the data was analyzed with MODFIT.
Determination of DNA strand breaks with single-cell gel electrophoresis comet assay
The alkaline comet assay was done as general guidance that previously described (37) with minor modifications. Briefly, HT-29 cells treated with different concentrations of DHA for 24hr were washed with and suspended in PBS buffer before they were embedded in 0.75% low-melting point agarose on the top of the three layer comet slide. Cells treated with 9 Gy of gamma radiation after embedding were used as positive controls (38). The cells on the slides were lysed for three hours in lysis buffer (pH 10) and then were transferred to ice-cold electrophoresis buffer (pH 13; 300 mM NaOH, 1mM EDTA) for 40 min. The single cell DNA was separated with electrophoresis at the current of 300 mA, 1.3V/cm for 45 min. After electrophoresis, the slides were neutralized by 0.4 M Tris buffer (pH 7.4) and fixed by methanol for 10 min and washed twice by water. The slides were stained with ethidium bromide and analyzed using an Olympus BX-51 fluorescence microscope equipped with ND50 and ND25 neutral density filters and a cooled QImaging Micropublisher 5.0 RTV 5-megapixel digital video camera. Fifty comets on each slide, duplicate slides for each dose, coded and blindly scored, were acquired using the LAI HCSA (v. 2.3.5) automatic image analysis system (Loats Associates, Inc.). Percentage of DNA in tail was used as the measure of DNA damage (38).
Results
Apoptosis induced in HT-29 cells treated with DHA, AA and LA
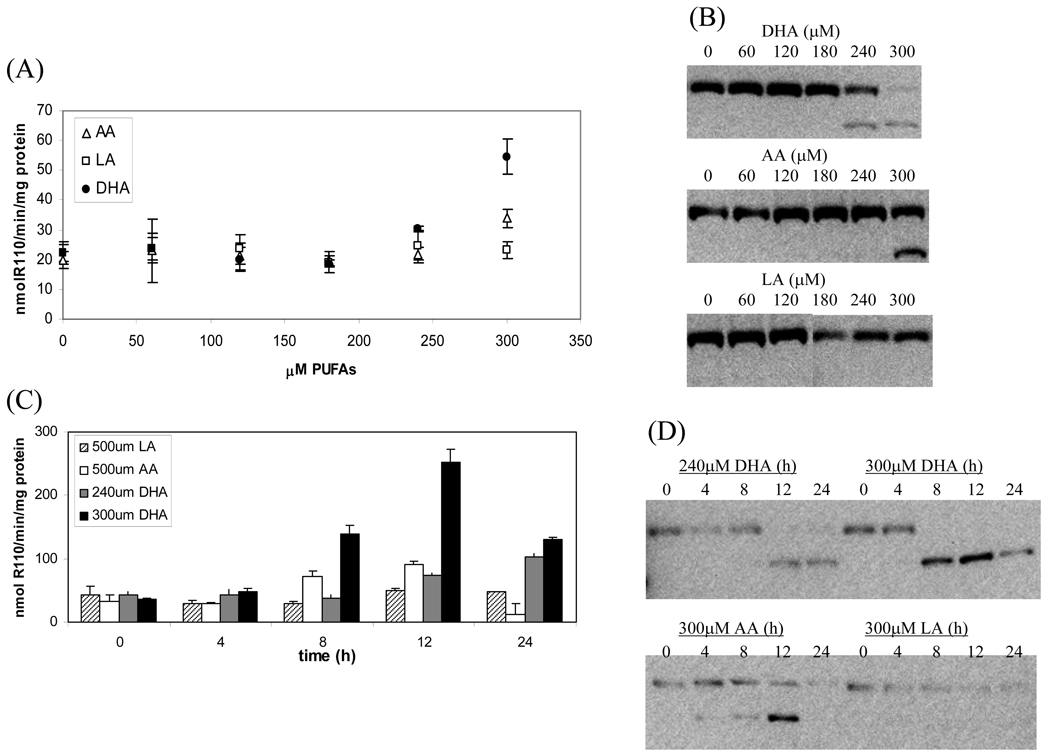
To study the apoptosis induction by PUFAs, HT-29 cells were treated with different concentrations (0, 60, 120, 180, 240 and 300µM) of DHA, AA and LA for 24 h. The apoptosis was measured by caspase-3 activity and PARP cleavage using western blot. As shown in Figure 2A, the caspase-3 activities remained at the basal level with concentrations of DHA up to 200µM. A slight, but significant, increase was observed at 240µM (p=0.0004 DHA vs AA and p=0.04 DHA vs LA) and the increase became highly significant at 300µM (p=0.0004 DHA vs AA and p=0.00005 DHA vs LA). The induction of apoptosis was confirmed by PARP cleavage assay (Figure 2B). Similar but a lesser degree effects were observed with AA at 300µM (p=0.001 AA vs. LA). Neither caspase-3 activity nor PARP cleavage were increased in cells treated with LA up to 300µM (Figure 2A and 2B). LA did not induce apoptosis until its concentration reached 600µM (data not shown).
Fig. 2.
Apoptosis induction in HT-29 cells by DHA, AA or LA. In A and B, HT-29 cells were treated with different concentrations of PUFAs for 24h. The apoptotic responses were measured using caspase-3 activities (A) and PARP cleavage (B). In C and D, cells were treated with indicated concentrations of DHA, AA or LA for 0,4,8,12 and 24h and the apoptotic responses were measured by caspase-3 activities (C) and PARP cleavage (D).
A time-dependent study was carried out to investigate the apoptosis response in HT-29 cells treated with DHA, AA and LA (Figure 2C and 2D). At 300 µM DHA induced caspase-3 activity by three-fold at 8h (p=0.015 for DHA vs. AA and p=0.004 DHA vs. LA) and six-fold at 12h (p=0.004 for DHA vs. AA and p=0.003 DHA vs. LA) after incubation. The activity decreased at 24h because of considerable cell death. At 240µM DHA induced caspase-3 activity up to 24h without significant cell death. Because AA and LA were weaker inducers of apoptosis than DHA, higher concentrations were used. At 500µM AA induced caspase-3 activity at 8h and 12h, followed by a decrease at 24h. LA had no significant effect on caspase-3 activities throughout the 24h incubation even at 500µM. The PARP cleavage assay shown in Figure 2D also demonstrated similar kinetics in the apoptotic responses of HT-29 cells treated with DHA, AA and LA. These results clearly demonstrated that the induction of apoptosis follows an order of DHA>AA>LA in HT-29 cells.
Formation of Acr-dG and 8-oxo-dG in cells treated DHA, AA and LA
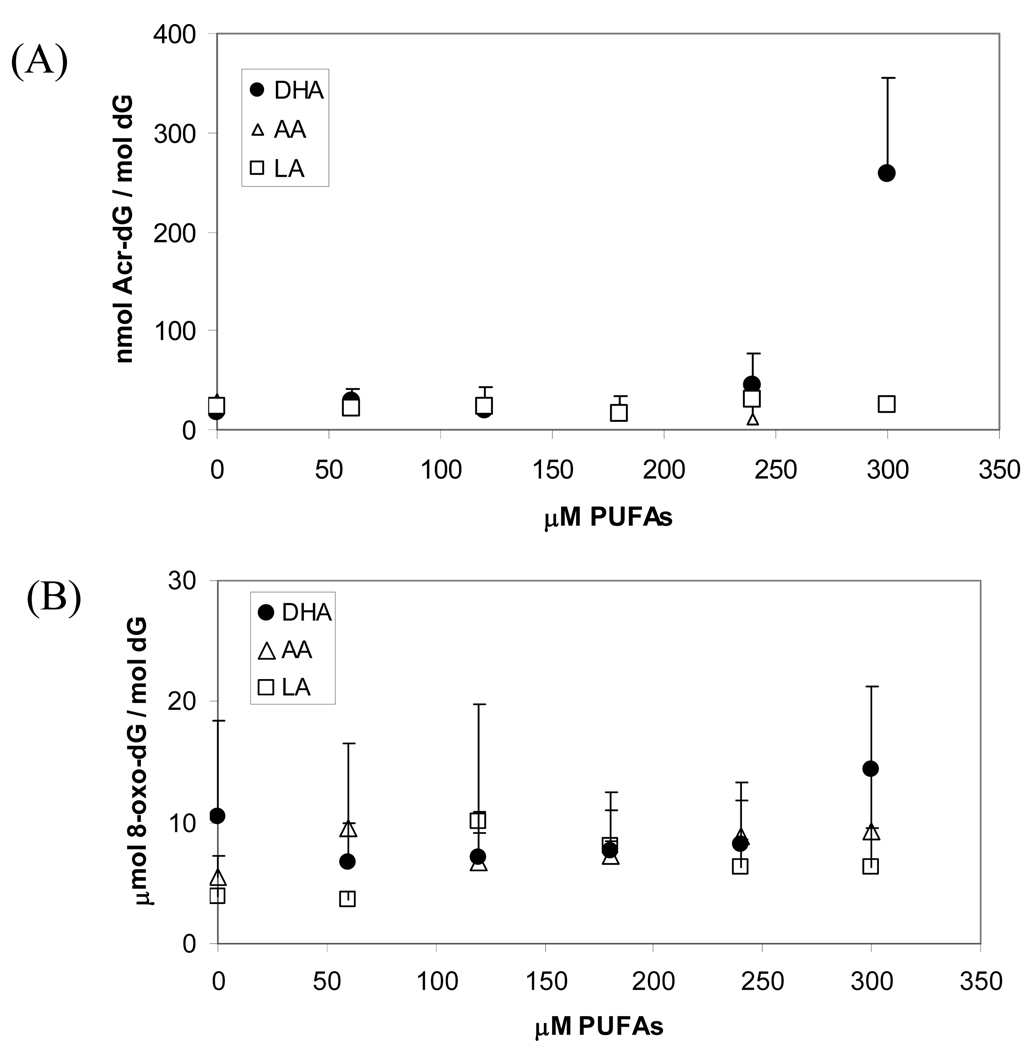
To see whether Acr-dG adducts formation in DNA is correlated with apoptosis, we analyzed Acr-dG in HT-29 cells treated with PUFAs. DNA isolated from the cells treated with DHA, AA or LA from 0 to 300µM was analyzed by the previously described SPE/HPLC-based 32P-postlabeling method (34). The results (Figure 3A) showed that although Acr-dG levels were increased from ~20 to 45.3nmol/mol dG by the treatment with DHA at 240µM for 24h, the increase was not statistically significant due to the large assay variability (p=0.15 for DHA vs AA and p=0.13 DHA vs LA). Remarkably, treatment with DHA at 300µM sharply raised the levels of Acr-dG to 220 nmol/mol dG, a more than ten-fold increase from the untreated controls. Treatment with AA and LA at the same concentration did not lead to significant Acr-dG level changes (p=0.01 for both DHA vs. AA and DHA vs. LA). Contrary to the results of DHA treatment, no significant differences were observed in Acr-dG formation in DNA of cells treated with up to 300µM AA or LA for 24h with Acr-dG levels to be 27.3 or 24.2 nmol/mol dG, respectively. These results showed an excellent agreement between Acr-dG formation and apoptosis induction in cells treated with these PUFAs.
Fig. 3.
Acr-dG and 8-oxo-dG levels in HT-29 cells treated with different concentrations of PUFAs. Acr-dG adduct levels were measured with the 32P/SPE/HPLC-based postlabeling assay (A) and 8-oxo-dG levels were measured with an HPLC/electrochemical assay (B).
We also measured 8-oxo-dG, another important oxidative DNA damage marker, in the same DNA. The results (Figure 3B) showed that, unlike Acr-dG, 8-oxo-dG levels were not significantly changed in cells treated with DHA, AA or LA in the same concentrations.
BSA decreased apoptosis and Acr-dG formation in HT-29 cells treated with DHA
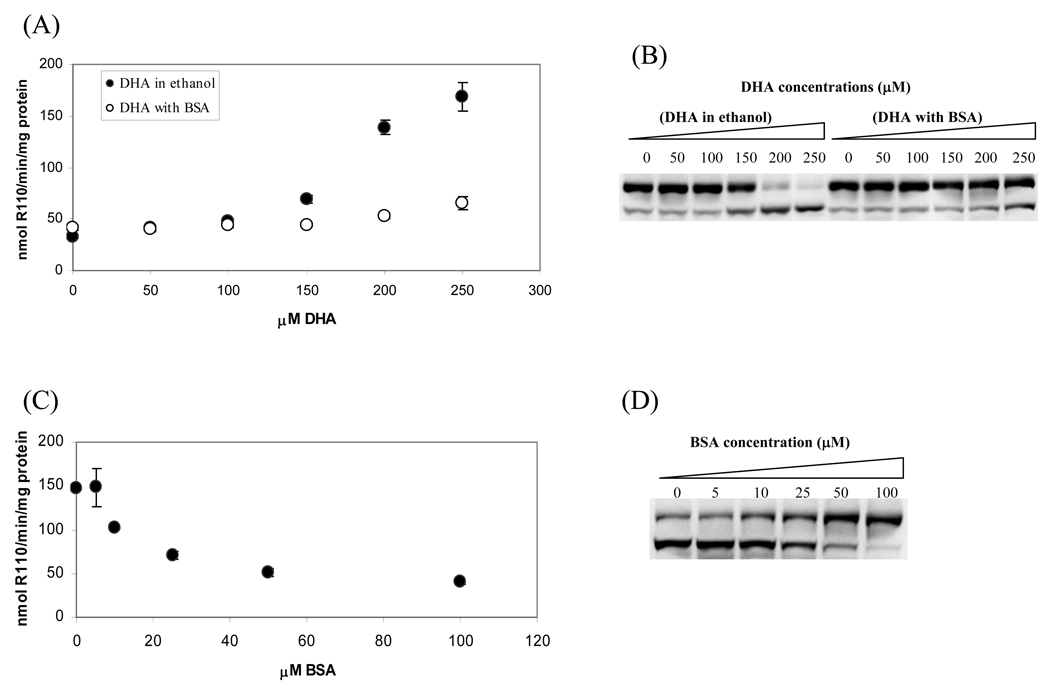
Because proteins, like DNA, can also be modified by enals (36,39), therefore, they may compete with DNA toward reactions with enals. We examined whether the addition of BSA to the cell culture media will change the levels of Acr-dG adduct formation in the DNA of cells treated with PUFAs and how the level changes will affect apoptosis. The cell culture media containing DHA-BSA complex were prepared by adding different amounts of DHA to the DMEM media containing 100µM BSA with the final concentrations of DHA 0, 50, 100, 150, 200 and 250µM. As controls, the same amounts of DHA were dissolved in ethanol and added into the DMEM media without BSA. Cells were harvested after 24h incubations for caspase-3 activity and PARP cleavage assays (Figure 4A and B). When HT-29 cells were treated with DHA in the presence of ethanol, both caspase-3 activity and PARP cleavage were increased in a dose-dependent manner up to 250µM. In contrast, there were little or no significant changes in apoptotic activities in cells treated with the same concentrations of DHA delivered as the DHA-BSA complex up to 250µM. To determine whether these changes are indeed related to BSA, in a separate experiment we treated the HT-29 cells with 200µM DHA complexed with different amounts of BSA (0, 5, 10, 25, 50 and 100µM). As shown in Figure 4C and 4D, as the concentrations of BSA increased, the caspase-3 activities and the PARP cleavage both decreased in a dose-dependent manner. These results indicate that the addition of BSA can dampen the apoptosis response in cells treated with DHA.
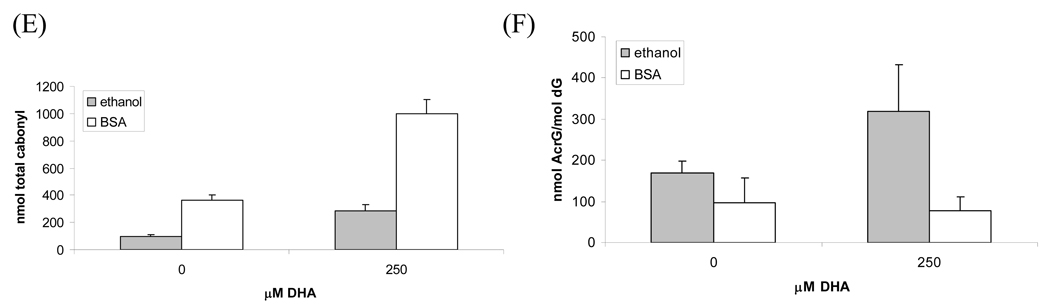
Fig. 4.
Comparison of apoptotic responses of HT-29 cells treated with DHA delivered with ethanol or BSA. HT-29 cells were treated with different concentrations of DHA delivered with and without BSA for 24h, then caspase-3 activities (A) and PARP cleavage (B) were measured. In another experiment, cells were treated with 200µM DHA in the presence of different concentrations of BSA for 24h, the dose-dependent effect of BSA on caspase-3 activities (C) and PARP cleavage (D) were examined. To investigate the oxidative effects of DHA on protein and DNA in the presence of BSA, the cells were treated with 0 or 250µM of DHA with and without 100µM BSA. The total protein carbonyl formation in media (E) and Acr-dG in DNA were measured (F).
Since BSA can also serve as a target for oxidation metabolites of DHA, next we examined the total protein carbonyls in the culture media from control and 250µM DHA treatment using the DNPH assay (36). As shown in Figure 4E, the presence of 100µM BSA in the media increased the total carbonyls by about three times in both control and DHA treatment media. These results showed that the significantly higher carbonyls in the DHA-BSA system than the corresponding DHA-ethanol system can be attributed to trapping of DHA aldehyde metabolites by BSA in the cell culture media. The Acr-dG levels in the DNA of cells treated with under these conditions were also measured. As expected, the increase in protein carbonyls in the incubation mixture was accompanied by a concomitant decrease of Acr-dG formation in the cells treated with DHA/BSA. Taken together, these results showed that the decrease in the DHA induced apoptosis in HT-29 cells after adding BSA to the cultural media that correlated well with Acr-dG formation is likely due to the reaction of BSA toward the DHA oxidation generated enals.
Acr-dG formation and apoptosis in HT-29 cells treated with Acr
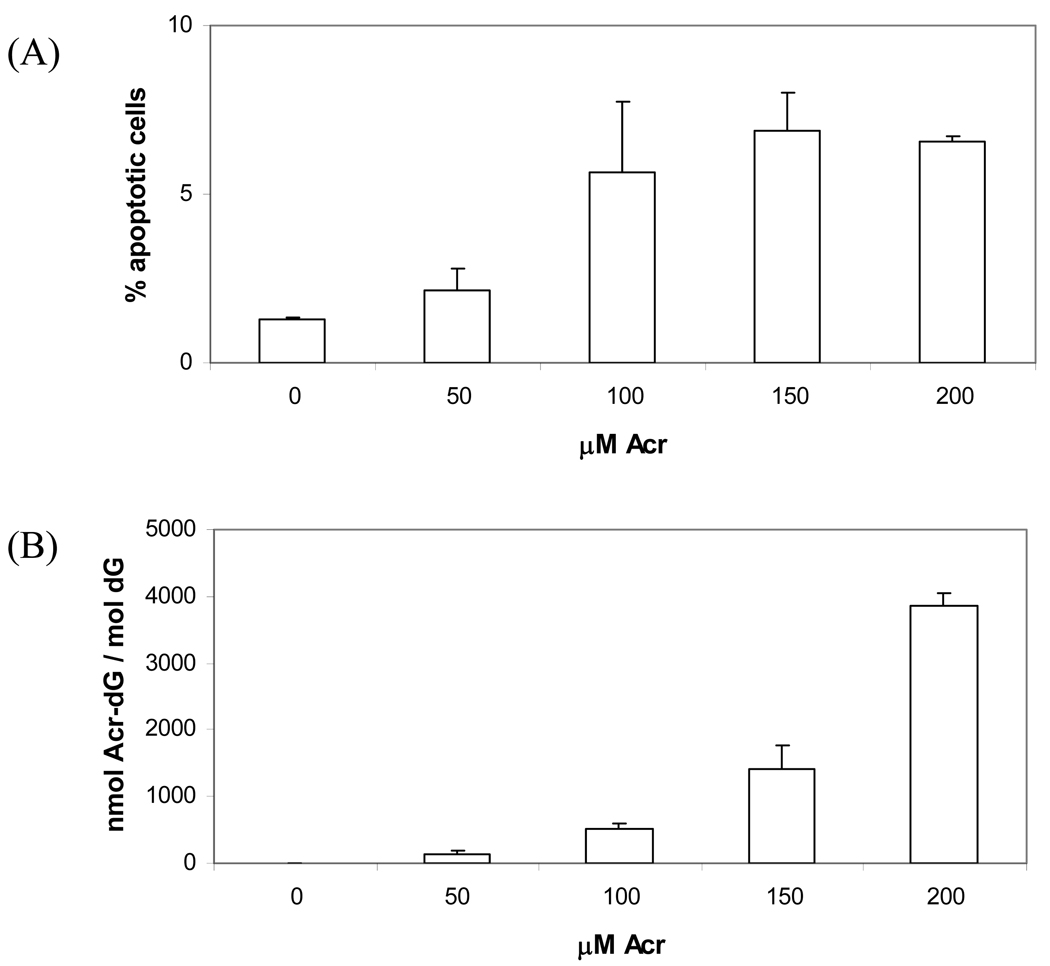
PUFA induced Acr-dG adducts is the major focus of this study, so although Acr is highly reactive towards proteins, DNA and therefore can affect or initiate various signal transduction pathways, it is still interesting to investigate the apoptosis and Acr-dG formation in HT-29 cells treated with Acr and compare the results with those from DHA study. Cells treated with different concentrations (0, 50, 100, 150, and 200µM) of Acr were used to measure apoptosis with Annexin V staining assay, the Acr-dG levels were also measured. As shown in Figure 5A, apoptotic cells population significantly increased when Acr concentrations reached 100µM and maintained at about the same levels when Acr concentrations were 150µM and 200µM. For Acr-dG formation (Figure 5B), Acr-dG levels increased steadily in a dose-dependent manner.
Fig. 5.
Comparison of apoptosis and Acr-dG formation of HT-29 cells treated with Acr. Cells were incubated with from 0 to 200µM of Acr for 24h. Then, the Annexin V-FITC / PI assay were used to measure apoptosis (A), and Acr-dG adduct levels were measured (B).
DHA caused DNA strand breaks
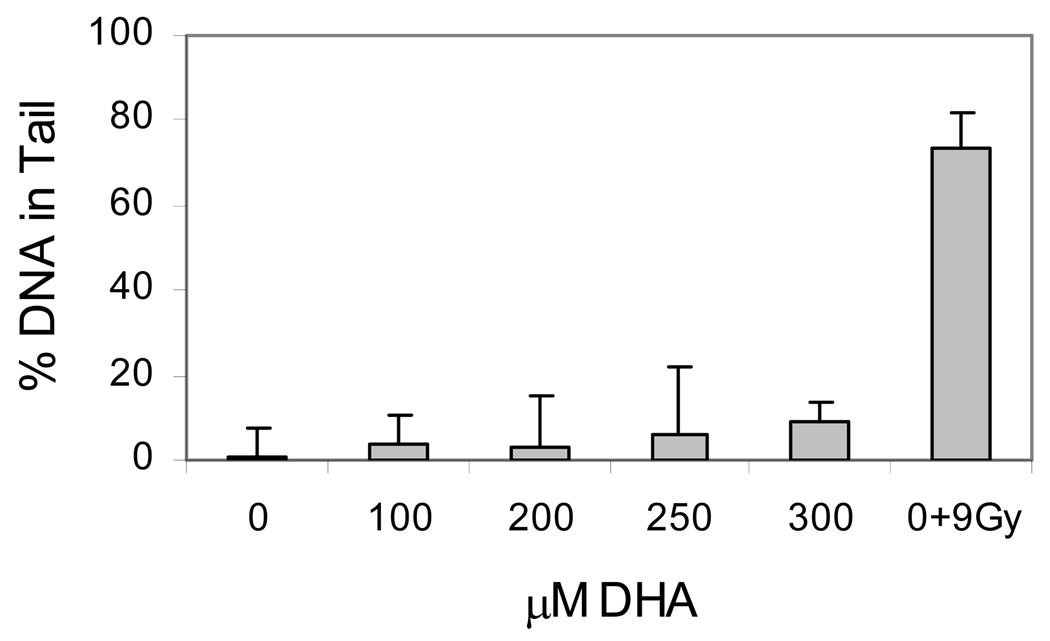
DNA strand breaks have been shown to be involved in initiation of apoptosis (23,24). The strand breaks in DNA can be generated by at least two mechanisms; either direct attack by ROS or repair of DNA lesions, such as Acr-dG. To determine whether DNA strand-breakage is formed by DHA treatment, we used the single-cell electrophoresis based comet assay to measure DNA strand breaks in HT-29 cells treated with different concentrations of DHA (Figure 6). The results indicate that DHA treated cells have higher percentage of DNA in tails (p<0.01 for all concentrations) compared with the control. The relatively large increase of DNA strand breaks at 300µM (p<0.002) coincided with the apoptosis induced by DHA at this concentration.
Fig. 6.
DNA strand breaks in HT-29 cells treated with DHA. Cells were treated with different concentrations of DHA and DNA strand breaks were measured with an electrophoresis-based comet assay. Irradiation of 9Gy was to treated cells as positive control.
DHA induced cell cycle arrest at G1 phase
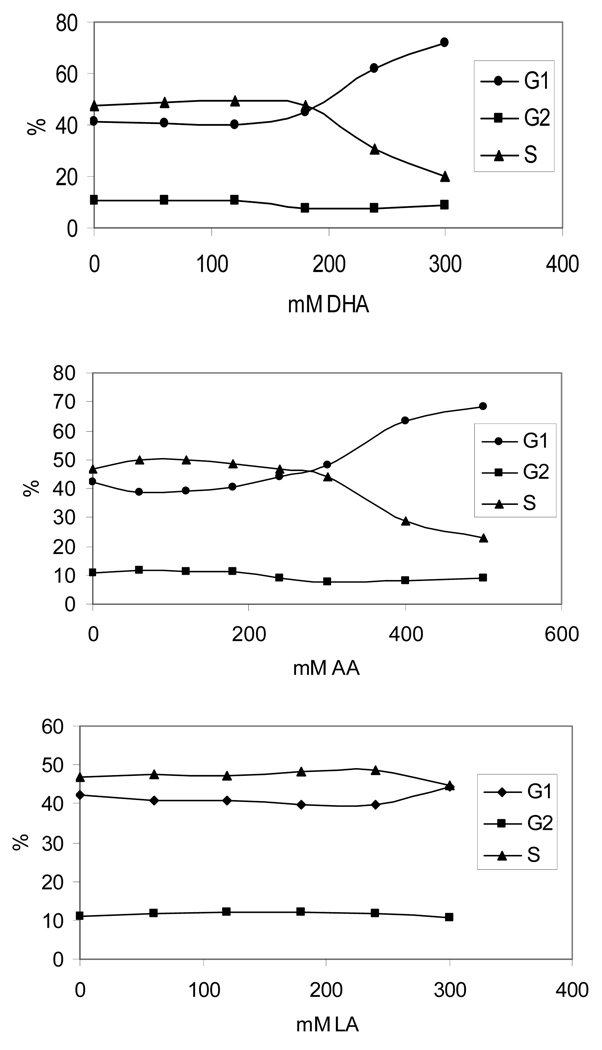
Previous study reported that DHA can inhibit cell growth progression and arrest cells at G1 phase in colon cancer cells (40). Although it is not the focus of this study, cell cycle analysis was performed to provide information about the possible checkpoints and pathways that are activated by DHA. The PI stain-based cell cycle analysis showed that cells were arrested at G1 phase when treated with 180µM DHA and the percentage of G1 cell population increased in a dose-dependent manner. Similar results can be observed in cells treated with AA and LA, but the cell cycle arrest was observed only at higher concentrations of more than 300µM (Figure 7). The induction of G1 arrest follows the same order: DHA>AA>LA.
Fig. 7.
Cell cycle arrest analysis of HT-29 cells treated with PUFAs. Cells treated with different concentrations of DHA, AA, or LA were harvested and stained with propidium iodide for flow-cytometry-based FACT analysis.
Discussion
Evidence from animal as well as epidemiological studies has shown that DHA and other ω-3 PUFAs possess cancer chemopreventive effects, yet earlier studies have shown that ω-3 PUFAs generate more DNA damage, such as Acr-dG, than ω-6 PUFAs or saturated fatty acids (13). A possible explanation that we propose and study here is that specific DNA damage may contribute to the initiation of DHA induced apoptosis and cell cycle arrest, and thus protect the cells against carcinogenesis. Several other possible mechanisms have been suggested for the apoptotic effects of DHA and other ω-3 PUFAs, including prostaglandin metabolism by cyclooxgenase and lipooxygenase, nuclear receptors such as peroxisome prolifirator-activated receptors and PUFAs’ membrane altering effects (6,41). Protein modification by enals and other PUFA oxidative products has also been shown to play a role in ω-3 PUFAs induced apoptosis (42,43). It is conceivable that more than one mechanisms are likely involved in ω-3 PUFAs induced apoptosis.
Although extensively studied, the mutagenic potential of Acr-dG is uncertain due to conflicting results (17–21, 44–46). Other related cyclic DNA adducts from ω-6 PUFAs, specifically HNE-dG and the etheno adducts, have been shown to be mutagenic (47–51). Because of its relatively high levels of modification induced by ω-3 PUFAs, Acr-dG formation in cellular DNA could be one of the triggers in apoptosis. Here we investigated whether Acr-dG and 8-oxo-dG, two major in vivo DNA lesions induced by DHA and other ω-3 PUFAs, play a role in this process. This study showed that DHA is a potent inducer of apoptosis in HT-29 cells compared with AA and LA. We also found that the apoptotic responses in cells treated with various concentrations of DHA, AA or LA are in parallel with Acr-dG formation, but not with 8-oxo-dG, in DNA. Furthermore, we demonstrated that BSA can block the apoptosis in cells treated with DHA with simultaneous decrease in Acr-dG formation, by trapping Acr to form protein carbonyls before it reacts with nuclear DNA. Additionally, our results showed that incubation of Acr induced apoptosis and Acr-dG formation in HT-29 cells. Together, the results established a close correlation between apoptosis and Acr-dG formation in HT-29 cells, suggesting a potential role of Acr-dG in the apoptosis induced by DHA.
Our data showed the same patterns of dose-response in apoptosis induction and Acr-dG formation in cells treated with DHA, suggesting that the apoptotic response could be initiated only when Acr-dG adduct levels in cells reach a threshold of around 300nmol Acr-dG/mol dG, which coincide with the data from the Acr treatment experiments where the apoptosis was initiated when Acr-dG levels reached and passed around 500nmol Acr-dG/mol dG. The lack of dose-response in Acr-dG formation with the lower concentrations of DHA (Figure 3A) may be explained by efficient repair of Acr-dG. Acr-dG can be readily removed through the nucleotide excision repair (NER)-mediated pathway similar to that for HNE-dG (52,53). DNA repair can act as genome ‘caretakers’ to repair and remove DNA damages, thus maintain the genomic integrity. However, new evidence shows that they can also act as ‘gatekeepers’ reacting to DNA damage by themselves or through interacting with other molecules to initiate cell cycle arrest and apoptosis pathways when certain DNA damage reaches a threshold level (26,27,54). NER has been shown to be involved in transcription-coupled DNA damage sensing and signaling, whereas mismatch repair in DNA replication-related signaling recognition (24,25,54,55).
Our data suggest that when the Acr-dG and perhaps DNA strand breaks induced by DHA reached a certain threshold level beyond the capability of the NER and other pathways, cells may undergo cell cycle arrest or initiate the apoptosis signaling pathways. More studies are needed in order to determine the roles of the NER repair in DHA induced apoptosis. Nevertheless, our results suggest that NER repair and NER engaged transcription-coupled DNA damage signaling pathway may be involved in the apoptotic response to the Acr-dG formation and other possible oxidative DNA damage caused by DHA. In addition, the apoptosis induced by DHA is likely via a p53-independent pathway because HT-29 cells express mutant p53. Our preliminary experiments with HCT116 and p53 knocked out HCT116 cells also showed that there is no difference in apoptosis responses from these two cell lines treated with DHA (data not shown).
It should be noted that the concentrations of fatty acids used in this study are considerably higher than the in vivo intracellular free fatty acid concentration generally reported (<10µM) (56). However, the concentrations may still be relevant with the consideration of the following factors: 1) once in the cell, most fatty acids non-covalently bind to various fatty acid binding proteins, they are enzymatically converted into fatty acyl-CoA-thioesters (FA-CoA). FA-CoA level is highly variable and dependent on the cell examined. For example, in liver FA-CoA can range from 110~152µM (56); 2) the serum lipid level is relatively high as the average free fatty acids concentration in the post-absorptive stage is reported to be 0.7mM, and this level can be much higher during the absorptive phase following ingestion of a meal rich in fat. Other factors including age, gender, diet and genetic background can also affect the expression and regulation of genes involved in fatty acid metabolism and, therefore, its concentrations (57); and 3) DHA in the culture media may be oxidized and its oxidative products can readily react with proteins in the media resulting in the reduced effective intracellular DHA concentration.
It was interesting to observe that the addition of BSA to the cell culture media led to decrease of DNA adduct formation and apoptosis, since BSA and other fatty acid binding proteins have been widely used to facilitate fatty acid absorption and transportation by cells. Our results suggest that because BSA and other fatty acid binding proteins can readily react with certain reactive oxidative species, the oxidative effects of PUFAs on DNA and other proteins in cells may be altered. Caution needs to be exercised to interpret the data on oxidation when BSA and other fatty acid binding proteins were used.
A recent study showed that the accumulation of 8-oxo-dG in nuclear and mitochondrial DNAs can lead to a buildup of the mismatch repair mediated DNA single strand breaks, that trigger two distinctive apoptotic pathways (58). While our study showed that Acr-dG may play an important role in apoptosis induced by DHA, it is possible that 8-oxo-dG and maybe other types of DNA damage including the etheno adducts in cells treated with DHA could contribute to DNA strand breakage due to effective repair. It is therefore reasonable to assume that a variety of DNA damage could collectively be responsible for DHA-induced apoptosis. Nonetheless, our studies demonstrated a clear correlation between Acr-dG formation and apoptotic response in human colon HT-29 cells, and thus shed light on a potential role of Acr-dG adduct formation in the protective effect of DHA against tumorigenesis.
Acknowledgement
The authors thank Dr. Karen Creswell and Ms. Michelle Lombard for conducting the flowcytometry experiments. This work was supported by NCI grant CA043159 to FLC and in part by NCI grant CA115625-01A2 and Flight Attendant Medical Research Institute award 052444 to RG.
Abbreviations
- PUFA
polyunsaturated fatty acids
- DHA
docosahexaenoic acid
- EPA
eicosapentaenoic acid
- AA
arachidonic acid
- LA
linoleic acid
- Acr
acrolein
- HNE
4-hydroxy-2-nonenal
- dG
deoxyguanosine
- PdG
1,N2-propanodeoxyguanosine
- Acr-dG
acrolein-derived 1,N2-propanodeoxyguanosine
- 8-oxo-dG
8-oxo-deoxyguanosine
- SPE
solid phase extraction
- DNPH
2,4-dinitrophenylhydrazine
- PI
propidium iodide
- BSA
bovine serum album
- NER
nucleotide excision repair
References
- 1.Deschner EE, Lytle JS, Wong G, Ruperto JF, Newmark HL. The effect of dietary omega-3-fatty-acids (fish oil) on azoxymethanol-induced focal areas of dysplasia and colon-tumor incidence. Cancer. 1990;66(11):2350–2356. doi: 10.1002/1097-0142(19901201)66:11<2350::aid-cncr2820661117>3.0.co;2-6. [DOI] [PubMed] [Google Scholar]
- 2.Chang WCL, Chapkin RS, Lupton JR. Fish oil blocks azoxymethane-induced rat colon tumorigenesis by increasing cell differentiation and apoptosis rather than decreasing cell proliferation. Journal of Nutrition. 1998;128(3):491–497. doi: 10.1093/jn/128.3.491. [DOI] [PubMed] [Google Scholar]
- 3.Fernandez E, Chatenoud L, La Vecchia C, Negri E, Franceschi S. Fish consumption and cancer risk. American Journal of Clinical Nutrition. 1999;70(1):85–90. doi: 10.1093/ajcn/70.1.85. [DOI] [PubMed] [Google Scholar]
- 4.Rao CV, Hirose Y, Indranie C, Reddy BS. Modulation of experimental colon tumorigenesis by types and amounts of dietary fatty acids. Cancer Research. 2001;61(5):1927–1933. [PubMed] [Google Scholar]
- 5.Kimura Y, Kono S, Toyomura K, Nagano J, Mizoue T, Moore MA, Mibu R, Tanaka M, Kakeji Y, Maehara Y, Okamura T, Ikejiri K, Futami K, Yasunami Y, Maekawa T, Takenaka K, Ichimiya H, Imaizumi N. Meat, fish and fat intake in relation to subsite-specific risk of colorectal cancer: The Fukuoka Colorectal Cancer Study. Cancer Science. 2007;98(4):590–597. doi: 10.1111/j.1349-7006.2007.00425.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chapkin RS, McMurray DN, Lupton JR. Colon cancer, fatty acids and anti-inflammatory compounds. Current Opinion in Gastroenterology. 2007;23(1):48–54. doi: 10.1097/MOG.0b013e32801145d7. [DOI] [PubMed] [Google Scholar]
- 7.Dupertuis YM, Meguid MM, Pichard C. Colon cancer therapy: new perspectives of nutritional manipulations using polyunsaturated fatty acids. Current Opinion in Clinical Nutrition and Metabolic Care. 2007;10(4):427–432. doi: 10.1097/MCO.0b013e3281e2c9d4. [DOI] [PubMed] [Google Scholar]
- 8.Nath RG, Chung FL. Detection of exocyclic 1,N2-propanodeoxyguanosine adducts as common DNA lesions in rodents and humans. Proceedings of the National Academy of Sciences of the United States of America. 1994;91(16):7491–7495. doi: 10.1073/pnas.91.16.7491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chaudhary AK, Nokubo M, Reddy GR, Yeola SN, Morrow JD, Blair IA, Marnett LJ. Detection of Endogenous Malondialdehyde-Deoxyguanosine Adducts in Human Liver. Science. 1994;265(5178):1580–1582. doi: 10.1126/science.8079172. [DOI] [PubMed] [Google Scholar]
- 10.Chung FL, Chen HJC, Nath RG. Lipid peroxidation as a potential endogenous source for the formation of exocyclic DNA adducts. Carcinogenesis. 1996;17(10):2105–2111. doi: 10.1093/carcin/17.10.2105. [DOI] [PubMed] [Google Scholar]
- 11.Nair J, Barbin A, Velic I, Bartsch H. Etheno DNA-base adducts from endogenous reactive species. Mutation Research-Fundamental and Molecular Mechanisms of Mutagenesis. 1999;424(1–2):59–69. doi: 10.1016/s0027-5107(99)00008-1. [DOI] [PubMed] [Google Scholar]
- 12.Marnett LJ. Oxy radicals, lipid peroxidation and DNA damage. Toxicology. 2002;181–182:219–222. doi: 10.1016/s0300-483x(02)00448-1. [DOI] [PubMed] [Google Scholar]
- 13.Pan J, Chung FL. Formation of cyclic deoxyguanosine adducts from ω-3 and ω-6 polyunsaturated fatty acids under oxidative conditions. Chemical Research in Toxicology. 2002;15(3):367–372. doi: 10.1021/tx010136q. [DOI] [PubMed] [Google Scholar]
- 14.Chung FL, Nath RG, Ocando J, Nishikawa A, Zhang L. Deoxyguanosine adducts of t-4-hydroxy-2-nonenal are endogenous DNA lesions in rodents and humans: Detection and potential sources. Cancer Research. 2000;60(6):1507–1511. [PubMed] [Google Scholar]
- 15.Zhang SY, Villalta PW, Wang MY, Hecht SS. Detection and quantitation of acrolein-derived 1,N2-propanodeoxyguanosine adducts in human lung by liquid chromatography-electrospray ionization-tandem mass spectrometry. Chemical Research in Toxicology. 2007;20(4):565–571. doi: 10.1021/tx700023z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chung FL, Pan JS, Choudhury S, Roy R, Hu WW, Tang MS. Formation of trans-4-hydroxy-2-nonenal-and other enal-derived cyclic DNA adducts from ω-3 and ω-6 polyunsaturated fatty acids and their roles in DNA repair and human p53 gene mutation. Mutation Research-Fundamental and Molecular Mechanisms of Mutagenesis. 2003;531(1–2):25–36. doi: 10.1016/j.mrfmmm.2003.07.001. [DOI] [PubMed] [Google Scholar]
- 17.Kawanishi M, Matsuda T, Nakayama A, Takebe H, Matsui S, Yagi T. Molecular analysis of mutations induced by acrolein in human fibroblast cells using supF shuttle vector plasmids. Mutation Research. 1998;417(2–3):65–73. doi: 10.1016/s1383-5718(98)00093-x. [DOI] [PubMed] [Google Scholar]
- 18.VanderVeen LA, Hashim MF, Nechev LV, Harris TM, Harris CM, Marnett LJ. Evaluation of the mutagenic potential of the principal DNA adduct of acrolein. Journal of Biological Chemistry. 2001;276(12):9066–9070. doi: 10.1074/jbc.M008900200. [DOI] [PubMed] [Google Scholar]
- 19.Sanchez AM, Minko IG, Kurtz AJ, Kanuri M, Moriya M, Lloyd RS. Comparative evaluation of the bioreactivity and mutagenic spectra of acrolein-derived α-HOPdG and γ-HOPdG regioisomeric deoxyguanosine adducts. Chemical Research in Toxicology. 2003;16(8):1019–1028. doi: 10.1021/tx034066u. [DOI] [PubMed] [Google Scholar]
- 20.Kim SI, Pfeifer GP, Besaratinia A. Lack of mutagenicity of acrolein-induced DNA adducts in mouse and human cells. Cancer Research. 2007;67(24):11640–11647. doi: 10.1158/0008-5472.CAN-07-2528. [DOI] [PubMed] [Google Scholar]
- 21.Feng ZH, Hu WW, Hu Y, Tang MS. Acrolein is a major cigarette-related lung cancer agent: Preferential binding at p53 mutational hotspots and inhibition of DNA repair. Proceedings of the National Academy of Sciences of the United States of America. 2006;103(42):15404–15409. doi: 10.1073/pnas.0607031103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Branzei D, Foiani M. The DNA damage response during DNA replication. Current Opinion in Cell Biology. 2005;17(6):568–575. doi: 10.1016/j.ceb.2005.09.003. [DOI] [PubMed] [Google Scholar]
- 23.Zou L, Elledge SJ. Sensing DNA damage through ATRIP recognition of RPA-ssDNA complexes. Science. 2003;300(5625):1542–1548. doi: 10.1126/science.1083430. [DOI] [PubMed] [Google Scholar]
- 24.Ljungman M, Lane DP. Opinion - Transcription-guarding the genome by sensing DNA damage. Nature Reviews Cancer. 2004;4(9):727–737. doi: 10.1038/nrc1435. [DOI] [PubMed] [Google Scholar]
- 25.Hickman MJ, Samson LD. Role of DNA mismatch repair and p53 in signaling induction of apoptosis by alkylating agents. Proceedings of the National Academy of Sciences of the United States of America. 1999;96(19):10764–10769. doi: 10.1073/pnas.96.19.10764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Colton SL, Xu XXS, Wang YA, Wang G. The involvement of ataxia-telangiectasia mutated protein activation in nucleotide excision repair-facilitated cell survival with cisplatin treatment. Journal of Biological Chemistry. 2006;281(37):27117–27125. doi: 10.1074/jbc.M602826200. [DOI] [PubMed] [Google Scholar]
- 27.Branzei D, Foiani M. Regulation of DNA repair throughout the cell cycle. Nature Reviews Molecular Cell Biology. 2008;9(4):297–308. doi: 10.1038/nrm2351. [DOI] [PubMed] [Google Scholar]
- 28.Engelward BP, Allan JM, Dreslin AJ, Kelly JD, Wu MM, Gold B, Samson LD. A chemical and genetic approach together define the biological consequences of 3-methyladenine lesions in the mammalian genome. Journal of Biological Chemistry. 1998;273(9):5412–5418. doi: 10.1074/jbc.273.9.5412. [DOI] [PubMed] [Google Scholar]
- 29.Roos WP, Kaina B. DNA damage-induced cell death by apoptosis. Trends in Molecular Medicine. 2006;12(9):440–450. doi: 10.1016/j.molmed.2006.07.007. [DOI] [PubMed] [Google Scholar]
- 30.Chung FL, Young R, Hecht SS. A Study of Chemical Carcinogenesis .61. Formation of Cyclic 1,N2-Propanodeoxyguanosine Adducts in DNA Upon Reaction with Acrolein Or Crotonaldehyde. Cancer Research. 1984;44(3):990–995. [PubMed] [Google Scholar]
- 31.Mahoney EM, Hamill AL, Scott WA, Cohn ZA. Response of endocytosis to altered fatty acyl composition of macrophage phospholipids. Proc Natl Acad Sci U. S A. 1977;74(11):4895–4899. doi: 10.1073/pnas.74.11.4895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Edwards IJ, Berquin IM, Sun HG, O'Flaherty JT, Daniel LW, Thomas MJ, Rudel LL, Wykle RL, Chen YQ. Differential effects of delivery of omega-3 fatty acids to human cancer cells by low-density lipoproteins versus albumin. Clinical Cancer Research. 2004;10(24):8275–8283. doi: 10.1158/1078-0432.CCR-04-1357. [DOI] [PubMed] [Google Scholar]
- 33.Marmur J. Procedure for the Isolation of Deoxyribonucleic Acid from Microorganisms. J. Mol. Biol. 1961;3:208–218. [Google Scholar]
- 34.Emami A, Dyba M, Cheema AK, Pan J, Nath RG, Chung FL. Detection of the acrolein-derived cyclic DNA adduct by a quantitative 32P-postlabeling/solid-phase extraction/HPLC method: Blocking its artifact formation with glutathione. Analytical Biochemistry. 2008;374(1):163–172. doi: 10.1016/j.ab.2007.10.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Fiala ES, Conaway CC, Mathis JE. Oxidative DNA and RNA damage in the livers of Sprague-Dawley rats treated with the hepatocarcinogen 2-nitropropane. Cancer Research. 1989;49:5518–5522. [PubMed] [Google Scholar]
- 36.Refsgaard HHF, Tsai L, Stadtman ER. Modifications of proteins by polyunsaturated fatty acid peroxidation products. Proceedings of the National Academy of Sciences of the United States of America. 2000;97(2):611–616. doi: 10.1073/pnas.97.2.611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tice RR, Agurell E, Anderson D, Burlinson B, Hartmann A, Kobayashi H, Miyamae Y, Rojas E, Ryu JC, Sasaki YF. Single cell gel/comet assay: guidelines for in vitro and in vivo genetic toxicology testing. Environ. Mol. Mutagen. 2000;35(3):206–221. doi: 10.1002/(sici)1098-2280(2000)35:3<206::aid-em8>3.0.co;2-j. [DOI] [PubMed] [Google Scholar]
- 38.Saha DT, Davidson BJ, Wang A, Pollock AJ, Orden RA, Goldman R. Quantification of DNA repair capacity in whole blood of patients with head and neck cancer and healthy donors by comet assay. Mutation Research-Genetic Toxicology and Environmental Mutagenesis. 2008;650(1):55–62. doi: 10.1016/j.mrgentox.2007.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yamada S, Funada T, Shibata N, Kobayashi M, Kawai Y, Tatsuda E, Furuhata A, Uchida K. Protein-bound 4-hydroxy-2-hexenal as a marker of oxidized n-3 polyunsaturated fatty acids. Journal of Lipid Research. 2004;45(4):626–634. doi: 10.1194/jlr.M300376-JLR200. [DOI] [PubMed] [Google Scholar]
- 40.Danbara N, Yuri T, Tsujita-Kyutoku M, Sato M, Senzaki H, Takada H, Hada T, Miyazawa T, Okazaki K, Tsubura A. Conjugated docosahexaenoic acid is a potent inducer of cell cycle arrest and apoptosis and inhibits growth of colo 201 human colon cancer cells. Nutrition and Cancer. 2004;50(1):71–79. doi: 10.1207/s15327914nc5001_10. [DOI] [PubMed] [Google Scholar]
- 41.Jump DB, Clarke SD. Regulation of gene expression by dietary fat. Annual Review of Nutrition. 1999;19:63–90. doi: 10.1146/annurev.nutr.19.1.63. [DOI] [PubMed] [Google Scholar]
- 42.Gao L, Wang J, Sekhar KR, Yin H, Yared NF, Schneider SN, Sasi S, Dalton TP, Anderson ME, Chan JY, Morrow JD, Freeman ML. Novel n-3 fatty acid oxidation products activate Nrf2 by destabilizing the association between Keap1 and Cullin3. Journal of Biological Chemistry. 2007;282(4):2529–3257. doi: 10.1074/jbc.M607622200. [DOI] [PubMed] [Google Scholar]
- 43.Jacobs AT, Marnett LJ. Heat shock factor 1 attenuates 4-Hydroxynonenal-mediated apoptosis: critical role for heat shock protein 70 induction and stabilization of Bcl-XL. Journal of Biological Chemistry. 2007;282(4):33412–33420. doi: 10.1074/jbc.M706799200. [DOI] [PubMed] [Google Scholar]
- 44.Minko IG, Washington MT, Kanuri M, Prakash L, Prakash S, Lloyd RS. Translesion synthesis past acrolein-derived DNA adduct, γ-hydroxypropanodeoxyguanosine, by yeast and human DNA polymerase eta. Journal of Biological Chemistry. 2003;278(2):784–790. doi: 10.1074/jbc.M207774200. [DOI] [PubMed] [Google Scholar]
- 45.Yang IY, Miller H, Wang ZG, Frank EG, Ohmori H, Hanaoka F, Moriya M. Mammalian translesion DNA synthesis across an acrolein-derived deoxyguanosine adduct - Participation of DNA polymerase eta in error-prone synthesis in human cells. Journal of Biological Chemistry. 2003;278(16):13989–13994. doi: 10.1074/jbc.M212535200. [DOI] [PubMed] [Google Scholar]
- 46.Minko IG, Kozekov ID, Kozekova A, Harris TA, Rizzo CJ, Lloyd RS. Mutagenic potential of DNA-peptide crosslinks mediated by acrolein-derived DNA adducts. Mutation Research-Fundamental and Molecular Mechanisms of Mutagenesis. 2008;637(1–2):161–172. doi: 10.1016/j.mrfmmm.2007.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Cajelli E, Ferraris A, Brambilla G. Mutagenicity of 4-Hydroxynonenal in V79 Chinese-Hamster Cells. Mutation Research. 1987;190(2):169–171. doi: 10.1016/0165-7992(87)90050-9. [DOI] [PubMed] [Google Scholar]
- 48.Feng ZH, Hu WW, Amin S, Tang MS. Mutational spectrum and genotoxicity of the major lipid peroxidation product, trans-4-hydroxy-2-nonenal, induced DNA adducts in nucleotide excision repair-proficient and -deficient human cells. Biochemistry. 2003;42(25):7848–7854. doi: 10.1021/bi034431g. [DOI] [PubMed] [Google Scholar]
- 49.Choi JY, Zang H, Angel KC, Kozekov ID, Goodenough AK, Rizzo CJ, Guengerich FP. Translesion synthesis across 1,N2-ethenoguanine by human DNA polymerases. Chemical Research in Toxicology. 2006;19(6):879–886. doi: 10.1021/tx060051v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zang H, Goodenough AK, Choi JY, Irimia A, Loukachevitch LV, Kozekov ID, Angel KC, Rizzo CJ, Egli M, Guengerich FP. DNA adduct bypass polymerization by Sulfolobus solfataricus DNA polymerase Dpo4 - Analysis and crystal structures of multiple base pair substitution and frameshift products with the adduct 1,N2-ethenoguanine. Journal of Biological Chemistry. 2005;280(33):29750–29764. doi: 10.1074/jbc.M504756200. [DOI] [PubMed] [Google Scholar]
- 51.Hussain SP, Raja K, Amstad PA, Sawyer M, Trudel LJ, Wogan GN, Hofseth LJ, Shields PG, Billiar TR, Trautwein C, Hohler T, Galle PR, Phillips DH, Markin R, Marrogi AJ, Harris CC. Increased p53 mutation load in nontumorous human liver of Wilson disease and hemochromatosis: Oxyradical overload diseases. Proceedings of the National Academy of Sciences of the United States of America. 2000;97(23):12770–12775. doi: 10.1073/pnas.220416097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Choudhury S, Pan J, Amin S, Chung FL, Roy R. Repair kinetics of trans-4-hydroxynonenal-induced cyclic 1,N2-propanodeoxyguanine DNA adducts by human cell nuclear extracts. Biochemistry. 2004;43(23):7514–7521. doi: 10.1021/bi049877r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Johnson KA, Fink SP, Marnett LJ. Repair of propanodeoxyguanosine by nucleotide excision repair in vivo and in vitro. Journal of Biological Chemistry. 1997;272:11434–11438. doi: 10.1074/jbc.272.17.11434. [DOI] [PubMed] [Google Scholar]
- 54.Fousteri M, Mullenders LH. Transcription-coupled nucleotide excision repair in mammalian cells: molecular mechanisms and biological effects. Cell Research. 2008;18(1):73–84. doi: 10.1038/cr.2008.6. [DOI] [PubMed] [Google Scholar]
- 55.Wang JYJ, Edelmann W. Mismatch repair proteins as sensors of alkylation DNA damage. Cancer Cell. 2006;9(6):417–418. doi: 10.1016/j.ccr.2006.05.013. [DOI] [PubMed] [Google Scholar]
- 56.Gossett RE, Frolov AA, Roths JB, Behnke WD, Kier AB, Schroeder F. Acyl-CoA binding proteins: Multiplicity and function. Lipids. 1996;31(9):895–918. doi: 10.1007/BF02522684. [DOI] [PubMed] [Google Scholar]
- 57.Calviello G, Serini S, Piccioni E. n-3 polyunsaturated fatty acids and the prevention of colorectal cancer: Molecular mechanisms involved. Current Medicinal Chemistry. 2007;14(29):3059–3069. doi: 10.2174/092986707782793934. [DOI] [PubMed] [Google Scholar]
- 58.Oka S, Ohno M, Tsuchimoto D, Sakumi K, Furuichi M, Nakabeppu Y. Two distinct pathways of cell death triggered by oxidative damage to nuclear and mitochondrial DNAs. EMBO Journal. 2008;27(2):421–432. doi: 10.1038/sj.emboj.7601975. [DOI] [PMC free article] [PubMed] [Google Scholar]