Abstract
Circular bacterial chromosomes have highly polarized nucleotide composition in the two replichores, and this genomic strand asymmetry can be visualized using GC skew graphs. Here we propose and discuss the GC skew index (GCSI) for the quantification of genomic compositional skew, which combines a normalized measure of fast Fourier transform to capture the shape of the skew graph and Euclidean distance between the two vertices in a cumulative skew graph to represent the degree of skew. We calculated GCSI for all available bacterial genomes, and GCSI correlated well with the visibility of GC skew. This novel index is useful for estimating confidence levels for the prediction of replication origin and terminus by methods based on GC skew and for measuring the strength of replicational selection in a genome.
Keywords: GC skew, DNA replication, replicational selection, fast Fourier transforms, bioinformatics, GCSI
Introduction
In circular bacterial chromosomes, the replication process starts from a finite replication origin (ori) and continues bidirectionally along the two arms (i.e. the replichores) until the replication complex reaches the replication terminus (ter), located directly opposite of ori (Rocha, 2004a; Rocha, 2004b). Replication is obviously the most fundamental and essential process in the cell cycle of bacteria, and replication also exerts genome-wide mutational and selection pressure, shaping genomic polarity with asymmetrically biased nucleotide composition in leading and lagging strands (Lobry and Louarn, 2003; Lobry and Sueoka, 2002). This compositional skew can be easily observed by plotting the normalized excess of guanine (G) over cytosine (C) content in a subgenomic region with sliding windows along the complete genome sequence (Lobry, 1996). Such a GC skew graph segregates the genome into two regions: one with an excess of G over C corresponding to the leading strand, and the other with an excess of C over G corresponding to the lagging strand. Moreover, the shift points of the GC skew graphs are reportedly correlated with the loci of ori and ter (Frank and Lobry, 1999). GC skew is observed in many bacterial species with circular chromosomes, although with varying clarity of the shift points, and GC skew is usually not detectable in symbionts and bacteria with linear chromosomes (Worning et al. 2006) or in archaeal genomes, which employ different machinery for the replication process (Grabowski and Kelman, 2003; Lopez et al. 1999; Myllykallio et al. 2000). GC skew is also observed in local genomic regions primarily introduced by RNA synthesis (Fujimori et al. 2005), but the overall genomic polarity due to replication is present regardless of these local effects, and the GC skew is thus observed in intergenic regions as well as in the third nucleotide positions in codons. Although the underlying causes for GC skew is not completely understood, hydrolytic deamination of cytosine in the leading strand in single-stranded state during replication, is suggested as the major contributing factor (Rocha, 2004b).
Because only a few ori and ter positions had been identified by experimental means, analysis of GC skew was first utilized for the computational prediction of ori and ter positions by examining available genome sequences (Frank and Lobry, 2000). Similar method using nucleotide gradients of T/C and A/G is utilized for the detection of unidirectional replication in mitochondria (Krishnan et al. 2004; Seligmann et al. 2006). To improve the accuracy of prediction, cumulative diagrams are commonly employed to balance out the noise in sequence composition and to eliminate the requirement for window slides (Grigoriev, 1998), coupled with purine and keto excesses and GC skew (Freeman et al. 1998). However, predictions based on these methods are less accurate in genomes where GC skew cannot be strongly observed (Zawilak et al. 2001). To observe the control of replicational selection on the various genomic properties, genomic compositional skews are also used in conjunction with other genomic features such as the gene orientation (McLean et al. 1998), the distribution of RAG oligomers recognized by the FtsK translocase (Hendrickson and Lawrence, 2006), and the codon bias of genes along the genome (Daubin and Perriere, 2003). To our knowledge, however, no method to quantify the strength of GC skew has been proposed; therefore, it is difficult to compare the effects of replicational selection across bacterial genomes.
In this work, we present the GC skew index (GCSI), which quantifies the strength of GC skew of a given genome by combining Fourier power spectral analysis with the Euclidean distance between the maximum and minimum of the cumulative skew vector. Spectral analysis using fast Fourier transform (FFT) is able to identify the frequency components contributing to a given signal, and it has been applied successfully to the field of bioinformatics (Dodin et al. 2000; Katoh et al. 2002; Yin and Yau, 2005). Because GC skew emerges from the mutational selection in the two replichores, the greatest contributing frequency component of GC skew should be at 1 Hz, with two clear shift points. This observation of a 1-Hz signal combined with the degree of skew calculated by the distance measure between the two vertices of a cumulative skew diagram effectively quantifies the skew of genomic compositional asymmetry.
Materials and Methods
Sequences and software
Complete circular chromosomal sequences of 303 bacteria and complete genome sequences of 29 archaeal genomes in GenBank format were selected and obtained from the NCBI RefSeq FTP repository (ftp://ftp.ncbi.nih.gov/genomes/Bacteria/). All analyses were conducted using the G-language Genome Analysis Environment version 1.6.11 (Arakawa et al. 2003; Arakawa and Tomita, 2006). The positional coordinate system for the genomic sequence used in this work was set to originate at 0, unlike that of GenBank, which uses 1 for the position of the first base.
Calculation of GC skew
GC skew was defined as the normalized excess of C over G in a given sequence, (C − G)/(C + G), which is calculated with sliding windows along the genome. GC skew is defined to be 0 when the amount of C equals that of G. To eliminate the use of window slides, cumulative skew can be calculated as the cumulative sum of the walker graph score at each nucleotide position along the genome, with scores A = 0, T = 0, G = 1, and C = −1. In this work, however, the cumulative GC skew was calculated by taking the cumulative sum of the GC skew in each of the windows, to normalize the cumulative skew strength without it being affected by the length of the genome.
Fast fourier transform
FFT is the computationally optimized derivation of discrete Fourier transform (DFT) for the number of sampling units in the power of two. FFT transforms a given signal in the time domain to reveal the frequency components comprising the input signal. GC skew can be thought of as a signal along the continuous axis of genomic position, which was used in place of the time domain in this work. DFT F(k) of a signal of length N, f (n), where n = 0, 1, …, N − 1, at frequency k was calculated as follows:
| (1) |
where i = √ 1̅. The power spectrum PS(k) of F(k) was further defined as
| (2) |
at each frequency k. In this power spectrum, GC skew shows the greatest contributing component at 1-Hz frequency, corresponding to the two replichores shifting between two regions of opposite polarity as in a sine curve (Arakawa et al. 2007). The Math:FFT module of Perl (http://search.cpan.org/~rkobes/Math-FFT-1.28/FFT.pm) was used for FFT calculation. To level the effects of genome size when comparing the diverse bacterial species, all genome sequences were divided into 4096 windows, and then the GC skew used as the initial signal, the cumulative GC skew, and the power spectra were calculated in these windows. Number of windows must be the power of two for effective FFT calculation, and here 212 = 4096 windows were used to take account of the effects of gene positioning, since this window size roughly corresponds the size of genes (about 1kbp) in bacterial genomes. This window size also eliminates other local mutational factors including those within genes, generated by functional requirements in RNA synthesis and translation.
GC skew index
Because cumulative skew should remain around zero under conditions of no strand bias and inversely increase its value in both positive and negative directions where bias is strong, Euclidean distance between the maximal and minimal vertices can be used as a measure of skew. The limitation of this approach and the central challenge for the quantification of genomic compositional skews, however, reside in the mathematical assessment of the skew structure to have exactly two regions physically balanced in length but with opposite polarity of nucleotide content. FFT is a good method for such a purpose, because it is able to reveal the contributing frequency components. Therefore, we used FFT to assess the fitness of the skew to the replicational selection model and combined this with the Euclidean distance between the two vertices of cumulative skew to calculate the GCSI. The GCSI is defined as the normalized average of the Euclidean distance between the two vertices of cumulative skew (dist) and the ratio of spectral strength at 1 Hz and the average strength of spectra in frequency regions 2 Hz or above (SR). Because the replicational selection is the single most dominant factor for GC strand bias, the ratio of spectral strength at 1-Hz frequency and that of all other spectra or their average must be greater than 1. SR was normalized by division with the rounded maximal SR of all bacterial genomes, which we defined here as 6000. Likewise, dist was normalized by 600.
Statistical assessment of the significance of GCSI
Significance of the GCSI values is tested using the distribution of GCSI calculated using two sets of randomized data: GCSI calculated using shuffled GC skew, where the window order is randomized using the GC skew values calculated with the original genome sequence, and GCSI calculated using shuffled genome, where the entire nucleotide sequence of the genome is shuffled while conserving the original nucleotide content. Due to calculation costs, statistical test was conducted using 1000 shuffled GC skew and 100 shuffled genome data sets. Distribution of the resulting GCSI values for the randomized data set was firstly tested for its normality using Kolmogorov-Smirnov Lilliefors test, and the significance of the original GCSI value is calculated using the z-score in the distribution of the randomized data set.
Results
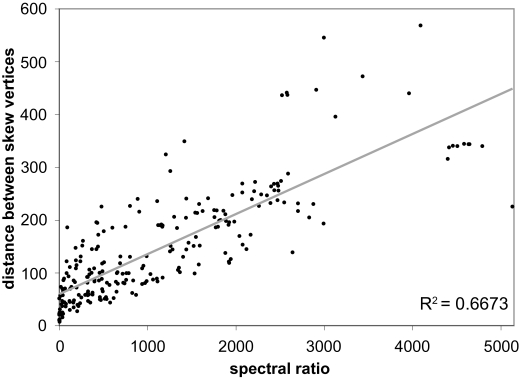
To test the applicability of GCSI for the quantification of GC skew strength, we first assessed the correlation between the Euclidean distance of the two vertices of cumulative GC skew, dist, and the Fourier power spectrum ratio, SR, using all genomes (Fig. 1). The two measures correlated with an R2 value of 0.6673, showing that the predominance of the 1-Hz frequency component leads to a stronger degree of skew.
Figure 1.
Scatter plot of spectral ratio RS against the Euclidean distance between the two vertices in cumulative graph dist. RS measures the goodness-of-fit of the “shape” of the overall GC skew to be partitioned into two segments corresponding the two replichores, by calculating the relative predominance of the spectral strength of the 1-Hz frequency component over other frequencies upon applying Fast Fourier Transform. dist measures the degree of bias in the leading and lagging strands, by calculating the Euclidean distance between the average GC skew in the two replichores. RS is generally correlated with dist, therefore combination of these two measures as GCSI should correctly represent both the shape of the graph and the degree of skew.
Using the measures dist and SR, GCSI was calculated for 304 bacterial genomes; 50 selected species are shown in Table 1 (see supplementary information for comprehensive listings). From the comprehensive list, nine genomes were further selected to illustrate the GC skew graphs plotted with 500 windows at various GCSI values (Fig. 2). As a control, GCSI was also calculated for 29 archaeal genomes, most of which showed no GC skew (Table 2). Because GCSI was normalized by the rounded maximum values of SR and dist, it ranged from 0 to 1. GCSI in bacterial genomes ranged from 0.006 for Gloeobacter violaceus to 0.815 for Clostridium perfringens (mean, 0.207; median, 0.145; SD, 0.173). The majority of archaeal genomes had GCSI <0.05, and the highest GCSI among archaeal genomes (0.122 of Halobacterium sp.) was low compared to those of bacterial genomes. GC skew was not clearly observable in species with GCSI <0.05, but it showed clear shift points when GCSI >0.10. Due to the limited number of iterations, normality test for the statistical assessment using shuffled genome sequence did not score well, but that using shuffled GC skew passed the test in all genome analyzed. The z-score was generally low and therefore not significant when GCSI <0.05 (especially <0.02), where the GCSI values may not be accurate. On the other hand, GCSI >0.05 scored extremely high z-scores, and therefore these values accurately depict the polarity of the genomes.
Table 1.
GCSI, spectral ratio RS, and the Euclidean distance between the two vertices in cumulative graph dist for randomly selected 50 bacterial chromosomes. Significance was calculated using 1) 1000 samples by shuffling GC skew windows, and 2) 100 samples by shuffling the entire nucleotide sequence of the genome while conserving the nucleotide composition, and the p-value from the normality test and the significance of the original GCSI value using the distribution of randomized samples was given as the z-score.
| shuffled GC skew | shuffled genome | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| species | accession | GCSI | SR | dist | mean | SD | z-Score | p-value (Lillefors) | mean | SD | z-Score | p-value (Lillefors) |
| Gloeobacter violaceus PCC 7421 | NC_005125 | 0.006 | 2.006 | 7.103 | 0.006002 | 7.88E–05 | 1 | 8.08E–62 | 0.002587 | 0.000671 | 5 | 0.014 |
| Synechocystis sp. PCC 6803 | NC_000911 | 0.009 | 0.296 | 10.443 | 0.008784 | 8.10E–05 | 0 | 1.12E–68 | 0.005882 | 0.000706 | 4 | 0.000 |
| Mycoplasma hyopneumoniae 232 | NC_006360 | 0.019 | 0.149 | 22.387 | 0.018741 | 9.19E–05 | 0 | 3.39E–86 | 0.008964 | 0.002092 | 4 | 0.015 |
| Synechococcus elongatus PCC 7942 | NC_007604 | 0.024 | 35.953 | 24.632 | 0.020612 | 8.28E–05 | 35 | 8.91E–63 | 0.005043 | 0.000938 | 19 | 0.022 |
| Shigella boydii Sb227 | NC_007613 | 0.035 | 120.160 | 29.756 | 0.024878 | 7.99E–05 | 124 | 7.41E–67 | 0.009254 | 0.000518 | 49 | 0.000 |
| Frankia alni ACN14a | NC_008278 | 0.040 | 125.894 | 35.514 | 0.029676 | 7.46E–05 | 139 | 1.95E–54 | 0.005153 | 0.000263 | 132 | 0.006 |
| Thiobacillus denitrificans ATCC 25259 | NC_007404 | 0.042 | 161.970 | 34.606 | 0.028922 | 8.35E–05 | 160 | 2.43E–69 | 0.003939 | 0.000703 | 54 | 0.923 |
| Tropheryma whipplei str. Twist | NC_004572 | 0.048 | 9.110 | 56.698 | 0.047333 | 8.84E–05 | 7 | 1.78E–79 | 0.02598 | 0.000883 | 24 | 0.063 |
| Geobacter sulfurreducens PCA | NC_002939 | 0.054 | 167.963 | 47.796 | 0.039911 | 8.49E–05 | 163 | 1.80E–80 | 0.007165 | 0.000401 | 116 | 0.024 |
| Rhodopseudomonas palustris BisB5 | NC_007958 | 0.064 | 365.258 | 40.691 | 0.033991 | 8.07E–05 | 376 | 1.32E–66 | 0.011625 | 0.000239 | 220 | 0.000 |
| Polaromonas sp. JS666 | NC_007948 | 0.071 | 424.069 | 43.091 | 0.035995 | 8.13E–05 | 433 | 7.59E–58 | 0.00298 | 0.000621 | 109 | 0.003 |
| Shigella flexneri 2a str. 2457T | NC_004741 | 0.077 | 320.650 | 60.344 | 0.050368 | 8.16E–05 | 326 | 4.89E–71 | 0.008386 | 0.000448 | 153 | 0.000 |
| Haemophilus influenzae 86–028NP | NC_007146 | 0.084 | 177.860 | 82.588 | 0.068907 | 8.17E–05 | 180 | 2.15E–63 | 0.00523 | 0.001235 | 63 | 0.143 |
| Mycoplasma genitalium G37 | NC_000908 | 0.086 | 103.679 | 93.248 | 0.077794 | 9.01E–05 | 94 | 3.10E–78 | 0.018262 | 0.002466 | 27 | 0.007 |
| Buchnera aphidicola str. APS (Acyrthosiphon pisum) | NC_002528 | 0.090 | 84.815 | 99.584 | 0.083067 | 8.06E–05 | 86 | 1.40E–71 | 0.022281 | 0.002142 | 31 | 0.114 |
| Helicobacter pylori HPAG1 | NC_008086 | 0.097 | 182.961 | 97.720 | 0.081517 | 8.68E–05 | 174 | 1.75E–77 | 0.00802 | 0.001226 | 72 | 0.043 |
| Escherichia coli K12 | NC_000913 | 0.098 | 486.480 | 69.038 | 0.057613 | 8.13E–05 | 497 | 1.84E–69 | 0.004953 | 0.00069 | 134 | 0.000 |
| Corynebacterium glutamicum ATCC 13032 | NC_006958 | 0.104 | 321.871 | 92.134 | 0.076862 | 8.03E–05 | 333 | 1.69E–61 | 0.012113 | 0.000354 | 258 | 0.044 |
| Helicobacter pylori J99 | NC_000921 | 0.106 | 187.288 | 108.770 | 0.090726 | 8.31E–05 | 186 | 2.02E–66 | 0.018694 | 0.000701 | 124 | 0.008 |
| Rhodospirillum rubrum ATCC 11170 | NC_007643 | 0.113 | 763.008 | 59.247 | 0.049457 | 8.74E–05 | 726 | 4.72E–77 | 0.003081 | 0.000736 | 149 | 0.017 |
| Helicobacter acinonychis str. Sheeba | NC_008229 | 0.119 | 239.480 | 118.460 | 0.098801 | 8.17E–05 | 243 | 1.12E–62 | 0.007669 | 0.001377 | 80 | 0.477 |
| Escherichia coli O157:H7 str. Sakai | NC_002695 | 0.121 | 662.307 | 79.123 | 0.06602 | 8.36E–05 | 658 | 3.89E–68 | 0.008232 | 0.000316 | 356 | 0.068 |
| Dehalococcoides sp. CBDB1 | NC_007356 | 0.127 | 490.612 | 102.952 | 0.085878 | 8.61E–05 | 473 | 1.69E–72 | 0.010229 | 0.0009 | 129 | 0.144 |
| Neisseria meningitidis Z2491 | NC_003116 | 0.138 | 484.060 | 117.689 | 0.098156 | 8.08E–05 | 498 | 7.08E–66 | 0.004362 | 0.000929 | 144 | 0.129 |
| Neisseria gonorrhoeae FA 1090 | NC_002946 | 0.142 | 508.853 | 119.737 | 0.099867 | 8.29E–05 | 510 | 2.72E–60 | 0.006794 | 0.001039 | 130 | 0.122 |
| Yersinia pestis KIM | NC_004088 | 0.148 | 785.937 | 99.533 | 0.083032 | 8.47E–05 | 772 | 4.61E–63 | 0.017378 | 0.00027 | 485 | 0.002 |
| Rickettsia typhi str. Wilmington | NC_006142 | 0.161 | 437.431 | 149.404 | 0.124586 | 8.32E–05 | 437 | 9.13E–71 | 0.048368 | 0.000704 | 159 | 0.533 |
| Salmonella typhimurium LT2 | NC_003197 | 0.167 | 1107.149 | 89.390 | 0.074569 | 7.57E–05 | 1217 | 4.49E–65 | 0.002732 | 0.000705 | 232 | 0.001 |
| Yersinia pseudotuberculosis IP 32953 | NC_006155 | 0.174 | 951.989 | 113.617 | 0.094763 | 8.49E–05 | 933 | 8.63E–77 | 0.017276 | 0.000232 | 676 | 0.054 |
| Dechloromonas aromatica RCB | NC_007298 | 0.199 | 1366.887 | 101.487 | 0.084655 | 8.43E–05 | 1350 | 3.78E–73 | 0.002758 | 0.000606 | 322 | 0.260 |
| Chromohalobacter salexigens DSM 3043 | NC_007963 | 0.220 | 1568.376 | 106.845 | 0.089121 | 8.36E–05 | 1561 | 1.67E–69 | 0.02062 | 0.000186 | 1070 | 0.000 |
| Shewanella denitrificans OS217 | NC_007954 | 0.231 | 1611.284 | 116.055 | 0.096797 | 8.05E–05 | 1666 | 3.01E–60 | 0.009546 | 0.000429 | 515 | 0.005 |
| Bacteroides fragilis YCH46 | NC_006347 | 0.253 | 1464.908 | 157.144 | 0.13104 | 8.95E–05 | 1362 | 3.16E–75 | 0.003523 | 0.000726 | 343 | 0.046 |
| Chlamydophila pneumoniae AR39 | NC_002179 | 0.256 | 1169.869 | 189.710 | 0.158179 | 8.84E–05 | 1101 | 3.49E–72 | 0.007879 | 0.00137 | 180 | 0.311 |
| Methylobacillus flagellatus KT | NC_007947 | 0.267 | 1921.680 | 127.706 | 0.106502 | 7.90E–05 | 2026 | 5.99E–66 | 0.005942 | 0.000789 | 330 | 0.057 |
| Bacillus licheniformis ATCC 14580 | NC_006270 | 0.271 | 1879.614 | 137.460 | 0.114632 | 8.87E–05 | 1764 | 3.54E–88 | 0.003241 | 0.000753 | 355 | 0.299 |
| Desulfotalea psychrophila LSv54 | NC_006138 | 0.298 | 2118.652 | 145.143 | 0.12104 | 8.74E–05 | 2018 | 1.04E–70 | 0.034761 | 0.000188 | 1393 | 0.002 |
| Bacillus subtilis subsp. subtilis str. 168 | NC_000964 | 0.312 | 2041.257 | 169.785 | 0.141571 | 8.43E–05 | 2017 | 1.59E–71 | 0.008504 | 0.000492 | 615 | 0.067 |
| Streptococcus thermophilus LMG 18311 | NC_006448 | 0.319 | 1827.666 | 200.247 | 0.166958 | 8.66E–05 | 1758 | 6.60E–73 | 0.015441 | 0.000843 | 360 | 0.245 |
| Lactococcus lactis subsp. lactis Il1403 | NC_002662 | 0.329 | 1753.562 | 218.954 | 0.182543 | 8.30E–05 | 1759 | 1.10E–73 | 0.022 | 0.000455 | 673 | 0.000 |
| Streptococcus pyogenes SSI-1 | NC_004606 | 0.332 | 1881.246 | 210.648 | 0.175623 | 8.53E–05 | 1836 | 5.66E–74 | 0.040869 | 0.000363 | 802 | 0.269 |
| Streptococcus mutans UA159 | NC_004350 | 0.378 | 2260.651 | 226.932 | 0.189192 | 8.33E–05 | 2259 | 6.68E–72 | 0.023889 | 0.000534 | 662 | 0.000 |
| Bacillus halodurans C-125 | NC_002570 | 0.406 | 2702.537 | 216.866 | 0.180803 | 7.92E–05 | 2841 | 1.12E–62 | 0.028966 | 0.000211 | 1788 | 0.643 |
| Buchnera aphidicola str. Bp (Baizongia pistaciae) | NC_004545 | 0.409 | 1415.720 | 348.910 | 0.290845 | 8.67E–05 | 1359 | 3.03E–70 | 0.011699 | 0.002443 | 162 | 0.005 |
| Staphylococcus aureus subsp. aureus COL | NC_002951 | 0.420 | 2398.078 | 263.652 | 0.219797 | 8.72E–05 | 2290 | 2.05E–70 | 0.02288 | 0.000463 | 857 | 0.124 |
| Lactobacillus johnsonii NCC 533 | NC_005362 | 0.438 | 2512.013 | 274.215 | 0.228599 | 8.63E–05 | 2425 | 8.56E–71 | 0.038523 | 0.000349 | 1142 | 0.001 |
| Ehrlichia ruminantium str. Welgevonden | NC_005295 | 0.579 | 2583.741 | 436.829 | 0.364108 | 7.99E–05 | 2694 | 6.11E–61 | 0.047406 | 0.000605 | 879 | 0.837 |
| Lactobacillus plantarum WCFS1 | NC_004567 | 0.615 | 5130.119 | 225.397 | 0.187914 | 8.59E–05 | 4977 | 4.07E–76 | 0.008182 | 0.000715 | 848 | 0.001 |
| Bacillus anthracis str. Sterne | NC_005945 | 0.669 | 4584.878 | 344.506 | 0.287173 | 8.28E–05 | 4611 | 1.26E–62 | 0.020516 | 0.000228 | 2840 | 0.271 |
| Clostridium perfringens str. 13 | NC_003366 | 0.815 | 4092.849 | 568.720 | 0.474015 | 7.93E–05 | 4301 | 1.73E–62 | 0.108625 | 0.000249 | 2832 | 0.153 |
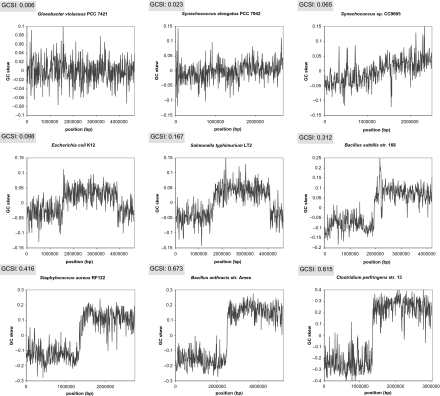
Figure 2.
GC skew graphs plotted with 500 windows for nine bacteria at different levels of GCSI. GC skew is not observable for the first two species at GCSI <0.05, and becomes evident at GCSI >0.08. At GCSI >0.1, graphs increase their skewness and the shift points and two replichores can be clearly discerned from the graph. Note that the range of Y-axis extends as GCSI values increase. Overall, GCSI correlates with and correctly captures the degree of skew.
Table 2.
GCSI, spectral ratio RS, and the Euclidean distance between the two vertices in cumulative graph dist for 29 archaeal chromosomes. See Table 1 legend for the details of the test of significance.
| shuffled GC skew | shuffled genome | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| species | accession | GCSI | SR | dist | mean | SD | z-Score | p-value (Lillefors) | mean | SD | z-Score | p-value (Lillefors) |
| Thermotoga maritima MSB8 | NC_000853 | 0.079 | 55.630 | 89.074 | 0.07431 | 8.28E–05 | 55 | 7.84E–73 | 0.053342 | 0.000293 | 87 | 0.6718442 |
| Aeropyrum pernix K1 | NC_000854 | 0.040 | 3.985 | 47.125 | 0.039355 | 8.41E–05 | 2 | 9.80E–70 | 0.024235 | 0.000362 | 42 | 0.0041011 |
| Pyrococcus abyssi GE5 | NC_000868 | 0.045 | 26.443 | 51.771 | 0.043226 | 8.29E–05 | 25 | 4.59E–69 | 0.012958 | 0.000897 | 36 | 0.172579 |
| Methanocaldococcus jannaschii DSM 2661 | NC_000909 | 0.087 | 14.536 | 102.413 | 0.085427 | 8.30E–05 | 13 | 4.54E–70 | 0.039171 | 0.000559 | 84 | 0.5234979 |
| Methanothermobacter thermautotrophicus str. Delta H | NC_000916 | 0.045 | 67.648 | 47.539 | 0.039698 | 7.85E–05 | 70 | 8.45E–61 | 0.007545 | 0.001076 | 35 | 0.0004862 |
| Archaeoglobus fulgidus DSM 4304 | NC_000917 | 0.020 | 3.695 | 23.137 | 0.019366 | 8.57E–05 | 2 | 1.63E–72 | 0.012813 | 0.000474 | 14 | 0.0639122 |
| Pyrococcus horikoshii OT3 | NC_000961 | 0.105 | 75.244 | 117.845 | 0.098287 | 8.08E–05 | 76 | 1.23E–64 | 0.043455 | 0.000316 | 193 | 0.5840531 |
| Thermoplasma acidophilum DSM 1728 | NC_002578 | 0.046 | 43.474 | 50.894 | 0.042499 | 8.71E–05 | 40 | 2.08E–69 | 0.014096 | 0.000693 | 46 | 0.0003436 |
| Halobacterium sp. NRC-1 | NC_002607 | 0.122 | 617.687 | 84.760 | 0.070717 | 8.43E–05 | 609 | 1.26E–70 | 0.006544 | 0.000784 | 147 | 0.3059486 |
| Thermoplasma volcanium GSS1 | NC_002689 | 0.044 | 39.413 | 49.277 | 0.041153 | 9.40E–05 | 33 | 2.58E–83 | 0.010706 | 0.001065 | 31 | 0.0006178 |
| Sulfolobus solfataricus P2 | NC_002754 | 0.042 | 17.321 | 48.204 | 0.040254 | 8.11E–05 | 16 | 5.41E–62 | 0.009386 | 0.000742 | 43 | 0.0113624 |
| Sulfolobus tokodaii str. 7 | NC_003106 | 0.033 | 1.862 | 39.909 | 0.033345 | 8.61E–05 | 0 | 2.92E–68 | 0.023109 | 0.000452 | 22 | 0.0383977 |
| Pyrobaculum aerophilum str. IM2 | NC_003364 | 0.038 | 6.535 | 44.690 | 0.037329 | 8.94E–05 | 5 | 3.60E–75 | 0.035224 | 0.000278 | 9 | 0.1593346 |
| Pyrococcus furiosus DSM 3638 | NC_003413 | 0.025 | 0.167 | 29.587 | 0.024742 | 8.62E–05 | 0 | 1.43E–69 | 0.005934 | 0.00129 | 14 | 0.3798953 |
| Methanopyrus kandleri AV19 | NC_003551 | 0.023 | 9.398 | 26.361 | 0.022049 | 8.20E–05 | 8 | 1.48E–70 | 0.01687 | 0.000478 | 12 | 0.0002658 |
| Methanosarcina acetivorans C2A | NC_003552 | 0.012 | 2.813 | 13.871 | 0.011641 | 7.88E–05 | 1 | 7.19E–61 | 0.003945 | 0.000539 | 14 | 0.1568932 |
| Methanosarcina mazei Go1 | NC_003901 | 0.015 | 2.817 | 17.149 | 0.014374 | 8.09E–05 | 1 | 4.87E–64 | 0.004366 | 0.000714 | 14 | 0.0536714 |
| Nanoarchaeum equitans Kin4-M | NC_005213 | 0.034 | 3.562 | 40.488 | 0.033818 | 7.74E–05 | 2 | 1.39E–66 | 0.012129 | 0.002861 | 7 | 0.0388642 |
| Methanococcus maripaludis S2 | NC_005791 | 0.041 | 21.214 | 46.718 | 0.039014 | 7.67E–05 | 21 | 4.53E–55 | 0.01073 | 0.001159 | 25 | 0.0074146 |
| Picrophilus torridus DSM 9790 | NC_005877 | 0.032 | 2.862 | 37.785 | 0.031568 | 7.96E–05 | 1 | 6.30E–68 | 0.027789 | 0.000466 | 8 | 0.1271154 |
| Haloarcula marismortui ATCC 43049 | NC_006396 | 0.007 | 5.473 | 8.170 | 0.006896 | 8.40E–05 | 4 | 4.38E–62 | 0.003083 | 0.000636 | 6 | 0.0262528 |
| Haloarcula marismortui ATCC 43049 | NC_006397 | 0.027 | 8.093 | 31.154 | 0.026042 | 7.79E–05 | 7 | 7.89E–64 | 0.017283 | 0.002185 | 4 | 5.38E–05 |
| Thermococcus kodakarensis KOD1 | NC_006624 | 0.023 | 12.060 | 26.681 | 0.022315 | 7.73E–05 | 11 | 1.29E–58 | 0.007225 | 0.000882 | 18 | 0.0146136 |
| Sulfolobus acidocaldarius DSM 639 | NC_007181 | 0.036 | 11.514 | 41.648 | 0.034791 | 8.63E–05 | 10 | 9.33E–75 | 0.006642 | 0.001216 | 23 | 0.2043022 |
| Methanosarcina barkeri str. fusaro | NC_007355 | 0.014 | 11.210 | 15.497 | 0.012995 | 7.86E–05 | 10 | 6.92E–63 | 0.013765 | 0.000319 | 0 | 8.15E–05 |
| Natronomonas pharaonis DSM 2160 | NC_007426 | 0.027 | 96.855 | 22.716 | 0.019012 | 8.05E–05 | 99 | 3.44E–67 | 0.004615 | 0.000762 | 29 | 1.68E–05 |
| Methanosphaera stadtmanae DSM 3091 | NC_007681 | 0.087 | 85.030 | 96.236 | 0.08028 | 8.38E–05 | 83 | 6.57E–71 | 0.023236 | 0.000748 | 85 | 0.2877452 |
| Methanospirillum hungatei JF-1 | NC_007796 | 0.027 | 17.105 | 30.715 | 0.025674 | 7.94E–05 | 16 | 3.41E–72 | 0.014578 | 0.000342 | 36 | 0.0096521 |
| Methanococcoides burtonii DSM 6242 | NC_007955 | 0.044 | 44.608 | 48.355 | 0.040381 | 8.44E–05 | 43 | 1.67E–66 | 0.029311 | 0.000319 | 46 | 0.0895566 |
As can be seen from the GC skew graphs in Figure 2, the degree of skew correlates with GCSI. No skew was observable for G. violaceus and Synechococcus elongatus PCC 7942, with GCSI values of 0.006 and 0.023, respectively, but a gradual rise from negative values to positive values was observed for Synechococcus sp. CC9605, with a GCSI of 0.065, although the skew was not well defined. GC skew became visible at a GCSI of 0.098 in Escherichia coli K12, and the clarity was increased in correlation with the GCSI values for scores greater than 1, as represented by the increasing range of the y-axis from ±0.15 at GCSI values around 1 to ±0.4 at a GCSI of 0.815.
Discussion
The nucleotide sequence of a genome is structured and controlled by a myriad of selection pressures, especially in subgenomic regions, as typified by the fact that coding regions are shaped by the essential order and usage of codons. In addition to such requirements in the subgenomic regions, circular bacterial chromosomes experience genome-wide selection through the replication process. The chiral nucleotide composition in the two replication arms is significant; however, with regard to the evolutionary aspects of replicational selection on bacterial chromosomes, no useful method to quantify the degree of genomic compositional asymmetry has been proposed, unlike the wealth of codon bias measures (Suzuki et al. 2005). This lack of indices for genomic compositional skews was likely due to the difficulty of mathematical formulation and detection of the skewing shape of GC skew graphs. To distinguish the degree of skew, we utilized FFT to observe the predominance of the 1-Hz frequency component, which corresponds to the replicational selection on the two replichores, over other frequency components. Combined with the Euclidean distance between the two vertices in cumulative skew graphs, the formulated GCSI captured the strength of GC skew in bacterial chromosomes, as shown by the above results. GCSI scores are diverse even within bacterial genomes with circular chromosomes, ranging from a number of genomes with extremely low values therefore implying the lack of observable GC skew in the genome, to groups of genomes with clear skews as can be seen in Bacilli.
The majority of the archaeal genomes had GCSI <0.05, at which point no noticeable skew is observed even in bacterial genomes. This is also confirmed by the z-score in the statistical test using randomized data, with low z-scores (therefore implying less significance) when GCSI is less than 0.05. Thus, 0.05 can be employed as a threshold value to determine whether GC skew is present in a genome and therefore whether replicational selection is acting on the organism. Because the GCSI values do not show a Gaussian distribution, however, it should be noted that the indices are not necessarily proportionate with each other. Therefore, GCSI values should not be compared in terms of ratios but in terms of their rank orders. For the direct comparison of quantitative degrees of skew calculated as the ratio of two values, the use of Euclidean distance may be more suitable. However, significant Euclidean distance between the two vertices of cumulative skew may not always result from the polarity exhibited by the GC skew graph; it could also result from local regions of highly biased nucleotide content. Therefore, to ascertain that the skews are controlled by replicational selection, genomes used for such analyses should be selected beforehand using GCSI or SR at sufficiently high thresholds (e.g. 0.07 for GCSI and 200 for SR, also noting the z-scores).
GCSI would be a useful index for the estimation of confidence levels for bioinformatics analyses using genomic compositional skews. Predictions of replication origin and terminus by the observation of shift points (i.e. vertices) of cumulative skew diagrams become erroneous when the GC skew is not well defined. However, the confidence level can be easily estimated by taking into account of the magnitude of the GCSI. In this work we have only described the index for GC skew, although the same method is applicable to purine and keto excesses or any other genomic compositional skews, given that the selection is on the two replichores. Similarly, for comparative studies of genomic features related to evolutionary pressures and replication machinery, GCSI can also be used as a measure of replicational selection.
Suppliment Material
Acknowledgments
We thank the members of MGSP at the Institute for Advanced Biosciences, Keio University, especially Nozomu Yachie and Ayako Kinoshita, for critical discussions. This research was supported by the Japan Society for the Promotion of Science (JSPS).
Footnotes
Please note that this article may not be used for commercial purposes. For further information please refer to the copyright statement at http://www.la-press.com/copyright.htm
References
- Arakawa K, Mori K, Ikeda K, et al. G-language Genome Analysis Environment: a workbench for nucleotide sequence data mining. Bioinformatics. 2003;19:305–306. doi: 10.1093/bioinformatics/19.2.305. [DOI] [PubMed] [Google Scholar]
- Arakawa K, Saito R, Tomita M. Noise-reduction filtering for accurate detection of replication termini in bacterial genomes. FEBS Lett. 2007;581:253–258. doi: 10.1016/j.febslet.2006.12.021. [DOI] [PubMed] [Google Scholar]
- Arakawa K, Tomita M. G-language System as a platform for large-scale analysis of high-throughput omics data. Journal of Pesticide Science. 2006;31:282–288. [Google Scholar]
- Daubin V, Perriere G. G + C3 structuring along the genome: a common feature in prokaryotes. Mol. Biol. Evol. 2003;20:471–483. doi: 10.1093/molbev/msg022. [DOI] [PubMed] [Google Scholar]
- Dodin G, Vandergheynst P, Levoir P, et al. Fourier and wavelet transform analysis, a tool for visualizing regular patterns in DNA sequences. J. Theor. Biol. 2000;206:323–326. doi: 10.1006/jtbi.2000.2127. [DOI] [PubMed] [Google Scholar]
- Frank AC, Lobry JR. Asymmetric substitution patterns: a review of possible underlying mutational or selective mechanisms. Gene. 1999;238:65–77. doi: 10.1016/s0378-1119(99)00297-8. [DOI] [PubMed] [Google Scholar]
- Frank AC, Lobry JR. Oriloc: prediction of replication boundaries in unannotated bacterial chromosomes. Bioinformatics. 2000;16:560–561. doi: 10.1093/bioinformatics/16.6.560. [DOI] [PubMed] [Google Scholar]
- Freeman JM, Plasterer TN, Smith TF, et al. Patterns of Genome Organization in Bacteria. Science. 1998;279:1827a. [Google Scholar]
- Fujimori S, Washio T, Tomita M. GC-compositional strand bias around transcription start sites in plants and fungi. BMC Genomics. 2005;6:26. doi: 10.1186/1471-2164-6-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grabowski B, Kelman Z. Archeal DNA replication: eukaryal proteins in a bacterial context. Annu. Rev. Microbiol. 2003;57:487–516. doi: 10.1146/annurev.micro.57.030502.090709. [DOI] [PubMed] [Google Scholar]
- Grigoriev A. Analyzing genomes with cumulative skew diagrams. Nucleic Acids Res. 1998;26:2286–2290. doi: 10.1093/nar/26.10.2286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hendrickson H, Lawrence JG. Selection for chromosome architecture in bacteria. J. Mol. Evol. 2006;62:615–629. doi: 10.1007/s00239-005-0192-2. [DOI] [PubMed] [Google Scholar]
- Katoh K, Misawa K, Kuma K, et al. MAFFT: a novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Res. 2002;30:3059–3066. doi: 10.1093/nar/gkf436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krishnan NM, Seligmann H, Raina SZ, et al. Detecting gradients of asymmetry in site-specific substitutions in mitochondrial genomes. DNA Cell Biol. 2004;23:707–714. doi: 10.1089/1044549042476901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lobry JR. Asymmetric substitution patterns in the two DNA strands of bacteria. Mol. Biol. Evol. 1996;13:660–665. doi: 10.1093/oxfordjournals.molbev.a025626. [DOI] [PubMed] [Google Scholar]
- Lobry JR, Louarn JM. Polarisation of prokaryotic chromosomes. Curr. Opin. Microbiol. 2003;6:101–108. doi: 10.1016/s1369-5274(03)00024-9. [DOI] [PubMed] [Google Scholar]
- Lobry JR, Sueoka N.2002Asymmetric directional mutation pressures in bacteria Genome Biol 3RESEARCH0058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez P, Philippe H, Myllykallio H, et al. Identification of putative chromosomal origins of replication in Archaea. Mol. Microbiol. 1999;32:883–886. doi: 10.1046/j.1365-2958.1999.01370.x. [DOI] [PubMed] [Google Scholar]
- McLean MJ, Wolfe KH, Devine KM. Base composition skews, replication orientation, and gene orientation in 12 prokaryote genomes. J. Mol. Evol. 1998;47:691–696. doi: 10.1007/pl00006428. [DOI] [PubMed] [Google Scholar]
- Myllykallio H, Lopez P, Lopez-Garcia P, et al. Bacterial mode of replication with eukaryotic-like machinery in a hyperthermophilic archaeon. Science. 2000;288:2212–2215. doi: 10.1126/science.288.5474.2212. [DOI] [PubMed] [Google Scholar]
- Rocha EP. Order and disorder in bacterial genomes. Curr. Opin. Microbiol. 2004a;7:519–527. doi: 10.1016/j.mib.2004.08.006. [DOI] [PubMed] [Google Scholar]
- Rocha EP. The replication-related organization of bacterial genomes. Microbiology. 2004b;150:1609–1627. doi: 10.1099/mic.0.26974-0. [DOI] [PubMed] [Google Scholar]
- Seligmann H, Krishnan NM, Rao BJ. Possible multiple origins of replication in primate mitochondria: Alternative role of tRNA sequences. J. Theor. Biol. 2006;241:321–332. doi: 10.1016/j.jtbi.2005.11.035. [DOI] [PubMed] [Google Scholar]
- Suzuki H, Saito R, Tomita M. A problem in multivariate analysis of codon usage data and a possible solution. FEBS Lett. 2005;579:6499–6504. doi: 10.1016/j.febslet.2005.10.032. [DOI] [PubMed] [Google Scholar]
- Worning P, Jensen LJ, Hallin PF, et al. Origin of replication in circular prokaryotic chromosomes. Environ. Microbiol. 2006;8:353–361. doi: 10.1111/j.1462-2920.2005.00917.x. [DOI] [PubMed] [Google Scholar]
- Yin C, Yau SS. A Fourier characteristic of coding sequences: origins and a non-Fourier approximation. J. Comput. Biol. 2005;12:1153–1165. doi: 10.1089/cmb.2005.12.1153. [DOI] [PubMed] [Google Scholar]
- Zawilak A, Cebrat S, Mackiewicz P, et al. Identification of a putative chromosomal replication origin from Helicobacter pylori and its interaction with the initiator protein DnaA. Nucleic Acids Res. 2001;29:2251–2259. doi: 10.1093/nar/29.11.2251. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.