Abstract
Proliferation of mammalian cells requires the coordinated function of many proteins to accurately divide a cell into two daughter cells. Several RNAi screens have identified previously uncharacterised genes that are implicated in mammalian cell division. The molecular function for these genes needs to be investigated to place them into pathways. Phenotypic profiling is a useful method to assign putative functions to uncharacterised genes. Here, we show that the analysis of protein localisation is useful to refine a phenotypic profile. We show the utility of this approach by defining a function of the previously uncharacterised gene C13orf3 during cell division. C13orf3 localises to centrosomes, the mitotic spindle, kinetochores, spindle midzone, and the cleavage furrow during cell division and is specifically phosphorylated during mitosis. Furthermore, C13orf3 is required for centrosome integrity and anaphase onset. Depletion by RNAi leads to mitotic arrest in metaphase with an activation of the spindle assembly checkpoint and loss of sister chromatid cohesion. Proteomic analyses identify C13orf3 (Ska3) as a new component of the Ska complex and show a direct interaction with a regulatory subunit of the protein phosphatase PP2A. All together, these data identify C13orf3 as an important factor for metaphase to anaphase progression and highlight the potential of combined RNAi screening and protein localisation analyses.
Keywords: esiRNA, kinetochore, shugoshin, Ska, spindle checkpoint
Introduction
Cell division and mitosis of eukaryotic somatic cells require the coordinated function of many proteins in a temporally and spatially well-orchestrated process (Nigg, 2001; Nasmyth, 2002; Varetti and Musacchio, 2008). Mitosis can be subdivided into different phases mainly depending on morphological features. The purpose of early mitosis from prophase to metaphase is the establishment of a bipolar spindle with all kinetochores attached amphitelic to spindle microtubules (Musacchio and Salmon, 2007). Proper attachment and the creation of tension at the kinetochores are believed to be the key factors for silencing of the spindle assembly checkpoint (SAC). Silencing of the SAC leads to the activation of the E3 ubiquitin–protein ligase, APC/C (Sullivan and Morgan, 2007). The APC/C is a multiprotein complex composed of at least 12 subunits, of which, among others, the subunits Cdc16 and Cdc27 are crucial for its activity (Peters, 2006; Thornton et al, 2006); it promotes execution of anaphase by polyubiquitylation of its main substrates cyclin B1 and securin, thereby targeting them for destruction by the proteasome. Degradation of cyclin B1 results in a decrease in Cdk1 activity that is required for entry into the late phases of mitosis. Loss of securin allows activation of separase required for sister chromatid separation (Sullivan and Morgan, 2007). In vertebrate cells, arm cohesion is largely lost during prophase and prometaphase in a separase-independent pathway requiring polo-like kinase 1 (Plk1) and aurora kinase B (AurkB) activity to facilitate sister chromatid resolution (Waizenegger et al, 2000; Watanabe, 2005). In contrast, centromeric cohesion is preserved until anaphase by the protein shugoshin-like 1 (SGOL1), which recruits protein phosphatase 2A (PP2A) to centromeric cohesion, thereby counter-acting phosphorylation by Plk1.
Although parts of the genes and the mechanisms that guard mammalian cell division have been identified, others remain elusive. The complexity of mammalian cell division calls for a systems-level approach to understand the sophisticated interaction and regulation of proteins involved (Kittler et al, 2008). Loss-of-function screening by RNAi is a valuable strategy for the systematic analysis of genes implicated in cell-cycle regulation, and different methods to carry out RNAi experiments in mammalian cells are available (Sachse and Echeverri, 2004). We and others have developed and successfully utilised endoribonuclease-prepared short interfering RNAs (esiRNAs) as mediator for RNAi (Yang et al, 2002; Kittler et al, 2004, 2007a; Galvez et al, 2007; Fazzio et al, 2008). As esiRNAs are highly specific, they are well suited for RNAi experiments, especially for large-scale RNAi screens (Kittler et al, 2007b). We conducted previously a genome-scale esiRNA screen on cell-cycle progression in mammalian cells (Kittler et al, 2007a), which identified many previously uncharacterised genes implicated in this process. The use of multiparametric analysis in combination with hierarchical clustering allowed the placement of some of these genes into pathways (Kittler et al, 2007a). However, it remains challenging to interpret multiparametric phenotypic data to build valid biological hypotheses, and for many uncharacterised genes the molecular role during cell division remains elusive.
Localisation is an independent indicator of gene function (Wang et al, 2008b), which may provide valuable information in addition to loss-of-function data. Antibodies are useful to determine the localisation pattern of proteins, and are commercially available for many known cell-cycle proteins. However, generating antibodies for uncharacterised genes is time-consuming and cost-intensive. Tagging of genes with fluorescent proteins is a rapid and cost-effective alternative to antibodies, which also allows the dynamic localisation of proteins in living cells, and collections of tagged genes based on cDNA constructs have been assembled (Pepperkok and Ellenberg, 2006). However, expression of tagged genes from cDNA constructs can be problematic because the genomic context of the gene is not preserved. As a consequence, the gene is often expressed at nonphysiological levels, which can lead to mislocalisation of the protein. Recently, the TransgeneOmics approach has been developed to allow rapid tagging of many genes that preserves the genomic context (Poser et al, 2008). Using recombineering technology (Muyrers et al, 2001) to tag genes, encoded on a bacterial artificial chromosome (BAC), allows the expression of genes close to the endogenous level. As a BAC usually contains all cis-regulatory elements of the promoter, 3′-UTR, and the coding region, the transgene maintains its physiological expression levels and splicing pattern (Poser et al, 2008), a feature especially interesting for genes implicated in cell-cycle control.
To test whether localisation data help to refine phenotypic profiles, we exemplified the analysis of a subcluster derived from two RNAi screens that were enriched for known regulator proteins of mitosis by using the TransgeneOmics approach. We nominated the previously uncharacterised protein, C13orf3 (also known as Rama1), to exhibit a similar localisation pattern and phenotypic features to the protein Ska1 (spindle and kinetochore associated protein 1). A detailed characterisation identified a direct interaction of C13orf3 with members of the Ska complex, described as a two-component complex, composed of Ska1 (C18orf24) and Ska2 (Fam33a), with a critical role in the maintenance of the metaphase plate and progression through mitosis (Hanisch et al, 2006). We show here that C13orf3 (Ska3) is an integral part of the Ska complex and localises to the mitotic spindle, kinetochores, and cleavage furrow during mitosis. In addition, we show that C13orf3 is required for the maintenance of a bipolar spindle. Depletion of C13orf3 leads to an arrest in a metaphase-like state with an activation of the SAC and sister chromatid separation. These findings underline the importance of C13orf3 in the mitotic progression of mammalian cells and show that the combination of phenotypic profiling and localisation data improves the predictive power helping to identify pathways for genes with important roles during cell division.
Results and discussion
Combining phenotypic profiling with protein localisation
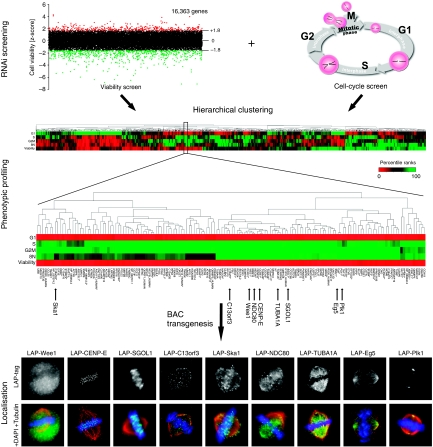
To predict functions of previously uncharacterised genes, we started with a data set from a cell-cycle esiRNA screen carried out previously in our laboratory (Kittler et al, 2007a). In this screen, a genome-wide analysis of genes implicated in cell-cycle progression was carried out using DNA content analysis combined with laser-scanning cytometry providing statistical scores (z-scores) for cell-cycle progression phenotypes (i.e., cells in G1, S, G2/M phases and 8 N) for each knockdown (Kittler et al, 2007a). To refine this data set, we carried out a genome-wide RNAi viability screen in the same cell line providing z-scores for viability of 16,363 genes (Figure 1 and Supplementary Table S1). Both data sets were combined for hierarchical clustering. All z-scores were converted into percentile ranks and these variables were used to build a hierarchical tree with an Euclidean metric, sorting the phenotypic data by similarity (Figure 1). We chose a subcluster from this phenotypic tree, which was enriched for genes with well-established mitotic functions for further analysis (Figure 1). This subcluster included known genes with important mitotic functions such as polo-like kinase 1 (Plk1), wee1-like protein kinase (Wee1), shugoshin-like protein 1 (SGOL1), centromere protein E (CENP-E), kinetochore associated protein 2 (Kntc2, NDC80), kinesin family member 11 (Eg5), spindle and kinetochore associated protein 1 (Ska1), and several tubulins, such as tubulin alpha 3 (TUBA1A) (Supplementary Table S2). The enrichment of mitosis-associated genes suggested that uncharacterised genes within this cluster might have an important role during cell division. To further refine the profile within this cluster, we determined the protein localisation in mitotic cells for known cell-cycle genes and previously uncharacterised genes using the BAC-based TransgeneOmics approach (Kittler et al, 2005b; Poser et al, 2008). BAC constructs maintain the genomic context of a gene and usually contain all cis-regulatory elements for gene expression. Furthermore, they typically integrate at low copy number, thereby allowing physiological expression of the tagged proteins (Kittler et al, 2005b; Poser et al, 2008). A modified version of the ‘localisation and affinity purification' (LAP) tag (Cheeseman and Desai, 2005) consisting of an EGFP and an S-peptide sequence was used. The analysis of images from these BAC-transgenic HeLa cell lines during mitosis confirmed the known localisation of cell-cycle-relevant proteins such as CENP-E (kinetochores), SGOL1 (centromeres), Eg5 (centrosomes, mitotic spindle), and Ska1 (kinetochores, mitotic spindle) (Figure 1), showing the utility of the TransgeneOmics approach. To improve the phenotypic profiling, we determined the localisation of 52 known and uncharacterised BAC-tagged proteins (Supplementary Table S2) from the subcluster comparing their localisation with each other to identify proteins with similar localisation patterns. Interestingly, the analysis of the uncharacterised protein, C13orf3, showed a spindle and kinetochore localisation during mitosis, most similar to the localisation pattern of Ska1 in the subcluster (Figure 1) (Hanisch et al, 2006; Rines et al, 2008). Although the phenotypic data alone were insufficient to link C13orf3 to Ska1, the combination with the localisation data predicted that Ska1 and C13orf3 might physically and/or genetically interact. To test this hypothesis, C13orf3 was selected for an in-depth analysis. Comparative sequence analysis identified putative C13orf3 orthologues in mammals, birds, amphibians, and bony fish (e.g., mouse: ENSMUSG00000021965, chicken: ENSGALG00000017128, frog: ENSXETG00000009595, and zebrafish: ENSDARG00000067746, respectively), but not in invertebrates. Structure-based bioinformatics analyses (Sippl and Flockner, 1996; Godzik, 2003) identified a Gle2-binding sequence motif (GLEBS motif) at the C-terminal region of C13orf3 (aa 345–382) (Supplementary Figure S3). Interestingly, an additional putative GLEBS motif has been proposed in the N-terminal region (aa 17–51) of C13orf3 by Gaitanos et al (E Nigg, personal communication, 2008). GLEBS motifs are present in Bub1 and BubR1 and have been structurally characterised to mediate binding to Bub3 (Larsen et al, 2007), substantiating a potential role of C13orf3 in mitosis.
Figure 1.
Strategy of comparative genomics. Genome-scale RNAi data from a cell cycle and a viability screen were combined for multiparametric hierarchical clustering. z-score values for five parameters, that is, cells in phases G1, S, G2/M, and aneuploidic cells (8N), as well as cell viability (decreased: red, increased: green), were converted in percentile ranks and used to build a hierarchical tree sorting the genes by similarity of their phenotype. A subcluster enriched with genes with important mitotic functions is shown in the blow-up. Selected examples of imaged BAC transgenic cell lines stained for α-tubulin (red), DNA (blue), and LAP-tag (green) are shown in the bottom panel.
C13orf3 localises to prominent structures during mitosis
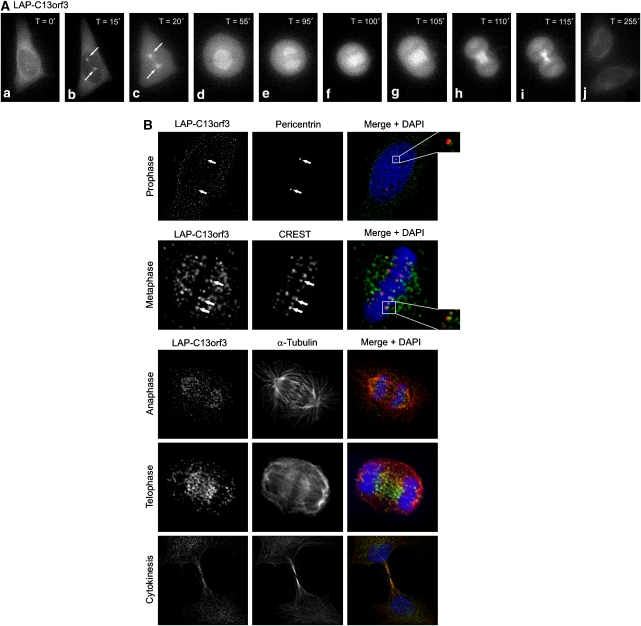
To analyse the dynamic localisation of C13orf3 during cell division and to substantiate the overlapping localisation with Ska1 observed in the profiling study (Figure 1), we used the C13orf3 BAC-transgenic HeLa cell line and imaged dividing cells using fluorescence time-lapse microscopy (Supplementary Movie S4). The protein was predominantly cytoplasmatic during interphase, with a noticeable concentration around the nuclear envelope (Figure 2Aa, j). In prophase, just before the nuclear envelope breakdown, an accumulation of C13orf3 at the centrosomes was readily detectable (Figure 2Ab and B). During prometaphase and metaphase, C13orf3 localised to the mitotic spindle as well as to the kinetochores (Figure 2Ac, d and B). Upon entry into anaphase, the fusion protein was enriched at the spindle (Figure 2Ae and B) and at the spindle midzone in late anaphase and telophase (Figure 2Af, g, and B). During cytokinesis, C13orf3 was found at the cleavage furrow (Figure 2Ah, i, and B). The localisation to important mitotic structures and the overlap with the localisation pattern of Ska1 (Hanisch et al, 2006) underlines a potential interaction of these proteins.
Figure 2.
Localisation pattern of C13orf3 in HeLa cells. (A) Selected frames from fluorescence time-lapse microscopy of HeLa cells expressing LAP-tagged C13orf3 at indicated time points are shown in interphase (a, j), prophase (b), prometaphase (c), metaphase (d), early anaphase (e), late anaphase (f), telophase (g), early cytokinesis (h), and late cytokinesis (i). (B) Immunofluorescence microscopy of LAP-C13orf3 co-stained with pericentrin, CREST, and α-tubulin antibodies during indicated cell cycle phases. Arrows and blow-ups point to areas of colocalisation.
C13orf3 is required for anaphase onset
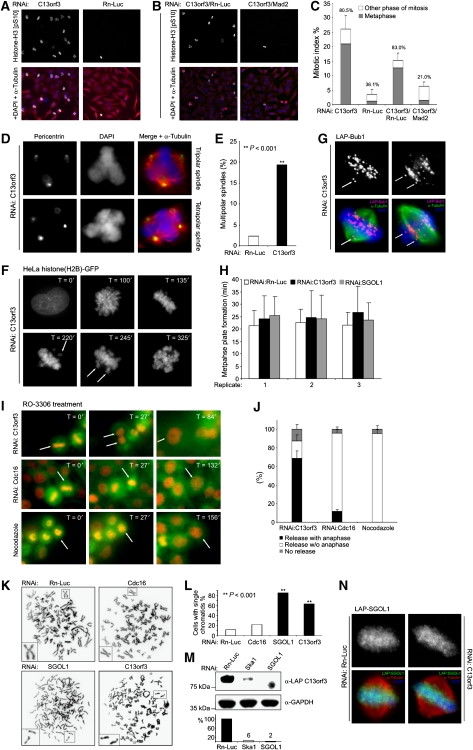
The prominent localisation of C13orf3 prompted us to carry out a more detailed phenotypic analysis. To confirm and validate findings from the genome-wide RNAi screens (Figure 1), we first repeated the DNA content analysis by flow cytometry with two independent esiRNAs targeting C13orf3 (Supplementary Table S5). For both esiRNAs, a significant cell-cycle arrest in the G2/M phase was observed 42 h post transfection (Supplementary Figure S6A and data not shown). To ensure efficient knockdown of the intended target genes by RNAi, we conducted Q-PCR and western blot analyses. For all esiRNAs, a knockdown of at least 80% was achieved at the mRNA level 24 h post transfection and a knockdown of at least 85% at the protein level 42 h post transfection, showing the efficacy of the employed esiRNAs (Supplementary Figure S7A and B). As the DNA content for cells in G2 phase and mitosis is 4 N, it is not possible to distinguish these two phases by DNA content measurements. To distinguish between a G2- and M-phase arrest, we depleted C13orf3 by RNAi and determined the mitotic index 42 h post transfection by staining for the phosphorylation of serine 10 of histone H3, a mitotic marker for chromatin condensation (Goto et al, 1999). Immunofluorescence microscopy showed an increase in the mitotic index to 26.2% upon C13orf3 depletion (3.6% for mock control), of which 80.5% of the cells were arrested in a metaphase-like state (36.1% for mock control) (Figure 3A and C). These data suggest that C13orf3 is required for anaphase onset. In addition to the metaphase arrest, we observed a significant increase in cells with tripolar and tetrapolar spindles (19 versus 2% for mock-transfected cells) (Figure 3D and E), indicating that C13orf3 might also have a function in centrosome duplication or maintenance. To obtain a dynamic description of the phenotypic consequences upon C13orf3 depletion, we carried out time-lapse microscopy analyses in a cell line expressing a histone(H2B)–GFP fusion protein. These analyses showed an apparently normal chromosome congression and a proper establishment of the metaphase plate (Figure 3F, H and Supplementary Movies S9, S10, S11, and S23). However, as early as 30 h post transfection, cells failed to maintain the metaphase plate, with individual chromosomes exiting from the aligned chromosomes (Figure 3F and Supplementary Movie S9, S10) causing a mitotic arrest, which ultimately led to cell death through caspase-dependent apoptosis (Supplementary Figure S12). Closer analysis of the first mitosis after C13orf3 depletion in the histone(H2B)-GFP cell line showed that affected cells first form a straight metaphase plate, indicative of a bipolar spindle with two centrosomes. At later stages, the metaphase suddenly became kinked, indicating that the bipolar spindles had reverted to a tripolar spindle, likely through fragmentation of one centrosome (Supplementary Movie S10). This observation confirms the results from the phospho-histone H3 (pS10) and anti-pericentrin immunofluorescence stains (Figure 3A, C, D, and E) and provides a possible explanation for the frequent appearance of tripolar and tetrapolar spindles. Staining of C13orf3-depleted cells with antibodies against the mitotic checkpoint proteins, Bub1 (Figure 3G) and Mad2 (data not shown), showed that the detached kinetochores were positive for both proteins, suggesting that the mitotic arrest is caused by the activation of the SAC. Other kinetochore proteins such as CASC5, Mis12, or NDC80 did not loose their localisation, showing that the overall structure of the kinetochores is not affected upon C13orf3 depletion (Supplementary Figure S13). Co-depletion of Mad2 together with C13orf3 rescued the mitotic arrest (Figure 3B, C, and Supplementary Figure S6B), showing that the activation of the SAC is indeed the primary cause of the mitotic arrest. Together, the comparison of the RNAi phenotype with the described Ska1 RNAi phenotype, that is, mitotic arrest in metaphase, SAC activation (Hanisch et al, 2006), and mitotic centrosome fragmentation (Supplementary Movie S14 and S15 (Rines et al, 2008)), substantiated a possible functional link between these two proteins.
Figure 3.
RNAi phenotypes upon C13orf3 depletion. (A) C13orf3-depleted cells arrest in metaphase. Anti-phospho-histone H3 (pS10) (green), DAPI (blue), and α-tubulin (red) stains are shown of HeLa cells treated with esiRNAs as indicated. Representative images of three independent experiments are shown. (B) Metaphase arrest upon C13orf3 depletion is dependent on spindle-assembly checkpoint integrity. Anti-phospho-histone H3 (pS10) (green), DAPI (blue), and α-tubulin (red) stains of HeLa cells treated with indicated mixtures of esiRNAs are shown. (C) C13orf3 depletion results in a SAC-dependent metaphase arrest with high statistical significance (P<0.001). Quantitative evaluation of phospho-histone H3 (pS10) stains (as in panels A and B) are shown. The mitotic index is shown with the percentages of cells arrested in metaphase indicated above the bars. At least 200 cells were counted for each experiment. Error bars indicate the standard deviation of three independent experiments. Significance tests were carried out with differences calculated between mitotic and metaphase indices of cells treated with C13orf3 or C13orf3/Mad2 esiRNAs versus controls by a two-tailed t-test. (D) Cells arrested by C13orf3 depletion show an increased frequency of multipolar spindles compared with control cells. Representative cells stained with antibodies against α-tubulin (red), pericentrin (green), and DAPI (blue) are shown. (E) Depletion of C13orf3 increases the number of multipolar spindles with high statistical significance (P<0.001). Quantitative evaluations of pericentrin stains (as in panel D) are shown. At least 80 metaphases were evaluated for each value. Significance tests were carried out with differences calculated between percentages of multipolar spindles of cells treated with C13orf3- versus Rn-Luc esiRNAs by a Pearson's χ2 test with Yates' correction for continuity (**). (F) C13orf3 is necessary for metaphase plate maintenance. HeLa cells stably expressing histone(H2B)-GFP depleted of C13orf3 followed by fluorescence time-lapse microscopy for the indicated time periods are shown. Arrows indicate unaligned chromosomes. Representative images from a total of 45 cells filmed by time-lapse microscopy are presented. (G) Immunofluorescence staining of LAP-Bub1 in mitotic HeLa cells after depletion of C13orf3 by RNAi. The single arrow (left panel) points to detached paired sister kinetochores, and the two arrows (right panel) point to separated sister kinetochores. Representative images are depicted from a total of 56 mitotic cells evaluated from two independent experiments. (H) C13orf3 depletion leaves the timing of metaphase plate formation unaltered. Measurement of the time from nuclear envelope breakdown to metaphase plate formation is shown. For every RNAi treatment, the average time and standard deviation of 15 cells are shown. The experiment was repeated three times independently. Average and standard deviations are given for every replicate separately. (I) Cdk1 inhibition promotes anaphase entry in arrested cells after C13orf3 depletion. HeLa cells stably expressing Cherry-histone(H2B) and GFP-tubulin arrested by RNAi against C13orf3, Cdc16, or nocodazole are shown (T=0). Selected frames from time-lapse microscopy after the addition of the Cdk1 inhibitor, RO-3306, are presented (T=27–156). Arrows track cells exiting from mitosis. (J) Statistical quantification shows significant differences in mitotic exit upon C13orf3-RNAi, Cdc16-RNAi, or nocodazole treatments. Cells arrested in mitosis after indicated treatments were analysed with respect to mitotic release by RO-3306 with progression to anaphase (black) or without anaphase (white). Arrested cells that could not be released by RO-3306 are shown in grey. Error bars indicate standard deviation for 200 evaluated cells from three independent experiments. (K) Sister chromatids are separated after mitotic arrest by C13orf3 or SGOL1 depletion. Representative chromosome preparations for HeLa cells treated with indicated esiRNAs are shown. Cells treated with the negative control Rn-Luc were arrested by nocodazole treatment before harvesting. RNAi against the APC/C component, Cdc16, was used as control for metaphase-arrested cells with X-shaped chromosomes. Individual chromosomes are shown as blow-ups. (L) Quantitative evaluation of metaphase spreads shows significant increase in single chromatids upon treatment with esiRNA for SGOL1 and C13orf3 compared with Cdc16 or Rn-Luc RNAi (P<0.001). For each treatment, 40–60 metaphases from two independent experiments were evaluated. Significance tests were carried out with differences calculated between the percentage of cells showing single chromatids treated with C13orf3, Cdc16, or SGOL1 and Rn-Luc esiRNA by a Pearson's χ2 test with Yates' correction for continuity (**). (M) C13orf3 protein stability is dependent on Ska1 and SGOL1. Western blot analysis of extracts from mitotic cells transfected with indicated esiRNAs and stained with indicated antibodies are shown. Size standards are depicted on the left. Quantifications of the band intensities by densitometry are shown below the western blot. Numbers indicate the protein levels in percent after RNAi treatment, normalised to GAPDH. The intensity for the mock control (i.e., Rn-Luc esiRNA) was used as reference. (N) SGOL1 protein stability and localisation is not dependent on C13orf3. Immunofluorescence stains of LAP-SGOL1 in mitotic HeLa cells after treatment with esiRNAs against C13orf3 or Rn-Luc are shown. Representative images are depicted from a total of 48 mitotic cells evaluated from two independent experiments.
Next, we wanted to investigate whether C13orf3 is required for chromosome segregation. Onset from metaphase to anaphase with chromosome segregation during anaphase requires, among others, the inhibition of the kinase activity of Cdk1 (Sullivan and Morgan, 2007). Furthermore, Cdk1 activity is necessary for mitotic entry and maintenance of the mitotic state in early mitosis (Vassilev et al, 2006). Consequently, an inhibition of Cdk1 kinase activity during early mitosis, for example, by the small molecular inhibitor, RO-3306, results in mitotic exit (Supplementary Movie S16 and S17) (Vassilev et al, 2006). In contrast, inhibition of Cdk1 activity in interphase prevents mitosis entry and induces an arrest in the G2 phase (Supplementary Movie S16). Accordingly, mitotic arrest in prometaphase by nocodazole could be released by RO-3306 treatment, leading to mitotic exit without chromosome segregation (Supplementary Movie S17 (Vassilev et al, 2006)). To investigate possible differences of RO-3306-induced mitotic exit in metaphase, we arrested cells through RNAi against the APC/C subunits Cdc16 and C13orf3. After treatment with RO-3306, we imaged the exiting cells by fluorescence time-lapse microscopy. Although Cdc16 depletion leads to a metaphase arrest with the formation of a metaphase plate and a bipolar spindle, the release by RO-3306 treatment does not result in chromosome segregation (Figure 3I and Supplementary Movie S18), inter alia because sister chromatids are still held together at the centromeres by cohesin. In contrast, a release from the metaphase arrest after C13orf3 depletion by RO-3306 resulted in anaphase onset and chromosome segregation with high statistical significance (Figure 3I, J, and Supplementary Movie S19). Consequently, C13orf3 is not per se required for anaphase execution or chromosome segregation. To exclude effects caused by tagging with GFP and Cherry, we repeated these assays with unlabelled cells leading to the same conclusion (Supplementary Movies S16 and S20, S21, S22).
Together, these data identify C13orf3 as an essential protein to satisfy the SAC but not for chromosome segregation during anaphase. Beside the inhibition of Cdk1 activity, the separation of sister chromatids is an important step before execution of anaphase (Sullivan and Morgan, 2007; Yamagishi et al, 2008). Therefore, we asked whether cells arrested in metaphase after C13orf3 depletion have intact sister chromatid cohesion. The protein SGOL1 is implicated in the protection of centromeric sister chromatid cohesion during early mitosis mainly by recruiting PP2A to the centromeric region of chromosomes. Hence, knockdown of SGOL1 leads to mitotic arrest due to premature sister chromatid separation (Waizenegger et al, 2000; Riedel et al, 2006). We analysed chromosome spreads prepared from cells arrested by nocodazole treatment or RNAi against Cdc16, C13orf3, and SGOL1. As expected, X-shaped chromosomes were observed in the cases of Cdc16 RNAi and nocodazole treatment (Figure 3K), indicating that the sister chromatid cohesion is still intact. In contrast, single sister chromatids were observed with high statistical significance for RNAi of C13orf3 and SGOL1 (Figure 3K and L), indicating that the phenotypic consequences of the depletion of these two proteins may be similar. Interestingly, on closer inspection of the RNAi phenotype of SGOL1, other striking similarities to the C13orf3 depletion were seen, for example, exiting chromosomes from the metaphase plate and centrosome fragmentation without alteration on timing of early mitosis (Nakajima et al, 2007; Wang et al, 2008a) (Figure 3H, Supplementary Movies S23). Similar to the C13orf3 knockdown, the RNAi depletion of SGOL1 does not alter the localisation of kinetochore proteins such as CASC5, Mis12, or NDC80, showing that the overall kinetochore structure stays intact (Supplementary Figure S13). To further investigate a potential connection of SGOL1 and Ska complex, we depleted Ska1 and SGOL1 in the C13orf3 BAC-tagged HeLa cell line and monitored possible changes in C13orf3 protein levels by western blot analysis. Strikingly, both knockdowns greatly reduced protein levels of C13orf3 in mitotic cells (Figure 3M), corroborating the link between C13orf3, Ska1 and SGOL1 and indicating that C13orf3 requires Ska1 and SGOL1 for its stability. To test whether the loss of sister chromatid cohesion phenotype upon C13orf3 depletion (Figure 3K) might be due to loss of SGOL1 protein or mislocalisation, we depleted C13orf3 in a BAC transgenic HeLa cell line expressing LAP-tagged SGOL1. No significant loss in SGOL1 protein level or mislocalisation was observed upon C13orf3 depletion compared with the mock control (Figure 3N). These results suggest that there is no mutual dependency of C13orf3 and SGOL1 protein levels and places C13orf3 downstream of SGOL1.
C13orf3 is differentially phosphorylated during mitosis
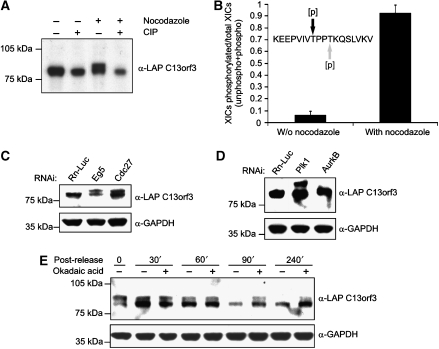
Western blot analysis of lysates isolated from asynchronously growing LAP-tagged C13orf3 cells identified a single band of the predicted size. However, cell extracts prepared from mitotic cells showed an additional band of higher molecular weight (Figure 4A), suggesting that C13orf3 is modified in mitosis. Treatment of mitotic extracts with calf intestine phosphatase led to the disappearance of the slower migrating band (Figure 4A), showing that C13orf3 is phosphorylated during mitosis. Furthermore, the mass spectrometry analysis identified a C13orf3 peptide that was specifically phosphorylated at threonine 190 or 193 during mitosis (Figure 4B and Supplementary Figure S24A and B). To study phosphorylation of C13orf3 during mitosis in more detail, we depleted the protein Eg5 (kinesin 11, depletion produces monopoles and causes a prometaphase arrest) and the APC/C subunit Cdc27 (depletion of important APC/C subunits cause a metaphase arrest) by RNAi. Protein extracts isolated from these cells showed that C13orf3 is phosphorylated during prometa- and metaphase (Figure 4C). To identify potential kinases implicated in C13orf3 phosphorylation, we depleted selected kinases with prominent roles during mitosis by RNAi in the LAP-tagged C13orf3 BAC-transgenic HeLa cells and analysed mitotic cell lysates by western blot. This analysis showed that depletion of AurkB, but not Plk1, abolished phosphorylation of C13orf3 (Figure 4D), indicating that C13orf3 phosphorylation is AurkB-dependent. Interestingly, in addition to the reduced protein levels, no band of higher molecular weight was visible in mitotic extracts upon Ska1 RNAi treatment (Figure 3M), indicating that C13orf3 phosphorylation was also dependent on Ska1. Given the differential phosphorylation of C13orf3 in interphase and mitosis, it appeared likely that C13orf3 is dephosphorylated by a protein phosphatase at the end of mitosis. To test this hypothesis, we released HeLa cells stably expressing LAP-tagged C13orf3 from nocodazole arrest and treated the cells with okadaic acid, an inhibitor of the phosphatase activity of PP2A and PP1 (Mailhes et al, 2003). The phosphorylation of C13orf3 persisted in the presence of okadaic acid (Figure 4E), indication that either PP2A or PP1 activity is required to remove C13orf3 phosphorylation at the end of mitosis. We conclude that C13orf3 is differentially phosphorylated during the cell cycle, with AurkB and PP2A or PP1 being potential candidates that phosphorylate and dephosphorylate the protein, respectively.
Figure 4.
C13orf3 is phosphorylated during mitosis. (A) C13orf3 is phosphorylated in mitotic cells but not in interphase. Extracts from cells stably expressing LAP-tagged C13orf3 and treated with indicated reagents were analysed by western blotting with an anti-GFP antibody (CIP, calf intestine phosphatase). (B) Identification of a cell-cycle-dependent phosphorylation site in C13orf3. HeLa cells expressing LAP-C13orf3 were treated with nocodazole and harvested for analysis by mass spectrometry. Asynchronously growing cells served as reference. The mono-phosphorylated peptide detected and quantified by mass spectrometry is shown (either position T190 (black) or T193 (grey) is phosphorylated). Peak areas from the ion chromatogram for the phosphorylated and unphosphorylated peptides were used for given ratios. Error bars represent the standard deviation of three independent replicates. (C) C13orf3 is phosphorylated in different phases of mitosis. BAC-transgenic HeLa cells expressing LAP-C13orf3 arrested in prometa- (RNAi: Eg5) or metaphase (RNAi: Cdc27) analysed by western blotting are presented. (D) C13orf3 is not phosphorylated in AurkB-depleted cells. HeLa cells expressing LAP-C13orf3 treated with esiRNA against Plk1 or AurkB are shown. Mitotic cells were harvested by mechanical shake-off and analysed by western blot. (E) C13orf3 phosphorylation persists in the presence of okadaic acid. BAC-transgenic HeLa cells expressing LAP-C13orf3 released from nocodazole arrest and treated with okadaic acid for indicated time periods are shown. Cell lysates were analysed by western blotting with indicated antibodies. Markers are shown in the left of western blots.
C13orf3 forms a complex with Ska1, Ska2 and PPP2R2B
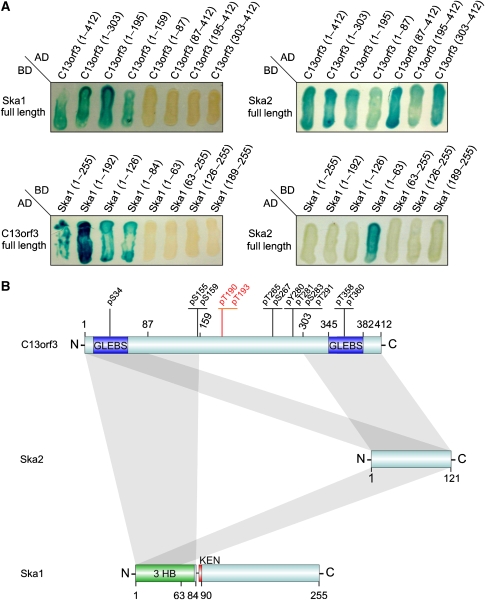
To identify protein interaction partners of C13orf3 we performed immunoprecipitation assays followed by mass spectrometry using the LAP-tagged C13orf3 HeLa cell line. These analyses showed interactions of C13orf3 with the Ska complex proteins Ska2 (Fam33A) and Ska1 (C18orf24) (Table I). Mass spectrometry analyses utilising LAP-tagged BAC-transgenic Ska1 and Ska2 cell lines validated the physical interaction of these three proteins (Table I). Hence, these studies identify C13orf3 as a new member of the Ska complex. Based on this data we propose to rename C13orf3 into Ska3. In contrast to the Ska protein interactions, we did not detect a direct interaction of C13orf3 with SGOL1 by mass spectrometry. However, a global proteomic study with subunits of PP2A showed a physical interaction of C13orf3 and Ska1/-2 with PPP2R2B (Glatter et al, 2009) (Table I). The interaction with PPP2R2B indicates that PP2A is the phosphatase that dephosphorylates C13orf3 at the end of mitosis (Figure 4E) and also provides a possible link to SGOL1 through the regulation of PP2A activity. To map the interaction domains of Ska proteins, we tested different protein fragments in yeast two-hybrid assays (Boxem et al, 2008). These analyses showed an interaction of the N-terminal part of C13orf3 (aa 1–159) with the N-terminal part of Ska1 (aa 1–84) (Figure 5A and B), as well as with Ska2 at the two terminal regions of C13orf3 (aa 1–87 and aa 303–412). In addition, these assays defined a minimal motif for binding of Ska1 and Ska2 at the N-terminal side of Ska1 (aa 1–63) (Figure 5A and B). Hence, all Ska complex proteins interact directly with each other (Figure 5B). Bioinformatic analysis (threading) of Ska1 predicts a three-helical bundle at the N-terminus that is structurally homologous to a Spectrin repeat-like fold (SCOP-ID: 46965), followed by a KEN box (Figure 5B and Supplementary Figure S25), a short motif found in many proteins involved in cell-cycle regulation and mitosis (Michael et al, 2008). Three helical bundles are known to mediate protein–protein interactions (Fridmann-Sirkis et al, 2006). Hence, we propose that the observed interaction of Ska1 with C13orf3 might be mediated through the three-helical bundle region (Supplementary Figure S25). In summary, the two-hybrid assays validate the observed interaction of C13orf3 with Ska1 and Ska2, and define the minimal motifs for these interactions (Figure 5A and B).
Table 1.
Results of pulldown assays and mass spectrometry
| Bait | Interaction partner | Ensembl-ID | International protein index | Detected peptides: unique for protein/coverage (%) |
|---|---|---|---|---|
| C13orf3 | — | ENSG00000165480 | IPI00333014 | 20/46 |
| C13orf3 | Ska1 | ENSG00000154839 | IPI00059912 | 13/40 |
| C13orf3 | Ska2 | ENSG00000182628 | IPI00103149 | 10/66 |
| Ska1 | — | ENSG00000154839 | IPI00059912 | 13/41 |
| Ska1 | C13orf3 | ENSG00000165480 | IPI00333014 | 14/39 |
| Ska1 | Ska2 | ENSG00000182628 | IPI00103149 | 9/58 |
| Ska2 | — | ENSG00000182628 | IPI00789882 | 4/58 |
| Ska2 | C13orf3 | ENSG00000165480 | IPI00333014 | 10/29 |
| Ska2 | Ska1 | ENSG00000154839 | IPI00059912 | 6/21 |
| PPP2R2B | — | ENSG00000156475 | IPI00020850 | 70/55 |
| PPP2R2B | C13orf3 | ENSG00000165480 | IPI00333014 | 10/28 |
| PPP2R2B | Ska1 | ENSG00000154839 | IPI00059912 | 2/20 |
| PPP2R2B | Ska2 | ENSG00000182628 | IPI00103149 | 4/29 |
| Baits are shown in bold characters. | ||||
Figure 5.
Identification of minimal binding motifs and model of interaction. (A) Yeast two-hybrid analysis of Ska complex protein interactions. Ska proteins were shortened from both termini (fragment sizes are given as numbers of amino acids) and combined for yeast two-hybrid analyses (AD, activation domain; BD, binding domain). (B) Model for Ska complex interactions. Minimal binding motifs derived from panel A are indicated as grey bars. Phosphorylated residues are indicated (Brill et al, 2004; Rush et al, 2005; Nousiainen et al, 2006; Cantin et al, 2008). Newly identified phosphorylation at threonine 190 or threonine 193 are highlighted in red. Predicted GLEBS motifs, three-helical bundle (3 HB), or KEN-box motifs are shown in blue, green, and red, respectively. Numbers indicate the amino acid positions.
In conclusion, our data on C13orf3 show that the combination of phenotypic profiling with protein localisation data is a useful approach to predict functions of uncharacterised genes. Large-scale tagging of proteins at endogenous expression levels is possible in yeast, and comprehensive protein localisation (Huh et al, 2003) and protein interaction network studies (Gavin et al, 2006; Krogan et al, 2006) have been carried out in this organism. To broaden this approach, systematic BAC tagging (Poser et al, 2008) of most proteins would allow similar studies in mammalian cells. In a test study, applying this technology in small scale, we identified C13orf3 as a new interaction partner of the Ska complex. This study provides a first link between the Ska complex and regulation of sister chromatid cohesion possibly through SGOL1 and PP2A pathways.
Materials and methods
Phenotypic profiling
esiRNAs were synthesised as described previously (Kittler et al, 2005a, 2007b) and, after normalisation, arrayed into 384-well plates. Briefly, esiRNAs are silencing triggers for RNAi in mammalian cells prepared by enzymatic digestion (bacterial RNase III) of long dsRNA (300–600 bp, derived from target mRNA). esiRNAs were chosen in this study in favour of chemically synthesised siRNAs because they were shown to produce less off-target effects (Kittler et al, 2007b). Sequences of the esiRNAs especially relevant to this study are given in Supplementary Table S5. All esiRNAs used in this study were designed to target all splicing variants of their target genes, respectively. For the genome-scale viability screen, esiRNAs (15 ng each) were reversely transfected into HeLa cells with Oligofectamine (Invitrogen) in black, tissue culture plates (Greiner) and incubated for 72 h. For viability analysis, the cell culture medium was supplemented with AlamarBlue dye (Serotec) and after 3 h incubation the fluorescence intensities (excitation: 535 nm; emission: 590 nm) were measured using a plate reader (GENiosPro, Tecan). The resulting fluorescence intensities were transformed to z-scores by using plate mean and standard deviation. Hierarchical clustering analysis was carried out with R statistical package (R Development Core Team). As a data source, we used 16,363 phenotypic profiles from the viability and previously conducted cell-cycle RNAi screen (Kittler et al, 2007a). The input variables were z-scores of cells in G1, S, G2/M phase, 8 N cells, and viability. To normalise variables, all scores were converted into percentile ranks. The percentile rank of a score is the percentage of scores, which are lower or equal to it; they were chosen for hierarchical clustering because they are more robust towards skewed distributions. Distances between genes were calculated with a weighted Euclidean metric. Weights were chosen to ensure equal influence of both experiments in the calculated distance (1/2 for viability variable and 1/8 for each of the four cell-cycle variables). The final tree was constructed using clustering with the Unweighted Pair Group Method with Arithmetic Mean (UPGMA).
BAC TransgeneOmics
BACs harbouring the genes of interest were obtained from the BACPAC Resource Center (http://bacpac.chori.org; BAC-IDs see Supplementary Table S2). A LAP cassette (Cheeseman and Desai, 2005) was inserted as a C-terminal fusion using recombineering (Zhang et al, 2000) (Gene Bridges). Isolated BAC DNA was transfected and selected for stable integration as described (Poser et al, 2008).
Immunofluorescence microscopy
Cells were grown on coverslips in 12-well plates, transfected with 300ng esiRNAs, fixed in cold methanol at −20°C for 8 min, and blocked with 0.2% gelatin from cold-water fish skin (Sigma) in phosphate buffered saline (PBS) (PBS/FSG) for 10 min, 36–48 h post transfection for Ska complex esiRNAs and SGOL1 esiRNA. Staining was carried out by incubation with the following primary antibodies for 20 min in PBS/FSG: goat anti-GFP (1:4000, MPI-CBG Antibody Facility), mouse anti-α-tubulin (1:2000, MPI-CBG Antibody Facility), human anti-CREST (1:500, Cortex Biochem), rabbit anti-pericentrin (1:5000, Abcam). After washes with PBS/FSG, the cells were incubated with fluorescently labelled secondary antibodies (donkey anti-mouse Alexa594, Molecular Probes; donkey anti-goat FITC, Molecular Probes; donkey anti-rabbit Alexa594, Molecular Probes; and goat anti-human Alexa594, Invitrogen). After washing with PBS/FSG, the coverslips were mounted on glass slides by inverting them in the mounting solution containing 4′,6-diamidino-2-phenylindole (DAPI, ProLong Gold antifade, Invitrogen). Images were taken on an Axioplan II Microscope (Zeiss) operated by MetaMorph (Molecular Devices) or on Olympus IX70 (Olympus) equipped with the imaging system DeltaVision RT using × 40/1.00 or × 63/1.40 Plan-Apochromat oil immersion objectives. Z-stacks (0.2 μm optical sections) were collected, deconvolved, and projected into one picture using softWoRx software (Applied Precision). Acquired images were cropped and contrast adjusted in Adobe Photoshop 8.0 (Adobe Systems) and then sized and placed in figures using Corel Draw 11.633 (Corel Corporation).
Live-cell imaging
HeLa cells stably expressing LAP-tagged proteins or histone(H2B)-GFP or Cherry-histone(H2B) and GFP-tubulin were grown in 96-well tissue culture plates and transfected with 40ng esiRNAs. Images were obtained 12–36 h after transfection with a ScanR system (Olympus) placed in a heated chamber (37°C) with 5% CO2 and filmed for 1–36 h as indicated. If appropriate, cells were arrested by treatment with 50 ng/ml nocodazole (Sigma) for 12 h and/or treated with 9 μM of RO-3306 (Merck Biosciences). For high-resolution time-lapse imaging, the cells were grown on eight-well LabTek chambered coverglasses (Nalge Nunc). Before imaging, the medium was changed to CO2-independent medium (Invitrogen), and the cell culture chamber was placed onto a heated sample stage within a heated chamber (37°C). Images were acquired with an Olympus IX70 DeltaVision RT system (Olympus).
Cell-based assays
For cellular DNA content analysis, esiRNA-transfected cells were fixed and stained with propidium iodide (Molecular Probes) and scanned with a FACSCalibur flow cytometer (BD Biosciences) 42 h post transfection. The resulting DNA content histograms were manually gated to determine the percentage of cells in G2/M phase. For determining the mitotic index, esiRNA-transfected cells were fixed and incubated with mouse anti-α-tubulin (1:2000; MPI-CBG Antibody Facility) and rabbit anti-phospho-histone H3 Ser10 antibodies (1:10, conjugated to Alexa488, Cell Signaling Technologies) 42 h post transfection. Subsequently, the cells were incubated with fluorescently labelled donkey anti-mouse Alexa594 antibody and DAPI; images were obtained as described above. For chromosome preparations, HeLa cells were treated with esiRNA (42 h: C13orf3 and SGOL1; 96 h: Cdc16) and/or nocodazole (50 ng/ml for 14 h). After harvesting, the cells were resuspended in hypertonic solution (30 mM sodium citrate) and incubated for 35–45 min at 37°C. Subsequently, the cells were fixed with ethanol/acetic acid (3:1), spread onto a coverslip, rehydrated for 15 min with PBS and fixed again with formaldehyde (4%). After washing for three times with PBS, the cells were dehydrated by washing with stepwise increasing concentrations of ethanol (70–100% in four steps). After drying, the coverslips were mounted on glass slides by inverting them in the mounting solution containing DAPI (ProLong Gold antifade, Invitrogen). Images were taken on an Axioplan II Microscope (Zeiss) as described above.
Quantitative PCR and apoptosis assays
To ensure an efficient silencing for all prominent esiRNAs used in this study, we conducted mRNA quantification by quantitative-PCR (Q-PCR) (Supplementary Table S5). HeLa cells were transfected in 12-well cell culture dishes using 300 ng esiRNA and 4.2 μl Oligofectamine (Invitrogen) per well. After 24 h incubation, the cells were harvested and total mRNA was extracted using the RNeasy Mini Kit (Qiagen) including an on-column DNaseI digest as given in the manufacturer's manual. Subsequently, total mRNA was reverse transcribed using SuperScript III reverse transcriptase (Invitrogen) and oligo (dT)12–18 primers (Invitrogen). Quantification of the targeted mRNA was conducted using the Absolute qPCR SYBR Green Kit and an Mx3000p (Stratagene) real-time PCR machine. Primer sequences for quantitative PCR are provided in the Supplementary Table S8. For apoptosis assays, HeLa cells were transfected with esiRNA (30 ng) in 96-well cell culture dishes. After 24 h incubation, the caspase inhibitor, z-VAD-FMK (Merck Biosciences), was added to the cell culture supernatant (50 nM) or to 1% DMSO as vehicle control. The cells were harvested and stained 48 h after transfection with fluorescently labelled Annexin-V (APC-conjugated, Becton Dickinson) and propidium iodide (Molecular Probes) and analysed by flow cytometry on a FACSCalibur system (BD Biosciences). Cells that stained positive for Annexin-V but negative for propidium iodide were considered apoptotic.
Western blot analysis
Whole-cell lysates or mitotic cells (mechanical shake-off) stably transfected with different BAC constructs and treated with esiRNAs, nocodazole (50 ng/ml, Sigma), or okadaic acid (250 nM; Sigma) were subjected to SDS–PAGE (NuPage 4–12% Bis-Tris; Invitrogen), blotted to nitrocellulose (Protran, Schleicher & Schuell) and incubated with primary antibodies (mouse anti-GFP, 1:5000, Roche; or mouse anti-GAPDH, 1:20 000, Acris Antibodies). Subsequently, the membranes were incubated with goat anti-mouse antibody conjugated to horseradish peroxidase (1:4000, Bio-Rad); bands were visualised with enhanced chemiluminescence Western Blotting Detection Reagents (GE Healthcare). As a molecular weight standard, the Full-Range Rainbow ladder 10–250 kDa (GE Healthcare) was used. Films were scanned and images were cropped and contrast adjusted in Adobe Photoshop 8.0 (Adobe Systems) and then sized and placed in figures using Corel Draw 11.633 (Corel Corporation). For phosphatase assays, nocodazole-arrested cells were harvested by mechanical shake-off, and lysates were incubated with calf intestinal phosphatase (New England Biolabs) for 15 min at 37°C or left untreated. Subsequently, all lysates were analysed by western blotting as described above. All band intensities of western blot images were quantified with ImageJ 1.40 g (National Institutes of Health).
Immunoprecipitation and mass spectrometry
Transgenic cells expressing LAP-tagged or Strep-HA-tagged versions of the proteins of interest were harvested and, after lysis, cleared from insoluble material by ultracentrifugation (100 000 g for 20 min at 2°C). Immunoprecipitation was carried out by incubation with goat anti-GFP antibody (MPI-CBG Antibody Facility, 1 h at 4°C) immobilised on G-protein sepharose (FastFlow, GE Healthcare, 200 μg antibody per 100 μl matrix) or on 200 μl Strep-Tactin beads (IBA TAGnologies). Specificity of the goat anti-GFP antibody in immunoprecipitation assays was extensively validated (Poser et al, 2008). After washing, elution from the affinity beads was carried out with 100 μl of glycine (100 mM, pH 2.0), which was subsequently neutralised with 1.5 M Tris at pH 8.0 or 100 mM NH4HCO3. Strep-HA-tagged proteins were eluted with TNN-HS buffer with 2 mM biotin and immunoprecipitated with 100 μl anti-HA agarose (Sigma) before glycine elution. Modified porcine trypsin (Promega) was added (16 ng/μl) and proteins were digested overnight. The tryptic peptides were analysed by mass spectrometry (detailed protocol S26, see Supplementary data).
Quantification of phosphorylation
Peak areas of extracted ion chromatograms (XICs) corresponding phosphorylated and non-phosphorylated precursors at 2+/3+ charge states were determined using Xcalibur 2.0 software (Thermo Fisher Scientific), assuming better than 10 ppm mass accuracy and <1 min retention time variation within multiple runs. Survey spectra were examined to make sure no peaks of the same mass were co-eluted with the quantified peptides. Phosphorylation was calculated as a ratio of the peak areas of phosphorylated precursors to the sum of peak areas of phosphorylated and non-phosphorylated forms and averaged between four repetitive runs for arrested cells and duplicated samples for non-arrested cells. When calculating phosphorylation of the peptide presented in Supplementary Figure S24, the peak areas of the partially miscleaved form (both in phosphorylated and in non-phosphorylated states) were considered.
Yeast two-hybrid analysis
Yeast two-hybrid analysis was carried out using a system described previously (Vidal et al, 1996). Full-length proteins and fragments of Ska1, Ska2, and C13orf3 were fused to Gal4 DNA-binding domain (BD, aa 1–147) or Gal4 activation domain (AD, aa 768–881) as indicated in Figure 5A. Clones growing on media lacking uracil were streaked out on selective media lacking histidine and containing 50 mM 3-amino-1,2,4-triaziole (MP Biomedicals). Positive clones were further analysed for β-galactosidase activity.
Computational sequence analysis and comparative modelling
Sequence analyses were carried out with the ELM server (Puntervoll et al, 2003). Secondary structure predictions were carried out with Jpred (Cole et al, 2008). Threading analysis of the Ska protein sequences was carried out with the program Prohit (Proceryon GmbH) and a fold library containing 20008 chains from the Brookhaven Protein Data Bank (PDB). GO terms according to the observed RNAi phenotype were used to distinguish possible true hits from false positives in the fold hit list. The N-terminal region of Ska1 was modelled as a Spectrin repeat-like fold using the sequence–structure alignment obtained from threading and the X-ray structure of SNARE Tlg1 at 2.05 Å resolution as template (PDBId 2c5k, chain T; Fridmann-Sirkis et al, 2006). The C-terminal region of Ska3 was modelled as a GLEBS motif using the X-ray structure of the yeast Bub1-GLEBS/Bub3 at 1.9 Å resolution as template (PDBId 2i3s, chain B; Larsen et al, 2007). The human Bub3 was modelled by homology based on the chain A of the yeast complex X-ray structure. The Discovery Studio package (v1.7, Accelrys, San Diego, CA) was used for model construction and refinement. The RosettaDock Server (Lyskov and Gray, 2008) was used to dock the modelled human Ska3-GLEBS and Bub3 structures, and the complex with the highest score was selected after visual inspection.
Supplementary Material
Supplementary Movie S4
Supplementary Movie S9
Supplementary Movie S10
Supplementary Movie S11
Supplementary Movie S14
Supplementary Movie S15
Supplementary Movie S16
Supplementary Movie S17
Supplementary Movie S18
Supplementary Movie S19
Supplementary Movie S20
Supplementary Movie S21
Supplementary Movie S22
Supplementary Movie S23
Supplementary Information
Supplementary Table S1
Supplementary Table S2
Supplementary Table S3
Supplementary Legends
Acknowledgments
We thank E Krausz and J Wagner for support with automated transfection; W Zachariae for critical reading of the paper; M Augsburg, M Biesold, A Ssykor, S Rose, and A Weise for technical assistance; I Nüsslein for support with FACS analysis; A Bird for support with DeltaVision imaging; Y Toyoda, Z Maliga, and J Ellenberg for providing cell lines and all members of the F Buchholz laboratory for discussions. This study was funded by the Max Planck Society, the Bundesministerium für Bildung und Forschung grands Go-Bio [0315105] and NGFN-Plus [01GS0859] and by the 6th Framework Program of the European Union, Integrated Project ‘MitoCheck' (LSHG-CT-2004-503464).
References
- Boxem M, Maliga Z, Klitgord N, Li N, Lemmens I, Mana M, de Lichtervelde L, Mul JD, van de Peut D, Devos M, Simonis N, Yildirim MA, Cokol M, Kao HL, de Smet AS, Wang H, Schlaitz AL, Hao T, Milstein S, Fan C et al. (2008) A protein domain-based interactome network for C. elegans early embryogenesis. Cell 134: 534–545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brill LM, Salomon AR, Ficarro SB, Mukherji M, Stettler-Gill M, Peters EC (2004) Robust phosphoproteomic profiling of tyrosine phosphorylation sites from human T cells using immobilized metal affinity chromatography and tandem mass spectrometry. Anal Chem 76: 2763–2772 [DOI] [PubMed] [Google Scholar]
- Cantin GT, Yi W, Lu B, Park SK, Xu T, Lee JD, Yates III JR (2008) Combining protein-based IMAC, peptide-based IMAC, and MudPIT for efficient phosphoproteomic analysis. J Proteome Res 7: 1346–1351 [DOI] [PubMed] [Google Scholar]
- Cheeseman IM, Desai A (2005) A combined approach for the localization and tandem affinity purification of protein complexes from metazoans. Sci STKE 2005: pl1. [DOI] [PubMed] [Google Scholar]
- Cole C, Barber JD, Barton GJ (2008) The Jpred 3 secondary structure prediction server. Nucleic Acids Res 36 (Web Server issue): W197–W201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fazzio TG, Huff JT, Panning B (2008) An RNAi screen of chromatin proteins identifies Tip60-p400 as a regulator of embryonic stem cell identity. Cell 134: 162–174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fridmann-Sirkis Y, Kent HM, Lewis MJ, Evans PR, Pelham HR (2006) Structural analysis of the interaction between the SNARE Tlg1 and Vps51. Traffic 7: 182–190 [DOI] [PubMed] [Google Scholar]
- Galvez T, Teruel MN, Heo WD, Jones JT, Kim ML, Liou J, Myers JW, Meyer T (2007) siRNA screen of the human signaling proteome identifies the PtdIns(3,4,5)P3-mTOR signaling pathway as a primary regulator of transferrin uptake. Genome Biol 8: R142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gavin AC, Aloy P, Grandi P, Krause R, Boesche M, Marzioch M, Rau C, Jensen LJ, Bastuck S, Dumpelfeld B, Edelmann A, Heurtier MA, Hoffman V, Hoefert C, Klein K, Hudak M, Michon AM, Schelder M, Schirle M, Remor M et al. (2006) Proteome survey reveals modularity of the yeast cell machinery. Nature 440: 631–636 [DOI] [PubMed] [Google Scholar]
- Glatter T, Wepf A, Aebersold R, Gstaiger M (2009) An integrated workflow for charting the human interaction proteome: insights into the PP2A system. Mol Syst Biol 5: 237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Godzik A (2003) Fold recognition methods. Methods Biochem Anal 44: 525–546 [DOI] [PubMed] [Google Scholar]
- Goto H, Tomono Y, Ajiro K, Kosako H, Fujita M, Sakurai M, Okawa K, Iwamatsu A, Okigaki T, Takahashi T, Inagaki M (1999) Identification of a novel phosphorylation site on histone H3 coupled with mitotic chromosome condensation. J Biol Chem 274: 25543–25549 [DOI] [PubMed] [Google Scholar]
- Hanisch A, Sillje HH, Nigg EA (2006) Timely anaphase onset requires a novel spindle and kinetochore complex comprising Ska1 and Ska2. EMBO J 25: 5504–5515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huh WK, Falvo JV, Gerke LC, Carroll AS, Howson RW, Weissman JS, O'Shea EK (2003) Global analysis of protein localization in budding yeast. Nature 425: 686–691 [DOI] [PubMed] [Google Scholar]
- Kittler R, Heninger AK, Franke K, Habermann B, Buchholz F (2005a) Production of endoribonuclease-prepared short interfering RNAs for gene silencing in mammalian cells. Nat Methods 2: 779–784 [DOI] [PubMed] [Google Scholar]
- Kittler R, Pelletier L, Buchholz F (2008) Systems biology of mammalian cell division. Cell Cycle 7: 2123–2128 [DOI] [PubMed] [Google Scholar]
- Kittler R, Pelletier L, Heninger AK, Slabicki M, Theis M, Miroslaw L, Poser I, Lawo S, Grabner H, Kozak K, Wagner J, Surendranath V, Richter C, Bowen W, Jackson AL, Habermann B, Hyman AA, Buchholz F (2007a) Genome-scale RNAi profiling of cell division in human tissue culture cells. Nat Cell Biol 9: 1401–1412 [DOI] [PubMed] [Google Scholar]
- Kittler R, Pelletier L, Ma C, Poser I, Fischer S, Hyman AA, Buchholz F (2005b) RNA interference rescue by bacterial artificial chromosome transgenesis in mammalian tissue culture cells. Proc Natl Acad Sci USA 102: 2396–2401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kittler R, Putz G, Pelletier L, Poser I, Heninger AK, Drechsel D, Fischer S, Konstantinova I, Habermann B, Grabner H, Yaspo ML, Himmelbauer H, Korn B, Neugebauer K, Pisabarro MT, Buchholz F (2004) An endoribonuclease-prepared siRNA screen in human cells identifies genes essential for cell division. Nature 432: 1036–1040 [DOI] [PubMed] [Google Scholar]
- Kittler R, Surendranath V, Heninger AK, Slabicki M, Theis M, Putz G, Franke K, Caldarelli A, Grabner H, Kozak K, Wagner J, Rees E, Korn B, Frenzel C, Sachse C, Sonnichsen B, Guo J, Schelter J, Burchard J, Linsley PS et al. (2007b) Genome-wide resources of endoribonuclease-prepared short interfering RNAs for specific loss-of-function studies. Nat Methods 4: 337–344 [DOI] [PubMed] [Google Scholar]
- Krogan NJ, Cagney G, Yu H, Zhong G, Guo X, Ignatchenko A, Li J, Pu S, Datta N, Tikuisis AP, Punna T, Peregrin-Alvarez JM, Shales M, Zhang X, Davey M, Robinson MD, Paccanaro A, Bray JE, Sheung A, Beattie B et al. (2006) Global landscape of protein complexes in the yeast Saccharomyces cerevisiae. Nature 440: 637–643 [DOI] [PubMed] [Google Scholar]
- Larsen NA, Al-Bassam J, Wei RR, Harrison SC (2007) Structural analysis of Bub3 interactions in the mitotic spindle checkpoint. Proc Natl Acad Sci USA 104: 1201–1206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyskov S, Gray JJ (2008) The RosettaDock server for local protein–protein docking. Nucleic Acids Res 36 (Web Server issue): W233–W238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mailhes JB, Hilliard C, Fuseler JW, London SN (2003) Okadaic acid, an inhibitor of protein phosphatase 1 and 2A, induces premature separation of sister chromatids during meiosis I and aneuploidy in mouse oocytes in vitro. Chromosome Res 11: 619–631 [DOI] [PubMed] [Google Scholar]
- Michael S, Trave G, Ramu C, Chica C, Gibson TJ (2008) Discovery of candidate KEN-box motifs using cell cycle keyword enrichment combined with native disorder prediction and motif conservation. Bioinformatics 24: 453–457 [DOI] [PubMed] [Google Scholar]
- Musacchio A, Salmon ED (2007) The spindle-assembly checkpoint in space and time. Nat Rev Mol Cell Biol 8: 379–393 [DOI] [PubMed] [Google Scholar]
- Muyrers JP, Zhang Y, Stewart AF (2001) Techniques: recombinogenic engineering—new options for cloning and manipulating DNA. Trends Biochem Sci 26: 325–331 [DOI] [PubMed] [Google Scholar]
- Nakajima M, Kumada K, Hatakeyama K, Noda T, Peters JM, Hirota T (2007) The complete removal of cohesin from chromosome arms depends on separase. J Cell Sci 120(Part 23): 4188–4196 [DOI] [PubMed] [Google Scholar]
- Nasmyth K (2002) Segregating sister genomes: the molecular biology of chromosome separation. Science 297: 559–565 [DOI] [PubMed] [Google Scholar]
- Nigg EA (2001) Mitotic kinases as regulators of cell division and its checkpoints. Nat Rev Mol Cell Biol 2: 21–32 [DOI] [PubMed] [Google Scholar]
- Nousiainen M, Sillje HH, Sauer G, Nigg EA, Korner R (2006) Phosphoproteome analysis of the human mitotic spindle. Proc Natl Acad Sci USA 103: 5391–5396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pepperkok R, Ellenberg J (2006) High-throughput fluorescence microscopy for systems biology. Nat Rev Mol Cell Biol 7: 690–696 [DOI] [PubMed] [Google Scholar]
- Peters JM (2006) The anaphase promoting complex/cyclosome: a machine designed to destroy. Nat Rev Mol Cell Biol 7: 644–656 [DOI] [PubMed] [Google Scholar]
- Poser I, Sarov M, Hutchins JR, Heriche JK, Toyoda Y, Pozniakovsky A, Weigl D, Nitzsche A, Hegemann B, Bird AW, Pelletier L, Kittler R, Hua S, Naumann R, Augsburg M, Sykora MM, Hofemeister H, Zhang Y, Nasmyth K, White KP et al. (2008) BAC TransgeneOmics: a high-throughput method for exploration of protein function in mammals. Nat Methods 5: 409–415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puntervoll P, Linding R, Gemund C, Chabanis-Davidson S, Mattingsdal M, Cameron S, Martin DM, Ausiello G, Brannetti B, Costantini A, Ferre F, Maselli V, Via A, Cesareni G, Diella F, Superti-Furga G, Wyrwicz L, Ramu C, McGuigan C, Gudavalli R et al. (2003) ELM server: a new resource for investigating short functional sites in modular eukaryotic proteins. Nucleic Acids Res 31: 3625–3630 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riedel CG, Katis VL, Katou Y, Mori S, Itoh T, Helmhart W, Galova M, Petronczki M, Gregan J, Cetin B, Mudrak I, Ogris E, Mechtler K, Pelletier L, Buchholz F, Shirahige K, Nasmyth K (2006) Protein phosphatase 2A protects centromeric sister chromatid cohesion during meiosis I. Nature 441: 53–61 [DOI] [PubMed] [Google Scholar]
- Rines DR, Gomez-Ferreria MA, Zhou Y, DeJesus P, Grob S, Batalov S, Labow M, Huesken D, Mickanin C, Hall J, Reinhardt M, Natt F, Lange J, Sharp DJ, Chanda SK, Caldwell JS (2008) Whole genome functional analysis identifies novel components required for mitotic spindle integrity in human cells. Genome Biol 9: R44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rush J, Moritz A, Lee KA, Guo A, Goss VL, Spek EJ, Zhang H, Zha XM, Polakiewicz RD, Comb MJ (2005) Immunoaffinity profiling of tyrosine phosphorylation in cancer cells. Nat Biotechnol 23: 94–101 [DOI] [PubMed] [Google Scholar]
- Sachse C, Echeverri CJ (2004) Oncology studies using siRNA libraries: the dawn of RNAi-based genomics. Oncogene 23: 8384–8391 [DOI] [PubMed] [Google Scholar]
- Sippl MJ, Flockner H (1996) Threading thrills and threats. Structure 4: 15–19 [DOI] [PubMed] [Google Scholar]
- Sullivan M, Morgan DO (2007) Finishing mitosis, one step at a time. Nat Rev Mol Cell Biol 8: 894–903 [DOI] [PubMed] [Google Scholar]
- Thornton BR, Ng TM, Matyskiela ME, Carroll CW, Morgan DO, Toczyski DP (2006) An architectural map of the anaphase-promoting complex. Genes Dev 20: 449–460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varetti G, Musacchio A (2008) The spindle assembly checkpoint. Curr Biol 18: R591–R595 [DOI] [PubMed] [Google Scholar]
- Vassilev LT, Tovar C, Chen S, Knezevic D, Zhao X, Sun H, Heimbrook DC, Chen L (2006) Selective small-molecule inhibitor reveals critical mitotic functions of human CDK1. Proc Natl Acad Sci USA 103: 10660–10665 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vidal M, Braun P, Chen E, Boeke JD, Harlow E (1996) Genetic characterization of a mammalian protein–protein interaction domain by using a yeast reverse two-hybrid system. Proc Natl Acad Sci USA 93: 10321–10326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waizenegger IC, Hauf S, Meinke A, Peters JM (2000) Two distinct pathways remove mammalian cohesion from chromosome arms in prophase and from centromeres in anaphase. Cell 103: 399–410 [DOI] [PubMed] [Google Scholar]
- Wang X, Yang Y, Duan Q, Jiang N, Huang Y, Darzynkiewicz Z, Dai W (2008a) sSgo1, a major splice variant of Sgo1, functions in centriole cohesion where it is regulated by Plk1. Dev Cell 14: 331–341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Shyy JY, Chien S (2008b) Fluorescence proteins, live-cell imaging, and mechanobiology: seeing is believing. Annu Rev Biomed Eng 10: 1–38 [DOI] [PubMed] [Google Scholar]
- Watanabe Y (2005) Shugoshin: guardian spirit at the centromere. Curr Opin Cell Biol 17: 590–595 [DOI] [PubMed] [Google Scholar]
- Yamagishi Y, Sakuno T, Shimura M, Watanabe Y (2008) Heterochromatin links to centromeric protection by recruiting shugoshin. Nature 455: 251–255 [DOI] [PubMed] [Google Scholar]
- Yang D, Buchholz F, Huang Z, Goga A, Chen CY, Brodsky FM, Bishop JM (2002) Short RNA duplexes produced by hydrolysis with Escherichia coli RNase III mediate effective RNA interference in mammalian cells. Proc Natl Acad Sci USA 99: 9942–9947 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Muyrers JP, Testa G, Stewart AF (2000) DNA cloning by homologous recombination in Escherichia coli. Nat Biotechnol 18: 1314–1317 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Movie S4
Supplementary Movie S9
Supplementary Movie S10
Supplementary Movie S11
Supplementary Movie S14
Supplementary Movie S15
Supplementary Movie S16
Supplementary Movie S17
Supplementary Movie S18
Supplementary Movie S19
Supplementary Movie S20
Supplementary Movie S21
Supplementary Movie S22
Supplementary Movie S23
Supplementary Information
Supplementary Table S1
Supplementary Table S2
Supplementary Table S3
Supplementary Legends