Abstract
Rationale: Little is known about the genetic regulation of granulomatous inflammation in sarcoidosis.
Objectives: To determine if tissue gene array analysis would identify novel genes engaged in inflammation and lung remodeling in patients with sarcoidosis.
Methods: Gene expression analysis was performed on tissues obtained from patients with sarcoidosis at the time of diagnosis (untreated) (n = 6) compared with normal lung tissue (n = 6). Expression of select genes was further confirmed in lung tissue from a second series of patients with sarcoidosis and disease-free control subjects (n = 11 per group) by semi-quantitative RT-PCR. Interactive gene networks were identified in patients with sarcoidosis using Ingenuity Pathway Analysis (Ingenuity Systems, Inc., Redwood, CA) software. The expression of proteins corresponding to selected overexpressed genes was determined using fluorokine multiplex analysis, and immunohistochemistry. Selected genes and proteins were then analyzed in bronchoalveolar lavage fluid in an independent series of patients with sarcoidosis (n = 36) and control subjects (n = 12).
Measurements and Main Results: A gene network engaged in Th1-type responses was most significantly overexpressed in the sarcoidosis lung tissues, including genes not previously reported in the context of sarcoidosis (e.g., IL-7). MMP-12 and ADAMDEC1 transcripts were most highly expressed (> 25-fold) in sarcoidosis lung tissues, corresponding with increased protein expression by immunohistochemistry. MMP-12 and ADAMDEC1 gene and protein expression were increased in bronchoalveolar lavage samples from patients with sarcoidosis, correlating with disease severity.
Conclusions: Tissue gene expression analyses provide novel insights into the pathogenesis of pulmonary sarcoidosis. MMP-12 and ADAMDEC1 emerge as likely mediators of lung damage and/or remodeling and may serve as markers of disease activity.
Keywords: genetic, gene array, granuloma, lung, BAL
AT A GLANCE COMMENTARY
Scientific Knowledge on the Subject
The pathogenesis of sarcoidosis remains poorly understood, including the genetic factors regulating granulomatous inflammation at the tissue level. Genome-wide gene expression profiling can provide novel insights into interactive gene networks involved in the pathogenesis of disease.
What This Study Adds to the Field
Using gene expression profiling, highly interactive gene networks were identified, including genes and gene products not previously linked to the pathogenesis of sarcoidosis. Among these, gene and protein expression of proteases MMP-12 and ADAMDEC1 were greatly increased at the tissue level and in bronchoalveolar fluid of patients with sarcoidosis and correlated with the severity of lung disease.
The primary cause of sarcoidosis remains a mystery, and the lack of an established environmental cause, together with clustering of cases within families and in certain ethnic groups, suggests that genetic factors strongly contribute to disease pathogenesis. A number of genetic variations are implicated as potentially disease-predisposing or disease-modifying factors. Germline polymorphisms of genes regulating Th1 immune responses, particularly those of major histocompatibility complex II (1, 2), IFN-γ (3), IL-18 (4), and signal transducer and activator of transcription 1 (STAT1) (5, 6), have been identified as potential disease susceptibility genes. However, little is known about the genetic regulation of granulomatous inflammation operating at the tissue level in patients with active pulmonary sarcoidosis.
Recent technological advances provide access to enormous genetic data sets with opportunities to discover novel disease mechanisms. This approach has been applied to diseases of unknown cause to create new hypotheses relating to disease pathogenesis (7, 8) and is shown to have prognostic (9) and diagnostic applications (10, 11).
Using the global genomic approach, we sought to identify novel gene transcripts and transcript networks engaging in common biological processes operating at the tissue level in pulmonary sarcoidosis. Tissue gene transcript profiling confirmed previous investigations generally linking Th1-type immune responses to sarcoidosis; however, the finding of highly interactive gene networks featuring IL-7 and IL-15, and the corresponding protein expression, has not been previously reported. Furthermore, a gene network regulating macrophage-derived proteases, including matrix metalloproteinase 12 (MMP-12), a potent elastase, and ADAM-like, decysin 1 (ADAMDEC1), a recently discovered member of the metalloproteinase family, were dramatically overexpressed in the lungs of patients with sarcoidosis. Expression of MMP-12 protein colocalized to macrophages within lung granulomas and was associated with local depletion of elastin fibers. Moreover, increased MMP-12 and ADAMDEC1 gene and protein expression in bronchoalveolar lavage (BAL) samples were associated with more advanced disease. Some of the results of these studies have been previously reported in abstract form (12, 13).
METHODS
Patient Recruitment
Gene expression analysis was performed on lung tissues from patients who met the operational diagnosis of sarcoidosis. Specifically, sarcoidosis samples displayed well-formed, nonnecrotizing epithelioid granuloma in the absence of identifiable infection or foreign objects, in accordance with the diagnostic criteria described in the American Thoracic Society's Joint Statement on Sarcoidosis (14). Samples exhibiting signs of infection and atypical pathological features, such as necrosis or fibrosis, were excluded from analysis. Disease-free lung tissues were obtained during surgical lung resections or in the immediate postmortem period from patients who had submitted for organ donation for the purposes of medical research. Each sample was verified to have normal lung histology by a certified pathologist. Informed consent was provided for the use of all tissues analyzed in these studies, which were performed in compliance with the universities' institutional review boards. In some cases, sarcoidosis lung tissues were obtained from diagnostic video-assisted thoracic surgery procedures. Presumably, these patients were referred to thoracic surgeons after nondiagnostic bronchoscopy or by physicians from smaller communities lacking pulmonology consultation services. For the BAL studies, subjects were recruited at the time of diagnostic bronchoscopy and were not using systemic medications for sarcoidosis. Healthy control individuals were all nonsmokers with no history of lung disease and were not on any medications at the time of the study. BAL was performed by instilling three 50-ml aliquots of warmed saline in a segmental bronchus. Processing of BAL specimens was done as previously reported (15). Viability of cells exceeded 95% as assessed by trypan blue exclusion. Total RNA was isolated from resuspended cells and stored at −80°C for further analysis.
Isolation and Validation of High-Quality RNA for Gene Chip Analysis and PCR
Total RNA was isolated from frozen lung tissue using TRIzol reagent (Invitrogen Corp., Carlsbad, CA) according to the manufacturer's protocol and as described in the online supplement. The integrity of purified total RNA samples was assessed qualitatively on an Agilent 2100 Bioanalyzer (Agilent Technologies, Santa Clara, CA) and verified by the presence of two discrete electropherogram peaks corresponding to the 28S and 18S rRNA at a ratio approaching 2:1. Housekeeping genes GAPDH and CAP1 were used to assess the quality of thesynthesized, labeled cDNA and to normalize PCR results. Samples were excluded from gene array analysis if the ratio between the 3′ and 5′ signals exceeded 4, with ideal values being between 1 and 2. RNA degradation was shown to be comparable in all samples used for Affymetrix analyses (see online supplement).
Gene Array
Samples were processed using high-density oligonucleotide HG-U133 Plus 2 arrays (Affymetrix, Inc., Santa Clara, CA), which analyze the expression levels of over 47,000 transcripts and variants including 38,500 well-characterized human genes. Gene expression levels from probe intensities were estimated using a robust multiarray analysis method with quantile normalization and background correction (16).
Statistical analysis comparing the sarcoidosis and control groups was performed using BRB Array Tools software developed by the Biometric Research Branch of the U.S. National Cancer Institute (http://linus.nci.nih.gov/BRB-ArrayTools). Array data were excluded if less than 20% of expression data had at least a twofold change in either direction from the gene's median value; 6,533 probe sets passed the filtering criteria. To identify differentially expressed genes between the two groups, a modified t test with random variance model was applied. The random variance model is an improvement over the standard t test because it permits sharing information among genes about within-class variation without assuming that all genes have the same variance (17). To adjust for multiple test comparisons, the Benjamin and Hochberg false discovery rate, which controls the proportion of genes expected in a list of genes by chance, was applied. Fold change between the two groups was estimated by the ratio of geometric means of intensities in each group. By first applying a t-distribution to obtain 95% confidence intervals for the difference in means of logarithmic intensities and then using an exponential transformation of these intervals, 95% confidence intervals for fold changes were obtained. A gene was identified as differentially expressed when the P value was < 0.005, the false discovery rate was <0.03, and the fold increase/decrease was ≥2.
Gene Network Analysis
Functional and network analyses of gene expression data derived from the Affymetrix HG-U133 Plus 2 chips was performed using Ingenuity Systems' IPA software (Ingenuity Systems Inc., Redwood, CA), which is described in greater detail in the Discussion. Briefly, gene expression changes are considered in the context of physical, transcriptional, or enzymatic interactions of the gene/gene products and are grouped according to interacting gene networks through the use of Ingenuity Pathways Analysis. The score assigned to any given gene network takes into account the total number of molecules in the data set, the size of the network, and the number of “network eligible” genes/molecules in the data set. The network score is based on the hypergeometric distribution and is calculated with the right-tailed Fisher's exact test. The network score is the negative log of this P value.
Semi-Quantitative RT-PCR and Real-Time PCR
Expression of select human genes (ADAMDEC1, MMP12, HLA-DRB1, STAT1, CXCL9, CCL5, IL-7, IL-15, CAP1, and GAPDH) was confirmed in an additional set of lung tissue samples using semi-quantitative reverse transcriptase–polymerase chain reaction (RT-PCR), as detailed in the online supplement. Isolated tissue RNA was reverse-transcribed to cDNA and amplified using RT-PCR after first determining the optimal number of cycles needed for appropriate PCR amplification. The PCR product corresponding to each gene for every sample was further examined by high-resolution gel electrophoresis and semi-quantitated by band intensity analysis normalized to that of the housekeeping genes.
Human gene expression for MMP12 and ADAMDEC1 was determined in cells derived from BAL using real-time PCR with normalization to that of GAPDH (see the online supplement).
Cytokine Analysis
Multiplex quantification of cytokine expression was conducted in lung tissue homogenates. Frozen lung samples (∼0.5 g) were processed into tissue homogenates, as described in the online supplement, and the concentration of each cytokine was analyzed from a 50-μl sample. A custom-made Bio-Plex Human Cytokine panel that included IL-1β, IL-1ra, IL-2, IL-6, IL-7, IL-8, IL-10, IL-12 (p40), IL-12 (p70), IL-13, IL-15, granulocyte macrophage colony-stimulating factor (GM-CSF), macrophage colony-stimulating factor (M-CSF), macrophage inflammatory protein-1 (MIP-1), and tumor necrosis factor (TNF)-α was used according to the manufacturer's recommendations using the Bio-Plex 200 Analysis System (Bio-Rad Laboratories, Inc., Hercules, CA). Concentrations determined from standards were normalized per mg tissue homogenate protein.
Immunohistochemistry
Frozen lung tissue samples obtained from patients with sarcoidosis and control subjects were processed and cut at 4 μm for slide preparation and immunohistochemical staining according to standard techniques. Immunoreactivity was evaluated using the following anti-human mouse monoclonal primary antibodies: anti–MMP-12 (1:150; R&D Systems, Inc., Minneapolis, MN), anti-CD68 (1:150; Dako North America, Inc., Carpinteria, CA), anti-ADAMDEC1 (1:150; Abnova Corp., Taipei City, Taiwan), and anti–TIMP-1 (1:150; R&D Systems, Inc.).
Tissue Elastin Staining
Unstained slides prepared from the frozen lung tissue samples as indicated above were routinely stained and evaluated for the presence of elastin fibers using Verhoeff's elastic stain and van Gieson's counterstain according to standard techniques (18).
MMP-12 and ADAMDEC1 Protein Expression
Bronchoalveolar lavage fluid (BALF) was concentrated tenfold (by volume) using Amicon Ultra-15 centrifugal filters (Millipore, Burlington, MA). The total protein content was determined with the bicinchoninic acid protein assay (Pierce Biotechnology, Inc., Rockford, IL). For Western blot analysis, 10 μg of total protein was loaded per lane as previously described (19), and protein detection was determined using an anti-human rabbit polyclonal antibody to MMP-12 (Abcam Inc., Cambridge, MA) and an anti-human mouse polyclonal antibody to ADAMDEC1 (Novus Biologicals, Inc., Littleton, CO). Densitometry was assessed using ImagePro software with between-group differences assessed by Tukey's test.
Characteristics of Patient Populations Tested
Whole-genome RNA expression in granulomatous pulmonary tissue was profiled in six patients with active pulmonary sarcoidosis, which was then compared with the profiles in normal lungs from six control subjects, as our training set. All of the patients included in the microarray experiments were recruited from the greater Columbus, Ohio area. Demographic data for each experimental group are summarized in Table 1 (Group 1). There was a trend toward younger age, with whites outnumbering African Americans in the sarcoidosis group, which is in keeping with regional trends. The control group included histologically normal lung tissue obtained from patients undergoing lung resection for benign tumors or noninflammatory infiltrates (n = 4) and samples obtained from organ donors (n = 2). The percentage of smokers in the control group was higher, although this did not achieve statistical significance. All patients were HIV negative. Genome-wide gene expression results were then validated for selected genes by semi-quantitative RT-PCR in a second series of subjects (patients with sarcoidosis [n = 11] and control subjects [n = 11]) who were recruited from the same demographic group (Table 1, Group 2). For the patients with sarcoidosis, the lung biopsy material was obtained by surgical or transbronchial biopsy, whereas disease-free lung tissues were obtained from organ donors or from those who had undergone resection of a lung tumor. With respect to the latter, histologically normal lung tissue was obtained at least 1 cm from the tumor.
TABLE 1.
PATIENT DEMOGRAPHICS FOR TISSUE GENE EXPRESSION ANALYSES
| Variable | Control (n = 6) | Sarcoidosis (n = 6) | Control (n = 11) | Sarcoidosis (n = 11) |
|---|---|---|---|---|
| Age, years* | 51 ± 10 | 41 ± 9 | 62 ± 8 | 47 ± 7 |
| Sex (male/female) | 3/3 | 2/4 | 6/5 | 4/7 |
| Race (white/black/other) | 4/2/0 | 4/2/0 | 8/3/0 | 6/5/0 |
| Smoking history (yes/no/unknown) | 5/1/0 | 3/2/1 | 7/3/1 | 5/4/2 |
| PFT data (% predicted)* | ||||
| FVC | 85 ± 13 | 83 ± 14 | ||
| TLC | 84 ± 11 | 89 ± 12 | ||
| DlCO | 71 ± 16 | 73 ± 20 | ||
| CXR stage | ||||
| 1 | 0 | 0 | ||
| 2 | 4 | 7 | ||
| 3 | 2 | 4 | ||
| 4 | 0 | 0 |
Definition of abbreviations: CXR = chest X-ray; PFT-pulmonary function test.
Mean ± SD.
Another independent group of patients with sarcoidosis (n = 36) and healthy control subjects (n = 12) underwent bronchoscopy and BAL. These patients were recruited by informed consent at the Cleveland Clinic Foundation (Cleveland, OH); all were nonsmokers, and none was using systemic immunosuppressants before bronchoscopy. The clinical characteristics of these patients are summarized in Table 2. Some subjects were not used for all studies due to availability of samples. The patients with sarcoidosis were prospectively stratified into “severe” and “nonsevere” phenotypes based on whether there was an indication for systemic treatment of sarcoidosis at the time of bronchoscopy or any time during follow-up (19, 20). Treatment decisions were made at the discretion of the treating physician; all samples were prospectively phenotyped before inclusion in these studies. Subjects with unclear treatment indications or indeterminant follow-up data were not included. In brief, the decision to treat for pulmonary disease was predicated upon the demonstration of deteriorating pulmonary physiology (i.e., 10% decline in serial FVC) or substantial pulmonary symptoms in the presence of significantly abnormal pulmonary function tests. All but one of the patients in the severe group had advanced Scadding chest X-ray (CXR) stages (II–IV). The median chest X-ray follow-up for these subjects was 29 months.
TABLE 2.
CHARACTERISTICS OF PATIENTS INCLUDED IN BAL ANALYSES
| Sarcoidosis
|
|||
|---|---|---|---|
| Variable | Control (n = 12) | Nonsevere (n = 18) | Severe (n = 18) |
| Age, years* | 34 ± 9 | 39 ± 7 | 48 ± 9 |
| Sex (male/female) | 6/6 | 14/4 | 10/8 |
| Race (white/black/other) | 7/4/1 | 16/1/1 | 8/10/0 |
| Smoking history (none/former/active) | 10/2/0 | 13/3/2 | 16/1/1 |
| BAL differential* | |||
| Lymphocytes, % | 5 ± 4 | 16 ± 13 | 19 ± 13 |
| PMNs, % | 1 ± 2 | 1 ± 1 | |
| CXR stage | |||
| 1 | 10 | 1 | |
| 2 | 7 | 8 | |
| 3 | 1 | 2 | |
| 4 | 0 | 7 | |
| FVC, % predicted | n/a | 94 ± 15 | 79 ± 22 |
| DlCO, % predicted* | n/a | 95 ± 17 | 77 ± 22 |
Definition of abbreviations: BAL = bronchoalveolar lavage; CXR = chest X-ray; PMN = polymorphonuclear leukocyte.
Mean ± SD.
RESULTS
General Characteristics of Differentially Expressed Genes in Lung Tissue from Patients with Sarcoidosis and the Control Group
We were interested in identifying functionally related genes that distinguished the patients with sarcoidosis from the control subjects. Using the gene expression cutoffs described above, it was observed that the permutation P values were < 0.02 for the patients with sarcoidosis versus the control subjects. There were 436 differentially expressed transcripts representing 319 unique genes (i.e., by at least twofold) in the patients with sarcoidosis compared with the control subjects. The false discovery rate was <3%. The gene list is available for review on the National Center for Biotechnology Information's Gene Expression Omnibus (http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc).
Interacting Gene Networks Overexpressed in Lungs of Patients with Sarcoidosis Relative to Control Subjects
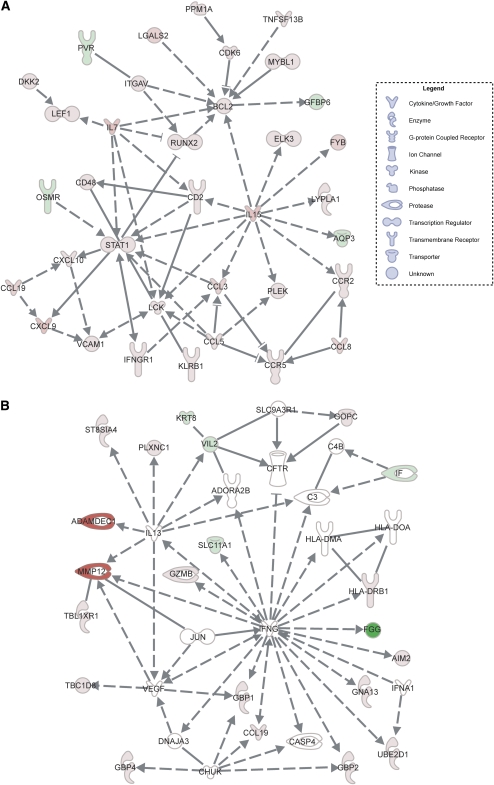
A unique feature of the IPA tool is the capacity to prioritize the gene expression data according to gene networks that are known to interact, as reflected by the capacity of the genes or gene products (e.g., enzymes) to influence the function of one another. This interacting network of genes, referred to as the “interactome,” potentially provides novel insight into the disease mechanism. For instance, interactive gene networks corresponding to cell movement and immune functions were most highly overexpressed in the lung tissues of patients with sarcoidosis (Figure 1A), among which STAT1, IL-7, IL-15, and specific chemokine genes (CCR5, CXCL9) are shown to be highly engaged. The network score was 61, indicating that there was a 10−61 chance of this many genes being present in the network by random chance (see Methods). MMP12 and ADAMDEC1, which are represented in a different gene network (network score >13), were among the genes most highly overexpressed (>25-fold) in the lungs of patients with sarcoidosis. These genes are regulated by IL-13 (Figure 1B).
Figure 1.
Interactive gene networks in lung tissues derived from patients with sarcoidosis. (A) The gene network most highly overrepresented in the sarcoidosis group relative to control subjects based on statistical analysis (see Methods) includes STAT1, IL7, and IL15. (B) The gene network in which the two most highly overexpressed genes (MMP12 and ADAMDEC1) in the sarcoidosis lung samples were found. Genes and gene products are represented by their common abbreviated names according to established direct (solid lines) or indirect (dashed lines) interactions and the specific nature of the interaction. These include binding (straight line), activation (arrow), inhibition (truncated line), or either activation or inhibition (truncated line with arrow). Genes that are significantly overexpressed in the sarcoidosis group relative to control subjects are highlighted in red; green denotes reduced expression, and white indicates no difference in expression level. The shape of each gene “node” denotes the function of the gene product.
Select Human Gene Expression in Lung Tissues by RT-PCR Confirms Genome-wide Affymetrix Results
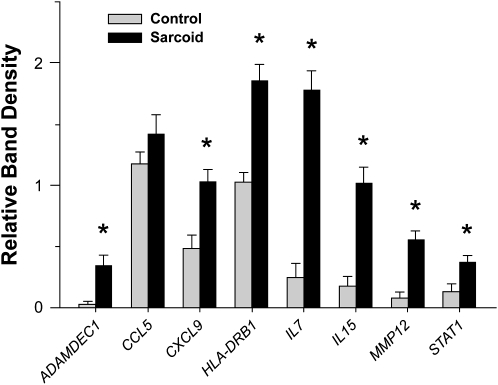
In a subsequent series of patients with sarcoidosis and control subjects, selected genes that were differentially expressed in sarcoidosis tissues in association with an interactive network (Figure 1; Table 3) were evaluated by semiquantitative RT-PCR analysis (Figure 2). The patients with sarcoidosis exhibited distinct increases in MMP12, ADAMDEC1, IL-7, and IL-15 expression levels compared with the control subjects, as was observed by Affymetrix.
TABLE 3.
STATISTICAL INFORMATION ON SELECT DIFFERENTIALLY EXPRESSED GENES AFTER GENE CHIP ANALYSIS
| Gene | Affymetrix Overall Transcript Rank | Fold Difference* (95% CI) | P Value | q Value |
|---|---|---|---|---|
| MMP12 | 1 | 29.7 (4.4–198.5) | 0.0003 | 0.0064 |
| ADAMDEC1 | 2 | 25.0 (4.3–147.8) | 0.0003 | 0.0058 |
| CXCL9 | 5 | 9.5 (2.7–33.5) | 0.0008 | 0.0103 |
| STAT1 | 19 | 4.2 (2.2–8.4) | 0.0002 | 0.0047 |
| CCL5 | 21 | 4.0 (2.2–5.7) | 0.0017 | 0.0157 |
| HLA-DRB1 | 35 | 3.3 (1.6–9.8) | 0.0001 | 0.0040 |
| IL-7 | 77 | 2.7 (1.5–4.8) | 0.0004 | 0.0077 |
| IL-15 | 170 | 2.2 (1.4–3.5) | 0.0011 | 0.0121 |
Definition of abbreviation: CI = confidence interval.
Relative to control subjects.
Figure 2.
Semi-quantitative reverse transcriptase–polymerase chain reaction (RT-PCR) analysis demonstrates enhanced select gene expression in lung tissues of patients with sarcoidosis. Semi-quantitative RT-PCR analysis was performed on lung samples obtained from in independent series of patients with sarcoidosis (n = 11) and control subjects (n = 11), the clinical characteristics of which are shown in Table 1 (mean ± SEM; *P < 0.05; Wilcoxon Rank-Sum test with a multiple comparison adjustment using the Bonferonni-Holm's procedure). See online supplement for actual RT-PCR images.
Cytokine Analysis in Lung Tissues of Patients with Sarcoidosis
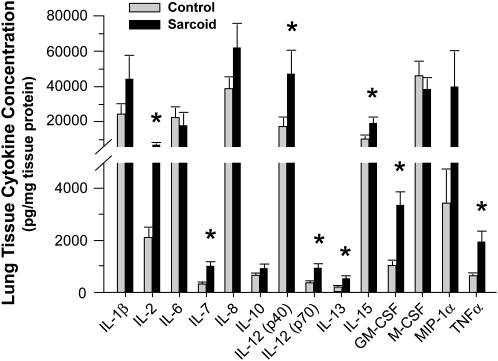
In keeping with the tissue gene expression data, quantitative cytokine protein analysis confirmed increased expression of IL-7 and IL-15 proteins in lung tissues relative to control subjects. In addition, IL-1ra (288.3 ± 40.0 vs. 113.0 ± 15.3 ng/mg tissue protein; P < 0.05), IL-2, IL-12 (p40), IL-12 (p70), IL-13, GM-CSF, and TNF-α protein expression were significantly increased in the patients with sarcoidosis (Figure 3), even though there was no significant change in their respective levels of mRNA expression. By contrast, and in keeping with gene expression results, tissue concentrations of IL-1β, IL-6, IL-8, IL-10, M-CSF, and MIP-1α were not statistically different in the patients with sarcoidosis relative to control subjects.
Figure 3.
Cytokine analysis in lung tissues of patients with sarcoidosis. Bio-Plex Human Cytokine Bead Array analysis of lung tissue samples identified a number of proinflammatory cytokine proteins that were significantly elevated in the lungs of patients with sarcoidosis relative to control subjects (mean ± SEM; *P < 0.05; Wilcoxon Rank-Sum test with a multiple comparison adjustment using the Bonferonni-Holm's procedure).
Lung MMP-12 and ADAMDEC1 Protein Expression Levels Are Highest in the Vicinity of Granulomatous Inflammation
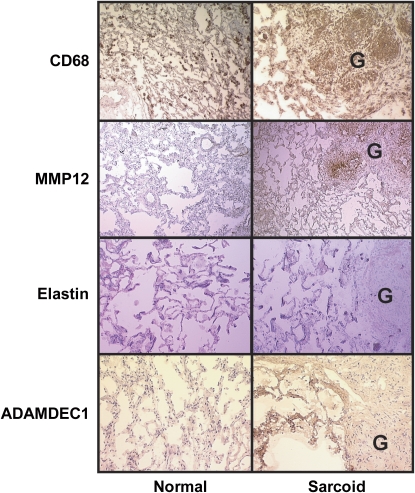
The expression of MMP-12 protein was greatest in the areas of active granulomatous inflammation and colocalized with CD68 expression (Figure 4), identifying macrophages, which are known to produce MMP-12 (21). Tissue inhibitor of metalloproteinase 1 (TIMP-1), an endogenous inhibitor of MMP-12 (22), was uniformly expressed throughout the lung tissue, including the granulomas, and the expression level did not differ among the two groups (not shown). To the extent that high levels of MMP-12 protein expression were not opposed by increases in TIMP-1, it follows that the target of MMP-12, elastin, would be diminished in areas of MMP-12 overexpression (Figure 4). In contrast to MMP-12, ADAMDEC1 expression was greatest in lung tissues adjacent to the granulomas (Figure 4).
Figure 4.
Immunohistochemical analyses of MMP-12 and ADAMDEC1 protein expression in pulmonary sarcoidosis. Immunohistochemical staining of frozen lung samples for CD68, MMP-12, elastin, and ADAMDEC1 from representative control subjects and patients with sarcoidosis. Granulomas labeled “G.” Comparison of anti-CD68 immunostaining with anti–MMP-12 showed colocalization within the granulomas of sarcoidosis tissues. Elastin staining was depleted in granulomas but was distributed uniformly throughout the lung tissue otherwise. Anti-ADAMDEC1 immunostaining was uniformly distributed at lower levels in control lungs but was expressed at high levels in tissue adjacent to granulomas in the lungs of patients with sarcoidosis.
MMP12 and ADAMDEC1 Gene Expression are Elevated in the BALF of Patients with Sarcoidosis and Correlate with Clinical Phenotype
The expression of MMP12 and ADAMDEC1 was determined in cells from BALF obtained from an independent series of patients with sarcoidosis and healthy control subjects (Figure 5). As in the lung tissue, higher MMP12 expression was observed in the BAL cells obtained from patients with active pulmonary sarcoidosis (3.03-fold higher in sarcoidosis; P < 0.004). A trend was also observed for elevations of ADAMDEC1 expression compared with healthy control subjects (2.38-fold increase; P = 0.07). Because macrophages are the major producers of MMP-12 and ADAMDEC1 (23), they are the likely source of these gene transcripts. In this regard, there was no significant correlation between the percentage of lymphocytes or neutrophils and the abundance of mRNA to either protein.
Figure 5.
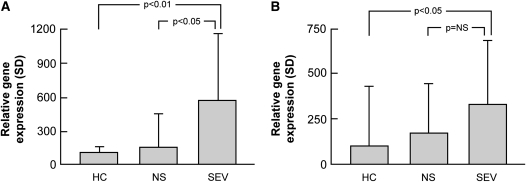
MMP12 and ADAMDEC1 are overexpressed in bronchoalveolar cells from patients with sarcoidosis. MMP12 and ADAMDEC1 (gene) expression were determined by real-time PCR in bronchoalveolar lavage cells obtained from an independent series of normal subjects and patients with sarcoidosis. RNA expression was normalized to housekeeping genes as described in Methods. Sarcoidosis was associated with (A) increases in MMP12 and (B) ADAMDEC1 expression in bronchoalveolar lavage cells, which was highest in those with severe disease (mean ± SD; Tukey's test). HC = healthy control subjects; NS = nonsevere; SEV = severe.
There was also a significant correlation between BAL MMP12 expression and sarcoidosis phenotype (Figure 5), and there was a nonsignificant trend (1.92-fold higher expression; P = 0.17) toward increased ADAMDEC1 expression in the BAL cells of patients with sarcoidosis with severe compared with nonsevere disease.
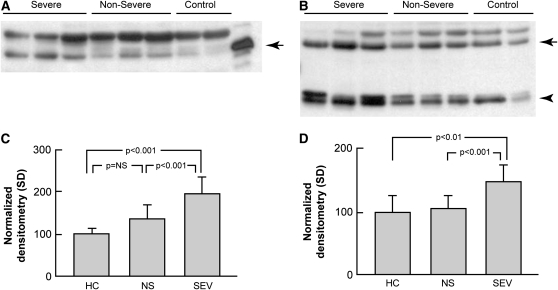
Similar to the gene expression data, Western blot analysis of acellular BALF from patients with sarcoidosis was enriched in MMP-12 (Figures 6A and 6C; P = 0.003 vs. control subjects) and ADAMDEC1 (Figures 6B and 6D; P = 0.056). There were differences apparent between sarcoidosis phenotypes, with more unbound MMP-12 (P < 0.001) and ADAMDEC1 (P < 0.001) in the BALF from severe versus nonsevere phenotypes.
Figure 6.
Soluble MMP-12 and ADAMDEC1 are elevated in acellular bronchoalveolar lavage (BAL) in sarcoidosis. Western blot analysis of concentrated BAL fluid from patients with sarcoidosis (n = 12) and healthy control subjects (n = 8). Gel loading was normalized to 10 μg total protein per lane using the bicinchoninic acid protein assay. Images are representative of two independent sample sets. (A) For MMP-12, recombinant pro–MMP-12 was used as a control with the expected band seen at 54 kD (arrow). (B) For ADAMDEC1, two bands were visualized: a species around the expected size (53 kD, arrow) and a dimer of approximately 28 kD that represents a cleavage product (arrowhead). The unlabeled bands likely represent nonspecific binding. (C and D) Densitometry results. (C) The MMP-12 level was higher overall in patients with sarcoidosis compared with healthy control subjects (1.65-fold; P = 0.003; Student's t test); (D) the increase was nearly significant for ADAMDEC1 (1.28-fold; P = 0.056). Stratification of the subjects by clinical phenotype revealed that MMP12 and ADAMDEC1 levels were elevated in severe versus nonsevere subjects (1.45-fold and 1.39-fold, respectively; P < 0.001 for both; Tukey's test). HC = healthy controls; NS = nonsevere; SEV = severe.
Lack of Correlation between MMP12 or ADAMDEC1 Expression with Age, Race, or Gender
Given the unequal distribution of the sarcoidosis and control populations with respect to age, race, and gender (see Tables 1 and 2) and the limited size of the groups, multivariate regression analyses of MMP12 and ADAMDEC1 expression with respect to these clinical variables was not feasible. However, a negative correlation by univariate regression analysis can identify genes that are unlikely to be influenced by a specific clinical variable (e.g., age) (24). In this regard, regression analysis of tissue MMP12 and ADAMDEC1 expression derived from 22 subjects, represented in Figure 2, did not correlate with age (r2 values of 0.06 and 0.13, respectively; NS). Despite a bias toward female gender and African American race in the patients with sarcoidosis, nonparametric analysis (Wilcoxon rank-sum) of tissue MMP12 and ADAMDEC1 expression with respect to race and gender yielded nonsignificant relationships (P > 0.2 for all comparisons). Similar findings were obtained when BAL results were analyzed. However, there was a poor but significant correlation between age and BAL cell MMP12 expression (r2 = 0.15; P = 0.025), which was eliminated after controlling for the presence of sarcoidosis and is consistent with the bias toward more advanced age in the patients with sarcoidosis relative to the control subjects (see Table 2). These observations should be considered preliminary due to the limited sample sizes.
DISCUSSION
This genome-wide expression analysis of lung tissue derived from patients with active pulmonary sarcoidosis identified interacting gene networks that are likely to be critical for the maintenance of granulomatous inflammation of the lung and associated lung damage. Network analyses tools, such as those used in these investigations, provide an unbiased interpretation of gene array data based upon established interactions between genes/gene products, thereby reducing user bias. Gene network analyses also provide a template for interpreting large datasets generated through genome-wide gene array analyses to identify relevant biological processes. Not surprisingly, many of the genes identified in the most over-represented network, such as STAT1 and CCL5 (RANTES) (see Figure 1A), are recognized to participate in Th1-type immune responses; however, there were several unexpected or previously unreported findings. For instance, IL-7 was not previously linked to pulmonary sarcoidosis but is known to play a central role in the development of granulomatous inflammation in the context of infections (25). Furthermore, the tissue expression of two macrophage-derived proteases, MMP-12 and ADAMDEC1, was dramatically increased in the setting of granulomatous lung inflammation and has not been previously reported. These findings were confirmed in an independent series of patients with sarcoidosis, wherein the expression of these proteases in the BALF was shown to correlate with disease severity.
Given the intense proinflammatory milieu attendant to pulmonary sarcoidosis and the results of previous analyses performed on BALF derived from patients with sarcoidosis, it is surprising that IL-7 and IL-15 were the only cytokine genes significantly overexpressed in patients with sarcoidosis relative to control subjects. The finding of increased IL-15 gene expression in the lungs of our patients with sarcoidosis is consistent with those of Muro and colleagues (26) and with experimental evidence linking IL-15 to macrophage–T-cell interactions in patients with active sarcoidosis (27). IL-15 expression in macrophages is regulated by IL-4 and GM-CSF, a mechanism favoring granuloma formation in the context of infections (28). IL-15 is highly expressed in alveolar macrophages derived from patients with sarcoidosis and is shown to induce the release of IFN-γ from T cells (27). Furthermore, IL-15 regulates the expression of CXCR3, one of the genes overexpressed in sarcoidosis lung tissues (see Figure 1), in T cells derived from the lungs of patients with sarcoidosis (29). Unlike IL-15, increased expression of IL-7 has not been previously described in sarcoidosis; however, IL-7 protein is shown to be essential for the expansion of T-cell populations (25) and is expressed in the stroma surrounding infectious granulomas (30), where it is thought to promote the clearance of intracellular pathogens (31). A recent investigation indicates that IL-7 is essential for CD4+ T-cell responses to viral infection and that IL-15 plays an accessory role in this process (32). It is unclear whether these cytokines are essential for pulmonary granuloma formation in the context of sarcoidosis, but their known functions and central positioning within the gene network that is most overrepresented in the lungs of patients with sarcoidosis predicts that they would.
Previous gene expression analyses have shown that gene and protein expression are not reliably linked, and our findings support this. Elevated levels of Th1-related cytokines, including IFN-γ and IL-2, have been previously reported in BALF from patients with sarcoidosis (31), and increased IL-2 gene expression has been observed in alveolar lymphocytes but does not occur in circulating lymphocytes of patients with sarcoidosis (32). Despite no significant increase in IL-2 gene expression, IL-2 protein levels were increased in the lung tissue of patients with sarcoidosis. Furthermore, MMP12 and ADAMDEC1, which were highly expressed in lung tissues, are regulated by IL-13 (33, 34). IL-13 was not overexpressed at the gene transcript level, whereas protein levels were significantly elevated in the patients with sarcoidosis. In this regard, recent studies have shown that protein expression is not directly related to mRNA expression due, in part, to the actions of microRNAs, which are capable of imperfectly base pairing with the target mRNA, leading to mRNA translational inhibition (35). The role of microRNAs and other mechanisms influencing gene translation in the lungs during sarcoidosis has not been considered but is likely to play a significant role in disease pathogenesis, including the regulation of inflammatory pathways.
Among the gene candidates identified by gene array that are most likely to directly contribute to lung pathology, MMP12 and ADAMDEC1 are of particular interest because both were highly overexpressed in lung tissue (>25-fold) and BAL cells (>2-fold) of patients with pulmonary sarcoidosis, and both could contribute to lung injury. MMP12, which encodes a potent tissue elastase, is generally expressed in macrophages, where it can be up-regulated by several mediators, including IL-13, and suppressed by TGF-β. It is known that MMP-12 participates in lung remodeling (36) and fibrosis responses (37), and there is strong evidence linking MMP-12 to alveolar destruction in tobacco-induced models of emphysema in mice (38). Macrophage-specific transgenic overexpression of MMP-12 in rabbits resulted in larger carrageenan-induced granuloma size (39). In cutaneous granulomas from patients with sarcoidosis, expression of MMP-12 has been reported (40); however, a specific role in pulmonary sarcoidosis has not been previously identified. We demonstrate for the first time that MMP-12 is overexpressed at the site of pulmonary granulomatous inflammation. Moreover, the gene and protein expression levels of TIMP-1, an important natural inhibitor of MMP-12 (37), were unchanged at the site of granulomatous inflammation, and elastin fibers in the vicinity of MMP-12 expression were depleted, suggesting that enzymatic actions of MMP-12 were increased. Together with the finding of increased expression of MMP-12 in the BALF in more severe cases of sarcoidosis, these findings suggest that MMP-12 plays an important role in facilitating lung remodeling, possibly by recruiting inflammatory cells (41) and promoting fibrosis (37).
The role of ADAMDEC1 in the pathogenesis of lung disease is less clear. ADAMDEC1 is a metalloprotease expressed in macrophages and dendritic cells (21) of which little is known. It is inducible by LPS (42) and is highly expressed in unstable carotid artery plaques (43). In contrast to MMP-12, which was most highly expressed within granulomas, ADAMDEC1 was highly expressed in lung tissue adjacent to the granulomas. It is possible that the ADAMDEC1 observed in the BALF was partially derived from stromal cells, rather than alveolar macrophages. Further investigations are needed to establish the roles of MMP-12 and ADAMDEC1 in the pathogenesis of lung injury and as biomarkers of disease activity in the context of sarcoidosis.
The relatively small size of the patient population included in the gene array analysis could be considered to be a limitation of this study; however, this concern is obviated by several factors. First, the IPA analysis approach, which assigns differentially expressed transcripts to the most biologically relevant networks based upon known molecular interactions and functions, increases the statistical power of the study. For example, the odds of a single gene transcript (e.g., STAT1) being identified by chance is reflected by a false discovery rate of <3% (see Methods). However, the probability that multiple differentially expressed transcripts within our data set would associate with a given gene network is proportionate to the total number of differentially expressed genes, the size of the gene network, and the number of genes that are associated with the gene network. This approach allows for the identification of biologically relevant genes in relatively small experimental groups (44, 45). Thus, despite the fact that the gene array data depicted in Figure 1 was derived from only six patients with sarcoidosis, the statistical probability that this particular cogent gene network would be found merely by chance is extremely low (e.g., 10−61 for the gene network shown in Figure 1A). This point is further substantiated by the confirmation of increased expression of selected genes in additional lung (11 patients) and BALF samples (36 patients).
In summary, this genome-wide oligonucleotide expression analysis provides a number of novel insights into the pathogenesis of pulmonary sarcoidosis. As expected, many of the genes or products of genes overexpressed in sarcoidosis participate in Th1-type antigen responses, including HLA-DRB1, which has been specifically linked to the pathogenesis of sarcoidosis (46, 47). The most significant finding of this analysis was the identification of highly interactive gene networks, which regulate the expression of granuloma-promoting signaling proteins (STAT1), inflammatory cytokines (IL-7, IL-15), and proteolytic enzymes (MMP-12, ADAMDEC1), that are operating within the lung compartment of patients with sarcoidosis. It is unclear whether the genetic forces driving pulmonary sarcoidosis are the same for other affected tissues. However, the expression levels of MMP-12 and ADAMDEC1 in the BALF were shown to correlate with disease severity. As such, it is interesting to speculate that manipulation of genes or gene products identified in these analyses would alter the course of the disease. Additional studies are needed to confirm whether tissue gene expression profiles have prognostic and diagnostic applications in the setting of pulmonary sarcoidosis.
Supplementary Material
Acknowledgments
The authors thank the Midwest Division of the Cooperative Human Tissue Network (CHTN), the Lifeline of Ohio, a state-supported organ and tissue procurement agency, and the Information Warehouse of the Ohio State University Medical Center for assisting with the procurement of tissue samples and related clinical data. The authors also thank Xiuping Liu of the Ohio State University Comprehensive Cancer Center Microarray Core Facility for help with the preparation of samples for gene array analysis; Susie Jones and the OSU Medical Center Pathology Core Facility for assistance with the tissue immunohistochemistry; and Timothy Eubanks for his help with data presentation.
Supported by NIH grants HL077466 (E.D.C. and C.E.), HL081538 (D.A.C.), and HL083468 (K.S.K.); the Foundation for Sarcoidosis Research (D.A.C. and E.D.C.); and Clarian Health Partners, Indianapolis, Indiana (K.S.K.). C.E. is a recipient of the Doris Duke Distinguished Clinical Scientist Award.
This article has an online supplement, which is available from the issue's table of contents at www.atsjournals.org
Originally Published in Press as DOI: 10.1164/rccm.200803-490OC on February 12, 2009
Conflict of Interest Statement: None of the authors has a financial relationship with a commercial entity that has an interest in the subject of this manuscript.
References
- 1.Sharma SK, Balamurugan A, Pandey RM, Saha PK, Mehra NK. Human leukocyte antigen-DR alleles influence the clinical course of pulmonary sarcoidosis in Asian Indians. Am J Respir Cell Mol Biol 2003;29:225–231. [DOI] [PubMed] [Google Scholar]
- 2.Mrazek F, Holla LI, Hutyrova B, Znojil V, Vasku A, Kolek V, Welsh KI, Vacha J, du Bois RM, Petrek M. Association of tumour necrosis factor-alpha, lymphotoxin-alpha and HLA-DRB1 gene polymorphisms with Lofgren's syndrome in Czech patients with sarcoidosis. Tissue Antigens 2005;65:163–171. [DOI] [PubMed] [Google Scholar]
- 3.Akahoshi M, Ishihara M, Remus N, Uno K, Miyake K, Hirota T, Nakashima K, Matsuda A, Kanda M, Enomoto T, et al. Association between IFNA genotype and the risk of sarcoidosis. Hum Genet 2004;114:503–509. [DOI] [PubMed] [Google Scholar]
- 4.Zhou Y, Yamaguchi E, Hizawa N, Nishimura M. Roles of functional polymorphisms in the interleukin-18 gene promoter in sarcoidosis. Sarcoidosis Vasc Diffuse Lung Dis 2005;22:105–113. [PubMed] [Google Scholar]
- 5.Vidal M, Ramana CV, Dusso AS. Stat1-vitamin D receptor interactions antagonize 1,25-dihydroxyvitamin D transcriptional activity and enhance stat1-mediated transcription. Mol Cell Biol 2002;22:2777–2787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tanaka G, Matsushita I, Ohashi J, Tsuchiya N, Ikushima S, Oritsu M, Hijikata M, Nagata T, Yamamoto K, Tokunaga K, et al. Evaluation of microsatellite markers in association studies: a search for an immune-related susceptibility gene in sarcoidosis. Immunogenetics 2005;56:861–870. [DOI] [PubMed] [Google Scholar]
- 7.Calvano SE, Xiao W, Richards DR, Felciano RM, Baker HV, Cho RJ, Chen RO, Brownstein BH, Cobb JP, Tschoeke SK, et al. Inflammation and Host Response to Injury Large Scale Collaborative Research Program. A network-based analysis of systemic inflammation in humans. Nature 2005;437:1032–1037. [DOI] [PubMed] [Google Scholar]
- 8.Ergun A, Lawrence CA, Kohanski MA, Brennan TA, Collins JJ. A network biology approach to prostate cancer. Mol Syst Biol 2007;3:82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Korkola JE, Baveri E, DeVries S, Moore DH II, Hwang ES, Chen YY, Estap AL, Chew KL, Jensen RH, Waldman FM. Identification of a robust gene signature that predicts breast cancer outcome in independent data sets. BMC Cancer 2007;7:61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Takahashi M, Yang XJ, McWhinney S, Sano N, Eng C, Kagawa S, Teh BT, Kanayama HO. cDNA microarray analysis assists in diagnosis of malignant intrarenal pheochromocytoma originally masquerading as a renal cell carcinoma. J Med Genet 2005;42:e48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Aldred MA, Huang Y, Liyanarachchi S, Pellagata NS, Gimm O, Jhiang S, Davuluri RV, de la Chapelle A, Eng C. Papillary and follicular thyroid carcinomas show distinctly different microarray expression profiles and can be distinguished by a minimum of five genes. J Clin Oncol 2004;22:3531–3539. [DOI] [PubMed] [Google Scholar]
- 12.Culver DA, Farver CF, Abraham S, Swaisgood C, Moeller S, Barna BP, Thomassen MJ, Bonfield TL. Nuclear receptor corepressor and PPAR gamma in severe sarcoidosis [abstract]. Am J Respir Crit Care Med 2007;175:A186. [Google Scholar]
- 13.Crouser ED, Huff JE, Wewers MD, Knox KS, Ross P, Abbas A, Eng C. Analysis of lung gene expression in sarcoidosis and granulomatous infections [abstract]. Am J Respir Crit Care Med 2007;175:A185. [Google Scholar]
- 14.American Thoracic Society. Statement on sarcoidosis. Am J Respir Crit Care Med 1999;160:736–755. [DOI] [PubMed] [Google Scholar]
- 15.Culver DA, Barna BP, Raychaudhuri B, Bonfield TL, Abraham S, Malur A, Farver CF, Kavuru MS, Thomassen MJ. Peroxisome proliferator-activated receptor gamma activity is deficient in alveolar macrophages in pulmonary sarcoidosis. Am J Respir Cell Mol Biol 2004;30:1–5. [DOI] [PubMed] [Google Scholar]
- 16.Irizarry RA, Hobbs B, Collin F, Beazer-Barclay YD, Antonellis KJ, Scherf U, Speed TP. Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics 2003;4:249–264. [DOI] [PubMed] [Google Scholar]
- 17.Wright GW, Simon R. A random variance model for detection of differential gene expression in small microarray experiments. Bioinformatics 2003;19:2448–2455. [DOI] [PubMed] [Google Scholar]
- 18.Carson FL. Histotechnology: a self instructional text. Chicago: ASCP Press; 1990. p. 147–149.
- 19.Barna BP, Culver DA, Abraham S, Malur A, Bonfield TL, John N, Farver CF, Drazba JA, Raychaudhuri B, Kavuru MS, et al. Depressed peroxisome proliferator-activated receptor gamma is indicative of severe pulmonary sarcoidosis: possible involvement of interferon gamma. Sarcoidosis Vasc Diffuse Lung Dis 2006;23:93–100. [PubMed] [Google Scholar]
- 20.Baughman RP, Judson MA, Teirstein A, Yeager H, Rossman M, Knatterud GL, Thompson B. Presenting characteristics as predictors of duration of treatment of sarcoidosis. QJM 2006;99:307–315. [DOI] [PubMed] [Google Scholar]
- 21.Shapiro SD. Proteolysis in the lung. Eur Respir J 2003;22:30S–32S. [DOI] [PubMed] [Google Scholar]
- 22.Roberts AB. Medicine: smoke signals for lung disease. Nature 2003;422:130–131. [DOI] [PubMed] [Google Scholar]
- 23.Mueller CG, Rissoan MC, Salinas B, Ait-Yahia S, Ravel O, Bridon JM, Briere F, Lebacque S, Liu YJ. Polymerase chain reaction selects a novel disintegrin proteinase from CD40-activated germinal center dendritic cells. J Exp Med 1997;186:655–663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zapala MA, Schork NJ. Multivariate regression analysis of distance matrices for testing associations between gene expression patterns and related variables. Proc Natl Acad Sci USA 2006;103:19430–19435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Guimond M, Fry TJ, Mackall CL. Cytokine signals in T-cell homeostasis. J Immunother 2005;28:289–294. [DOI] [PubMed] [Google Scholar]
- 26.Muro S, Taha R, Tsicopoulos A, Olivenstein R, Tonnel AB, Christodoulopoulos P, Wallaert B, Hamid Q. Expression of IL-15 in inflammatory pulmonary diseases. J Allergy Clin Immunol 2001;108:970–975. [DOI] [PubMed] [Google Scholar]
- 27.Agostini C, Trentin L, Perin A, Facco M, Siviero M, Piazzo F, Basso U, Adami F, Zambello R, Semenzato G. Regulation of alveolar macrophage-T cell interactions during sarcoid inflammatory process. Am J Physiol 1999;277:L240–L250. [DOI] [PubMed] [Google Scholar]
- 28.Maeurer M, Selinger B, Trinder P, Gerdes J, Seitzer U. Interleukin-15 in mycobacterial infection of antigen-presenting cells. Scand J Immunol 1999;50:280–288. [DOI] [PubMed] [Google Scholar]
- 29.Agostini C, Cabrelle A, Calabrese F, Bortoli M, Scquizzato E, Carraro S, Miorin M, Beghe B, Trentin L, Zambello R, et al. Role for CXCR6 and its ligand CXCL16 in the pathogenesis of T-cell alveolitis in sarcoidosis. Am J Respir Crit Care Med 2005;172:1290–1298. [DOI] [PubMed] [Google Scholar]
- 30.Rossi MI, Dutra HS, El-Cheikh MC, Bonomo A, Borojevic R. Extramedullar B lymphopoiesis in liver schistosomal granulomas: presence of the early stages and inhibition of the full B cell differentiation. Int Immunol 1999;11:509–518. [DOI] [PubMed] [Google Scholar]
- 31.Maeurer MJ, Trinder P, Hommel G, Walter W, Freitag K, Atkins D, Storkel S. Interleukin-7 or interleukin-15 enhances survival of Mycobacterium tuberculosis-infected mice. Infect Immun 2000;68:2962–2970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lenz DC, Kurz SK, Lemmens E, Schoenberger SP, Sprent J, Oldstone MB, Homann D. IL-7 regulates basal homeostatic proliferation of antiviral CD+T cell memory. Proc Natl Acad Sci USA 2004;101:9357–9362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pouladi MA, Robbins CS, Swirski FK, Cundall M, McKenzie ANJ, Jordana M, Shapiro SD, Stampfli MR. Interleukin-13-dependent expression of matrix metalloproteinase-12 is required for the development of airway eosinophilia in mice. Am J Respir Cell Mol Biol 2004;30:84–90. [DOI] [PubMed] [Google Scholar]
- 34.Scotton CJ, Martinez FO, Smelt MJ, Sironi M, Locati M, Mantovani A, Sozzani S. Transcriptional profiling reveals complex regulation of the monocyte IL-1β system by IL-13. J Immunol 2005;174:834–845. [DOI] [PubMed] [Google Scholar]
- 35.Shyu AB, Wilkinson MF, van Hoof A. Messenger RNA regulation: to translate or to degrade. EMBO J 2008;27:471–481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lanone S, Zheng T, Zhu Z, Liu W, Lee CG, Ma B, Chen Q, Homer RJ, Wand J, Rabach LA, et al. Overlapping and enzyme-specific contributions of matrix metalloproteinases-9 and -12 in IL-13-induced inflammation and remodeling. J Clin Invest 2002;110:463–474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Matute-Bello G, Wurfel MM, Lee JS, Park DR, Frevert CW, Madtes DK, Shapiro SD, Martin TR. Essential role of MMP-12 in Fas-induced lung fibrosis. Am J Respir Cell Mol Biol 2007;37:210–221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Morris DG, Huang X, Kaminski N, Wang Y, Shapiro SD, Dolganov G, Glick A, Sheppard D. Loss of integrin alpha(v)beta6-mediated TGF-beta activation causes MMP12-dependent emphysema. Nature 2003;422:169–173. [DOI] [PubMed] [Google Scholar]
- 39.Fan J, Wang X, Wu L, Matsumoto SI, Liang J, Koike T, Ichikawa T, Sun H, Shikama H, Sasaguri Y, et al. Macrophage-specific overexpression of human matrix metalloproteinase-12 in transgenic rabbits. Transgenic Res 2004;13:261–269. [DOI] [PubMed] [Google Scholar]
- 40.Vaalamo M, Kariniemi AL, Shapiro SD, Saarialho-Kere U. Enhanced expression of human metalloelastase (MMP-12) in cutaneous granulomas and macrophage migration. J Invest Dermatol 1999;112:499–505. [DOI] [PubMed] [Google Scholar]
- 41.Houghton AM, Quintero PA, Perkins DL, Kobayashi DK, Kelley DG, Marconcini LA, Mecham RP, Senior RM, Shapiro SD. Elastin fragments drive disease progression in a murine model of emphysema. J Clin Invest 2006;116:753–759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Fritsche J, Muller A, Hausmann M, Rogler G, Andreesen R, Kreutz M. Inverse regulation of the ADAM-family members, decysin and MADDAM/ADAM19 during monocyte differentiation. Immunology 2003;110:450–457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Papaspyridonos M, Smith A, Burnand KG, Yaylor P, Padayachee S, Suckling KE, James CH, Greaves DR, Patel L. Novel candidate genes in unstable areas of human atherosclerotic plaques. Arterioscler Thromb Vasc Biol 2006;26:1837–1844. [DOI] [PubMed] [Google Scholar]
- 44.Yamashiro K, Myokai F, Hiratsuka K, Yamamoto T, Senoo K, Arai H, Nishimura F, Abiko Y, Takashiba S. Oligonucleotide array analysis of cyclic tension-responsive genes in human periodontal ligament fibroblasts. Int J Biochem Cell Biol 2007;39:910–921. [DOI] [PubMed] [Google Scholar]
- 45.Keller P, Vollaard N, Babraj J, Ball D, Sewell DA, Timmons JA. Using systems biology to define the essential biological networks responsible for adaptation to endurance exercise training. Biochem Soc Trans 2007;35:1306–1309. [DOI] [PubMed] [Google Scholar]
- 46.Voorter CE, Drent M, van den Berg-Loonen EM. Severe pulmonary sarcoidosis is strongly associated with the haplotype HLA-DQB1*0602–DRB1*150101. Hum Immunol 2005;66:826–835. [DOI] [PubMed] [Google Scholar]
- 47.Rossman MD, Thompson B, Frederick M, Maliarik M, Iannuzzi MC, Rybicki BA, Pandey JP, Newman LS, Magira E, Beznik-Cizman B, et al. ACCESS Group. HLA-DRB1*1101: a significant risk factor for sarcoidosis in blacks and whites. Am J Hum Genet 2003;73:720–735. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.