Abstract
Skeletal muscle atrophy is a severe consequence of ageing, neurological disorders and chronic disease. Identifying the intracellular signalling pathways controlling changes in skeletal muscle size and function is vital for the future development of potential therapeutic interventions. Striated activator of Rho signalling (STARS), an actin-binding protein, has been implicated in rodent cardiac hypertrophy; however its role in human skeletal muscle has not been determined. This study aimed to establish if STARS, as well as its downstream signalling targets, RhoA, myocardin-related transcription factors A and B (MRTF-A/B) and serum response factor (SRF), were increased and decreased respectively, in human quadriceps muscle biopsies taken after 8 weeks of both hypertrophy-stimulating resistance training and atrophy-stimulating de-training. The mRNA levels of the SRF target genes involved in muscle structure, function and growth, such as α-actin, myosin heavy chain IIa (MHCIIa) and insulin-like growth factor-1 (IGF-1), were also measured. Following resistance training, STARS, MRTF-A, MRTF-B, SRF, α-actin, MHCIIa and IGF-1 mRNA, as well as RhoA and nuclear SRF protein levels were all significantly increased by between 1.25-and 3.6-fold. Following the de-training period all measured targets, except for RhoA, which remained elevated, returned to base-line. Our results show that the STARS signalling pathway is responsive to changes in skeletal muscle loading and appears to play a role in both human skeletal muscle hypertrophy and atrophy.
Skeletal muscle is a unique tissue that it is characterized by its strong capacity to alter its size as well as its fibre composition in response to changes in mechanical loading (Fluck & Hoppeler, 2003). A reduction in skeletal muscle size (atrophy), integrity and contractile activity is associated with denervation, neurological disorders, disease, orthopaedic trauma and a reduction in physical activity (Jagoe et al. 2002; Glass, 2003). In contrast, an increase in muscle size (hypertrophy) and contractile activity can be stimulated by resistance training (Widrick et al. 2002; Leger et al. 2006a), while electrical stimulation can rescue the loss of muscle mass in spinal cord injured patients (Baldi et al. 1998). Changes in skeletal muscle size, structure and function are initiated by extracellular signals (Schiaffino et al. 2007). These signals result in the intracellular activation of transcriptional co-activators and transcription factors with a consequent increase in their target genes that encode proteins mediating contractile, metabolic and structural functions of the skeletal muscle. The intracellular signals influencing human skeletal muscle size, structure and function are not well understood and elucidating these pathways will assist with combating the debilitating effects of muscle wasting.
Striated muscle activator of Rho signalling (STARS) (also known as ms1) is a novel actin-binding protein specifically expressed in cardiac and skeletal muscle (Arai et al. 2002; Mahadeva et al. 2002). In cardiac tissue STARS mRNA is upregulated in response to pressure overload (Arai et al. 2002; Mahadeva et al. 2002). Forced overexpression of STARS in mouse heart tissue results in an increased sensitivity to overload resulting in cardiac hypertrophy (Kuwahara et al. 2007). It has been shown that STARS increases actin polymerization, partly in collaboration with RhoA (Arai et al. 2002). By reducing the cytoplasmic concentration of monomeric G-actin, STARS promotes the nuclear translocation of the serum response factor (SRF) transcriptional co-activators myocardin-related transcription factor-A and-B (MRTF-A and-B), resulting in an increase in SRF-mediated gene transcription (Kuwahara et al. 2005).
Several targets downstream of STARS, including RhoA and SRF, have previously been linked with skeletal muscle development and remodelling. Rho is involved in skeletal muscle differentiation (Takano et al. 1998) and regulates the expression of the myogenic transcription factors MyoD and myogenin in myoblasts (Carnac et al. 1998; Cai et al. 2004). RhoA protein levels in skeletal muscle have been shown to increase and decrease respectively, following functional overload (McClung et al. 2003 2004; Sakuma et al. 2003) and hindlimb suspension in rats (McClung et al. 2004). RhoA, via increasing actin polymerization, has been shown to increase the nuclear localization of SRF (Liu et al. 2003). SRF activity and expression are increased during load-induced hypertrophy in rooster (Fluck et al. 1999) and rat (Sakuma et al. 2003) skeletal muscle. Gene deletion studies in mice reveal that SRF is required for skeletal muscle growth and maturation (Li et al. 2005). Known SRF target genes include the structural protein α-actin (Carson et al. 1996), the motor protein myosin heavy chain type IIa (MHCIIa) (Allen et al. 2001), and the insulin-like growth factor-1 (IGF-1) (Charvet et al. 2006).
STARS signalling through a RhoA/MRTF/SRF pathway may play an important role in skeletal muscle remodelling and therefore may be an interesting target for therapeutic manipulation. However, the regulation of STARS and its downstream signalling targets in human skeletal muscle following hypertrophy and/or atrophy is unknown. Therefore, the aim of the present study was to measure the regulation of STARS, RhoA, MRTF-A/B and SRF, as well as the expression of the SRF target genes, α-actin, MHCIIa and IGF-1. All measurements were made before and after a period of resistance training (hypertrophy) and again after a period of de-training (atrophy) in human skeletal muscle (Leger et al. 2006a).
Methods
Subjects details
Twenty-five healthy males participated in the study. It was approved by the canton of Valais medical ethics commission (Commission cantonale valaisanne d'éthique médicale; CCVEM) and conformed with the Declaration of Helsinki. All participants gave their informed consent and agreed to muscle biopsies and physiological testing. The subjects were physically active but had not participated in a resistance training programme for more than 12 months. The subjects were divided into two resistance training groups (low repetitions or high repetitions) that were matched for age, height, weight,  and for strength and endurance for each of the exercises completed. Physiological characteristics of the subjects are summarized in Table 1.
and for strength and endurance for each of the exercises completed. Physiological characteristics of the subjects are summarized in Table 1.
Table 1.
Characteristics of the subjects in the strength and endurance training groups
| Parameter | Strength group | Endurance group |
|---|---|---|
| Age (years) | 36.8 ± 5.5 | 32.8 ± 2.5 |
| Height (cm) | 177 ± 7 | 180 ± 7 |
| Weight (kg) | 80 ± 13 | 77 ± 13 |
 (ml kg−1 min−1) (ml kg−1 min−1) |
45.1 ± 6.3 | 47.7 ± 8.0 |
Muscle biopsies
Skeletal muscle samples were obtained under local anaesthesia (Rapidocaine, 1% plain) from the belly of the vastus lateralis muscle using a percutaneous needle biopsy technique (Pro-Mag, Medical Device Technologies Inc., Gainsville, FL, USA). A single incision was made in the skin and three individual muscle samples were taken. The post-training muscle biopsies were taken 48–72 h after the last training session. The muscle samples were immediately frozen in liquid nitrogen and used for RNA and protein extraction. These muscle samples remained from a previously published study (Leger et al. 2006a).
Measurement of oxygen consumption
 was measured using a Quark B2 metabolic cart (Cosmed, Rome, Italy) while subjects were cycling on an ergometer (Ergoline 900, Sensor Medic, Bitz, Germany). The subjects began cycling at a power of 90 W. The power was increased by 30 W every 3 min until the subject could not maintain a minimal revolution of 75 r.p.m. At the end of each step, lactate concentration was obtained (Lactate Pro, Axon Lab AG, Reichenbach/Stuttgart, Germany). The duration of the test was between 20 and 30 min. Heart rate (Polar) and oxygen consumption were measured continually throughout the test.
was measured using a Quark B2 metabolic cart (Cosmed, Rome, Italy) while subjects were cycling on an ergometer (Ergoline 900, Sensor Medic, Bitz, Germany). The subjects began cycling at a power of 90 W. The power was increased by 30 W every 3 min until the subject could not maintain a minimal revolution of 75 r.p.m. At the end of each step, lactate concentration was obtained (Lactate Pro, Axon Lab AG, Reichenbach/Stuttgart, Germany). The duration of the test was between 20 and 30 min. Heart rate (Polar) and oxygen consumption were measured continually throughout the test.  was calculated as the highest value averaged over a 30 s period.
was calculated as the highest value averaged over a 30 s period.
Maximal power and endurance tests
The subjects were familiarized with the equipment (Technogym, Gambettola, Italy) and correct lifting technique for the exercises, which included leg press, squat and leg extension. Maximal strength load was estimated as the load (kg) which corresponded to the estimated hypothetical isometric maximal force determined from the force–velocity curve (Rahmani et al. 2001) using a Myotest accelerometer (Myotest SA, Sion, Switzerland). For the leg press the subjects started with a load of 60 kg which was increased by 20 kg per lift until the power–resistance graph demonstrated a decrease in power. For the squat and leg extensions the starting resistance was 25 and 40 kg, respectively, which were increased by 20 and 10 kg respective, per lift. Approximately 3 min rest was provided between each lift. Once the estimated maximal strength load was determined, 50% of this value was calculated for the local muscular endurance test. Following a 10 min rest the subjects completed the maximal number of repetitions possible when lifting 50% of the estimated maximal strength load.
Muscle cross sectional area
Computed tomography (GE Health Care) was used to determine the cross-sectional area (CSA) of the quadriceps. While in the supine position, an image was taken at a distance of 20 cm from the roof of the acetabulum. The CSA was determined by measurement of the surface area of the tissue by two observers (Leger et al. 2006a).
Training protocols
All subjects completed an 8 week resistance training programme as described previously (Campos et al. 2002). The training was performed 2 days per week for the first 4 weeks and 3 days per week for the final 4 weeks. Each session was supervised by a qualified instructor. The subjects were divided into two groups which performed either a LOW number (3–5) or HIGH number (20–28) of repetitions for each exercise, until fatigue. Therefore the resistance for the LOW group was considerably heavier that the resistance used by the HIGH group. The training programme was adapted from previous studies (Anderson & Kearney, 1982; Jackson et al. 1990; Campos et al. 2002) and designed to be approximately equal in volume (resistance × repetitions × sets) with the rest periods between sets adjusted according to the strength–endurance continuum (Fleck & Kraemer, 1997). For example, the LOW group performed three to five repetitions for four sets with 3 min rest between each set, while the HIGH group performed 20–28 repetitions for two sets with 1 min rest between each set (Campos et al. 2002). The exercises were performed in the fixed order of leg press, squat and leg extension. The resistance was continually increased during the training programme so that the ranges of repetitions could be respected.
RNA extraction and real time quantitative PCR
RNA from skeletal muscle (approximately 20 mg of muscle) was extracted using a commercially available preparation, peqGOLD Tri-Fast (Peqlab, Germany). Five micrograms of RNA was reverse transcribed to cDNA using Random Hexomer primers and a Stratascript enzyme (Stratagene, The Netherlands; De Bock et al. 2005). Real-Time PCR was performed using an MX3000p thermal cycler system and Brilliant® Multiplex QPCR Master Mix (Stratagene) (Leger et al. 2006b; Vergani et al. 2007). The PCR conditions consisted of one denaturing cycle at 90°C for 10 min, followed by 40 cycles consisting of denaturing at 90°C for 30 s and annealing at 60°C for 60 s. To control for any variations due to efficiencies of the reverse transcription and PCR, acidic ribosomal phosphoprotein PO (RPLPO; also known a 36B4) was used as an internal control for all PCR runs and remained stable throughout the training and de-training periods (Leger et al. 2008). All PCR runs were performed in triplicate. PCR primer sequences are provided in Table 2.
Table 2.
Human Primer sequences, and annealing temperatures used for the real-time PCR
| Gene | Sequence 5′–3′ | Temp (°C) |
|---|---|---|
| STARS | Sense TGG CTG TGG TCA GGA TCA AG | 60 |
| Anti CTT TGC AGT TGA GTT TCT CTG TAA ATC T | ||
| Probe TXS Red-CCC CTT GCC CTC CCA GGT AAA-BHQ2 | ||
| MRTF-A | Sense TCC ACC CCC ACA CTC ATT AAG | 60 |
| Anti TCT TGC TGC GCT GTG ACT TC | ||
| Probe 6-FAM-AAG CCA ACC CAA GTC TGC CAG TG-BHQ1 | ||
| MRTF-B | Sense TCG GAG TCG ACC AGA TCG TT | 60 |
| Anti GGG ATG GCT CTG CAA ATG TT | ||
| Probe TXS Red-TGA ACT TGT CAG GAT GCA CAT TTT A-BHQ2 | ||
| SRF | Sense TAC CAG GTG TCG GAG TCT GAC A | 60 |
| Anti GGC AGG TTG GTG ACT GTG AA | ||
| Probe AGA CCA AGG ACA CAC TGA AGC CGG C | ||
| α-Actin | Sense GTA GCT ACC CGC CCA GAA ACT | 60 |
| Anti AGG CCG GAG CCA TTG TC | ||
| Probe 6-FAM-ACA ATG TGC GAC GAA GAC GAG ACC AC-BHQ1 | ||
| MHCIIa | Sense GAT GGC ACA GAA GTT GCT GA | 57 |
| Anti CTT CTC GTA GAC GGC TTT GG | ||
| IGF-1 | Sense AGG TAT ACT GGA ATC CGATCT CTG A | 60 |
| Anti CTT GTT GTT TCC TGC ACT CCC TCT | ||
| RPLPO | Sense TCT ACA ACC CTG AAG TGC TTG ATA TC | 60 |
| Anti GCA GAC AGA CAC TGG CAA CAT T | ||
| Probe AGG AAA CTC TGC ATT CTC GCT TCC TGG AG |
Protein extraction and Western blotting
Cytosolic and nuclear proteins were extracted from approximately 20 mg of skeletal muscle using the NE-PER kit (Pierce Biotechnology, Rockford, USA) with the addition of 2 μl phosphatase inhibitor cocktails I, II and III and protease inhibitor cocktail (Sigma), as published previously (Leger et al. 2006b; Doucet et al. 2007). The BCA Protein Assay was used to determine protein concentration (Pierce Biotechnology). Electrophoresis and protein transfer were performed as published previously (Leger et al. 2006b; Doucet et al. 2007). Antibodies used for the detection of SRF (sc-335; 1: 2000) in the nuclear protein fractions and RhoA (sc-119): 1: 2000) in the cytoplasmic protein fractions were from Santa Cruz Biotechnology, CA, USA. Following incubation at room temperature for 60 min with a horseradish peroxidase-conjugated goat anti-rabbit IgG antibody (Cell Signaling Technology, Inc., Beverly, MA, USA) at a dilution of 1: 5,000, the membranes were washed for 5 × 5 min in 0.1% Tween 20–Tris-buffered saline (TBS) and treated for 5 min with a chemiluminescence substrate (ECL-plus, Amersham). All blots were normalized against the tubulin (cytosolic proteins) and lamin A proteins (nuclear proteins) as published previously (Leger et al. 2006b; Doucet et al. 2007).
Statistics
A two-way ANOVA was used to test for the interaction between time and training intensity. Following this a one-way ANOVA with repeated measures was used to test for differences between groups. The level of significance was set at P < 0.05.
Results
There was no significant effect of training intensity on any of the dependent variables. Therefore the two groups of subjects were pooled for all analyses. As shown previously (Leger et al. 2006a) the resistance training performed in this study resulted in a 20% and 15% increase in maximal strength, respectively, for the leg extension and squat exercises (P < 0.01). Muscular endurance also increased by 30%, 100% and 60%, respectively, for the leg extension, leg press and squat exercises (P < 0.01). These functional outcomes were associated with a 10% increase in quadriceps cross-sectional area after training when compared to the pre-training levels (P < 0.01). After 8 weeks of de-training a muscle atrophy of 5% was observed when compared with the post-training hypertrophy (P < 0.05). Muscle size following de-training was not significantly different from the pre-training size (P= 0.1).
Following 8 weeks of resistance training a 3.4-fold increase in STARS mRNA was observed in the post-training group (P < 0.05). STARS mRNA returned to basal levels after 8 weeks of detraining (Fig. 1). The increase in STARS mRNA was also paralleled by approximately a 2-fold increase in RhoA protein content following training which remained after the de-training period (Fig. 1).
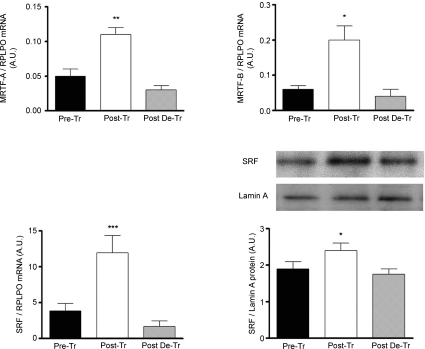
Figure 1. Effect of 8 weeks of resistance training (Post-Tr) and 8 weeks of de-training (Post-DeTr) on STARS mRNA and RhoA protein content.
Significantly different from pre-training and 8 weeks post-training levels: *P < 0.05; **P < 0.01.
The mRNA levels for MRTF-A and MRTF-B were increased by 2.5-and 3.6-fold, respectively, following the training period and both returned to basal levels following de-training (Fig. 2.) The skeletal muscle transcription factor SRF showed an increase of 3-fold, at the mRNA level, after 8 weeks of resistance training in the post-training group (P < 0.005). Following 8 weeks of detraining, SRF mRNA decreased 6.9-fold from post-training. SRF nuclear protein content was also increased by 1.25-fold post-training and returned to basal levels following the de-training period (Fig. 2).
Figure 2. Effect of 8 weeks of resistance training (Post-Tr) and 8 weeks of de-training (Post-DeTr) on MRTF-A, MRTF-B and SRF mRNA expression as well as on SRF protein levels.
Significantly different from pre-training and 8 weeks post-training levels: *P < 0.05; **P < 0.01; ***P < 0.005.
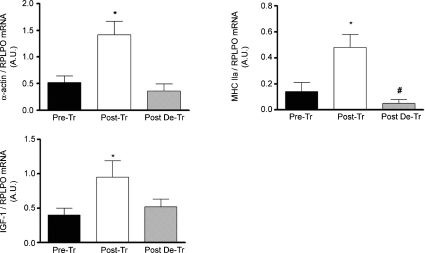
Following 8 weeks of resistance training, a 2.7-, 2.4-and 2.0-fold increase, respectively, in the SRF target genes, α-actin, MHCIIa and IGF-1, was observed (P < 0.05). α-Actin and IGF-1 mRNA returned to basal levels following de-training, while MHCIIa was 65% lower that the pre-training levels (Fig. 3).
Figure 3. Regulation of α-actin, MHCIIa and IGF-1 mRNA expression following 8 weeks of resistance training (Post-Tr) and 8 weeks of de-training (Post-DeTr).
*Significantly different from pre-training levels: P < 0.05; #significantly different from post-training levels: P < 0.05.
Discussion
Identifying the intracellular signalling pathways which control skeletal muscle integrity, growth and function are vital for developing therapeutic strategies aimed at reducing the devastating effects of muscle wasting, as seen in chronic diseases, specific neuromuscular disorders and following musculo-skeletal trauma. At present, the molecular targets and signalling pathways influencing human muscle hypertrophy and atrophy are not well understood. The results from the present study demonstrate, for the first time in skeletal muscle, the up-regulation of key members of the novel STARS signalling pathway following resistance exercise-induced muscle hypertrophy, as well as their down-regulation following inactivity-induced muscle unloading. Several novel and important findings were observed. Firstly, following 8 weeks of progressive overloading of the upper leg muscles, via resistance training, increases in the mRNA levels of STARS, MRTF-A, MRTF-B and SRF, as well as RhoA protein and nuclear SRF protein were observed. These increases were associated with hypertrophy of the upper leg. Secondly, increases in the SRF target genes, α-actin, MHCIIa and IGF-1, were also observed. Thirdly, these observations were all reversed, except for RhoA, during the period of inactivity-induced muscle unloading. It should be noted that muscle hypertrophy was achieved in both the high weight–low repetition, as well as the in the low weight–high repetition training regimes, the latter being contrary to previous observations in young men (Campos et al. 2002). A possible explanation is that the subjects used in the present study were on average 14 years older than those in the study by Campos et al. (2002) and may have been significantly less trained. Therefore any type of resistance training may have provided significant functional overload to induce an increase in muscle size.
The adaptation of skeletal muscle to external mechanical stress, such as increased loading or muscle contraction, requires the initial sensing of the stress, followed by the transduction of the stress into signals that will generate the appropriate physiological response. Previous studies have shown that STARS mRNA levels increase in the left ventricle in response to pressure overload caused by aortic banding (Mahadeva et al. 2002; Kuwahara et al. 2007) and that STARS increases F-actin polymerization, MRTF-A nuclear translocation and consequently, SRF-dependent gene transcription (Kuwahara et al. 2005 2007). Therefore, STARS has been suggested to provide an important link between the transduction of external stress to intracellular signalling which controls genes involved in the maintenance of cytoskeletal integrity and muscle function. In the present study the observed increase in STARS in skeletal muscle, following load-induced skeletal muscle hypertrophy, extends previous findings in cardiac tissue and supports the notion that STARS plays an a role in positively regulating muscle growth.
In addition to STARS, several members of its downstream signalling pathway were also increased in hypertrophied human skeletal muscle following resistance training. For the first time, the present study shows an increase in RhoA protein and SRF mRNA and SRF nuclear protein contents in hypertrophied human skeletal muscle, findings supporting several observations made previously in rat and rooster skeletal muscle. In these studies, RhoA protein and mRNA levels were increased in rat plantaris muscle following 3 and 21 days of functional overload, stimulated by surgical ablation of the distal third of the lateral and medial gastrocnemius muscle (McClung et al. 2003). SRF mRNA and nuclear protein content (Fluck et al. 1999) as well as SRF activity (Fluck et al. 2000) have been shown to increase in rooster anterior latissimus dorsi (ALD) muscle following 7 and 13 days of load-induced hypertrophy. SRF levels were also observed to increase in soleus muscle following 1 day and in plantaris muscle following 8 days of muscle overload-induced stretch (Gordon et al. 2001). SRF is a known transcriptional regulator of genes involved in cytoskeletal structure, growth and contraction (Miano et al. 2007) and therefore is seen as an important regulator of muscle integrity, growth and function. In the present study, the increase in nuclear SRF protein levels as well as SRF mRNA was associated with an increase in the SRF target genes, α-actin (Carson et al. 1996), MHCIIa (Allen et al. 2005), and IGF-1 (Charvet et al. 2006). It has previously been shown that SRF binding to its response element sequence on the α-actin promoter is increased in hypertrophied chicken ALD muscles (Carson et al. 1996). It should be noted that the parallel changes in nuclear SRF protein content and the SRF target genes, α-actin, MHCIIa, and IGF-1, are associations and no causal relationship can be drawn. It cannot be ruled out that other transcription factors, known to bind to the promoter regions of α-actin, MHCIIa, and IGF-1 in muscle tissue and muscle cells, may have been involved in the regulation of these genes. These include the androgen receptor (Hong et al. 2008) and Myc-associated zinc finger protein (MAZ) (Himeda et al. 2008) for α-actin, MEF2 and NFAT (Allen et al. 2001) for MHCIIa, and STAT5b (Woelfle & Rotwein, 2004) for IGF-1.
It has been well established that SRF transcriptional activity is enhanced by the transcriptional co-activator MRTF-A (Wang et al. 2002; Miralles et al. 2003; Cen et al. 2004) and that STARS overexpression contributes to the nuclear translocation of MRTF-A and MRTF-B (Kuwahara et al. 2005 2007). The increase in MRTF-A and MRTF-B mRNA observed in the present study is the first demonstration that MRTF levels are sensitive to the signals induced by increased loading of skeletal muscle. These results suggest that the MRTFs play a positive role in skeletal muscle adaptation to resistance exercise.
As members of the STARS signalling pathway were increased following a period of increased skeletal muscle loading, it was of interest to determine if unloading, stimulated by reduced physical activity, would reverse these increases. Following 8 weeks of reduced physical activity or de-training, STARS and MRTF-A/B mRNA, as well as SRF mRNA and nuclear protein levels returned to pre-training levels. In parallel, the SRF target genes α-actin, MHCIIa and IGF-1, also returned to pre-training levels following the period of reduced muscle loading. These results are the first to show that, in human skeletal muscle, members of the STARS signalling pathway are negatively influenced by reductions in muscle loading. A recent study observed that STARS, MRTF-A and SRF are lower in the skeletal muscle from sarcopenic 24-month-old mice, when compared with 3-month-old mice (Sakuma et al. 2008), suggesting a role for the STARS signalling pathway in age-induced skeletal muscle atrophy. Along similar lines, mice lacking muscle SRF and expressing dominant negative MRTF-A in skeletal muscle presented muscle atrophy, significant fibrosis as well as reduced levels of α-actin mRNA, a phenotype often observed in myopathies and muscle damage. In contrast to the other members of the STARS signalling pathway, RhoA remained elevated following the period of reduced physical activity. This is in contrast to the inactivity model of hindlimb suspension which reduced both muscle mass and RhoA in rat plantaris muscle (McClung et al. 2004). However, along similar lines to our observation, RhoA was observed to be elevated in atrophied skeletal muscle of aged, when compared with younger, mice (Sakuma et al. 2008). As STARS activation of transcription is only partially attenuated following RhoA inhibition (Arai et al. 2002), it appears that STARS signalling can be regulated independently of RhoA.
The regulation of SRF as well as MRTF-A and MRTF-B mRNA in the present study suggests that changes in skeletal muscle loading can regulate a transcriptional programme which may alter the transcription or stability of SRF, MRTF-A and MRTF-B mRNA. This response may, in part, be involved in increasing SRF, MRTF-A and MRTF-B protein levels and therefore provide the muscle with a larger pool of these proteins to be translocated to the nucleus upon stimulation. However, post-translational modifications may also be involved in changes in their protein levels. In the present study MRTF-A and MRTF-B protein levels were not measured; however work by Sakuma et al. (2008) has shown that reduced MRTF-A mRNA expression is paralleled by reduced MRTF-A protein levels in both cytoplasmic and nuclear fractions in older when compared with younger mice. It should be noted that while STARS activation has been shown to increase the transcriptional activity of SRF, via MRTF-A nuclear translocation, it has not been established if STARS, and/or MRTF-A, is involved in SRF nuclear translocation. These are areas for future investigation.
In conclusion, our results show that the STARS signalling pathway is up-and downregulated, respectively, in human skeletal muscle hypertrophy and atrophy. During muscle hypertrophy, induced through increased loading via resistance training, STARS and its downstream targets RhoA, MRTF-A/B and SRF were increased, as well as several SRF target genes involved in skeletal muscle structure, function and growth. The response of these targets was reversed after a period of atrophy-inducing unloading or de-training, when compared to the post-training levels. These findings show that, in human skeletal muscle, STARS signalling is responsive to changes in muscle loading and supports the notion that STARS signalling plays a role in muscle growth and function.
Acknowledgments
This work was supported by the Fonds National Suisse de la Recherche Scientifique (no. 3200B0-105936) and a Faculty Research Development Grant (A.P.R.). Dr Aaron Russell is supported by a NH&MRC Biomedical Career Development Award (479536).
Glossary
Abbreviations
- IGF-1
insulin-like growth factor-1
- MRTF-A and-B
myocardin-related transcription factor-A and-B
- MHCIIa
myosin heavy chain isoform IIa
- RPLPO
acidic ribosomal phosphoprotein PO
- SRF
serum response factor
- STARS
striated activator of Rho signalling
Author contributions
S.L., M.A.W. and B.L. contributed to the analysis and interpretation of the data, and to revising the manuscript; they approved the final version. A.P.R. was responsible for the conception and design of the study, analysis and interpretation of the data and drafting the article, and approved the final submission. Exercise testing and training were performed at the Clinique Romande de Réadaptation, Sion, Switzerland. The analysis of gene and protein expression was performed at the Institut de recherche en réadaptation-réinsertion, Sion, Switzerland and at the School of Exercise and Nutrition Sciences, Deakin University, Burwood, Australia.
References
- Allen DL, Sartorius CA, Sycuro LK, Leinwand LA. Different pathways regulate expression of the skeletal myosin heavy chain genes. J Biol Chem. 2001;276:43524–43533. doi: 10.1074/jbc.M108017200. [DOI] [PubMed] [Google Scholar]
- Allen DL, Weber JN, Sycuro LK, Leinwand LA. Myocyte enhancer factor-2 and serum response factor binding elements regulate fast Myosin heavy chain transcription in vivo. J Biol Chem. 2005;280:17126–17134. doi: 10.1074/jbc.M501207200. [DOI] [PubMed] [Google Scholar]
- Anderson T, Kearney JT. Effects of three resistance training programs on muscular strength and absolute and relative endurance. Res Q Exerc Sport. 1982;53:1–7. doi: 10.1080/02701367.1982.10605218. [DOI] [PubMed] [Google Scholar]
- Arai A, Spencer JA, Olson EN. STARS, a striated muscle activator of Rho signaling and serum response factor-dependent transcription. J Biol Chem. 2002;277:24453–24459. doi: 10.1074/jbc.M202216200. [DOI] [PubMed] [Google Scholar]
- Baldi JC, Jackson RD, Moraille R, Mysiw WJ. Muscle atrophy is prevented in patients with acute spinal cord injury using functional electrical stimulation. Spinal Cord. 1998;36:463–469. doi: 10.1038/sj.sc.3100679. [DOI] [PubMed] [Google Scholar]
- Cai D, Frantz JD, Tawa NE, Melendez PA, Oh B-C, Lidov HGW, Hasselgren P-O, Frontera WR, Lee J, Glass DJ, Shoelson SE. IKK[beta]/NF-[kappa]B activation causes severe muscle wasting in mice. Cell. 2004;119:285–298. doi: 10.1016/j.cell.2004.09.027. [DOI] [PubMed] [Google Scholar]
- Campos GE, Luecke TJ, Wendeln HK, Toma K, Hagerman FC, Murray TF, Ragg KE, Ratamess NA, Kraemer WJ, Staron RS. Muscular adaptations in response to three different resistance-training regimens: specificity of repetition maximum training zones. Eur J Appl Physiol. 2002;88:50–60. doi: 10.1007/s00421-002-0681-6. [DOI] [PubMed] [Google Scholar]
- Carnac G, Primig M, Kitzmann M, Chafey P, Tuil D, Lamb N, Fernandez A. RhoA GTPase and serum response factor control selectively the expression of MyoD without affecting Myf5 in mouse myoblasts. Mol Biol Cell. 1998;9:1891–1902. doi: 10.1091/mbc.9.7.1891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carson JA, Schwartz RJ, Booth FW. SRF and TEF-1 control of chicken skeletal alpha-actin gene during slow-muscle hypertrophy. Am J Physiol Cell Physiol. 1996;270:C1624–1633. doi: 10.1152/ajpcell.1996.270.6.C1624. [DOI] [PubMed] [Google Scholar]
- Cen B, Selvaraj A, Prywes R. Myocardin/MKL family of SRF coactivators: key regulators of immediate early and muscle specific gene expression. J Cell Biochem. 2004;93:74–82. doi: 10.1002/jcb.20199. [DOI] [PubMed] [Google Scholar]
- Charvet C, Houbron C, Parlakian A, Giordani J, Lahoute C, Bertrand A, Sotiropoulos A, Renou L, Schmitt A, Melki J, Li Z, Daegelen D, Tuil D. New role for serum response factor in postnatal skeletal muscle growth and regeneration via the interleukin 4 and insulin-like growth factor 1 pathways. Mol Cell Biol. 2006;26:6664–6674. doi: 10.1128/MCB.00138-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Bock K, Richter EA, Russell AP, Eijnde BO, Derave W, Ramaekers M, Koninckx E, Leger B, Verhaeghe J, Hespel P. Exercise in the fasted state facilitates fibre type-specific intramyocellular lipid breakdown and stimulates glycogen resynthesis in humans. J Physiol. 2005;564:649–660. doi: 10.1113/jphysiol.2005.083170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doucet M, Russell AP, Leger B, Debigare R, Joanisse DR, Caron MA, Leblanc P, Maltais F. Muscle Atrophy and Hypertrophy Signalling in Patients with Chronic Obstructive Pulmonary Disease. Am J Respir Crit Care Med. 2007;176:261–269. doi: 10.1164/rccm.200605-704OC. [DOI] [PubMed] [Google Scholar]
- Fleck S, Kraemer W. Designing Resistance Training Programs. Champaign: Human Kinetics; 1997. IL USA. [Google Scholar]
- Fluck M, Carson JA, Schwartz RJ, Booth FW. SRF protein is upregulated during stretch-induced hypertrophy of rooster ALD muscle. J Appl Physiol. 1999;86:1793–1799. doi: 10.1152/jappl.1999.86.6.1793. [DOI] [PubMed] [Google Scholar]
- Fluck M, Hoppeler H. Molecular basis of skeletal muscle plasticity–from gene to form and function. Rev Physiol Biochem Pharmacol. 2003;146:159–216. doi: 10.1007/s10254-002-0004-7. [DOI] [PubMed] [Google Scholar]
- Fluck M, Waxham MN, Hamilton MT, Booth FW. Skeletal muscle Ca(2+)-independent kinase activity increases during either hypertrophy or running. J Appl Physiol. 2000;88:352–358. doi: 10.1152/jappl.2000.88.1.352. [DOI] [PubMed] [Google Scholar]
- Glass DJ. Signalling pathways that mediate skeletal muscle hypertrophy and atrophy. Nat Cell Biol. 2003;5:87–90. doi: 10.1038/ncb0203-87. [DOI] [PubMed] [Google Scholar]
- Gordon SE, Fluck M, Booth FW. Selected Contribution: Skeletal muscle focal adhesion kinase, paxillin, and serum response factor are loading dependent. J Appl Physiol. 2001;90:1174–1183. doi: 10.1152/jappl.2001.90.3.1174. [DOI] [PubMed] [Google Scholar]
- Himeda CL, Ranish JA, Hauschka SD. Quantitative proteomic identification of MAZ as a transcriptional regulator of muscle-specific genes in skeletal and cardiac myocytes. Mol Cell Biol. 2008;28:6521–6535. doi: 10.1128/MCB.00306-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong MH, Sun H, Jin CH, Chapman M, Hu J, Chang W, Burnett K, Rosen J, Negro-Vilar A, Miner JN. Cell-specific activation of the human skeletal alpha-actin by androgens. Endocrinology. 2008;149:1103–1112. doi: 10.1210/en.2007-0530. [DOI] [PubMed] [Google Scholar]
- Jackson CG, Dickinson AL, Ringel SP. Skeletal muscle fiber area alterations in two opposing modes of resistance-exercise training in the same individual. Eur J Appl Physiol Occup Physiol. 1990;61:37–41. doi: 10.1007/BF00236691. [DOI] [PubMed] [Google Scholar]
- Jagoe RT, Lecker SH, Gomes M, Goldberg AL. Patterns of gene expression in atrophying skeletal muscles: response to food deprivation. FASEB J. 2002;16:1697–1712. doi: 10.1096/fj.02-0312com. [DOI] [PubMed] [Google Scholar]
- Kuwahara K, Barrientos T, Pipes GC, Li S, Olson EN. Muscle-specific signaling mechanism that links actin dynamics to serum response factor. Mol Cell Biol. 2005;25:3173–3181. doi: 10.1128/MCB.25.8.3173-3181.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuwahara K, Teg Pipes GC, McAnally J, Richardson JA, Hill JA, Bassel-Duby R, Olson EN. Modulation of adverse cardiac remodeling by STARS, a mediator of MEF2 signaling and SRF activity. J Clin Invest. 2007;117:1324–1334. doi: 10.1172/JCI31240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leger B, Cartoni R, Praz M, Lamon S, Deriaz O, Crettenand A, Gobelet C, Rohmer P, Konzelmann M, Luthi F, Russell AP. Akt signalling through GSK-3{beta}, mTOR and Foxo1 is involved in human skeletal muscle hypertrophy and atrophy. J Physiol. 2006a;576:923–933. doi: 10.1113/jphysiol.2006.116715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leger B, Derave W, De Bock K, Hespel P, Russell AP. Human Sarcopenia Reveals an Increase in SOCS-3 and Myostatin and a Reduced Efficiency of Akt Phosphorylation. Rejuvenation Res. 2008;11:163–175B. doi: 10.1089/rej.2007.0588. [DOI] [PubMed] [Google Scholar]
- Leger B, Vergani L, Soraru G, Hespel P, Derave W, Gobelet C, D'Ascenzio C, Angelini C, Russell AP. Human skeletal muscle atrophy in amyotrophic lateral sclerosis reveals a reduction in Akt and an increase in atrogin-1. FASEB J. 2006b;20:583–585. doi: 10.1096/fj.05-5249fje. [DOI] [PubMed] [Google Scholar]
- Li S, Czubryt MP, McAnally J, Bassel-Duby R, Richardson JA, Wiebel FF, Nordheim A, Olson EN. Requirement for serum response factor for skeletal muscle growth and maturation revealed by tissue-specific gene deletion in mice. Proc Natl Acad Sci U S A. 2005;102:1082–1087. doi: 10.1073/pnas.0409103102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu HW, Halayko AJ, Fernandes DJ, Harmon GS, McCauley JA, Kocieniewski P, McConville J, Fu Y, Forsythe SM, Kogut P, Bellam S, Dowell M, Churchill J, Lesso H, Kassiri K, Mitchell RW, Hershenson MB, Camoretti-Mercado B, Solway J. The RhoA/Rho kinase pathway regulates nuclear localization of serum response factor. Am J Respir Cell Mol Biol. 2003;29:39–47. doi: 10.1165/rcmb.2002-0206OC. [DOI] [PubMed] [Google Scholar]
- Mahadeva H, Brooks G, Lodwick D, Chong NW, Samani NJ. ms1, a novel stress-responsive, muscle-specific gene that is up-regulated in the early stages of pressure overload-induced left ventricular hypertrophy. FEBS Lett. 2002;521:100–104. doi: 10.1016/s0014-5793(02)02833-8. [DOI] [PubMed] [Google Scholar]
- McClung JM, Lee WJ, Thompson RW, Lowe LL, Carson JA. RhoA induction by functional overload and nandrolone decanoate administration in rat skeletal muscle. Pflugers Arch. 2003;447:345–355. doi: 10.1007/s00424-003-1151-7. [DOI] [PubMed] [Google Scholar]
- McClung JM, Thompson RW, Lowe LL, Carson JA. RhoA expression during recovery from skeletal muscle disuse. J Appl Physiol. 2004;96:1341–1348. doi: 10.1152/japplphysiol.01015.2003. [DOI] [PubMed] [Google Scholar]
- Miano JM, Long X, Fujiwara K. Serum Response Factor: Master Regulator of the Actin Cytoskeleton and Contractile Apparatus. Am J Physiol Cell Physiol. 2007;292:C70–81. doi: 10.1152/ajpcell.00386.2006. [DOI] [PubMed] [Google Scholar]
- Miralles F, Posern G, Zaromytidou AI, Treisman R. Actin dynamics control SRF activity by regulation of its coactivator MAL. Cell. 2003;113:329–342. doi: 10.1016/s0092-8674(03)00278-2. [DOI] [PubMed] [Google Scholar]
- Rahmani A, Viale F, Dalleau G, Lacour JR. Force/velocity and power/velocity relationships in squat exercise. Eur J Appl Physiol. 2001;84:227–232. doi: 10.1007/PL00007956. [DOI] [PubMed] [Google Scholar]
- Sakuma K, Akiho M, Nakashima H, Akima H, Yasuhara M. Age-related reductions in expression of serum response factor and myocardin-related transcription factor A in mouse skeletal muscles. Biochim Biophys Acta. 2008;1782:453–461. doi: 10.1016/j.bbadis.2008.03.008. [DOI] [PubMed] [Google Scholar]
- Sakuma K, Nishikawa J, Nakao R, Nakano H, Sano M, Yasuhara M. Serum response factor plays an important role in the mechanically overloaded plantaris muscle of rats. Histochem Cell Biol. 2003;119:149–160. doi: 10.1007/s00418-003-0499-2. [DOI] [PubMed] [Google Scholar]
- Schiaffino S, Sandri M, Murgia M. Activity-dependent signaling pathways controlling muscle diversity and plasticity. Physiology (Bethesda) 2007;22:269–278. doi: 10.1152/physiol.00009.2007. [DOI] [PubMed] [Google Scholar]
- Takano H, Komuro I, Oka T, Shiojima I, Hiroi Y, Mizuno T, Yazaki Y. The Rho family G proteins play a critical role in muscle differentiation. Mol Cell Biol. 1998;18:1580–1589. doi: 10.1128/mcb.18.3.1580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vergani L, Malena A, Sabatelli P, Loro E, Cavallini L, Magalhaes P, Valente L, Bragantini F, Carrara F, Leger B, Poulton J, Russell AP, Holt IJ. Cultured muscle cells display defects of mitochondrial myopathy ameliorated by anti-oxidants. Brain. 2007;130:2715–2724. doi: 10.1093/brain/awm151. [DOI] [PubMed] [Google Scholar]
- Wang DZ, Li S, Hockemeyer D, Sutherland L, Wang Z, Schratt G, Richardson JA, Nordheim A, Olson EN. Potentiation of serum response factor activity by a family of myocardin-related transcription factors. Proc Natl Acad Sci U S A. 2002;99:14855–14860. doi: 10.1073/pnas.222561499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Widrick JJ, Stelzer JE, Shoepe TC, Garner DP. Functional properties of human muscle fibers after short-term resistance exercise training. Am J Physiol Regul Integr Comp Physiol. 2002;283:R408–416. doi: 10.1152/ajpregu.00120.2002. [DOI] [PubMed] [Google Scholar]
- Woelfle J, Rotwein P. In vivo regulation of growth hormone-stimulated gene transcription by STAT5b. Am J Physiol Endocrinol Metab. 2004;286:E393–401. doi: 10.1152/ajpendo.00389.2003. [DOI] [PubMed] [Google Scholar]