Abstract
Nondistorting C4′ backbone adducts serve as molecular tools to analyze the strategy by which a limited number of human nucleotide excision repair (NER) factors recognize an infinite variety of DNA lesions. We have constructed composite DNA substrates containing a noncomplementary site adjacent to a nondistorting C4′ adduct to show that the loss of hydrogen bonding contacts between partner strands is an essential signal for the recruitment of NER enzymes. This specific conformational requirement for excision is mediated by the affinity of xeroderma pigmentosum group A (XPA) protein for nonhybridizing sites in duplex DNA. XPA recognizes defective Watson–Crick base pair conformations even in the absence of DNA adducts or other covalent modifications, apparently through detection of hydrophobic base components that are abnormally exposed to the double helical surface. This recognition function of XPA is enhanced by replication protein A (RPA) such that, in combination, XPA and RPA constitute a potent molecular sensor of denatured base pairs. Our results indicate that the XPA–RPA complex may promote damage recognition by monitoring Watson–Crick base pair integrity, thereby recruiting the human NER system preferentially to sites where hybridization between complementary strands is weakened or entirely disrupted.
Hypersensitivity to sunlight, skin cancer, and neurodegeneration are major endpoints of a nucleotide excision repair (NER) deficiency in xeroderma pigmentosum patients (1, 2). This versatile DNA repair system normally protects the genome by removing bulky base adducts imposed by UV radiation or other environmental carcinogens (3–5). The same pathway also is implicated in the repair of many nonbulky base lesions generated by intracellular genotoxic processes or spontaneous decay (6, 7). Although a minimal set of about 25 core subunits necessary for the complete NER reaction in humans has been defined (8, 9), the strategy by which these factors locate a broad range of chemically unrelated DNA defects remains to be established.
Several findings suggest that human NER enzymes operate preferentially on DNA lesions that disturb hydrogen bonding between complementary strands. In a previous study, we observed considerable differences in the efficiency by which human NER factors are recruited to helix-destabilizing or helix-stabilizing adducts (10). Among the helix-stabilizing lesions, for example, monofunctional psoralen adducts are processed to some extent by NER enzymes (11). However, quantitative comparisons using a repair competition assay revealed that the human NER system recognizes psoralen monoadducts up to three orders of magnitude less efficiently than acetylaminofluorene adducts or other helix-destabilizing lesions (10). In a parallel report, we found that fast excision of benzo[a]pyrene (B[a]P) adducts correlates with disruption of Watson–Crick hydrogen bonds, whereas the presence of partially normal interactions between the modified base and its complementary partner results in slow excision of B[a]P adducts (12). Similarly, the human NER system processes l-deoxyribonucleosides (the mirror images of natural d-deoxyribonucleosides) only when these stereochemical variants are accompanied by local base-pairing defects (13). Enhanced excision repair at sites of reduced hydrogen bond formation also was noted by other authors when cyclobutane pyrimidine dimers (14) or cisplatin adducts (15) were combined with base mismatches. To uncouple covalent DNA modifications from such losses of hydrogen-bonding capabilities, we designed nondistorting lesions by adding functional groups such as the pivaloyl moiety depicted in Fig. 1A to carbon C4′ of a single deoxyribose residue. On incorporation into duplex DNA, these C4′ backbone adducts maintain standard Watson–Crick hydrogen bonding between complementary bases and are refractory to human NER activity (16). Here, we exploited a nondistorting C4′ pivaloyl adduct to construct composite substrates in which the site of defective Watson–Crick strand pairing is physically dissociated from the site of covalent modification. Probing of NER activity with such composite substrates and subsequent binding assays with purified components led us to propose that the key structural feature of DNA, i.e., its intermolecular association by base pairing, constitutes the critical determinant by which at least one NER subunit, the xeroderma pigmentosum group A–replication protein A (XPA–RPA) complex, recruits the NER system to damaged substrates. This discrimination function recognizes improper base pairing conformations but is independent of the particular adduct chemistry and, hence, accounts for the versatility of NER responses.
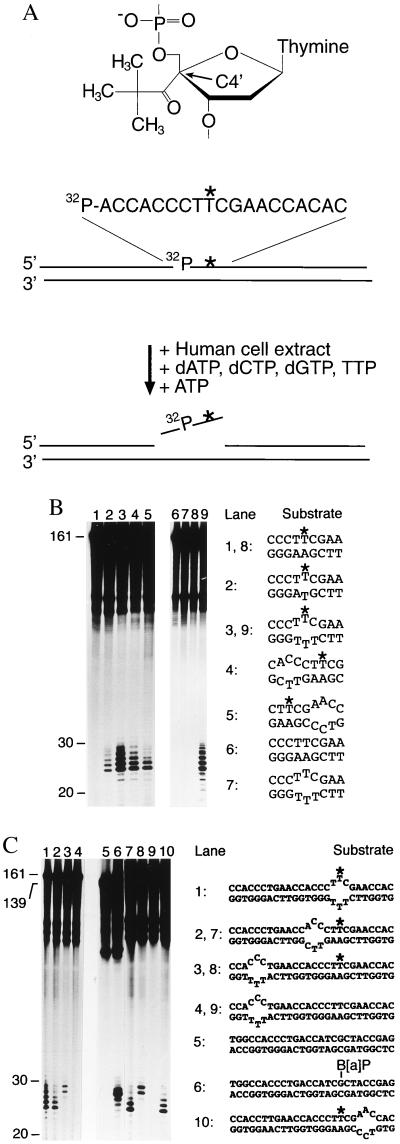
Figure 1.
Recruitment of human NER activity depends on defective hybridization. (A) Structure of the nondistorting C4′ pivaloyl adduct (Top), basic design of the 161-mer substrates (Middle) and oligonucleotide excision assay (Bottom). The C4′ pivaloyl modification is indicated by ∗, and a 32P-labeled residue is situated on the 5′ side of this lesion. (B) Determination of human NER activity in HeLa cell extract. After incubations of 40 min at 30°C, excision products were visualized by denaturing gel electrophoresis and autoradiography. All reactions contained the same amount of radioactive substrate (5 fmol), and oligonucleotide lengths were estimated by using appropriate markers. These incubations in cell extract also generate nonspecific degradation products, visible in the upper part of each lane, which do not interfere with detection of excised oligomers. The sequence environment in the central portion of each substrate is indicated, with the asterisks denoting the C4′ pivaloyl adduct. (C) Probing of NER activity using composite substrates. The central portion of each double-stranded fragment is outlined, again with ∗ denoting the C4′ modification. Lane 6 shows a control reaction with a (−)-cis-B[a]P-modified substrate of 139 bp (12).
MATERIALS AND METHODS
DNA Substrates. The 19-mer oligonucleotide 5′-ACCACCCTT∗CGAACCACAC-3′ containing a C4′ pivaloyl adduct (indicated by ∗) was provided by M. Petretta and B. Giese (University of Basel); 19-mer oligonucleotides with 3-nitropyrrole or 5-nitroindole residues were obtained from MedProbe (Oslo). The 11-mer oligonucleotides (5′-CCATCGCTACC-3′) containing a single guanine adduct with either (−)-trans-B[a]P, (−)-cis-B[a]P, or acetylaminofluorene were produced as described (12). Internally radiolabeled fragments of 161 or 139 bp were constructed by ligating six partially overlapping oligonucleotides as described (6, 17). B[a]P- and acetylaminofluorene-modified 11-mers, or the respective 11-mer control, were elongated to 19-mer oligonucleotides by annealing with an appropriate 43-mer (5′-GCTCTAGAATTCCACGGTAGCGATGGCTCGACGTCTGCAGTCG-3′). The single-stranded arms of the resulting partial duplex were hybridized with the 15-mer 5′-GTGGAATTCTAGAGC-3′ (on the 5′ side) and the 17-mer 5′-CGACTGCAGACGTCGAG-3′ (on the 3′ side), followed by ligation using T4 DNA ligase (Life Technologies, Grand Island, NY). After gel purification, the full-length 43-mer products were incubated with EcoRI (Life Technologies) and AatII (New England Biolabs) and repurified on denaturing 20% polyacrylamide gels to obtain 19-mer oligonucleotides of the sequence 5′-CGAGCCATCGCTACCGGTG-3′. Substrates of 43 bp were obtained by annealing the oligonucleotide 5′-CGACTGCAGACGTCGAGCCTTCGCTACCGTGGAATTCTAGAGC-3′ with either 5′-GCTCTAGAATTCCACGGTAGCGAAGGCTCGACGTCTGCAGTCG-3′ (to generate homoduplexes) or 5′GCTCTAGAATTCCACGGTAGCTTTGGCTCGACGTCTGCAGTCG-3′ (to generate heteroduplexes with three mismatches in the center).
Oligonucleotide Excision Assay.
Human NER activity was assayed in a HeLa cell extract, and excision products were analyzed on 10% polyacrylamide denaturing gels as reported (12, 13, 16).
Mobility Shift Assay.
Human XPA protein was expressed with an N-terminal polyhistidine tag in Escherichia coli strain BL21 (Invitrogen) and purified to homogeneity (18). Recombinant human RPA protein (19) was provided by R. Hindges and U. Hübscher (University of Zürich). The oligonucleotide substrates were 32P-labeled at the 5′ end and mixed with a 1.5-fold excess of unlabeled complementary oligonucleotides of the same length. Annealing was performed in 50 mM Tris⋅HCl (pH 7.4), 10 mM MgCl2, and 1 mM DTT by heating to 80°C for 10 min and then incubating for another 3 h at 25°C. 32P-labeled single-stranded or double-stranded 19-mer substrate (40 fmol), unlabeled 19-mer homoduplexes (1.4 pmol), and the indicated amounts of XPA protein were coincubated at 20°C in reactions of 20 μl containing 25 mM Hepes⋅KOH (pH 8.3), 30 mM KCl, 4 mM MgCl2, 1 mM EDTA, 0.9 mM DTT, 45 μg/ml BSA, and 10% glycerol. Reactions were stopped after 30 min by cooling the samples to 0°C. After addition of gel loading buffer (4 μl) containing 100 mM Tris⋅HCl (pH 8.3), 10% glycerol, and 0.05% Orange G, the extent of binding was determined on 5% native polyacrylamide gels. Electrophoresis was performed at 1.5 mA/cm for 50 min at 4°C, using 45 mM Tris⋅HCl (pH 8.3), 45 mM boric acid, and 1 mM EDTA as the running buffer. Gels were dried and subjected to autoradiography, and the radioactivity migrating with free and bound fractions was determined by densitometric analysis of appropriately exposed x-ray films. The linearity of each densitometric quantification was confirmed by counting Cerenkov radiations of representative gel slices. In the presence of RPA, mobility shift assays were performed as indicated before, but with substrates of 43 bp in length and without unlabeled competitor.
RESULTS
Excision of Composite Substrates.
Human NER activity was assessed by using linear DNA fragments of 161 bp that carry the site-directed C4′ pivaloyl in a central position. As control substrates, we used DNA fragments containing a site-directed B[a]P adduct (12). The modified strands were synthesized to include a 32P-labeled phosphate at the ninth phosphodiester bond on the 5′ side to the covalent lesion (Fig. 1A). Such internally radiolabeled substrates were mixed with a soluble HeLa cell extract containing the full repertoire of human NER factors (20, 21). On addition of deoxyribonucleotides and ATP, human NER enzymes promote DNA incision both 5′ and 3′ to the offending lesion, thereby releasing DNA damage as part of oligomeric segments of 24–32 nt in length (22). Because of their extension mainly on the 5′ side to each lesion, all excision products also include the incorporated radiolabel (Fig. 1A) and, therefore, can be detected by gel electrophoresis and autoradiography. As reported (16), this standard assay established that a single C4′ pivaloyl adduct is refractory to oligonucleotide excision when located within a fully complementary DNA duplex (Fig. 1B, lanes 1 and 8). Conversely, the same nondistorting pivaloyl adduct was able to induce substantial NER activity, resulting in 25- to 29-nt excision products, when Watson–Crick base pairing at the site of C4′ modification was artificially denatured by combination with a noncomplementary base (Fig. 1B, lane 2). NER activity was stimulated even more efficiently by three noncomplementary base pairs at or near the nondistorting C4′ modification (Fig. 1B, lanes 3–5 and 9). Mismatched bases on their own caused no oligonucleotide excision (Fig. 1 B, lane 7 and C, lanes 4 and 9), but the requirement of such artificially denatured sites for excision of the nondistorting pivaloyl adduct demonstrates that local disruption of Watson–Crick base pairing is an indispensable component of the molecular signal that attracts human NER enzymes to damaged DNA.
To dissect this complex recognition signal, the noncomplementary site that simulates base pair denaturation was dislocated from the C4′-modified residue and, surprisingly, NER activity was maintained even when the mismatches were moved by about 15 nt in the 5′ direction (Fig. 1C, lanes 3 and 8). This composite substrate involving dislocation of the nonhybridizing bases from the nondistorting adduct resulted in a slightly different excision pattern, but the observed repair products (composed mainly of 28- and 29-nt oligomers) remained within the characteristic 24- to 32-nt range and, in fact, were similar in length to the oligonucleotides caused by excision of a B[a]P adduct (Fig. 1C, lane 6). Irrespective of this limited variability in the final products, the reparability of such a composite substrate shows that nonhybridizing bases trigger the NER reaction even when located some distance from the nondistorting adduct. Thus, destabilization of Watson–Crick base pairing provides a molecular signal that can be dissociated from the site of aberrant chemistry, implying the involvement of a NER factor that recognizes defective base pairing conformations but is indifferent to the precise location or chemical composition of the accompanying lesion.
Binding of Human XPA Protein to Modified DNA.
We then searched for NER subunits that mediate the predicted damage-independent interaction with nonhybridizing double helical sites. For that purpose, the binding selectivity of known NER factors was tested by using 32P-labeled DNA fragments of 19 or 43 bp. Human XPA is a protein of 31 kDa that is thought to promote damage recognition (4, 5, 18) and coordinate protein–protein interactions on the DNA substrate (23–25). Electrophoretic mobility-shift assays confirmed that purified XPA protein forms nucleoprotein complexes preferentially with DNA duplexes carrying single carcinogen lesions such as a site-directed acetylaminofluorene or B[a]P adduct (Fig. 2A). Strong preferential binding was also observed in response to a site-directed cisplatin intrastrand crosslink (data not shown).
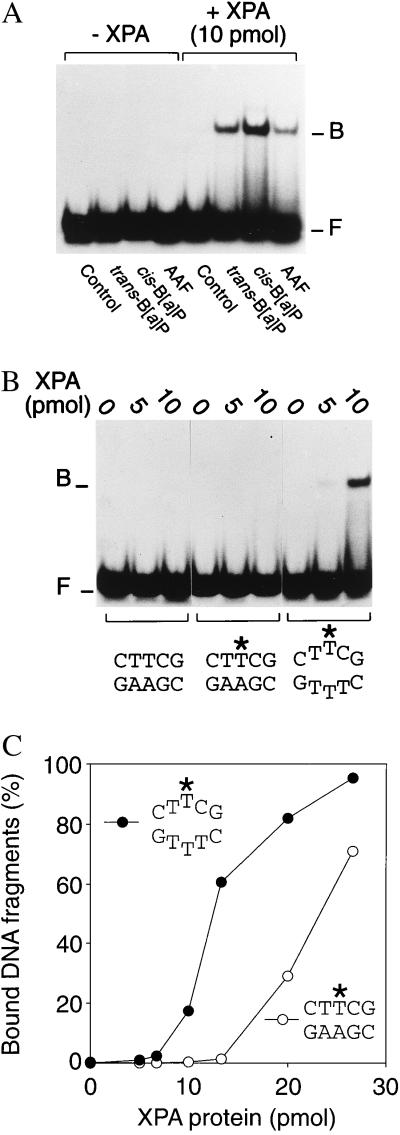
Figure 2.
Binding of human XPA protein to covalently modified DNA. The indicated amounts of XPA protein were coincubated with DNA duplexes of 19 residues (40 fmol per reaction), followed by analysis of protein–DNA interactions by electrophoretic mobility-shift assays. One strand of each substrate was 32P-labeled at its 5′ end. The positions of XPA–DNA complexes (B) and of free DNA fragments (F) in the gel autoradiographs are indicated. (A) DNA fragments were modified by a site-directed carcinogen–DNA adduct. (B) DNA fragments carried a C4′ pivaloyl adduct (denoted by ∗) in the center. (C) Mean percentages of bound DNA fragments obtained from quantitative densitometric scanning of 3–5 experiments.
After establishing that our XPA preparation displays the expected affinity for carcinogen-damaged duplexes, we examined its interaction with 32P-labeled fragments containing the C4′ pivaloyl lesion in a central position. To facilitate direct comparisons between substrate recognition and NER activity, the C4′ modification was located in exactly the same sequence environment that had already been used for the excision assays (Fig. 1). Under conditions of limiting protein (up to 10 pmol per reaction), XPA protein formed complexes with C4′ pivaloyl-modified DNA duplexes only when the nondistorting C4′ lesion was incorporated into a short sequence heterology that simulates base pair denaturation (Fig. 2 B and C). Binding to homoduplex controls was only detected when saturating amounts of XPA protein were added to the reactions (Fig. 2C). Thus, XPA protein shows a conformational requirement for interaction with C4′-modified DNA duplexes, i.e., disruption of hydrogen bonds mediating base pairing, that coincides with the local changes in DNA conformation required for excision of the same C4′ lesion.
Binding of XPA Protein to Artificially Denatured Base Pairs.
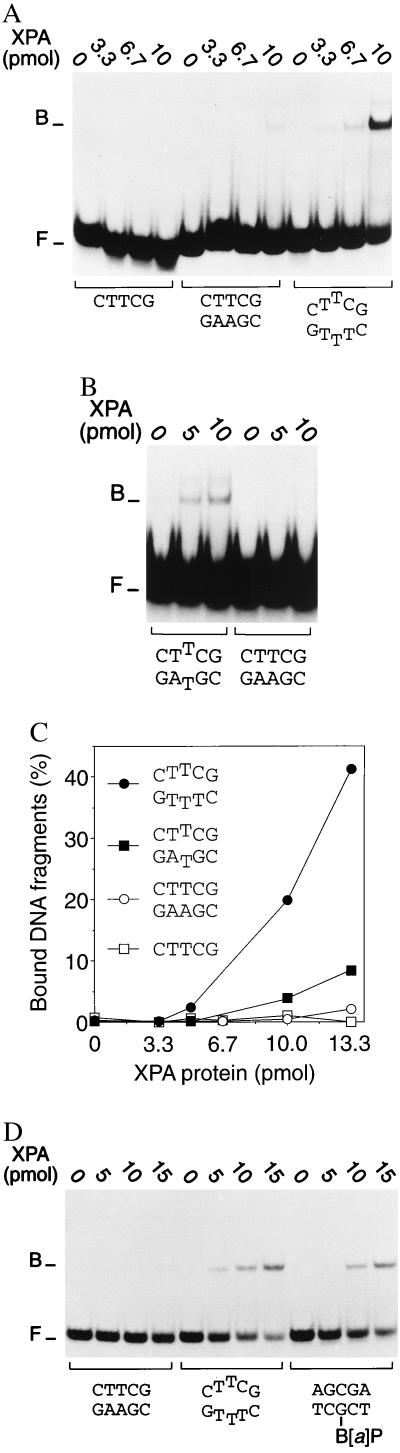
Prompted by our finding that noncomplementary base pairs stimulate oligonucleotide excision even when physically separated from the site of C4′ pivaloyl addition, we extended the binding studies to 32P-labeled duplexes containing mispaired bases but no covalent lesions. Again, under conditions of limiting protein (up to about 10 pmol per reaction), XPA retained a high level of affinity for heteroduplexes carrying three mispaired bases compared with the control homoduplex, even though the DNA substrate was devoid of covalent modifications (Fig. 3 A and C). A more moderate binding preference was observed in the presence of only one noncomplementary base pair (Fig. 3B and C) and, additionally, we found the same bias for artificially denatured sites when longer DNA fragments of 43 bp containing a different nucleotide sequence were tested (data not shown). We also found that XPA protein binds with considerably higher efficiency to partially duplex DNA than to single-stranded substrates of the same length (Fig. 3A). This latter observation rules out the simple possibility that the observed affinity for nonhybridizing base pairs results from extensive denaturation of the duplex and subsequent binding to single-stranded segments of the substrate. Finally, a direct comparison between different 19-mer substrates shows that XPA recognizes DNA duplexes with mispaired bases in the center at least as effectively as homoduplex substrates of the same length but containing a B[a]P carcinogen adduct (Fig. 3D).
Figure 3.
Recognition of nonhybridizing base pairs. Protein–DNA interactions were analyzed by mobility-shift assays, and the positions of XPA–DNA complexes (B) and free DNA fragments (F) are indicated. (A) Preferential interaction with 32P-labeled 19-mer DNA duplexes (40 fmol) containing artificially denatured sites that were generated by insertion of three consecutive mismatches. (B) Preferential binding to 19-mer duplexes containing a single mismatch. (C) Diagram representing the mean quantitative values (percentage of bound fragments) of at least 3 experiments. (D) Side-by-side comparison of XPA binding to homoduplex DNA, duplex DNA with three mispaired bases, or homoduplex DNA fragments containing a trans-B[a]P-modified guanine in a central position.
Probing of XPA Protein with Nonhybridizing Base Analogs.
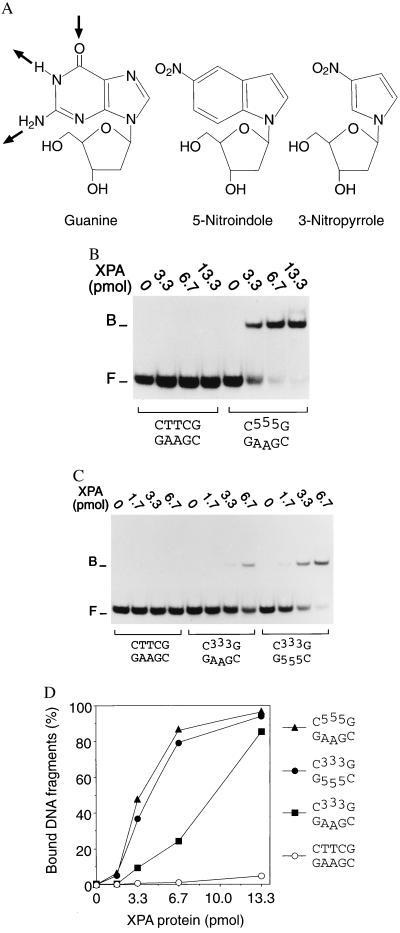
To identify the mechanism by which XPA protein discriminates between normal base pairs and nonhybridizing DNA constituents, we replaced natural bases by 5-nitroindole and 3-nitropyrrole analogs. These modifications lack hydrogen acceptor and donor groups for Watson–Crick hybridization (Fig. 4A). However, their aromatic ring structures have a similar electron distribution as natural bases and, as a consequence, retain at least in part the potential for hydrophobic interactions (26). When three consecutive 5-nitroindole (Fig. 4B) or three consecutive 3-nitropyrrole residues (Fig. 4C) were introduced in the center of one strand of 19-mer duplexes, the assembly of XPA–DNA complexes was stimulated even more efficiently than in the previous experiments, with three mismatches in the identical sequence context (compare with Fig. 3A). In the presence of only 6.7 pmol of protein, up to 80% of fragments containing the 5-nitroindole residues were complexed, whereas no XPA–DNA interactions could be detected when this low amount of protein was incubated with unmodified homoduplexes (Fig. 4D). To systematically remove all hydrogen acceptors and donors from the same site in the center of the duplex, 32P-labeled oligonucleotides carrying three consecutive 5-nitroindoles (used as purine analogs) were paired with complementary 19-mers containing three 3-nitropyrroles (used as pyrimidine analogs). Despite the complete absence of local hydrogen-bonding partners, XPA retained its strong affinity for the nonhybridizing site (Fig. 4 C and D). Thus, substitution of normal bases by 5-nitroindoles and 3-nitropyrroles demonstrates that unpaired hydrogen acceptors and donors are not needed for the preferential binding of XPA protein to nonhybridizing residues. Conversely, exposure of aromatic components by systematic removal of hydrogen donors and acceptors is sufficient to induce the characteristic substrate recognition function of XPA protein.
Figure 4.
XPA protein is guided to nonhybridizing sites by hydrophobic attractions. (A) Nucleoside analogs containing 5-nitroindole or 3-nitropyrrole preserve normal backbone structure, retain aromatic interactions but, in contrast to natural bases such as guanine, lack donor and acceptor groups for Watson–Crick hydrogen bonding. The arrows indicate the hydrogen acceptor and donors employed in Watson–Crick pairing between guanine and cytosine. (B) Preferential binding of XPA to 32P-labeled DNA duplexes (40 fmol) containing 5-nitroindole (“5”). (C) Preferential binding of XPA to DNA duplexes containing 3-nitropyrrole analogs (“3”), or both 5-nitroindoles in one strand and 3-nitropyrroles at the corresponding positions of the complementary strand. (D) Mean percentages of bound fragments obtained by densitometric scanning of three independent experiments. The composition in the center of each double-stranded 19-mer substrate is indicated.
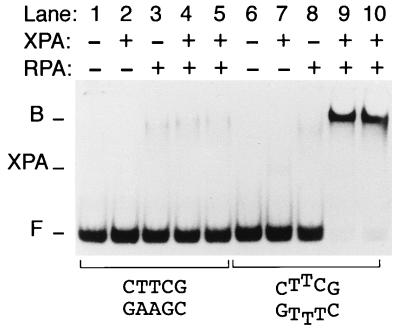
Cooperative Binding of XPA and RPA to Artificially Denatured Base Pairs.
XPA protein forms a tight stoichiometric complex with RPA, a heterotrimeric single-stranded DNA binding factor with a total molecular mass of 116 kDa that is critically involved in the human NER reaction (24, 25, 27). We first used mobility-shift assays to compare the individual behavior of XPA and RPA when incubated with different DNA species. As expected from its single-stranded DNA binding activity but unlike XPA, RPA interacts more efficiently with single-stranded oligonucleotides than with DNA duplexes of the same length and sequence (gel not shown). In the presence of RPA only, there is little or no preference for double-stranded fragments carrying three mismatched bases over the homoduplex control (Fig. 5, compare lanes 3 and 8), in agreement with earlier reports that RPA requires a minimum of 20–30 nucleotides of single-stranded DNA for its high-affinity binding (28, 29). Thus, XPA and RPA, although interacting with each other, exert different substrate-recognition functions: XPA binds preferentially to nonhybridizing base pairs in duplex DNA but displays little affinity for single strands, whereas RPA binds preferentially to single-stranded DNA but, even in the presence of a short nonhybridizing site, has a poor affinity for double-stranded DNA. The combination of XPA and RPA has been reported to stimulate the interaction of either factor alone with UV-irradiated or carcinogen-damaged DNA (30, 31). Here, we show that a very strong binding cooperativity is achieved in the absence of DNA adducts or any other covalent modification by incubating low equimolar amounts of XPA and RPA with DNA fragments containing just three mismatches in the center (Fig. 5, lanes 9 and 10). Thus, RPA potentiates the unique affinity of XPA protein for nonhybridizing sites in double-stranded DNA.
Figure 5.
Cooperative binding of XPA and RPA to nonhybridizing base pairs. XPA and RPA (2 pmol) were incubated with 43-mer DNA fragments (20 fmol), either homoduplexes (lanes 1–5) or partial duplexes containing three mismatches in the center (lanes 6–10), and protein–DNA interactions were analyzed by mobility shift gels. The positions of XPA–RPA–DNA complexes (B) and of free DNA fragments (F) is indicated. Additionally, the figure shows the position of complexes formed by XPA alone (XPA). Because of the low amount of XPA protein, these binary XPA–DNA complexes are visible in lane 7 only on overexposure of the gel.
DISCUSSION
In a previous report (16), we showed that human NER enzymes excise C4′ adducts only when these nondistorting backbone variants are incorporated into heterologous sequences that disrupt complementary strand pairing. In fact, neither a site of mispaired bases nor the tested C4′ lesions elicited NER activity, but the combination of these two substrate changes resulted in efficient recruitment of NER enzymes. These previous findings suggested a bipartite mode of substrate discrimination by which recognition of DNA damage depends on two different molecular determinants, i.e., destabilization of base pairing accompanied by a covalent modification of DNA (16). In this study, we further exploited the nondistorting C4′ pivaloyl adduct to dissect this damage recognition signal into its individual components. In fact, we found that human NER enzymes also process composite substrates where the mispaired bases are dislocated from the covalent adduct by an intervening sequence of about 15 nt. Because the NER system is active on this composite substrate despite physical dissociation of its two essential determinants, we concluded that the nonhybridizing site and the accompanying adduct may be recognized by two separate subunits. Also, processing of this composite substrate confirms that the presence of nonhybridizing bases constitutes a true molecular signal for the recruitment of human NER factors, arguing against the possibility that mismatches induce repair of a nondistorting adduct by mimicking an unwound reaction intermediate at the covalent modification.
XPA (18) and RPA (32) have been reported to display a preference for single-stranded DNA over double-stranded DNA and, hence, may detect the single-strand character of destabilized or disrupted helical sites. XPC also has a strong affinity for single-stranded DNA but, in contrast to XPA or RPA, this factor interacts with DNA duplexes as efficiently as with DNA single strands (33). Unexpectedly, we found in this study that XPA protein binds to duplexes containing short nonhybridizing sites even more efficiently than to single-stranded substrates of the same length and sequence. Also, we show that XPA recognizes unstable or disrupted base pairs regardless of whether local denaturation of the double helix results from covalent carcinogen adducts or is induced artificially by the insertion of mismatches. In contrast, RPA (the interaction partner of XPA) displays only a poor affinity for short nonhybridizing sites. Nevertheless, RPA stimulates the intrinsic affinity of XPA protein for locally denatured sites such that, in combination, XPA and RPA act as a potent sensor of defective base pairing conformations. These findings are reminiscent of the preferential binding of two prokaryotic NER proteins (UvrA and UvrB) to synthetic “bubble” or “loop” regions in DNA (34).
Most carcinogen–DNA adducts destabilize Watson–Crick hydrogen bonds, causing displacement of one or more bases relative to their standard intrahelical position (10, 12). Similarly, mismatches generate abnormal strand pairing conformations in which the affected bases are displaced from their intrahelical position (35). On the other hand, there are several examples of DNA-binding enzymes that are stimulated on introduction of mismatches in their target sites (36–38) and, in fact, a common property of these enzymes is that their metabolic action involves extrahelical displacement of bases. In the case of XPA protein, we observed preferential binding not only to carcinogen-damaged or mismatched substrates but, with higher efficiency, also to duplexes containing 5-nitroindole and 3-nitropyrrole residues. These aromatic base analogs lack hydrogen acceptors and donors for Watson–Crick strand pairing and, therefore, generate nonhybridizing sites characterized by increased hydrophobicity (26). Thus, the affinity of XPA for duplexes containing such aromatic analogs suggests that its strong bias for displaced pairing conformations is mediated by hydrophobic interactions with aromatic base components that are abnormally exposed to the helical surface.
It was generally believed that XPA protein is the primary DNA damage recognition subunit of the NER system (4, 5, 18, 23, 30, 31). However, our report indicates that XPA does not recognize the damage itself but adopts a more general function by probing the propensity of each nucleobase to undergo correct strand pairing with its complementary partner. This discrimination function, presumably mediated by the presence of abnormal hydrophobic attractions on the helical surface and without specificity for a particular type of lesion, allows recognition of all damaged sites that destabilize (including for example cyclobutane pyrimidine dimers; ref. 39) or disrupt Watson–Crick hydrogen bonds. Therefore, XPA protein (together with RPA) may use this versatile strategy to target the human NER system to a wide range of chemically diverse forms of DNA damage. Our report, indicating that derangement of base pairing constitutes an essential recognition signal and that the XPA–RPA complex may recruit the human NER system in response to this signal, is in apparent conflict with another in vitro study, where order-of-addition experiments led to the conclusion that XPC protein is the “initiator” of human NER activity (40). Additional studies are necessary to establish how XPC may contribute to the recognition of nonhybridizing base pairs.
Acknowledgments
We thank R. D. Wood for plasmid pET15b/XPA, M. Petretta, B. Giese, N. Luneva, and N. E. Geacintov for modified oligonucleotides, and R. Hindges and U. Hübscher for RPA protein. This work was supported by the Swiss National Science Foundation (Grant 31-50518.97).
ABBREVIATIONS
- B[a]P
benzo[a]pyrene
- NER
nucleotide excision repair
- RPA
replication protein A
- XPA
xeroderma pigmentosum group A
Footnotes
This paper was submitted directly (Track II) to the Proceedings Office.
References
- 1.Friedberg E C, Walker G C, Siede W. DNA Repair and Mutagenesis. Washington, DC: Am. Soc. Microbiol.; 1995. pp. 634–649. [Google Scholar]
- 2.Hoeijmakers J H J. Eur J Cancer. 1994;30:1912–1921. doi: 10.1016/0959-8049(94)00381-e. [DOI] [PubMed] [Google Scholar]
- 3.Hanawalt P C. Science. 1994;266:1957–1958. doi: 10.1126/science.7801121. [DOI] [PubMed] [Google Scholar]
- 4.Sancar A. Annu Rev Biochem. 1996;65:43–81. doi: 10.1146/annurev.bi.65.070196.000355. [DOI] [PubMed] [Google Scholar]
- 5.Wood R D. Annu Rev Biochem. 1996;65:135–167. doi: 10.1146/annurev.bi.65.070196.001031. [DOI] [PubMed] [Google Scholar]
- 6.Huang J-C, Hsu D S, Kazantsev A, Sancar A. Proc Natl Acad Sci USA. 1994;91:12213–12217. doi: 10.1073/pnas.91.25.12213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Reardon J T, Bessho T, Kung H C, Bolton P H, Sancar A. Proc Natl Acad Sci USA. 1997;94:9463–9468. doi: 10.1073/pnas.94.17.9463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mu D, Park C H, Matsunaga T, Hsu D S, Reardon J T, Sancar A. J Biol Chem. 1995;270:2415–2418. doi: 10.1074/jbc.270.6.2415. [DOI] [PubMed] [Google Scholar]
- 9.Aboussekhra A, Biggerstaff M, Shivji M K K, Vilpo J A, Moncollin V, Podust V N, Protic M, Hübscher U, Egly J-M, Wood R D. Cell. 1995;80:859–868. doi: 10.1016/0092-8674(95)90289-9. [DOI] [PubMed] [Google Scholar]
- 10.Gunz D, Hess M T, Naegeli H. J Biol Chem. 1996;271:25089–25098. doi: 10.1074/jbc.271.41.25089. [DOI] [PubMed] [Google Scholar]
- 11.Svoboda D L, Taylor J-S, Hearst J E, Sancar A. J Biol Chem. 1993;268:1931–1936. [PubMed] [Google Scholar]
- 12.Hess M T, Gunz D, Luneva N, Geacintov N E, Naegeli H. Mol Cell Biol. 1997;17:7069–7076. doi: 10.1128/mcb.17.12.7069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hess M T, Naegeli H, Capobianco M. J Biol Chem. 1998;273:27867–27872. doi: 10.1074/jbc.273.43.27867. [DOI] [PubMed] [Google Scholar]
- 14.Mu D, Tursun M, Duckett D R, Drummond J T, Modrich P, Sancar A. Mol Cell Biol. 1997;17:760–769. doi: 10.1128/mcb.17.2.760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Moggs J G, Szymkowski D E, Yamada M, Karran P, Wood R D. Nucleic Acids Res. 1997;25:480–490. doi: 10.1093/nar/25.3.480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hess M T, Schwitter U, Petretta M, Giese B, Naegeli H. Proc Natl Acad Sci USA. 1997;94:6664–6669. doi: 10.1073/pnas.94.13.6664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Matsunaga T, Mu M, Park C-H, Reardon J T, Sancar A. J Biol Chem. 1995;270:20862–20869. doi: 10.1074/jbc.270.35.20862. [DOI] [PubMed] [Google Scholar]
- 18.Jones C J, Wood R D. Biochemistry. 1993;32:12096–12104. doi: 10.1021/bi00096a021. [DOI] [PubMed] [Google Scholar]
- 19.Henricksen L A, Umbricht C B, Wold M S. J Biol Chem. 1994;269:11121–11132. [PubMed] [Google Scholar]
- 20.Sibghat-Ullah, Husain I, Carlton W, Sancar A. Nucleic Acids Res. 1989;17:4471–4484. doi: 10.1093/nar/17.12.4471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wood R D, Robins P, Lindahl T. Cell. 1988;53:97–106. doi: 10.1016/0092-8674(88)90491-6. [DOI] [PubMed] [Google Scholar]
- 22.Huang J-C, Svoboda D, Reardon J T, Sancar A. Proc Natl Acad Sci USA. 1992;89:3664–3668. doi: 10.1073/pnas.89.8.3664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cleaver J E, States J C. Biochem J. 1997;328:1–12. doi: 10.1042/bj3280001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Li L, Elledge S J, Peterson C A, Bales E S, Legerski R J. Proc Natl Acad Sci USA. 1994;91:5012–5016. doi: 10.1073/pnas.91.11.5012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Park C-H, Sancar A. Proc Natl Acad Sci USA. 1994;91:5017–5021. doi: 10.1073/pnas.91.11.5017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Loakes D, Brown D M, Linde S, Hill F. Nucleic Acids Res. 1995;23:2361–2366. doi: 10.1093/nar/23.13.2361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Coverley D, Kenny M, Munn M, Rupp W D, Lane D P, Wood R D. Nature (London) 1991;349:538–541. doi: 10.1038/349538a0. [DOI] [PubMed] [Google Scholar]
- 28.Kim C, Paulus B F, Wold M S. Biochemistry. 1994;33:14197–14206. doi: 10.1021/bi00251a031. [DOI] [PubMed] [Google Scholar]
- 29.Matsunaga T, Park C-H, Bessho T, Mu D, Sancar A. J Biol Chem. 1996;271:11047–11050. doi: 10.1074/jbc.271.19.11047. [DOI] [PubMed] [Google Scholar]
- 30.He Z, Henricksen L A, Wold M S, Ingles C J. Nature (London) 1995;374:566–569. doi: 10.1038/374566a0. (1995). [DOI] [PubMed] [Google Scholar]
- 31.Li L, Lu X, Peterson C A, Legerski R J. Mol Cell Biol. 1995;15:5396–5402. doi: 10.1128/mcb.15.10.5396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wold M S. Annu Rev Biochem. 1997;66:61–92. doi: 10.1146/annurev.biochem.66.1.61. [DOI] [PubMed] [Google Scholar]
- 33.Reardon J T, Mu D, Sancar A. J Biol Chem. 1996;271:19451–19456. doi: 10.1074/jbc.271.32.19451. [DOI] [PubMed] [Google Scholar]
- 34.Ahn B, Grossman L. J Biol Chem. 1996;271:21462–21470. doi: 10.1074/jbc.271.35.21462. [DOI] [PubMed] [Google Scholar]
- 35.Hunter W N, Brown T, Anand N N, Kennard O. Nature (London) 1986;320:552–555. doi: 10.1038/320552a0. [DOI] [PubMed] [Google Scholar]
- 36.Verri A, Mazzarello P, Spadari S, Focher F. Biochem J. 1992;287:1007–1010. doi: 10.1042/bj2871007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Klimasauskas S, Roberts R J. Nucleic Acids Res. 1995;23:1388–1395. doi: 10.1093/nar/23.8.1388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhao X, Liu J, Hsu D S, Zhao S, Taylor J-S, Sancar A. J Biol Chem. 1997;272:32580–32590. doi: 10.1074/jbc.272.51.32580. [DOI] [PubMed] [Google Scholar]
- 39.Jing Y, Kao J F-L, Taylor J-S. Nucleic Acids Res. 1998;26:3845–3853. doi: 10.1093/nar/26.16.3845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sugasawa K, Ng J M, Masutani C, Iwai S, van der Spek P J, Eker A P, Hanaoka F, Bootsma D, Hoeijmakers J H. Mol Cell. 1998;2:223–232. doi: 10.1016/s1097-2765(00)80132-x. [DOI] [PubMed] [Google Scholar]